Abstract
This study was designed to evaluate the immunogenicity and the protective efficacy of a divalent fusion DNA vaccine encoding both the Brucella abortus L7/L12 protein (ribosomal protein) and Omp16 protein (outer membrane lipoprotein), designated pcDNA3.1-L7/L12-Omp16. Intramuscular injection of this divalent DNA vaccine into BALB/c mice elicited markedly both humoral and cellular immune responses. The specific antibodies exhibited a dominance of immunoglobulin G2a (IgG2a) over IgG1. In addition, the dual-gene DNA vaccine elicited a strong T-cell proliferative response and induced a large amount of gamma interferon-producing T cells upon restimulation in vitro with recombinant fusion protein L7/L12-Omp16, suggesting the induction of a typical T-helper-1-dominated immune response in vivo. This divalent DNA vaccine could also induce a significant level of protection against challenge with the virulent strain B. abortus 544 in BALB/c mice. Furthermore, the protection level induced by the divalent DNA vaccine was significantly higher than that induced by the univalent DNA vaccines pcDNA3.1-L7/L12 or pcDNA3.1-Omp16. Taken together, the results of this study verify for the first time that the Omp16 gene can be a candidate target for a DNA vaccine against brucellosis. Additionally, a divalent genetic vaccine based on the L7/L12 and Omp16 genes can elicit a stronger cellular immune response and better immunoprotection than the relevant univalent vaccines can.
Brucella abortus is a facultative intracellular pathogen and one of the etiological agents of brucellosis that can infect humans and domestic animals (11). Like other intracellular bacterial pathogens, the host resistance to B. abortus depends mainly on acquired cell-mediated immunity (CMI) (40). The development of a Th1 subset of CD4+ lymphocytes secreting gamma interferon (IFN-γ), a crucial cytokine that can up-regulate the anti-Brucella activity of macrophages (14), and the development of CD8+ T lymphocytes secreting IFN-γ and lysing Brucella-infected cells (24) are the two main components of the protective response of the infected hosts. Live attenuated vaccines that can stimulate strong CMI responses are usually very effective against brucellosis. Attenuated strains such as Brucella melitensis Rev1 and B. abortus S19 and RB51 are being used to control brucellosis in domestic animals (21). However, no safe, effective vaccine is available for human use. The vaccine strains used for animals are considered too virulent; thus, they are not safe for human use. A vaccine that will be noninfectious to humans but effective in stimulating a broad protective immune response is needed to control brucellosis. To develop this type of Brucella vaccine, several research groups are pursuing different strategies, including development of subunit vaccines (25), utilization of bacterial vectors (28), and overexpression of protective homologous antigen (38).
Another new strategy for developing safe and efficacious vaccines is immunization with plasmid DNA encoding the protective antigen. The DNA vaccines seem to offer the best approach to activate both cellular components of the immune response (Th1 and CD8+ T cell), owing to the intrinsic feature of DNA vaccine to produce endogenous antigen in professional antigen-presenting cells (20). Furthermore, DNA vaccines also confer other advantages, such as posing no risk of infection, induction of a long-lived immune response, better stability than live attenuated vaccines, easy preparation, and low cost. Accordingly, it is reasonable to use a DNA vaccine to protect the host from infection of intracellular pathogens. Indeed, plasmid DNA vaccination has been validated to protect the host from many intracellular pathogen infections, such as viruses and parasites (5, 13, 39). As B. abortus is an intracellular pathogen, DNA vaccination should be a good countermeasure to protect the host from its infection. Actually, extensive research on a B. abortus DNA vaccine has been performed using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (32), bacterioferritin (1), P39 (1), GroEL heat shock gene (17), ribosome recycling factor-homologous protein (CP24) (4), superoxide dismutase (22, 27), and others. These vehicles illustrate that DNA vaccination should provide a good countermeasure to protect the host from B. abortus infection.
The B. abortus L7/L12 ribosomal protein has been identified as an immunodominant antigen from this pathogen (23). The recombinant L7/L12 protein and plasmid encoding the L7/L12 gene have demonstrated that they can elicit strong CMI and engender protection from Brucella infection in mice; however, the protective effect is much lower than what the live attenuated B. abortus vaccine S19 provides (16, 25, 26). Researchers have also observed the protective role of other types of L7/L12 based-vaccines utilizing different vectors such as vaccinia virus and Lactococcus lactis; nevertheless, the protection level is far less than that of the currently used live B. abortus vaccine (2, 30, 31). These results suggest that vaccines based on L7/L12 alone could not induce enough protection, regardless of the type of L7/L12 vaccine used.
However, other evidence shows that polyvalent vaccines, including protein vaccines and DNA vaccines, can engender more effective protection than univalent vaccines in some cases (15, 33). Thus, polyvalent vaccines combining L7/L12 with other immunogenic antigen(s) of Brucella will be a strategy to offer higher protection levels for Brucella infection.
Omp16, a 16.5-kDa Brucella outer membrane protein, is a lipoprotein, and it is expressed in all six species and known biovars of Brucella (34, 35). It has been confirmed to be one of the key mediators of the proinflammatory response elicited by heat-killed B. abortus (10), and the monoclonal antibody against Omp16 can protect mice against a Brucella ovis challenge (3), which indicates the important biological role of Omp16 in Brucella life and the immunogenicity of Omp16 (3, 10). Therefore, vaccines based upon Omp16 probably can elicit a cellular immune response and provide the host some protection from Brucella infection.
Thus, in this study, we constructed DNA vaccine pcDNA3.1-Omp16 to study the potentiality of an Omp16-based DNA vaccine in protection against Brucella infection. We also constructed a divalent fusion DNA vaccine containing both the L7/L12 and Omp16 genes (i.e., pcDNA3.1-L7/L12-Omp16) and compared the protective effects between the polyvalent DNA vaccine, the univalent vaccines (i.e., pcDNA3.1-L7/L12 or pcDNA3.1-Omp16), and the divalent protein vaccine (recombinant L7/L12-Omp16) in a mouse model to develop a more efficacious DNA vaccine against brucellosis.
MATERIALS AND METHODS
Animals and bacterial strains.
Female BALB/c mice (5 to 6 weeks old; obtained from the Animal Center of Institute of Microbiology and Epidemiology, Academy of Military Medical Science, Beijing, China) were randomly distributed into experimental groups. The mice were kept in conventional animal facilities, where they received water and food ad libitum. Approval for animal experiments was obtained from the institutional animal welfare committee.
Virulent Brucella strain B. abortus 544 and attenuated strain B. abortus RB51, on the basis of which vaccines have been developed (21, 36, 37), were obtained from the Biology Research Institute (Lanzhou, China). The bacterial cells were grown under aerobic conditions in tryptose-soy broth at 37°C. For inoculation, the bacterial suspensions were adjusted to an optical density at 600 nm (OD600) of 1, corresponding to 104 CFU of B. abortus 544 and 2 × 108 CFU of B. abortus RB51. All experiments with live Brucella were performed in biosafety level 2 facilities. Escherichia coli strain DH5α was used to prepare the plasmid constructs. The E. coli cultures were routinely grown at 37°C in Luria-Bertani broth or agar and were supplemented, when required, with 100 μg/ml ampicillin.
Construction and preparation of L7/L12-Omp16 DNA vaccine.
Full-length open reading frames of the L7/L12 gene and Omp16 gene were amplified with PCR from the genome of attenuated B. abortus strain RB51. The PCR primers were designed as shown in Table 1. The gene amplified with L7/L12 primers (FL and RL-1) and the gene amplified with Omp16 primers (FO and RO) were inserted into pcDNA3.1(+) vector (Invitrogen) at the EcoRV/XhoI and BamHI/XhoI sites to construct recombinant plasmids L7/L12-pcDNA3.1 and Omp16-pcDNA3.1, respectively. To construct the recombinant fusion plasmid L7/L12-Omp16-pcDNA3.1, the L7/L16 gene fragment was amplified with the L7/L16 PCR primers (FL and RL-2) first, which removed only the TAA stop codon from the L7/L16 gene. This PCR product and the amplified Omp16 gene above were digested with EcoRV/BamHI and BamHI/XhoI, respectively, and ligated with T4 ligase; the ligated product was then inserted into the pcDNA3.1(+) vector between the EcoRV and XhoI sites. This process kept the two genes together in one open reading frame.
TABLE 1.
PCR primers for DNA vacine construction
| Gene and primers | Primer sequencea | Restriction enzyme |
|---|---|---|
| L7/L12 | ||
| FL | 5′-GAGATATCATGGCTGATCTCG-3′ | EcoRV |
| RL-1 | 5′-GCCCTCGAGTTACTTGAGTTC-3′ | XhoI |
| RL-2 | 5′-TAGGATCCCTTGAGTTCAAC-3′ | BamHI |
| OMP16 | ||
| FO | 5′-TCGGATCCATGCGCCGTATCCAGTC-3′ | BamHI |
| RO | 5′-TATCTCGAGTTACCGTCCGGCCCCGTTG-3′ | XhoI |
Restriction sites are underlined, and initiation and termination codons are shown in boldface.
Large-scale plasmid DNA isolation was performed by using an EndoFree Plasmid Giga Kit (Sigma) according to the manufacturer's instructions. The plasmid DNA was finally resolved in phosphate-buffered saline (PBS; pH 7.0) at a concentration of 1 mg/ml. The expression of the correct proteins of the recombinant plasmids was verified by immunocytochemistry and Western blot assays.
Purification of rL7/L12-Omp16 fusion protein.
To prepare the recombinant L7/L12-Omp16 (rL7/L12-Omp16) fusion protein, the fusion gene was excised from the above plasmid L7/L12-Omp16-pcDNA3.1 by EcoRV/XhoI digestion and inserted into the same restriction sites of prokaryotic expression vector pET32a(+) (Novagen). The recombinant plasmids were then transformed into E. coli BL21(DE3) (Novagen), and the positive clones were selected. The recombinant protein was expressed in successfully transformed bacteria by induction with isopropyl-β-d-thiogalactopyranoside (IPTG) in Luria-Bertani medium and then purified with an Ni2+-HiTrap chelating 5-ml prepacked column (Amersham Pharmacia Biotech) by using imidazole as the elution reagent, according to the manufacturer's protocol. The lysates of transformed cells and the purified protein were identified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot assays. The purified protein was then stored at −70°C until use for enzyme-linked immunosorbent assay (ELISA) or for in vitro stimulation of splenocytes.
SDS-PAGE and Western blot assays.
The lysates of COS-7 cells transformed with pcDNA3.1-L7/L12-Omp16 or BL21(DE3) bacteria transformed with pET32a(+)-L7/L12-Omp16 were migrated in SDS-PAGE gels, and the protein bands were stained with Coomassie brilliant blue. For Western blot assays, the protein bands in the gels were electrotransferred to a nitrocellulose filter, and this step was followed by the antigen-antibody reactions. The rabbit anti-B. abortus RB51 hyperimmune sera were used as the detecting antibody, and horseradish peroxidase-goat anti-rabbit immunoglobulin G (IgG) served as the secondary antibody. Following the addition of diaminobenzidine substrate, the antibody-specific protein band was revealed.
Immunization.
The mice were anesthetized with methoxyflurane (Metofan; Mallinckrodt) and inoculated intramuscularly with 100 μg of pcDNA3.1-L7/L12-Omp16, pcDNA3.1-L7/L12, or pcDNA3.1-Omp16 in 100 μl of PBS (50 μl of the solution was injected into each tibialis anterior muscle). The control mice were infected with PBS or the expression vector alone (pcDNA3.1). Each mouse in another group was injected with 10 μg of rL7/L12-Omp16 in 100 μl PBS according to the same schedule. Each mouse was injected on weeks 0, 2, and 4. The mice used as positive controls were inoculated intraperitoneally on day 0 with 2 × 108 CFU of B. abortus strain RB51 in 0.2 ml of PBS. Two weeks after the last immunization, sera were obtained, and spleens were removed from the experimental mice for antibody detection, lymphocyte proliferation assays, and cytokine detection.
ELISA.
The presence of serum IgG and of the subtypes (IgG1 and IgG2a) specific to rL7/L12-Omp16 was determined by indirect ELISA on the 14th day after the final immunization. The purified rL7/L12-Omp16 was diluted to 3 μg/ml in carbonate buffer (pH 9.6) and used to coat the wells of a polystyrene plate (100 μl/well; Nunc-Immuno plate with MaxiSorp surface). After overnight incubation at 4°C, the plates were washed, blocked, and then incubated with serially diluted sera for 3 h at room temperature. Following another washing, IgG or isotype-specific rabbit anti-mouse horseradish peroxidase conjugates were added (50 μl/well) at the appropriate dilutions. After 30 min of incubation at room temperature, the plates were washed, and 100 μl of substrate solution (200 μmol of o-phenylenediamine and 0.04% H2O2) was added to each well. The enzymatic reaction was allowed to proceed for 20 min at room temperature, after which the reaction was stopped with the addition of 50 μl of 2 M sulfuric acid/well. The absorbance of the developed color was measured at 450 nm (A450). The cutoff value for the assay was calculated as the mean specific OD plus standard deviation (SD) for 10 serum samples, assayed at a dilution of 1:40, from nonimmunized mice. The titer of each serum was calculated as the reciprocal of the highest serum dilution yielding a specific OD higher than the cutoff value. All assays were performed in triplicate and repeated three times.
Splenocyte culture and lymphocyte proliferation.
Two weeks after the last immunization, mice were sacrificed, and their spleens were removed under aseptic conditions. Single-cell suspensions were prepared from the spleens, and the red blood cells were lysed with ACK (150 mM NH4Cl, 1 mM KHCO3, 0.1 mM Na2 · EDTA, pH 7.3) solution. Splenocytes were cultured at 37°C in 5% CO2 in a 96-well flat-bottom plate at a concentration of 4 × 105 cells/well in RPMI 1640 medium supplemented with 2 mM l-glutamine and 10% heat-inactivated fetal calf serum (Sigma), in the presence of 0.08 μg of purified rL7/L12-Omp16 protein or no additives (unstimulated control). The cells were cultured for 3 days and pulsed for 8 h with 0.4 μCi of [3H]thymidine (50 Ci/mmol; Amersham) per well. The radioactivity incorporated into the DNA was measured in a liquid scintillation counter. Cell proliferation was expressed as mean counts per minute (cpm) from five mice for each group. All assays were performed in triplicate and repeated three times.
ELISPOT assay.
To measure the amount of cytokine-producing T cells, after 48 h of stimulation in vitro with rL7/L12-Omp16 protein, spleen cells were collected and tested for the presence of IFN-γ by antigen-capture enzyme-linked immunospot (ELISPOT) assay using MultiScreen-IP Sterile Plate γ (DIACLONE Biosciences). All assays were performed in triplicate, and the experiments were repeated three times. The spot number corresponding to the IFN-γ-producing cells was calculated on an ELISPOT spot counter (Biorader 4000; Biosys, Germany).
Protective experiments.
Two weeks after the final vaccination, five mice from each group were challenged intraperitoneally according to published methods (36, 37), but a relatively higher dose of strain 544 (5 × 105 CFU) was used here. Four weeks postchallenge, the mice were killed by cervical dislocation, and their spleens were removed aseptically and weighed. Each spleen was homogenized in sterile PBS, serially diluted 10-fold, and plated in triplicate on tryptic-soy agar. B. abortus 544 colonies were counted after 3 days of incubation at 37°C with 10% CO2. The results were represented as the mean log CFU ± SD per group. This experiment was repeated three times. Statistical analyses were performed with a Student's paired t test. Log10 units of protection were calculated as the mean log10 numbers of CFU of the negative control group (PBS) minus the mean log10 numbers of CFU of the experimental group.
Statistical analysis of the data.
The CFU data were normalized by transformation and evaluated by one-way analysis of variance, followed by Dennett's post hoc test (InStat; GraphPad, San Diego, Calif.). The Kruskal-Wallis test and one-way analysis of variance were used to analyze antibody responses and cellular responses, respectively.
RESULTS
Construction and expression of pcDNA3.1-L7/L12-Omp16 DNA vaccine.
To compare the relative roles of the DNA vaccine pcDNA3.1-L7/L12-Omp16 and its univalent form (pcDNA3.1-L7/L12 and pcDNA3.1-Omp16) in inducing an immune response and protective immunity against brucellosis, we constructed these DNA vaccines. The recombinant plasmids were verified with restriction digestions and sequencing. To verify that the constructed DNA vaccines can be expressed in mammalian cells correctly, we transformed the recombinant plasmids into COS-7 cells. When the cells were examined with Western blot assays, a 14-kDa protein and a 19-kDa protein, corresponding to the molecular masses of L7/L12 and Omp16, respectively, could be detected in the lysate of the transformed COS-7 cells (Fig. 1). When transformed with the recombinant plasmid pcDNA3.1-L7/L12-Omp16, COS-7 cells could also correctly express the fusion protein L7/L12-Omp16 with a molecular mass of 33 kDa, corresponding to that of the fusion protein (Fig. 1).
FIG. 1.
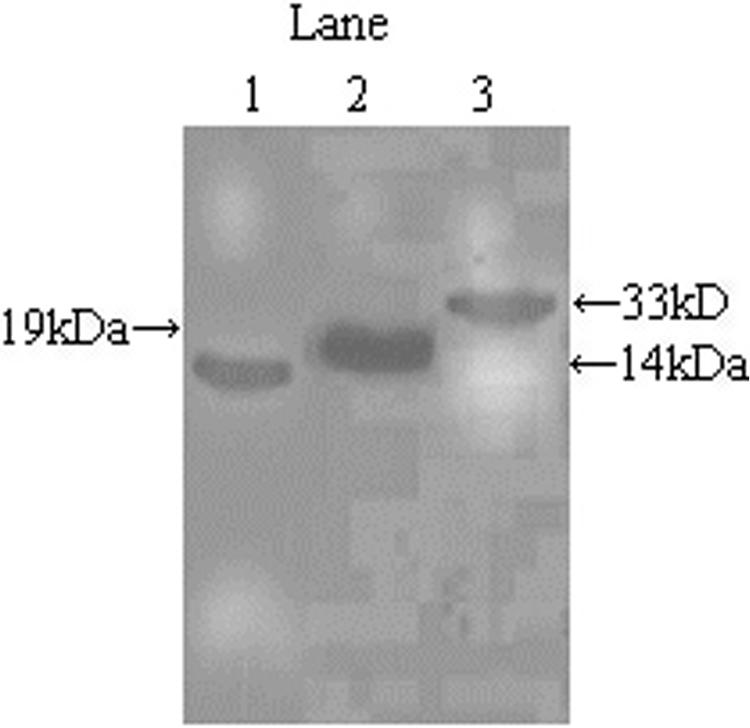
Expression of recombinant plasmid pcDNA3.1-L7/L12-Omp16. The lysates of COS-7 cells transformed with the different recombinant plasmids were analyzed for the respective target protein expression by Western blot assays. COS-7 cells were transformed with pcDNA3.1-L7/L12 (lane 1), pcDNA3.1-Omp16 (lane 2), or pcDNA3.1-L7/L12-Omp16 (lane 1). Molecular masses of the target proteins are indicated beside the figure.
Expression and purification of recombinant fusion protein L7/L12-Omp16.
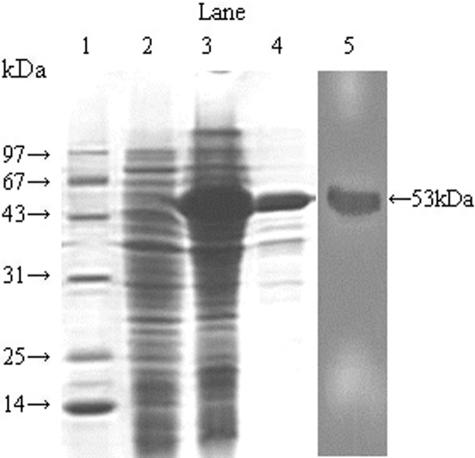
To compare the immunological effects of the divalent and univalent DNA vaccines with the effects of the divalent protein vaccine targeting the L7/L12 and/or Omp16 gene and to prepare the substrate required for ELISAs or in vitro splenocyte stimulation in the ELISPOT and the lymphocyte proliferation assays, we constructed the recombinant plasmid pET32a(+)-L7/L12-Omp16 and transformed it into E. coli BL21(DE3). Following the induction of target protein expression with IPTG, the His6-labeled target protein was purified with an Ni2+-chelating column and identified for further specificity. The lysates of transformed or nontransformed BL21(DE3) cells as well as the purified target protein migrated in the SDS-PAGE gel, and the staining results demonstrated that the L7/L12-Omp16 fusion protein could be expressed in BL21(DE3) cells correctly (Fig. 2). Additionally, the fusion protein could be purified specifically and efficiently because the purified protein had the expected molecular mass (Fig. 2). As the vector pET32a(+) itself had three tags with a molecular mass of 20 kDa, the molecular mass of the multiple fusion protein was about 53 kDa.
FIG. 2.
Expression and purification of recombinant protein L7/L12-Omp16. The expression of recombinant fusion protein L7/L12-Omp16 was analyzed by SDS-PAGE and Western blot assays. Lane 1, molecular size protein markers; lane 2, lysate of E. coli BL21(DE3) transformed with the vector pET32a(+); lane 3, lysate of E. coli BL21(DE3) transformed with pET32a(+)-L7/L12-Omp16; lane 4, the purified fusion protein; lane 5, immunoblotting assay for purified protein (molecular mass indicated at right).
Humoral immune response elicited by pcDNA3.1-L7/L12-Omp16 immunization.
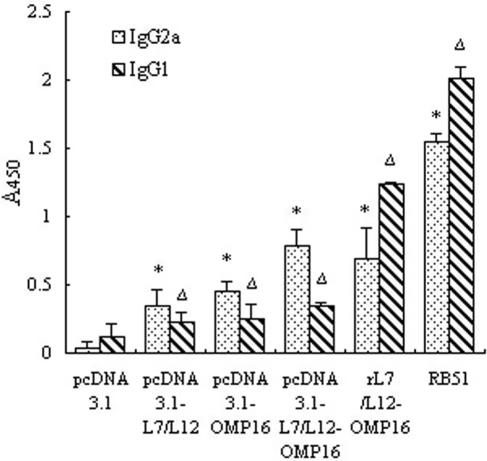
Sera collected 2 weeks after the last immunization were assayed for the presence of L7/L12- and/or Omp16-specific antibodies by ELISA. The results showed that the total IgG titer of the hyperimmune sera from mice immunized with pcDNA3.1-L7/L12-Omp16, pcDNA3.1-L7/L12, or pcDNA3.1-Omp16 reached 1:1,000, 1:800, or 1:600, respectively; immunization with rL7/L12 and live RB51 strain elicited much higher humoral immune responses in mice, with IgG titers reaching 1:25,600 and 1:50,000, respectively. To analyze the potential roles of IgG subtypes in the mechanism of preventing Brucella infection, we examined the proportion of Th1-associated IgG1 and Th2-associated IgG2a in the total serum IgG of each group with ELISAs by diluting the sera at 1:100. The analysis of IgG subtypes showed a significant increase in IgG1 and IgG2a from the DNA vaccine group, fusion protein vaccine group, and live RB51 group compared with the pcDNA3.1 vector control (P < 0.01) (Fig. 3). However, the ratio of the Th1-associated versus Th2-associated IgG subtype may reflect a substantial increase in certain IgG subtypes (12, 18, 29); thus, the IgG2a/IgG1 value for each group was calculated. Results showed that the ratios were 0.56 and 0.77 after rL7/L12-OMP16 and RB51 immunization, although the two types of antigen had higher IgG1 and/or IgG2a levels than the DNA vaccines. However, after the pcDNA3.1-L7/L12, pcDNA3.1-OMP16, and pcDNA3.1-L7/L12-OMP16 immunizations, the ratios were 1.5, 1.8, and 2.2, respectively, which suggested that the DNA vaccines had a more dominant Th1 response than other groups.
FIG. 3.
Antibody subtype profiles of mice immunized with various vaccines. Mice (five per group) were inoculated intramuscularly with various DNA vaccines, protein vaccine, and live Brucella strain RB51. Mice that received a pcDNA3.1 injection were negative controls. Two weeks after the last immunization, sera were collected from the experimental mice, and antibody titers were evaluated by ELISA. The results are represented as mean A450. Data are from three independent experiments. Each bar represents the mean A450 value of antibodies in the same group. * and ▵, P < 0.01, compared with the amount of IgG2a and IgG1, respectively, raised in the pcDNA 3.1-immunized group.
Th1 cellular immune response induced by pcDNA3.1-L7/L12-Omp16.
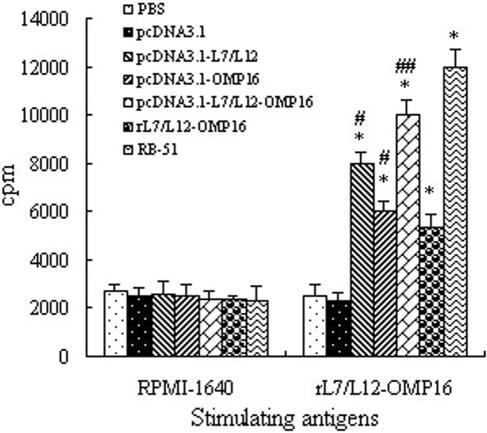
To further investigate the CMI response induced by the various DNA vaccines, we analyzed the proliferative T-cell response. As shown in Fig. 4, all three genetic vaccines (pcDNA3.1-L7/L12, pcDNA3.1-Omp-15, and pcDNA3.1-L7/L12-Omp16) induced significant and specific T-cell proliferation in immunized BALB/c mice in response to recombinant protein rL7/L12-Omp16, compared with PBS or pcDNA3.1 vector immunization (P < 0.01). Though the recombinant protein rL7/L12-Omp16 could prime a specific T-cell proliferative response significantly, this effect was much lower than that in the pcDNA3.1-L7/L12-Omp16 and pcDNA3.1-L7/L12 groups (P < 0.05) (Fig. 4). Among the three DNA vaccines, pcDNA3.1-L7/L12-Omp16 showed the strongest stimulant effect (P < 0.05); however, this effect was lower than that of the RB51 strain (P < 0.05) (Fig. 4). As a stimulus control, none of these vaccines, including DNA, protein, and RB51 vaccines, affected specific T-cell proliferation in response to RPMI 1640 medium.
FIG. 4.
Lymphocyte proliferation assay. BALB/c mice were immunized with DNA vaccines (pcDNA3.1-L7/L12, pcDNA3.1-Omp16, or pcDNA3.1-L7/L12-Omp16), rL7/L12-Omp16, or strain RB51, with PBS and expressing vector pcDNA3.1 as negative immunization controls. The T-cell proliferation response was measured 2 weeks after the last immunization. Splenocytes from each group were prepared from 4 × 105 cells per well in a 96-well flat-bottom plate and stimulated in vitro with purified rL7/L12-Omp16 (0.8 μg/ml) or the RPMI 1640 medium (control) as antigens. Each sample was assayed in quadruplicate wells. Data represent the mean cpm ± SD from five each group of five mice. *, P < 0.01, compared with PBS group; for ## versus #, P < 0.05.
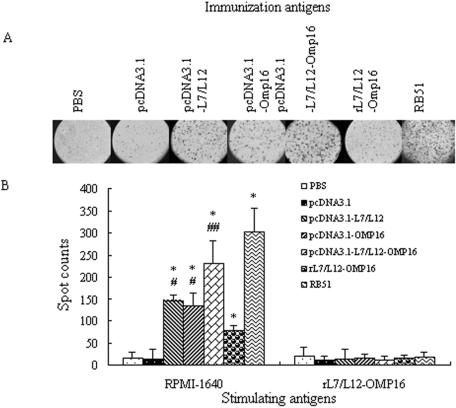
Two weeks after the last immunization, cytokine-producing T-cell profiles from splenocytes of five vaccinated mice per group were examined in in vitro stimulation with rL7/L12-Omp16 or with medium control (RPMI 1640). The spot number in each well was calculated on an ELISPOT spot counter. The typical results from the splenocytes of immunized mice are shown in Fig. 5A. After stimulation with rL7/L12-Omp16 in vitro, the number of spots in each vaccine group in this study was significantly more than that of the PBS or pcDNA3.1 group (P < 0.01) (Fig. 5B). However, the spot number in the rL7/L12 group was much lower than spot numbers in DNA vaccine groups (P < 0.05 versus the univalent DNA vaccines or P < 0.01 versus the divalent DNA vaccine) (Fig. 5B). In particular, although the number of spots in the pcDNA3.1-L7/L12-Omp16 group was lower than in the RB51 strain group (P < 0.05), it was much higher than in either the pcDNA3.1-L7/L12 group or the pcDNA3.1-Omp16 group (P < 0.01) (Fig. 5B). Nevertheless, the expressing vector pcDNA3.1 could not elicit more IFN-γ-producing cells than the PBS control did. In particular, as a stimulus control, splenocytes from each group could not produce IFN-γ while stimulated in vitro with RPMI 1640 medium (Fig. 5B).
FIG. 5.
Quantitative ELISPOT analysis of IFN-γ-producing lymphocytes upon in vitro stimulation with different antigens. Spleen cells (4 × 106 cells/ml) from mice inoculated with DNA vaccines, protein vaccines, live RB51 strain, or pcDNA3.1 were stimulated in vitro with rL7/L12-Omp16 or RPMI 1640 medium. (A) The typical ELISPOT assay results from the groups stimulated in vitro with rL7/L12-Omp16. (B) Statistics for data from each group. Each sample was assayed in quadruplicate wells. Data represent the mean spot number ±SD from each group of five mice. *, P < 0.01 compared with PBS immunization control group; ## versus #, P < 0.01.
Efficacy of pcDNA3.1-L7/L12-Omp16 immunization in generating protective immunity against B. abortus 544.
Two weeks after the last immunization, the vaccinated mice were challenged with intraperitoneal injection of virulent strain 544. Four weeks after the challenge, the level of infection in each mouse was evaluated by determining the CFU in the spleen. Data from three independent protection experiments demonstrated that immunization with any type of DNA vaccine resulted in a significantly higher degree of protection (1.25- to 2.05-log increase in protection) than the controls that received PBS (P < 0.01) (Table 2). Among the three recombinant DNA vaccines, the divalent DNA vaccine provided a higher protection level than either of the univalent DNA vaccines (P < 0.05) (Table 2). Immunization with the recombinant fusion protein rL7/L12 also created significant protection; nevertheless, this protective effect was significantly lower than that created by the immunization with the DNA vaccines (P < 0.05, versus pcDNA3.1-Omp16; P < 0.01, versus pcDNA3.1-L7/L12 and pcDNA3.1-L7/L12-Omp16). To compare the extent to which mice could be protected, we included the live B. abortus strain RB51 group for immunization, and it induced 2.25-log protection compared with the PBS group (P < 0.01). No reduction in the number of CFU was observed in animals injected with pcDNA3.1 compared to the PBS group (Table 2). These results indicated that pcDNA-L7/L12-Omp16 could confer a significant degree of protection against Brucella infection that was close to the level created by live B. abortus RB51.
TABLE 2.
Protection of mice against challenge with B. abortus 544 after immunization with various vaccines
| Vaccine | Log CFU/spleen (mean ± SD) | Log protectiona | Significance (P) |
|---|---|---|---|
| PBS | 4.35 ± 0.35 | 0.00 | |
| pcDNA3.1 | 4.40 ± 0.30 | 0.05 | >0.05 |
| pcDNA3.1-Omp16 | 3.22 ± 0.55 | 1.25* | <0.01 |
| pcDNA3.1-L7/L12 | 3.15 ± 0.60 | 1.59* | <0.01 |
| pcDNA3.1-Omp16-L7/L12 | 2.34 ± 0.57 | 2.05** | <0.01 |
| rL7/L12-Omp16 | 3.59 ± 0.35 | 0.81 | <0.05 |
| RB51 | 2.21 ± 0.13 | 2.25 | <0.01 |
** versus *, P < 0.05.
DISCUSSION
To develop a new generation of vaccines to prevent brucellosis, it is vital to overcome the three main drawbacks of the currently used live Brucella vaccines, i.e., causing abortion in pregnant animals, pathogenicity for humans, and inducing antibodies that interfere with the diagnosis of field infection in vaccinated animals (6, 7, 21). Improvements in the methods for gene cloning and protein purification have led to the use of purified recombinant proteins as cellular vaccines in experimental trials. These preparations as well as synthetic peptides are more convenient to use than attenuated vaccines, but they do not confer a high degree of protection or induce a strong CMI response compared with that induced by live Brucella vaccines (25).
Immunization with plasmid DNA coding for the immunogenic antigen represents a novel and promising method in vaccine research and development. A number of studies have demonstrated that after naked DNA immunization, the antigen is naturally processed and presented to T cells in the context of major histocompatibility complex class I and class II molecules, inducing a broad range of immune responses including antibody production and the activation of CD8+ cytotoxic T cells and CD4+ T helper cells (8, 9). However, immunization with an L7/L12-based DNA vaccine cannot elicit a strong enough cellular immune response to provide sufficient protection against Brucella infection compared with a live Brucella vaccine (16). Furthermore, other types of L7/L12-based vaccines cannot evoke a protective effect close to the level offered by the currently used live Brucella vaccines (2, 30, 31). Considering that polyvalent vaccines can induce a more intensive immune response than the univalent vaccine in some cases (15, 33), we constructed a divalent DNA vaccine (pcDNA3.1-L7/L12-Omp16) that combined the L7/L12 gene and another immunogenic B. abortus gene, Omp16, to investigate the efficacy of the divalent vaccine in inducing a cellular immune response.
Immunization with the divalent DNA vaccine (pcDNA3.1-L7/L12-Omp16) and its univalent forms (pcDNA3.1-L7/L12 and pcDNA3.1-Omp16) could induce remarkable titers of total IgG (1:1,000, 1:800, and 1:600, respectively); however, the titers were less than those induced by the recombinant fusion protein L7/L12-Omp16 and live RB51 strain (1:25,600 and 1:50,000, respectively). However, the IgG subtype assay demonstrated that the ratio of IgG2a/IgG1 in DNA vaccine groups was much higher than that in the protein or live RB51 immunization groups, suggesting that DNA vaccines elicited a stronger Th1-type cellular immune response.
Although the lymphocyte proliferation assays demonstrated that all vaccine types in this study could induce a significant T-cell response, they cannot reflect which subtypes of T cells were primed, because T cells include CD4+ and CD8+ subtypes that consist of T-helper cells (including Th1 and Th2 subtypes) and cytotoxic T cells. Th1 CD4+ T cells will activate CD8+ T cells and prime the cellar immune response, whereas Th2 CD4+ T cells will activate a humoral immune response. To investigate which subtype of T cells is predominant in mice immunized with a protein or DNA vaccine, we examined the amount of IFN-γ-producing T cells by ELISPOT assay. The results showed that the DNA vaccines could produce markedly more IFN-γ-producing T cells than protein vaccines could, thereby indicating that a DNA vaccine can induce a predominantly Th1-type T-cell immune response because IFN-γ is mainly secreted by Th1 and CD8+ T cells. The predominant Th1-type immune response in the DNA vaccine groups was validated by further protection experiments. In these experiments, DNA vaccines provided better protection against Brucella infection than the protein vaccine. Brucella is one kind of intracellular pathogen, and its clearance needs a strong cellular immune response, especially the production of IFN-γ that will activate macrophages to kill the pathogen. Thus, the results from the protection experiments confirmed the results from IFN-γ ELISPOT assay. The results indicate the importance of Th1-type cellular immune response in the clearance of B. abortus from the infected host, which has been observed by other researchers (24).
In this study, it was verified for the first time that a DNA vaccine based on the Omp16 gene induces significant humoral immunity and a Th1-type cellular immune response. The vaccine also conferred on the host remarkable protection against Brucella infection, indicating that the Omp16 gene can be used as a relatively efficacious target for genetic immunization. All of the DNA vaccines seemed to promote a better cellular immune response than protein vaccines in this study, even if the protein antigen was the fusion form of the L7/L12 and Omp16 proteins; this result may reflect the different processing and presenting mechanisms for endogenous antigen (e.g., DNA vaccine) and exogenous antigen (e.g., protein). Among the three DNA vaccines, the divalent DNA vaccine elicited the strongest Th1-type cellular response and provided the greatest protection against Brucella infection, which may indicate that more T-cell epitopes are present in the divalent DNA vaccine than in the univalent DNA vaccine. Thus, the former can promote a broader spectrum of T-cell clones than the latter, causing a difference in the levels of protection they offer from Brucella infection.
The RB51 strain conferred the best protection against Brucella infection of any groups in this study. The reason could be that RB51 can infect the host cells efficiently and produce endogenous antigens in antigen-presenting cells. These results are concordant with other observations (27). Although the divalent DNA vaccine pcDNA3.1-L7/L12-Omp16 stimulated a lower immune response than the RB51 strain, especially in terms of humoral immunity, it still conferred 2.05-log protection versus the 2.25-log protection conferred by RB51. Nevertheless, the two univalent DNA vaccines used in this study, pcDNA3.1-Omp16 and pcDNA3.1-L7/L12, only provided 1.25- and 1.59-log protection, respectively, which confirms Kurar's observation (16). The higher protection level for the divalent DNA vaccine pcDNA3.1-L7/L12-Omp16 may reflect the stacked effect of the two univalent DNA vaccines. Other published univalent DNA vaccines for Brucella based on the GAPDH, bacterioferritin, P39, GroEL, and CP24 genes could not provide protection above 1.0 log (1, 4, 17, 32), although univalent DNA vaccines based on the superoxide dismutase gene had protection above 1.5 log (22, 27). These findings suggest that the L7/L12 or Omp16 gene can be a potent target for DNA vaccines against Brucella.
To summarize, our univalent Brucella DNA vaccine can elicit a higher cellular immune response and provide greater protection for the host against Brucella infection. However, it can still provide less protection than the attenuated rough strain B. abortus RB51. In view of the deficient targeting uptake of the naked plasmids in vivo, it might be possible by using targeting delivery systems (2, 19) with the divalent DNA vaccine to obtain a protection level closer to that offered by the currently used live Brucella vaccines.
Acknowledgments
This work was supported by the Key Research Project of Natural Science Foundation of China (grant 30490240), Outstanding Youth Scientist Foundation of China (30325020), “863” Projects of China (2003AA208224, 2003AA208212, and 2003AA208218), and the National Natural Science Foundation of China (C03011403 and C30170853).
Editor: J. T. Barbieri
REFERENCES
- 1.Al-Mariri, A., A. Tibor, P. Mertens, X. DeBolle, P. Michel, J. Godfroid, K. Walravens, and J. -J. Letesson. 2001. Induction of immune response in BALB/c mice with a DNA vaccine encoding bacterioferritin or P39 of Brucella spp. Infect. Immun. 69:6264-6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baloglu, S., S. M. Boyle, R. Vemulapalli, N. Sriranganathan, G. G. Schurig, and T. E. Toth. 2005. Immune responses of mice to vaccinia virus recombinants expressing either Listeria monocytogenes partial listeriolysin or Brucella abortus ribosomal L7/L12 protein. Vet. Microbiol. 109:11-17. [DOI] [PubMed] [Google Scholar]
- 3.Bowden, R. A., S. M. Estein, M. S. Zygmunt, G. Dubray, and A. Cloeckaert. 2000. Identification of protective outer membrane antigens of Brucella ovis by passive immunization of mice with monoclonal antibodies. Microbes Infect. 2:481-488. [DOI] [PubMed] [Google Scholar]
- 4.Cassataro, J., C. A. Velikovsky, G. H. Giambartolomei, S. Estein, L. Bruno, A. Cloeckaert, R. A. Bowden, M. Spitz, and C. A. Fossati. 2002. Immunogenicity of the Brucella melitensis recombinant ribosome recycling factor-homologous protein and its cDNA. Vaccine 20:1660-1669. [DOI] [PubMed] [Google Scholar]
- 5.Castelruiz, Y., M. Blixenkrone-Moller, and B. Aasted. 2005. DNA vaccination with the Aleutian mink disease virus NS1 gene confers partial protection against disease. Vaccine 23:1225-1231. [DOI] [PubMed] [Google Scholar]
- 6.Cheville, N. F., M. G. Stevens, A. E. Jensen, F. M. Tatum, and S. M. Halling. 1993. Immune responses and protection against infection and abortion in cattle experimentally vaccinated with mutant strains of Brucella abortus. Am. J. Vet. Res. 54:1591-1597. [PubMed] [Google Scholar]
- 7.Corner, L. A., and G. G. Alton. 1981. Persistence of Brucella abortus strain 19 infection in adult cattle vaccinated with reduced doses. Res. Vet. Sci. 31:342-344. [PubMed] [Google Scholar]
- 8.Donnelly, J. J., J. B. Ulmer, J. W. Shiver, and M. A. Liu. 1997. DNA vaccines. Annu. Rev. Immunol. 15:617-648. [DOI] [PubMed] [Google Scholar]
- 9.Feltquate, D. M., S. Heaney, R. G. Webster, and H. L. Robinson. 1997. Different T helper cell types and antibody isotypes generated by saline and gene gun DNA immunization. J. Immunol. 158:2278-2284. [PubMed] [Google Scholar]
- 10.Giambartolomei, G. H., A. Zwerdling, J. Cassataro, L. Bruno, C. A. Fossati, and M. T. Philipp. 2004. Lipoproteins, not lipopolysaccharide, are the key mediators of the proinflammatory response elicited by heat-killed Brucella abortus. J. Immunol. 173:4635-4642. [DOI] [PubMed] [Google Scholar]
- 11.Godfroid, J., A. Cloeckaert, J. P. Liautard, S. Kohler, D. Fretin, K. Walravens, B. Garin-Bastuji, and J. J. Letesson. 2005. From the discovery of the Malta fever's agent to the discovery of a marine mammal reservoir, brucellosis has continuously been a re-emerging zoonosis. Vet. Res. 36:313-326. [DOI] [PubMed] [Google Scholar]
- 12.Hovden, A. O., R. J. Cox, and L. R. Haaheim. 2005. Whole influenza virus vaccine is more immunogenic than split influenza virus vaccine and induces primarily an IgG2a response in BALB/c mice. Scand. J. Immunol. 62:36-44. [DOI] [PubMed] [Google Scholar]
- 13.Jiang, C., D. M. Magee, and R. A. Cox. 1999. Coadministration of interleukin 12 expression vector with antigen 2 cDNA enhances induction of protective immunity against Coccidioides immitis. Infect. Immun. 67:5848-5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang, X., and C. L. Baldwin. 1993. Effects of cytokines on intracellular growth of Brucella abortus. Infect. Immun. 61:124-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnston, D., and J. C. Bystryn. 2005. Heterogeneous antibody response to polyvalent melanoma vaccines in syngeneic mice. Cancer Immunol. Immunother. 54:345-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurar, E., and G. A. Splitter. 1997. Nucleic acid vaccination of Brucella abortus ribosomal L7/L12 gene elicits immune response. Vaccine 15:1851-1857. [DOI] [PubMed] [Google Scholar]
- 17.Leclerq, S., J. S. Harms, G. M. Rosinha, V. Azevedo, and S. C. Oliveira. 2002. Induction of a th1-type of immune response but not protective immunity by intramuscular DNA immunisation with Brucella abortus GroEL heat-shock gene. J. Med. Microbiol. 51:20-26. [DOI] [PubMed] [Google Scholar]
- 18.Lefeber, D. J., B. Benaissa-Trouw, J. F. Vliegenthart, J. P. Kamerling, W. T. Jansen, K. Kraaijeveld, and H. Snippe. 2003. Th1-directing adjuvants increase the immunogenicity of oligosaccharide-protein conjugate vaccines related to Streptococcus pneumoniae type 3. Infect. Immun. 71:6915-6920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Little, S. R., D. M. Lynn, Q. Ge, D. G. Anderson, S. V. Puram, J. Chen, H. N. Eisen, and R. Langer. 2004. Poly-β amino ester-containing microparticles enhance the activity of nonviral genetic vaccines. Proc. Natl. Acad. Sci. USA 101:9534-9539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, M., B. Acres, J. M. Balloul, N. Bizouarne, S. Paul, P. Slos, and P. Squiban. 2004. Gene-based vaccines and immunotherapeutics. Proc. Natl. Acad. Sci. USA 101(Suppl. 2):14567-14571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moriyon, I., M. J. Grillo, D. Monreal, D. Gonzalez, C. Marin, I. Lopez-Goni, R. C. Mainar-Jaime, E. Moreno, and J. M. Blasco. 2004. Rough vaccines in animal brucellosis: structural and genetic basis and present status. Vet. Res. 35:1-38. [DOI] [PubMed] [Google Scholar]
- 22.Munoz-Montesino, C., E. Andrews, R. Rivers, A. Gonzalez-Smith, G. Moraga-Cid, H. Folch, S. Cespedes, and A. A. Onate. 2004. Intraspleen delivery of a DNA vaccine coding for superoxide dismutase (SOD) of Brucella abortus induces SOD-specific CD4+ and CD8+ T cells. Infect. Immun. 72:2081-2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oliveira, S. C., and G. A. Splitter. 1994. Subcloning and expression of the Brucella abortus L7/L12 ribosomal gene and T-lymphocyte recognition of the recombinant protein. Infect. Immun. 62:5201-5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oliveira, S. C., and G. A. Splitter. 1995. CD8+ type 1 CD44hi CD45 RBlo T lymphocytes control intracellular Brucella abortus infection as demonstrated in major histocompatibility complex class I- and class II-deficient mice. Eur. J. Immunol. 25:2551-2557. [DOI] [PubMed] [Google Scholar]
- 25.Oliveira, S. C., and G. A. Splitter. 1996. Immunization of mice with recombinant L7/L12 ribosomal protein confers protection against Brucella abortus infection. Vaccine 14:959-962. [DOI] [PubMed] [Google Scholar]
- 26.Oliveira, S. C., Y. Zhu, and G. A. Splitter. 1994. Recombinant L7/L12 ribosomal protein and gamma-irradiated Brucella abortus induce a T-helper 1 subset response from murine CD4+ T cells. Immunology 83:659-664. [PMC free article] [PubMed] [Google Scholar]
- 27.Onate, A. A., S. Cespedes, A. Cabrera, R. Rivers, A. Gonzalez, C. Munoz, H. Folch, and E. Andrews. 2003. A DNA vaccine encoding Cu, Zn superoxide dismutase of Brucella abortus induces protective immunity in BALB/c mice. Infect. Immun. 71:4857-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Onate, A. A., R. Vemulapalli, E. Andrews, G. G. Schurig, S. Boyle, and H. Folch. 1999. Vaccination with live Escherichia coli expressing Brucella abortus Cu/Zn superoxide dismutase protects mice against virulent B. abortus. Infect. Immun. 67:986-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paulovicova, E., S. Bystricky, J. Masarova, E. Machova, and D. Mislovicova. 2005. Immune response to Saccharomyces cerevisiae mannan conjugate in mice. Int. Immunopharmacol. 5:1693-1698. [DOI] [PubMed] [Google Scholar]
- 30.Pontes, D. S., F. A. Dorella, L. A. Ribeiro, A. Miyoshi, Y. Le Loir, A. Gruss, S. C. Oliveira, P. Langella, and V. Azevedo. 2003. Induction of partial protection in mice after oral administration of Lactococcus lactis producing Brucella abortus L7/L12 antigen. J. Drug Target. 11:489-493. [DOI] [PubMed] [Google Scholar]
- 31.Ribeiro, L. A., V. Azevedo, Y. Le Loir, S. C. Oliveira, Y. Dieye, J. C. Piard, A. Gruss, and P. Langella. 2002. Production and targeting of the Brucella abortus antigen L7/L12 in Lactococcus lactis: a first step towards food-grade live vaccines against brucellosis. Appl. Environ. Microbiol. 68:910-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosinha, G. M., A. Myioshi, V. Azevedo, G. A. Splitter, and S. C. Oliveira. 2002. Molecular and immunological characterisation of recombinant Brucella abortus glyceraldehyde-3-phosphate-dehydrogenase, a T- and B-cell reactive protein that induces partial protection when co-administered with an interleukin-12-expressing plasmid in a DNA vaccine formulation. J. Med. Microbiol. 51:661-671. [DOI] [PubMed] [Google Scholar]
- 33.Schirmbeck, R., M. Kwissa, N. Fissolo, S. Elkholy, P. Riedl, and J. Reimann. 2002. Priming polyvalent immunity by DNA vaccines expressing chimeric antigens with a stress protein-capturing, viral J-domain. FASEB J. 16:1108-1110. [DOI] [PubMed] [Google Scholar]
- 34.Tibor, A., B. Decelle, and J. J. Letesson. 1999. Outer membrane proteins Omp10, Omp16, and Omp19 of Brucella spp. are lipoproteins. Infect. Immun. 67:4960-4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tibor, A., V. Weynants, P. Denoel, B. Lichtfouse, X. De, B., E. Saman, J. N. Limet, and J. J. Letesson. 1994. Molecular cloning, nucleotide sequence, and occurrence of a 16.5-kilodalton outer membrane protein of Brucella abortus with similarity to pal lipoproteins. Infect. Immun. 62:3633-3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Velikovsky, C. A., J. Cassataro, G. H. Giambartolomei, F. A. Goldbaum, S. Estein, R. A. Bowden, L. Bruno, C. A. Fossati, and M. Spitz. 2002. A DNA vaccine encoding lumazine synthase from Brucella abortus induces protective immunity in BALB/c mice. Infect. Immun. 70:2507-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Velikovsky, C. A., F. A. Goldbaum, J. Cassataro, S. Estein, R. A. Bowden, L. Bruno, C. A. Fossati, and G. H. Giambartolomei. 2003. Brucella lumazine synthase elicits a mixed Th1-Th2 immune response and reduces infection in mice challenged with Brucella abortus 544 independently of the adjuvant formulation used. Infect. Immun. 71:5750-5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vemulapalli, R., Y. He, S. Cravero, N. Sriranganathan, S. M. Boyle, and G. G. Schurig. 2000. Overexpression of protective antigen as a novel approach to enhance vaccine efficacy of Brucella abortus strain RB51. Infect. Immun. 68:3286-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xue, T., E. Stavropoulos, M. Yang, S. Ragno, M. Vordermeier, M. Chambers, G. Hewinson, D. B. Lowrie, M. J. Colston, and R. E. Tascon. 2004. RNA encoding the MPT83 antigen induces protective immune responses against Mycobacterium tuberculosis infection. Infect. Immun. 72:6324-6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhan, Y., J. Yang, and C. Cheers. 1993. Cytokine response of T-cell subsets from Brucella abortus-infected mice to soluble Brucella proteins. Infect. Immun. 61:2841-2847. [DOI] [PMC free article] [PubMed] [Google Scholar]