Abstract
The ability of Candida albicans to transit between different cellular morphologies is believed to be important for virulence. Morphological transitions occur in response to a variety of environmental signals. One such signal is encountered when cells are grown in a semisolid matrix. An important regulator of cellular morphology is the putative transcription factor CZF1. Here we demonstrate that transcription of CZF1 is responsive to growth parameters such as the temperature, carbon source, growth phase of cells, and the physical environment. In wild-type cells, a CZF1 transcript of about 4 kb was expressed when cells were grown embedded in semisolid agar medium, as well as in late exponential phase when cells were grown in liquid medium. Deletion of EFG1, a key regulator of morphogenesis, abolished CZF1 expression. Overexpression of CZF1 revealed that this gene also autoregulates its expression. Efg1p and Czf1p were shown by chromatin immunoprecipitation to act by binding to the promoter of CZF1. The coupling of environmental cues to the expression of a morphogenetic transcription factor may allow C. albicans to coordinate morphogenesis in response to specific conditions encountered in the human host.
Candida albicans is an important pathogenic fungus of humans. Although it inhabits its mammalian host primarily as a commensal organism, it is capable of causing a variety of diseases and is the fourth-leading cause of nosocomial infections in the United States (54). C. albicans changes its morphology from the yeast form to filamentous forms in response to various environmental cues, and this ability is thought to be important for the success of the fungus both as a commensal organism and as an opportunistic pathogen (32).
Many growth conditions induce filamentous growth, including growth at 37°C, at neutral pH, in nutrient-poor media, or in media containing serum or being embedded within a semisolid matrix (for a review, see reference 15).
Deletion of the EFG1 and CPH1 genes, which encode transcriptional regulators, results in a mutant that fails to produce filaments under most laboratory conditions but retains the ability to form filaments when grown embedded within agar (17). In contrast, deletion of the CZF1 gene, encoding a putative transcriptional regulator, results in the converse phenotype, i.e., the strain is defective in filamentation under embedded conditions but not under other filament-inducing conditions (7). These findings indicate that there is a genetic program that functions during filamentous growth under embedded conditions and is distinct from the program that regulates filamentation under other conditions. This genetic program may contribute to disease pathogenesis by promoting filamentation during C. albicans growth within host tissue.
CZF1 regulates the morphogenetic response to growth within a semisolid matrix (7). CZF1 is predicted to encode a 388-amino-acid protein with a glutamine-rich central region and a C-terminal cysteine-rich region similar to zinc cluster elements of DNA-binding proteins (55).
EFG1 is an important regulator of several processes in C. albicans, including induction and repression of cell wall genes (39), white-phase-specific gene expression (43), filamentous growth (17, 28, 46), chlamydospore formation (41), and metabolism (14). EFG1 is a key positive regulator of filamentation under most inducing conditions (28, 41, 42, 45, 46). However, EFG1 acts as a negative regulator in cells embedded in semisolid medium; under these conditions, efg1 deletion mutants are more filamentous than wild-type cells (17, 41). One way that Czf1p acts to promote filamentous growth is by relieving Efg1p-mediated repression of filamentation during growth in a semisolid matrix (17).
To understand the conditions under which CZF1 contributes to the regulation of cellular morphogenesis, we undertook an analysis of CZF1 expression. We found that CZF1 is transcriptionally upregulated in response to several growth conditions, such as during embedded growth. CZF1 was highly expressed in post-exponential-phase cells in a temperature- and carbon source-dependent manner and was induced in cells subjected to osmotic shock. We also found that CZF1 expression is regulated through the binding of Efg1p and Czf1p to the CZF1 promoter.
MATERIALS AND METHODS
Strains.
The strains of C. albicans used in this study are listed in Table 1. Transformation of C. albicans was performed by the lithium acetate method (2). Escherichia coli strains used were XL-1 Blue (Stratagene), for plasmid propagation, and TOP10F′, for TOPO cloning (Invitrogen).
TABLE 1.
C. albicans strains used in this work
| Strain | Genotype | Source or reference |
|---|---|---|
| CAI-4 | SC5314 Δura3::imm434/Δura3::imm434 | 15a |
| CKY101 | CAI-4 ade2::pDBI52 | 7 |
| CKY230 | CAI-4 Δczf1::hisG/Δczf1::hisG ade2::pDBI52 | 7 |
| CKY136 | CAI-4 Δefg1::hisG/Δefg1::hisG ade2::pDBI52 | 17 |
| MVY200 | CAI-4(pYPB-ADH1pt) | This study |
| MVY201 | CAI-4(pYPB-ADH1pt-CZF1) | This study |
| MVY202 | CAI-4 Δefg1::hisG/Δefg1::hisG leu2::pBI-HAHYD | 42 |
| MVY101 | Δczf1::hisG/Δczf1::hisG ade2::pMV101 | This study |
| MVY102 | Δczf1::hisG/Δczf1::hisG ade2::pMV102 | This study |
| WO-1 ura3 | Δura3 strain derived from strain WO-1 | P. T. Magee |
| CHY101 | CAI-4 efg1::hisG/efg1::hisG czf1::hisG/czf1::hisG ade2::pMV101 | This study |
| CHY102 | CAI-4 efg1::hisG/efg1::hisG czf1::hisG/czf1::hisG ade2::pMV102 | This study |
Media and growth conditions.
C. albicans cells were routinely grown in YP medium (1% yeast extract, 2% Bacto peptone) with either 2% glucose (YPD) or 2% sucrose (YPS) added. For a selective medium, CM−Ura (complete medium lacking uridine and uracil) was used (2). For embedding experiments, 0.7% low-melting-point agarose (SeaPlaque; Cambrex) was added. Spider medium (27) and SCAA medium (42) were used for induction of the PCK1 promoter. Escherichia coli strains were cultured in Luria broth or on Luria plates (31), with ampicillin added to a concentration of 100 μg/ml.
For shock experiments, cells were grown at 25°C to an optical density at 600 (OD600) of 1.0 in YPS medium. Compounds were added to the following final concentrations: 0.3 M NaCl, 5 mM sorbate, 5 mM H2O2, 7.5% ethanol, and 10 mM caffeine. The final pH for the acid shock was 2.8. Cells were exposed to shocks for 20 min before being harvested. For osmotic shock, cells were grown overnight in YPS with 1 M sorbitol at 30°C. After dilution in the same medium, cells were grown at 25°C to an OD600 of 1.0. Ten milliliters of this culture was then added to 40 ml water (for hypo-osmotic shock), 1 M sorbitol (for isosmotic control), or 2.5 M sorbitol (for hyperosmotic shock). Cells were grown for 20 min and then harvested.
For experiments with cyclic AMP (cAMP), cells were grown overnight at 37°C in YPD. After dilution in the same medium, cultures were grown for 6 h, and dibutyryl cAMP (dbcAMP; Sigma) was added to a final concentration of 10 mM (water was added to the control). Ten milliliters of culture was harvested 4 h after the cAMP addition.
For isolation of white and opaque cells, cultures of strain WO-1 ura3 were streaked on YPD with phloxine B (50 μg/ml) and uridine (61.2 μg/ml) and then purified. Cultures were observed microscopically to confirm that they were composed of only one phase before being harvested. After being harvested, cells were frozen at −80°C in RNALater (Ambion) before being processed for RNA extraction.
Plasmids.
The plasmids used in this study are listed in Table 2. Vectors pDBI52 and pDB212 were described previously (7). Plasmid pYPB-ADH1pt (24) was kindly provided by Csilla Csank and Malcolm Whiteway, and pBI-HAHYD (42) was provided by Joachim F. Ernst. pYPB-ADH1pt-CZF1 was made by amplifying the CZF1 open reading frame (ORF) with primers CZVECF (GAAGATCTACAGAAGAAGAATAATAAGAACCACCCAACAG) and CZVECR (CCGCTCGAGTTTTTAAACGATATCCCTCCAACACA) (BglII and XhoI sites are underlined) and cloning the PCR product into BglII/XhoI-cut pYPB-ADH1pt.
TABLE 2.
Plasmids used in this work
pMV101 and pMV102 (hemagglutinin [HA]-tagged and wild-type [WT] Czf1, respectively, under the control of the maltase promoter) were made by first amplifying the HA epitope from pFA6a-His3MX6-PGAL1-3HA (29) with primers LIZHAF (ACAACAACCATTGTACCCAGCAATATCCGCCACAATCTGTAGGTTACCAAATGTCTTTAATTAACATCTTTTACCCAT) and LIZHAR (ATTTGCACATATTCTTGGTGTTGGGGATTGTACACATCACCCGCTAGGCACTGAGCAGCGTAATCTGGAAC). The resultant PCR product, containing the HA sequence fused in frame with CZF1 (CZF1 sequences are shown in italics), was transformed along with BstEII-cut pYEplac-CZF1 (2μm plasmid pYEplac195 carrying CZF1 [T. Volkert and C. A. Kumamoto, unpublished]) into Saccharomyces cerevisiae. Homologous recombination resulted in the HA tag being inserted into the CZF1 ORF near the BstEII site. The resulting plasmid, pYEplac-CZF1-HA, along with the parent plasmid pYEplac-CZF1, was used as a PCR template to amplify the entire CZF1 ORF. The HA-tagged CZF1 product retained biological function, as demonstrated by complementation of the czf1 deletion mutant grown embedded in YPS agarose. The primers used for this PCR were CZFStu (GCAGGCCTTATCTGTTCACCCCATTTCTTTCA) and CZFXba (GCTCTAGAATTTCGTTTTGCTGGTGCTGTG) (StuI and XbaI sites are underlined). Both the HA-tagged and WT CZF1 PCR products were cloned into EcoRV/XbaI-digested pDBI52 to produce pMV101 and pMV102, respectively.
DNA analysis.
PCR, restriction digestion, and gel electrophoresis were performed according to standard methods, as previously described (35). Automated DNA sequencing was performed by Michael Berne and coworkers at the Tufts University core facility. Sequences were analyzed using the BLAST program (1), and alignments were constructed using MegAlign software (DNASTAR).
Sequence analysis of CZF1 locus.
Sequences of the CZF1 locus were obtained from assembly 6 of the unfinished C. albicans genome sequence available at the Stanford Genome Technology Center (http://www-sequence.stanford.edu/group/candida/) and from the Ca19 annotation of the Canadian Bioinformatics Resource of the National Research Council of Canada (http://candida.bri.nrc.ca/candida/index.cfm). The cladogram in Fig. 1 showing phylogenetic relationships between species was drawn according to data from other studies (13, 16, 44). Sequence information for C. albicans, Candida glabrata, Debaryomyces hansenii, S. cerevisiae, Saccharomyces paradoxus, Ashbya gossypii, Kluyveromyces lactis, Yarrowia lipolytica, and Schizosaccharomyces pombe was obtained from published reports (5, 9, 12, 21, 26, 56) and public genome databases and search engines (Gènolevures, NCBI Entrez Gene, and Stanford Genome Center's Saccharomyces Genome Database and Candida Genome Database). Sequences were identified using the BLAST program or Entrez Gene database available at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/). Analysis of the 3′ region was carried out using a web-based tool provided by the Graber laboratory (http://harlequin.jax.org/polyA/).
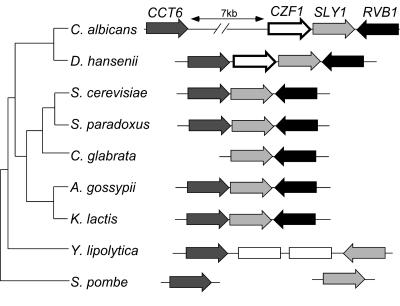
FIG. 1.
Features of CZF1 locus. The C. albicans CZF1 gene (white arrow) is depicted at the top along with neighboring genes. For comparison, related loci from several species of ascomycetes are shown below. A 7-kb region upstream of the C. albicans CZF1 ORF is devoid of significant ORFs. Orthologues of CCT6 are depicted by dark gray arrows, those of SLY1 are shown by light gray arrows, and those of RVB1 are shown by black arrows. White boxes represent genes not closely related to the four genes discussed here. S. pombe orthologues were not found close to each other in the genome. Genes and loci are drawn in the correct orientations but are not drawn to scale. The cladogram on the left shows phylogenetic relationships between species, determined as described in Materials and Methods.
Embedding and harvesting cells.
Embedding of cells in agarose medium was performed as previously described (7), with some modifications. For RNA preparation, cells were plated at a high density in YPS low-melting-point agarose (SeaPlaque), and after the agarose solidified, plates were overlaid with a thin layer of YPS low-melting-point agarose. Cells growing embedded within the agarose were harvested by mechanical disruption of the agarose in cold 1× phosphate-buffered saline and filtering of the slurry through nylon mesh (35 μm; Small Parts, Inc.). The filtrate containing cells was collected over ice, cells were harvested by centrifugation, and cell pellets were frozen at −80°C in RNALater (Ambion).
Northern analysis.
Total RNA from cells frozen in RNALater was extracted by mechanical disruption using glass zirconia beads, a Mini-BeadBeater (BioSpecs Products, Inc., Bartlesville, OK), and an RNeasy Midi kit (QIAGEN Inc.). Poly(A)+ mRNA was prepared by using an Oligotex spin column kit (QIAGEN Inc.).
Probes for the CZF1 locus were amplified by PCR with the following primers: TTCTCCCCTCAGCCTAACAG and GCTATAATTGCACCCCGTCTTT for the 603-bp probe A, GAAAAGTACCCACAAGCGACAAC and ACACAGCGTAAAAACATAAGG for the 928-bp probe B, CCTTCACCTCTAATGTTGTTAG and GTAATGTAAACTTAAGTTTCC for the 992-bp probe C, and GCCATGATGTCTGCCTCGACTC and CCCTCCAACACAGAGAAGC for the 659-bp probe D. Probe D was used in CZF1 expression studies. All probes for the CZF1 locus were demonstrated to hybridize normally by Southern blot analysis.
The 992-bp ACT1 probe used as a loading control was amplified by PCR using primers ACTFW (TATCATGGTTGGTATGGG) and ACTRV (TGTGGTGAACAATGGATG). The 18S rRNA probe used as an additional loading control in this study has been described previously (3).
Probes were labeled with [α-32P]dATP (NEN-Perkin-Elmer), using a Primer-It II random primer labeling kit (Stratagene).
Northern blot hybridization was performed according to previously described methods (35). Signals were recorded using a PhosphorImager gel scanner (Amersham), and values were calculated using ImageQuant software (Amersham). For experiments using total RNA, values were normalized to 18S rRNA levels, but for experiments using mRNA, it was not possible to use 18S rRNA, and therefore, ACT1 was used for normalization of these results.
5′ RACE analysis.
The transcription start site for CZF1 was determined using 5′ rapid amplification of cDNA ends (RACE) analysis with a BD SMART RACE cDNA amplification kit (BD Biosciences Clontech, Palo Alto, CA). Poly(A)+ mRNA from embedded cells was used as the template for cDNA synthesis. The CZF1-specific primer CGSP3 (AAGTATTTGTAGGGGTTGTCTGGGTAA) was used as a primer for reverse transcription cDNA synthesis. The CZF1-specific primer CGSP4 (GGGGATGGGGGAGTGAAAACAATAATAGGGTAGA) and a kit-supplied universal primer (UPM) were used for PCR amplification of the cDNA.
The PCR products were cloned into pCRII-TOPO vector (Invitrogen) and sequenced.
ChIP.
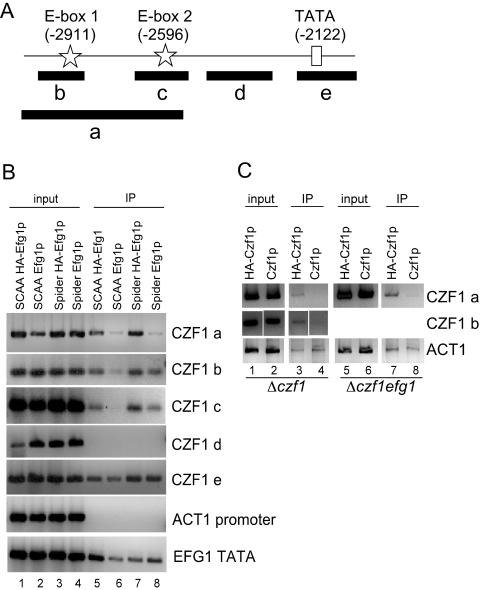
Strains carrying HA-tagged Efg1p (MVY202) and HA-tagged Czf1p (MVY101) were grown in promoter-inducing media (SCAA or Spider medium for PPCK1-HA-EFG1 and YPS for PMAL-HA-CZF1) to an OD600 of 1.0 to 1.5 and cross-linked with formaldehyde before the preparation of cell lysates as described in the standard chromatin immunoprecipitation (ChIP) protocol (47). Lysates were sonicated twice for 30 seconds each, using a microtip at 100% duty at level 5, resulting in an average DNA fragment size of about 800 bp. For input controls, a 1/100 volume of each lysate was saved. Immunoprecipitation was performed by the addition of 5 μl anti-HA monoclonal antibody (1:100; Babco, Richmond, CA) and 50 μl 50% protein G-Sepharose beads (Sigma, St. Louis, MO) to 500 μl lysate, followed by incubation for 4 h at 4°C. Cross-links were reversed by overnight incubation at 65°C. DNAs from immunoprecipitations and input controls were purified using a QIAquick PCR purification kit (QIAGEN), and aliquots of the eluates were used as templates for PCR. The primers used for the PCR products shown in Fig. 6A were as follows: for product a, CZF1AF (CAAAACAAGCCAGAATAAAAATA) and CZF1AR (ACCGGAAGTACCAAAAGAGTC); for product b, EBOX1F (TTATAGAACAACAACAACGAGAAA) and EBOX1R (TACGGAACACAATGGGAAGA); for product c, EBOX2F (ACTCATTAAATAGCCCATACTGC) and EBOX2R (ATCAACAAACCGACGAAGAA); for product d, INTERF (GCCTTATGTTTTTACGCTGTG) and INTERR (CATTAGAGGTGAAGGGAAAAAC); and for product e, TATAF (GGAAGTACAACGCAAATCAAAG) and TATAR (ATGGGGGAGTAAAAACAATAATAG).
FIG. 6.
In vivo binding of Efg1p and Czf1p to CZF1 promoter sequences. (A) Diagram of CZF1 upstream region and PCR products detected in ChIP experiments. Bars a to e denote the positions of PCR products used for ChiP analysis. The sizes of the PCR products were 565 bp (a), 125 bp (b), 147 bp (c), 245 bp (d), and 273 bp (e). (B) Strain MVY202, containing HA-tagged Efg1p, or CKY101, containing untagged Efg1p, was grown in medium that promotes strong induction (SCAA) or moderate induction (Spider) of the PCK1 promoter. ChIP of cellular extracts was performed, using anti-HA antibody to precipitate HA-tagged Efg1p. The presence of specific sequences from the CZF1 promoter was determined by PCRs using primers for fragments a to e, shown in panel A. An ACT1 promoter sequence was examined as a negative control, and a sequence containing the TATA box from the EFG1 promoter was examined as a positive control. (C) Strains MVY101 (HA-CZF1), MVY102 (CZF1), CHY101 (Δefg1/HA-CZF1), and CHY102 (Δefg1/CZF1) were grown in YPS medium, which induces the MAL2 promoter controlling HA-tagged or untagged CZF1. ChIP of cellular extracts was performed, using anti-HA antibody to precipitate HA-tagged Czf1p. The presence of specific sequences from the CZF1 promoter (fragment a or b in panel A) and from the negative control (ACT1 promoter) was examined by PCR.
Primers for amplifying the promoter sequences of ACT1 and EFG1 have been previously described (49).
RESULTS
CZF1 contains an unusually large 5′ untranslated region (UTR) transcribed from a 7-kb region lacking significant open reading frames.
Sequence analysis of CZF1 and the surrounding genomic region on chromosome 4 revealed a number of unusual features. First, there is a 7-kb region that separates CZF1 and its nearest upstream ORF (>300 bp). This large ORF-less region does not correspond to the centromeric DNA sequence for chromosome 4 (36). Second, sequence alignments revealed that CZF1 and the large upstream intergenic sequence are flanked by genes whose order and orientation are conserved among their homologues in S. cerevisiae and other Hemiascomycetes (Fig. 1). This was surprising, as there is little conservation of gene order between homologous genes of C. albicans and S. cerevisiae (37). Although this locus, composed of the genes CCT6, SLY1, and RVB1, has highly conserved synteny among several species of ascomycete fungi, the CZF1 gene is unique to one clade which includes C. albicans and close relatives. The halotolerant yeast Debaryomyces hansenii is a member of this clade and has a CZF1 homologue located within the syntenic block of genes, but this gene lacks the large noncoding upstream sequence. Homologues of CZF1 have also been found in two other Candida species, C. tropicalis (4) and C. dubliniensis (34), but their locations within their respective genomes and whether they have a large upstream ORF-less sequence remain to be determined.
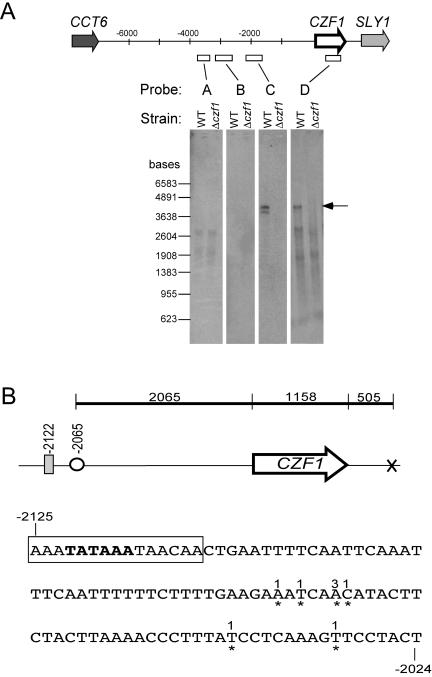
Northern analysis revealed a surprisingly large transcript for CZF1 (Fig. 2A). While the ORF is only 1.1 kb long, a probe for the ORF (probe D) detected a transcript of almost 4 kb (Fig. 2A). This CZF1 transcript was also detected using a probe for a sequence 2 kb upstream of the ORF (probe C; positions −2279 to −1287 from the ATG). However, a probe for a sequence 3 kb upstream (probe B; positions −3421 to −2501 from the ATG) did not yield a signal, nor did probes for sequences further upstream. No signal was detected in a Δczf1 strain with any of the probes tested. These results suggest that CZF1 transcription initiates approximately 2 kb upstream of the coding sequence. A smaller RNA was sometimes observed (for example, see Fig. 2A, probe C), which was more pronounced in mRNA than in total RNA preparations but was not consistently seen for any given probe, condition, or strain.
FIG. 2.
Large transcript of CZF1. (A) Northern analysis of the CZF1 transcript, using probes corresponding to the CZF1 ORF and upstream sequences. Each lane contains 3 μg mRNA from either wild-type strain CKY101 or czf1 mutant strain CKY230 grown embedded in agarose medium at 25°C. The CZF1 transcript is indicated by an arrow. The transcript size was estimated by measuring the electrophoretic mobility of the transcript and calibrating it to a standard curve of RNA markers of known sizes. A second smaller transcript was sometimes seen (for example, with probe C), but its appearance was not consistently observed. (B) CZF1 transcription start site determined by 5′ RACE analysis. The map diagrams the positions of the TATA box (gray bar), the transcription start site (open circle), and a putative polyadenylation site (×). The black line denotes the predicted span of the mRNA, including the large 5′ UTR, the ORF, and the 3′ UTR, with sizes indicated in base pairs. The sequence shown represents nucleotides −2125 to −2024 from the predicted translation start site (ATG) of CZF1. Start sites found by RACE are denoted by asterisks below the nucleotides. The numbers above the nucleotides indicate the numbers of clones with the indicated start sites. The putative TATA box is shown in bold. The boxed sequence represents a 15-bp sequence identical to the EFG1 promoter sequence that includes the TATA box for the major EFG1 transcript (49).
To confirm these results and identify the start point of transcription, RACE analysis was performed. Eight cloned products were sequenced, and the predicted start sites clustered in the region of bases −2070 to −2031 (Fig. 2B). Transcription from this region would result in the predicted 2-kb 5′ UTR.
The distance between TATA elements and transcription initiation sites in S. cerevisiae is typically between 40 and 120 bp (48). Consistent with this observation, there is a consensus TATA box (TATAAA) 50 bp upstream from the transcription start site for CZF1 determined by RACE. Interestingly, this TATA box and the surrounding 9 bp are identical to a 15-bp sequence containing the TATA box for one of the transcripts of the EFG1 gene (Fig. 2B, boxed sequence). This sequence has been shown by others to be required for EFG1 autoregulation (49).
Several potential transcription factor-binding sites can be found in the CZF1 promoter region. Four hundred and 700 bp upstream of the TATA box are two E boxes (CATTTG), DNA sequences known to be specifically recognized and bound by Efg1p (25). The TATA box of CZF1 is also about 800 bp downstream from a potential Tec1p-binding site (TCATTCT). The stress response element motif, CCCCT, is found numerous times throughout the region, both upstream and downstream of the TATA.
Computational analysis using a web-based tool (18) revealed consensus 3′ processing elements (52, 57) about 500 bp downstream of the CZF1 stop codon. These included a high-scoring efficiency element sequence (TATATA) 56 bp upstream of the putative poly(A) site, followed by a positioning element sequence (AATAAT) with a good score 21 bp upstream of the putative poly(A) site, followed by the putative polyadenylation site itself (TTATTA), which is flanked both upstream and downstream by T-rich elements (TGTTTG upstream and ATTGTT downstream). The location of a poly(A) site at this location is consistent with the calculated size of the transcript and the position of the transcription start site (Fig. 2B).
CZF1 is regulated in response to carbon source, temperature, growth phase, and physical environment.
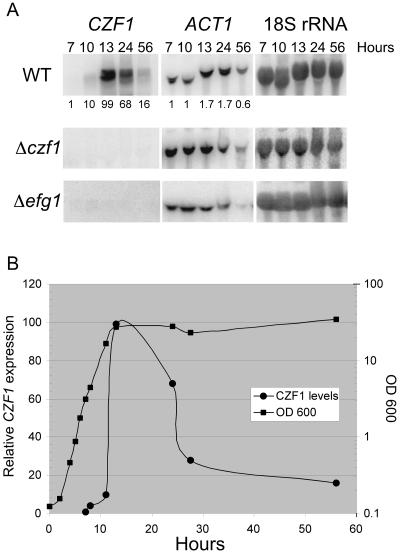
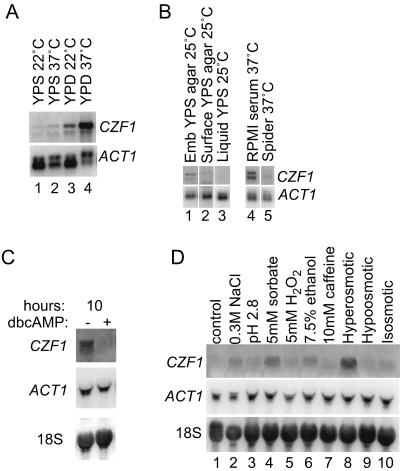
To determine conditions under which CZF1 was expressed, CZF1 transcription was studied during growth under a variety of conditions. In cells growing in liquid media, CZF1 was not expressed in log-phase cells but was most highly expressed in post-exponential-phase cells grown at 37°C in the presence of glucose (Fig. 3A, upper left panel). The use of sucrose as the carbon source or growth at a lower temperature resulted in less expression of CZF1 (Fig. 4A). Therefore, the carbon source, temperature, and growth phase all had strong effects on CZF1 expression.
FIG. 3.
Growth-phase-dependent expression of CZF1. (A) Northern blots using 20 μg total RNA from strains CKY101, czf1 mutant CKY230, and efg1 mutant CKY136 grown in rich liquid medium containing glucose as a carbon source (YPD) at 37°C for the indicated times. The blots were probed with a CZF1 ORF, ACT1, or 18S rRNA probe. Values underneath the blots indicate x-fold changes in transcript signal from expression at 7 h. All values were normalized to levels of 18S rRNA. (B) Expression profile of CZF1 during growth in YPD liquid medium at 37°C. Cell densities (squares) are given by the axis on the right, and relative expression levels of CZF1 (circles) are given by the axis on the left. Peak expression of CZF1 occurred during the diauxic shift.
FIG. 4.
CZF1 expression is responsive to carbon source, temperature, physical environment, and stress conditions. (A) Northern blots with RNAs from cells grown in liquid media with various carbon sources and temperatures, as indicated above the lanes. Each lane contained 1μg of mRNA extracted from wild-type (CKY101) cells. Blots were probed with CZF1 and ACT1 probes. Relative CZF1 expression levels were quantified and normalized to ACT1 levels. Values for induction were as follows: 1.0-fold (lane 1), 2.0-fold (lane 2), 5.6-fold (lane 3), and 87.9-fold (lane 4). (B) Northern blot showing expression of CZF1 under various growth conditions. The carbon sources for the conditions used were sucrose (YPS embedded, YPS surface, and YPS liquid), glucose (RPMI serum liquid), and mannitol (Spider liquid). Filamentous growth was induced during growth in YPS agar (embedded), RPMI serum liquid, and Spider liquid. One microgram of mRNA was run in each lane and probed with CZF1 and ACT1 probes. The values of CZF1 expression normalized to ACT1 levels were 1.0-fold (lane 1), 0.4-fold (lane 2), and 0.2-fold (lane 3). Lanes 1 to 3 are from one gel, and lanes 4 and 5 are from a separate gel, so the expression levels in lanes 4 and 5 cannot be compared quantitatively to those in lanes 1 to 3. (C) Northern blot showing cAMP-responsive CZF1 expression. Cells were grown for 10 h with or without dbcAMP, as described in Materials and Methods. Twenty micrograms of total RNA was loaded in each lane, and the blot was probed with CZF1, ACT1, and 18S rRNA probes. (D) Northern blot showing expression of CZF1 in response to stresses. Cells were grown in YPS liquid at 25°C to mid-log phase and then exposed to the indicated stress conditions for 20 min, as described in Materials and Methods. Twenty micrograms of total RNA was loaded in each lane, and the blot was probed with CZF1, ACT1, and 18S rRNA probes. Levels of CZF1 expression were normalized to 18S rRNA levels and yielded the following values: lane 1, 1.0-fold; lane 2, 3.7-fold; lane 3, 1.7-fold; lane 4, 4.4-fold; lane 5, 1.2-fold; lane 6, 2.6-fold; lane 7, 1.5-fold; lane 8, 9.4-fold; lane 9, 0.5-fold; and lane 10, 2.0-fold.
In order to test the effects of embedded growth on CZF1 expression, RNA was prepared from cells grown embedded in agar medium. Since CZF1 expression was very low in YPS liquid at a low temperature (Fig. 4A, lane 1), the embedding experiments were also done with YPS agarose at a low temperature to observe if there was any increase in CZF1 expression specifically during embedded growth. CZF1 expression was detected in cells embedded in YPS agar and grown at 25°C (Fig. 4B, lane 1). The expression level of CZF1 under embedded conditions was 2.5-fold higher than that in cells grown on the surface of YPS agar medium (Fig. 4B, lane 2) and 5-fold higher than that in cells grown in YPS liquid medium (Fig. 4B, lane 3). This observation is consistent with the previous finding that CZF1 regulates morphogenesis during embedded growth. CZF1 expression was also observed when cells were embedded and grown at 37°C in YPD agarose or YPS agarose, at levels that were slightly higher than the levels observed for cells embedded in YPS at 25°C (data not shown).
Cells under embedded conditions formed filaments, in contrast to cells grown on the surface of YPS agar or in YPS liquid medium. To determine whether CZF1 expression was associated with filamentation, cells were grown in RPMI serum (a glucose-containing medium) and Spider medium (a mannitol-containing medium). Cells produced filaments in both media, but RPMI serum-grown cells expressed CZF1 (Fig. 4B, lane 4), while Spider medium-grown cells did not (Fig. 4B, lane 5), demonstrating that CZF1 expression did not correlate with filamentous growth.
As noted above, CZF1 was expressed during the transition between exponential growth and diauxic growth (diauxic shift) (Fig. 3A and B). In S. cerevisiae, a decrease in 3′-5′ cAMP levels is important for the onset of the diauxic shift, and high levels of cAMP in growth media are known to antagonize changes in gene expression that occur at this time (6). To determine whether artificially high levels of cAMP would prevent CZF1 expression, dbcAMP (a membrane-permeative derivative of cAMP) was added to the medium of growing cells. The addition of dbcAMP resulted in a dramatic decrease in CZF1 expression at the diauxic transition (Fig. 4C). Therefore, as with genes expressed at the diauxic shift in S. cerevisiae, cAMP levels regulate the post-exponential-phase expression of CZF1.
To test the effects of stress conditions on CZF1 expression, cells were grown under various stress-inducing conditions, including exposure to 0.3 M NaCl, pH 2.8, 5 mM sorbate, 5 mM H2O2, 7.5% ethanol, 10 mM caffeine, hyperosmotic medium, and hypo-osmotic medium. The expression of CZF1 was analyzed after exposure of the cells to stress for 20 min. Cells subjected to hyperosmotic shock (20 min, 1 M sorbitol to 2 M sorbitol) expressed CZF1 at a high level (ninefold over the control level) (Fig. 4D, compare lane 8 to lane 1). The addition of sodium chloride, sorbate (a weak acid), and ethanol also resulted in some induction of CZF1 (3.7-fold, 4.4-fold, and 2.6-fold increases, respectively; Fig. 4D, lanes 2, 4, and 6).
These data indicate that CZF1 is transcriptionally regulated and that its expression is influenced by a variety of environmental parameters, including the carbon source, temperature, growth phase, osmotic conditions, and physical environment. At high temperatures, CZF1 expression occurred in post-exponential-phase cells growing in glucose-containing liquid medium. At low temperatures, CZF1 was poorly expressed unless the cells were growing within a matrix or under high-osmolarity conditions.
efg1 mutants are defective in CZF1 induction.
Because the sequence upstream of the CZF1 ORF contained putative binding sites for the morphogenesis regulatory protein Efg1p, we hypothesized that Efg1p plays a role in regulating CZF1 gene expression. To test this hypothesis, we analyzed CZF1 expression in an efg1 null mutant grown under conditions that normally lead to CZF1 expression.
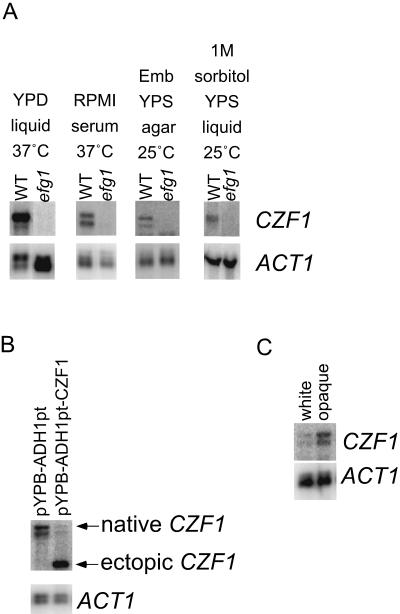
As shown in Fig. 5, CZF1 was expressed in cells grown in medium containing serum, in post-exponential-phase cells grown in YPD at 37°C, in cells grown embedded in agar medium, and in cells subjected to hyperosmotic shock. In contrast, efg1 mutant cells did not express CZF1 under any of these conditions (Fig. 5A). Therefore, Efg1p is required for the expression of CZF1 under all conditions tested.
FIG. 5.
CZF1 expression requires Efg1p and is negatively regulated by Czf1p. (A) Northern blot showing CZF1 expression under various conditions in WT or efg1 null mutant strain. One microgram of mRNA was used per lane (except for samples with 1 M sorbitol, which had 20 μg of total RNA). Blots were probed for CZF1 and ACT1. Emb, embedded. (B) Ectopic expression of CZF1 results in downregulation of the native CZF1 transcript. Strains carrying either empty vector (pYPB-ADH1pt) or vector with CZF1 under the control of the ADH1 promoter (pYPB-ADH1pt-CZF1) were analyzed by Northern blotting. All cells were grown in YPD liquid medium at 37°C for 13 h. One microgram of mRNA was used per lane. Blots were probed for CZF1 and ACT1. The positions of the large native transcript and the smaller CZF1 message transcribed from the ADH1 promoter are indicated. The smaller band seen below the native transcript was often, but not always, observed. Both lanes were adjacent on the same gel. (C) CZF1 expression in white and opaque cells. WO-1 ura3 cells in the white or opaque phase were grown to log phase in YPD+uridine at 25°C and observed microscopically before RNAs were prepared. One microgram of mRNA was loaded in each lane. The blot was hybridized with probes for CZF1 and ACT1.
CZF1 is autoregulated.
To ascertain whether overexpression of CZF1 affects expression from the native CZF1 promoter, a copy of the CZF1 ORF was cloned into plasmid pYPB-ADH1pt (24) so that expression of the gene was placed under the control of the constitutive ADH1 promoter. WT strains carrying this construct were grown in glucose-containing medium at 37°C to post-exponential phase, and RNAs were extracted. Northern blotting of these RNAs and probing with probe D (Fig. 2A) demonstrated the presence of an intense band of about 2.4 kb, corresponding to the transcript produced by the cloned copy of CZF1 (Fig. 5B). A large decrease in expression of the native 4-kb CZF1 transcript accompanied the threefold overexpression of the copy of CZF1 under the control of the ADH1 promoter (Fig. 5B). This result suggested that CZF1 negatively regulates its own expression.
CZF1 transcripts are the same size in opaque and white cells.
There are two transcripts produced from the EFG1 gene, and their abundances differ in different C. albicans cell types. Certain strains of C. albicans switch reversibly between two cell types, termed white and opaque (38). White cells appear to be the typical yeast cells, while opaque cells have a number of unique structural and physiological properties and are capable of mating at a high frequency. White cells preferentially express the 3.2-kb major EFG1 transcript, and opaque cells preferentially express the minor 2.2-kb transcript. Because the TATA box of CZF1 is identical to the TATA box of the EFG1 major transcript, the possibility that CZF1, like EFG1, exhibits differential transcript sizes in the two cell types was analyzed. Northern analysis of RNAs prepared from white and opaque cells grown at 25°C was performed using probes for the CZF1 ORF. As previously reported by others (51), CZF1 expression was higher in opaque cells than in white cells (Fig. 5C). However, the transcripts in both cell types appeared to be the same size (4 kb).
Efg1p and Czf1p bind to the CZF1 promoter.
The lack of CZF1 induction in efg1 mutants and the presence of putative Efg1p-binding sites suggested that Efg1p acts on the CZF1 promoter to regulate the expression of CZF1. The decrease in expression of CZF1 resulting from ectopic expression of CZF1 also suggested that Czf1p binds to its own promoter. To test whether this was the case, ChIP analyses were employed.
ChIP was performed using cell extracts from strains expressing either HA-tagged Efg1p under the control of the PCK1 promoter (expressed at high levels in SCAA medium and at moderate levels in Spider medium) or HA-tagged Czf1p under the control of the MAL2 promoter (expressed in YPS medium). Although CZF1 was not well expressed under these conditions (Fig. 4A and data not shown), growth in these media resulted in synthesis of the HA-tagged proteins, permitting analysis of their DNA-binding activities. As described in Materials and Methods, cell extracts were treated with formaldehyde and sonicated to shear the DNA to an average size of ∼800 bp. Immunoprecipitation was performed using anti-HA antibody, followed by reversal of cross-linking. The recovered DNAs were tested for the presence of sequences upstream of the CZF1 transcription start site (Fig. 6A) by PCR. As a control, strains with wild-type untagged EFG1 and CZF1 were used. Immunoprecipitation of HA-Efg1p using anti-HA antibody yielded enrichment for CZF1 promoter sequences compared to immunoprecipitation from a strain lacking HA-tagged Efg1p (Fig. 6B, fragments a to c, lanes 5 to 8). Enrichment was greater for upstream sequences that contained E boxes. This finding is consistent with the previous demonstration that in the absence of other C. albicans factors, Efg1p binds E-box-containing DNA fragments in vitro (25). The sequence containing the TATA box was only slightly enriched (Fig. 6B, fragment e, lanes 5 and 6). In contrast, neither another CZF1 sequence (Fig. 6A and B, fragment d) nor a control sequence from the ACT1 promoter (Fig. 6B, lanes 5 to 8) was enriched. As a positive control, enrichment of EFG1 promoter sequences after immunoprecipitation of HA-tagged Efg1p was shown, as previously reported (49). These results demonstrated that Efg1p bound the CZF1 promoter in vivo.
HA-tagged Czf1p also bound to CZF1 promoter sequences. Precipitation of HA-tagged Czf1p resulted in enrichment of CZF1 promoter sequences relative to precipitation from a strain lacking HA-tagged Czf1p (Fig. 6C, fragments a and b, lanes 3 and 4). In contrast, a negative control sequence from the ACT1 promoter did not show enrichment in the immunoprecipitated DNA from HA-Czf1p-expressing cells.
To determine whether CZF1 binding required the presence of Efg1p, HA-CZF1 was expressed in a strain lacking Efg1p. As shown in Fig. 6C (lanes 7 and 8), the CZF1 promoter fragment was detected in DNAs immunoprecipitated from extracts of this strain. Therefore, CZF1 was capable of binding to the CZF1 promoter in the absence of Efg1p.
The results of these ChIP experiments demonstrate that Efg1p and Czf1p bind to the CZF1 promoter in vivo. Thus, these proteins regulate the expression of CZF1 by binding to regulatory DNA sequences within the CZF1 promoter, either directly or as part of larger DNA-binding protein complexes.
DISCUSSION
Growth parameters influencing CZF1 expression.
In this report, we demonstrate that CZF1 is transcriptionally regulated in response to the carbon source, temperature, and growth phase. CZF1 was expressed most strongly in post-exponential-phase cells grown in glucose-containing medium at 37°C. This regulation suggests that CZF1 would be expressed by C. albicans cells growing within a human host. Glucose is the primary sugar found in human blood, and therefore, at least during bloodstream infection, glucose is probably the primary carbon source for C. albicans (10). Post-exponential-phase expression of virulence factors has been observed for several bacterial pathogens, suggesting that many microorganisms express host-damaging factors as they exit exponential growth phase within a host (33, 40, 50). CZF1 has also been shown to be transcriptionally upregulated in cells growing under alkaline (pH 8.0) conditions (22). This observation is noteworthy, as the pH of blood and certain organs ranges from neutral to slightly alkaline (11). Therefore, C. albicans growing within a host would be expected to express CZF1. The expression of CZF1 may help the cells to prepare for tissue invasion. When C. albicans encounters a surface and receives a contact-dependent cue, activation of filamentation through the function of Czf1p can readily occur, resulting in invasive growth into host tissue.
CZF1 was shown to be induced to high levels in liquid-grown cells during the diauxic transition, a period of transient growth arrest that occurs when exponentially growing cells exhaust a fermentable carbon source and switch to respiratory (oxidative) metabolism (20). Boy-Marcotte and colleagues proposed that a transient reduction of cAMP must occur after glucose exhaustion in order for post-log-phase genetic programs to be turned on (6). Consistent with this model, the prevention of cAMP reduction by artificially maintaining high levels of cAMP inhibits the expression of CZF1, suggesting that cAMP levels are important in regulating the diauxic transition in C. albicans, as in S. cerevisiae.
Exogenous cAMP may reduce the expression of CZF1 because cAMP promotes the repression activity of Efg1p. Overexpression of two protein kinase A (PKA) isoforms, Tpk1p and Tpk2p, members of the cAMP-PKA pathway, results in increased Efg1-mediated repression of the EFG1 promoter (49). Perhaps a repressive activity of Efg1p on the CZF1 promoter is activated by cAMP. Another possibility is that cAMP has Efg1p-independent effects on gene expression and that the effects of cAMP on CZF1 induction are mediated not through Efg1p, but rather through another, unidentified transcription factor. Indeed, results from DNA microarray studies indicate that the set of genes regulated by Efg1p is generally distinct from those regulated by other members of the cAMP-PKA pathway (19), indicating that cAMP may affect some gene expression through Efg1p-independent mechanisms. Studies by others also suggested that cAMP has a negative effect on filamentous growth under embedded conditions (8), consistent with the observation described in the present study that cAMP downregulates CZF1 expression.
The relevance of induction of CZF1 by hyperosmotic shock is unknown. One possibility is that C. albicans encounters hyperosmotic conditions in certain environmental niches of the host. One such location could be the skin, which is low in moisture and high in salt. Perhaps C. albicans cells encountering such conditions induce CZF1 to promote subsequent tissue invasion, thus bringing the cells to areas with more favorable conditions.
Effects of DNA-binding proteins on CZF1 expression.
The expression of CZF1 under all of the conditions described in this study was dependent on EFG1. Efg1p was shown to bind regions of the CZF1 promoter that contain two consensus E boxes (CATTTG), a type of sequence demonstrated to be bound by Efg1p (25). Binding of Efg1p to the TATA box region of CZF1 was poor. This was surprising, as the TATA box of CZF1 and the sequence immediately around it are identical to the TATA box region of the EFG1 white-phase-specific transcript, a sequence shown to be important for Efg1p binding and repression of its own promoter (49). Therefore, the sequence surrounding the TATA box region appears to influence Efg1p binding.
Under embedded conditions, the switch from yeast to filamentous growth appears to depend on downregulation of active Efg1p. EFG1 transcript levels also decline during serum-induced hyphal growth (46), and continued overexpression of EFG1 interferes with the formation of true hyphae (49). Thus, even under conditions where Efg1p is considered a positive regulator, this protein possesses some negative regulatory activity that must be lowered in order for morphogenesis to occur. Since Czf1p appears to antagonize the function of Efg1p (17), the expression of Czf1p may contribute to lowering the activity of Efg1p and favor the development of filamentous morphology.
Overexpression of CZF1 results in downregulation of the native CZF1 transcript, and ChIP analysis revealed binding of Czf1p to its own promoter within the same region where Efg1p binds. This finding, together with previous yeast two-hybrid results showing an interaction between Czf1p and Efg1p (17) and genetic evidence suggesting that Czf1p acts in opposition to Efg1p function (17), suggests that Czf1p may shut off expression at its own promoter by binding DNA near where Efg1p binds and antagonizing Efg1p-mediated transcription activation at this promoter. Inhibition of CZF1 expression would permit the level of Czf1p to decline so that Efg1p would regain activity. This cycle of CZF1 expression and inhibition may contribute to the regulation of the activity of Efg1p during morphogenesis.
Unusual features of the CZF1 locus and transcript.
The initiation of CZF1 transcription occurs far upstream of the ORF, and the resulting 5′ UTR is unusually large. Equally unusual is the size of the ORF-less region from which transcription of CZF1 is initiated. For C. albicans, the only other gene reported so far to have an unusually large 5′ UTR, EFG1, is located adjacent to a 10-kb ORF-less sequence. EFG1 has two developmentally regulated transcripts, the larger of which has a 1.2-kb 5′ UTR (23, 45). It is intriguing that both of these genes with unusually large intergenic regions are involved in filamentous growth. The results of this study suggest that CZF1 expression is responsive to a variety of growth parameters, and it may be that in order to integrate so many inputs, CZF1 requires a large and complex promoter.
The large 5′ UTR of CZF1 may serve a regulatory role. Developmentally regulated genes in vertebrates often have longer-than-average 5′ UTRs that control the fate of their mRNAs by a number of mechanisms (30). There are two small ORFs within the 5′ UTR that are in the same orientation as the CZF1 ORF. Such small upstream ORFs in 5′ UTRs are capable of regulating gene expression in a number of ways, including modulation of ribosome reinitiation at the main ORF, stalling of the scanning ribosome, and destabilization of mRNA caused by termination at the upstream ORF (53).
In conclusion, we have demonstrated that both Czf1p and Efg1p bind the CZF1 regulatory region and regulate the expression of CZF1. Transcription of CZF1 results in an unusually large message whose expression is subject to regulation in response to various environmental signals. Some of these signals correspond to conditions that would be encountered in the human host. Further investigation may reveal whether Czf1p is also regulated at the posttranslational level as well as shed light on the function of the large 5′ UTR and large upstream intergenic sequence.
Acknowledgments
We thank P. Riggle and members of the Sonenshein lab for assistance with gel shift assays; E. Kong for HA tagging of CZF1; C. Csank, J. Ernst, T. Volkert, and M. Whiteway for plasmids; P. T. Magee for strain WO-1 ura3; D. Dawson and R. Kamienecki for help with ChIP experiments; and A. L. Sonenshein, C. Moore, I. Bruzual, D. Nguyen, and P. Zucchi for discussions and for reading the manuscript.
This work was supported by grant AI038591 (to C.A.K.) from the National Institutes of Health. M.D.V. was the recipient of a Robert D. Watkins Minority Graduate Fellowship from the American Society for Microbiology.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F., R. Brent, R. Kingston, D. Moore, J. Seidman, J. Smith, and K. Struhl. 1989. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 3.Bahn, Y. S., and P. Sundstrom. 2001. CAP1, an adenylate cyclase-associated protein gene, regulates bud-hypha transitions, filamentous growth, and cyclic AMP levels and is required for virulence of Candida albicans. J. Bacteriol. 183:3211-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blandin, G., O. Ozier-Kalogeropoulos, P. Wincker, F. Artiguenave, and B. Dujon. 2000. Genomic exploration of the hemiascomycetous yeasts. 16. Candida tropicalis. FEBS Lett. 487:91-94. [DOI] [PubMed] [Google Scholar]
- 5.Bolotin-Fukuhara, M., C. Toffano-Nioche, F. Artiguenave, G. Duchateau-Nguyen, M. Lemaire, R. Marmeisse, R. Montrocher, C. Robert, M. Termier, P. Wincker, and M. Wesolowski-Louvel. 2000. Genomic exploration of the hemiascomycetous yeasts. 11. Kluyveromyces lactis. FEBS Lett. 487:66-70. [DOI] [PubMed] [Google Scholar]
- 6.Boy-Marcotte, E., D. Tadi, M. Perrot, H. Boucherie, and M. Jacquet. 1996. High cAMP levels antagonize the reprogramming of gene expression that occurs at the diauxic shift in Saccharomyces cerevisiae. Microbiology 142:459-467. [DOI] [PubMed] [Google Scholar]
- 7.Brown, D. H., Jr., A. D. Giusani, X. Chen, and C. A. Kumamoto. 1999. Filamentous growth of Candida albicans in response to physical environmental cues, and its regulation by the unique CZF1 gene. Mol. Microbiol. 34:651-662. [DOI] [PubMed] [Google Scholar]
- 8.Cao, F., S. Lane, P. P. Raniga, Y. Lu, Z. Zhou, K. Ramon, J. Chen, and H. Liu. 2006. The Flo8 transcription factor is essential for hyphal development and virulence in Candida albicans. Mol. Biol. Cell 17:295-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casaregola, S., C. Neuveglise, A. Lepingle, E. Bon, C. Feynerol, F. Artiguenave, P. Wincker, and C. Gaillardin. 2000. Genomic exploration of the hemiascomycetous yeasts. 17. Yarrowia lipolytica. FEBS Lett. 487:95-100. [DOI] [PubMed] [Google Scholar]
- 10.Cullen, P. J., and G. F. Sprague, Jr. 2000. Glucose depletion causes haploid invasive growth in yeast. Proc. Natl. Acad. Sci. USA 97:13619-13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis, D. 2003. Adaptation to environmental pH in Candida albicans and its relation to pathogenesis. Curr. Genet. 44:1-7. [DOI] [PubMed] [Google Scholar]
- 12.Dietrich, F. S., S. Voegeli, S. Brachat, A. Lerch, K. Gates, S. Steiner, C. Mohr, R. Pohlmann, P. Luedi, S. Choi, R. A. Wing, A. Flavier, T. D. Gaffney, and P. Philippsen. 2004. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science 304:304-307. [DOI] [PubMed] [Google Scholar]
- 13.Diezmann, S., C. J. Cox, G. Schonian, R. J. Vilgalys, and T. G. Mitchell. 2004. Phylogeny and evolution of medical species of Candida and related taxa: a multigenic analysis. J. Clin. Microbiol. 42:5624-5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doedt, T., S. Krishnamurthy, D. P. Bockmuhl, B. Tebarth, C. Stempel, C. L. Russell, A. J. Brown, and J. F. Ernst. 2004. APSES proteins regulate morphogenesis and metabolism in Candida albicans. Mol. Biol. Cell 15:3167-3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ernst, J. F. 2000. Transcription factors in Candida albicans—environmental control of morphogenesis. Microbiology 146:1763-1774. [DOI] [PubMed] [Google Scholar]
- 15a.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gasch, A. P., A. M. Moses, D. Y. Chiang, H. B. Fraser, M. Berardini, and M. B. Eisen. 2004. Conservation and evolution of cis-regulatory systems in ascomycete fungi. PLOS Biol. 2:e398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giusani, A. D., M. Vinces, and C. A. Kumamoto. 2002. Invasive filamentous growth of Candida albicans is promoted by Czf1p-dependent relief of Efg1p-mediated repression. Genetics 160:1749-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graber, J. H., G. D. McAllister, and T. F. Smith. 2002. Probabilistic prediction of Saccharomyces cerevisiae mRNA 3′-processing sites. Nucleic Acids Res. 30:1851-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harcus, D., A. Nantel, A. Marcil, T. Rigby, and M. Whiteway. 2004. Transcription profiling of cyclic AMP in Candida albicans. Mol. Biol. Cell 15:4490-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herman, P. K. 2002. Stationary phase in yeast. Curr. Opin. Microbiol. 5:602-607. [DOI] [PubMed] [Google Scholar]
- 21.Kellis, M., B. W. Birren, and E. S. Lander. 2004. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428:617-624. [DOI] [PubMed] [Google Scholar]
- 22.Kitanovic, A., M. Nguyen, G. Vogl, A. Hartmann, J. Gunther, R. Wurzner, W. Kunkel, S. Wolfl, and R. Eck. 2005. Phosphatidylinositol 3-kinase VPS34 of Candida albicans is involved in filamentous growth, secretion of aspartic proteases, and intracellular detoxification. FEMS Yeast Res. 5:431-439. [DOI] [PubMed] [Google Scholar]
- 23.Lachke, S. A., T. Srikantha, and D. R. Soll. 2003. The regulation of EFG1 in white-opaque switching in Candida albicans involves overlapping promoters. Mol. Microbiol. 48:523-536. [DOI] [PubMed] [Google Scholar]
- 24.Leberer, E., D. Harcus, I. Broadbent, K. L. Clark, D. Dignard, K. Ziegelbauer, A. Schmidt, N. A. R. Gow, A. J. P. Brown, and D. Y. Thomas. 1996. Signal transduction through homologs of the Ste20p and Ste7p protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans. Proc. Natl. Acad. Sci. USA 93:13217-13222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leng, P., P. R. Lee, H. Wu, and A. J. Brown. 2001. Efg1, a morphogenetic regulator in Candida albicans, is a sequence-specific DNA binding protein. J. Bacteriol. 183:4090-4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lepingle, A., S. Casaregola, C. Neuveglise, E. Bon, H. Nguyen, F. Artiguenave, P. Wincker, and C. Gaillardin. 2000. Genomic exploration of the hemiascomycetous yeasts. 14. Debaryomyces hansenii var. hansenii. FEBS Lett. 487:82-86. [DOI] [PubMed] [Google Scholar]
- 27.Liu, H., J. Köhler, and G. R. Fink. 1994. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266:1723-1726. [DOI] [PubMed] [Google Scholar]
- 28.Lo, H.-J., J. R. Köhler, B. DiDomenico, D. Loebenberg, A. Cacciapuoti, and G. R. Fink. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939-949. [DOI] [PubMed] [Google Scholar]
- 29.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 30.Mignone, F., C. Gissi, S. Liuni, and G. Pesole. 2002. Untranslated regions of mRNAs. Genome Biol. 3:REVIEWS0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Mitchell, A. P. 1998. Dimorphism and virulence in Candida albicans. Curr. Opin. Microbiol. 1:687-692. [DOI] [PubMed] [Google Scholar]
- 33.Molofsky, A. B., and M. S. Swanson. 2004. Differentiate to thrive: lessons from the Legionella pneumophila life cycle. Mol. Microbiol. 53:29-40. [DOI] [PubMed] [Google Scholar]
- 34.Moran, G., C. Stokes, S. Thewes, B. Hube, D. C. Coleman, and D. Sullivan. 2004. Comparative genomics using Candida albicans DNA microarrays reveals absence and divergence of virulence-associated genes in Candida dubliniensis. Microbiology 150:3363-3382. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 36.Sanyal, K., M. Baum, and J. Carbon. 2004. Centromeric DNA sequences in the pathogenic yeast Candida albicans are all different and unique. Proc. Natl. Acad. Sci. USA 101:11374-11379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seoighe, C., N. Federspiel, T. Jones, N. Hansen, V. Bivolarovic, R. Surzycki, R. Tamse, C. Komp, L. Huizar, R. W. Davis, S. Scherer, E. Tait, D. J. Shaw, D. Harris, L. Murphy, K. Oliver, K. Taylor, M. A. Rajandream, B. G. Barrell, and K. H. Wolfe. 2000. Prevalence of small inversions in yeast gene order evolution. Proc. Natl. Acad. Sci. USA 97:14433-14437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Slutsky, B., M. Staebell, J. Anderson, L. Risen, M. Pfaller, and D. R. Soll. 1987. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J. Bacteriol. 169:189-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sohn, K., C. Urban, H. Brunner, and S. Rupp. 2003. EFG1 is a major regulator of cell wall dynamics in Candida albicans as revealed by DNA microarrays. Mol. Microbiol. 47:89-102. [DOI] [PubMed] [Google Scholar]
- 40.Sonenshein, A. L. 2005. CodY, a global regulator of stationary phase and virulence in gram-positive bacteria. Curr. Opin. Microbiol. 8:203-207. [DOI] [PubMed] [Google Scholar]
- 41.Sonneborn, A., D. P. Bockmuhl, and J. F. Ernst. 1999. Chlamydospore formation in Candida albicans requires the Efg1p morphogenetic regulator. Infect. Immun. 67:5514-5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sonneborn, A., D. P. Bockmuhl, M. Gerads, K. Kurpanek, D. Sanglard, and J. F. Ernst. 2000. Protein kinase A encoded by TPK2 regulates dimorphism of Candida albicans. Mol. Microbiol. 35:386-396. [DOI] [PubMed] [Google Scholar]
- 43.Sonneborn, A., B. Tebarth, and J. F. Ernst. 1999. Control of white-opaque phenotypic switching in Candida albicans by the Efg1p morphogenetic regulator. Infect. Immun. 67:4655-4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Souciet, J., M. Aigle, F. Artiguenave, G. Blandin, M. Bolotin-Fukuhara, E. Bon, P. Brottier, S. Casaregola, J. de Montigny, B. Dujon, P. Durrens, C. Gaillardin, A. Lepingle, B. Llorente, A. Malpertuy, C. Neuveglise, O. Ozier-Kalogeropoulos, S. Potier, W. Saurin, F. Tekaia, C. Toffano-Nioche, M. Wesolowski-Louvel, P. Wincker, and J. Weissenbach. 2000. Genomic exploration of the hemiascomycetous yeasts. 1. A set of yeast species for molecular evolution studies. FEBS Lett. 487:3-12. [DOI] [PubMed] [Google Scholar]
- 45.Srikantha, T., L. K. Tsai, K. Daniels, and D. R. Soll. 2000. EFG1 null mutants of Candida albicans switch but cannot express the complete phenotype of white-phase budding cells. J. Bacteriol. 182:1580-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stoldt, V. R., A. Sonnenborn, C. E. Leuker, and J. F. Ernst. 1997. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 16:1982-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strahl-Bolsinger, S., A. Hecht, K. Luo, and M. Grunstein. 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 11:83-93. [DOI] [PubMed] [Google Scholar]
- 48.Struhl, K. 1987. Promoters, activator proteins, and the mechanism of transcriptional initiation in yeast. Cell 49:295-297. [DOI] [PubMed] [Google Scholar]
- 49.Tebarth, B., T. Doedt, S. Krishnamurthy, M. Weide, F. Monterola, A. Dominguez, and J. F. Ernst. 2003. Adaptation of the Efg1p morphogenetic pathway in Candida albicans by negative autoregulation and PKA-dependent repression of the EFG1 gene. J. Mol. Biol. 329:949-962. [DOI] [PubMed] [Google Scholar]
- 50.Thompson, L. J., D. S. Merrell, B. A. Neilan, H. Mitchell, A. Lee, and S. Falkow. 2003. Gene expression profiling of Helicobacter pylori reveals a growth-phase-dependent switch in virulence gene expression. Infect. Immun. 71:2643-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsong, A. E., M. G. Miller, R. M. Raisner, and A. D. Johnson. 2003. Evolution of a combinatorial transcriptional circuit: a case study in yeasts. Cell 115:389-399. [DOI] [PubMed] [Google Scholar]
- 52.van Helden, J., M. del Olmo, and J. E. Perez-Ortin. 2000. Statistical analysis of yeast genomic downstream sequences reveals putative polyadenylation signals. Nucleic Acids Res. 28:1000-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vilela, C., and J. E. McCarthy. 2003. Regulation of fungal gene expression via short open reading frames in the mRNA 5′ untranslated region. Mol. Microbiol. 49:859-867. [DOI] [PubMed] [Google Scholar]
- 54.Wenzel, R. P. 1995. Nosocomial candidemia: risk factors and attributable mortality. Clin. Infect. Dis. 20:1531-1534. [DOI] [PubMed] [Google Scholar]
- 55.Whiteway, M., D. Dignard, and D. Y. Thomas. 1992. Dominant negative selection of heterologous genes: isolation of Candida albicans genes that interfere with Saccharomyces cerevisiae mating factor-induced cell cycle arrest. Proc. Natl. Acad. Sci. USA 89:9410-9414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wood, V., R. Gwilliam, M. A. Rajandream, M. Lyne, R. Lyne, A. Stewart, J. Sgouros, N. Peat, J. Hayles, S. Baker, D. Basham, S. Bowman, K. Brooks, D. Brown, S. Brown, T. Chillingworth, C. Churcher, M. Collins, R. Connor, A. Cronin, P. Davis, T. Feltwell, A. Fraser, S. Gentles, A. Goble, N. Hamlin, D. Harris, J. Hidalgo, G. Hodgson, S. Holroyd, T. Hornsby, S. Howarth, E. J. Huckle, S. Hunt, K. Jagels, K. James, L. Jones, M. Jones, S. Leather, S. McDonald, J. McLean, P. Mooney, S. Moule, K. Mungall, L. Murphy, D. Niblett, C. Odell, K. Oliver, S. O'Neil, D. Pearson, M. A. Quail, E. Rabbinowitsch, K. Rutherford, S. Rutter, D. Saunders, K. Seeger, S. Sharp, J. Skelton, M. Simmonds, R. Squares, S. Squares, K. Stevens, K. Taylor, R. G. Taylor, A. Tivey, S. Walsh, T. Warren, S. Whitehead, J. Woodward, G. Volckaert, R. Aert, J. Robben, B. Grymonprez, I. Weltjens, E. Vanstreels, M. Rieger, M. Schafer, S. Muller-Auer, C. Gabel, M. Fuchs, A. Dusterhoft, C. Fritzc, E. Holzer, D. Moestl, H. Hilbert, K. Borzym, I. Langer, A. Beck, H. Lehrach, R. Reinhardt, T. M. Pohl, P. Eger, W. Zimmermann, H. Wedler, R. Wambutt, B. Purnelle, A. Goffeau, E. Cadieu, S. Dreano, S. Gloux, et al. 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415:871-880. [DOI] [PubMed] [Google Scholar]
- 57.Zhao, J., L. Hyman, and C. Moore. 1999. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63:405-445. [DOI] [PMC free article] [PubMed] [Google Scholar]