Abstract
The enhanced disease resistance 1 (edr1) mutation of Arabidopsis confers resistance to powdery mildew disease caused by the fungus Erysiphe cichoracearum. Resistance mediated by the edr1 mutation is correlated with induction of several defense responses, including host cell death. Double mutant analysis revealed that all edr1-associated phenotypes are suppressed by mutations that block salicylic acid (SA) perception (nim1) or reduce SA production (pad4 and eds1). The NahG transgene, which lowers endogenous SA levels, also suppressed edr1. In contrast, the ein2 mutation did not suppress edr1-mediated resistance and associated phenotypes, indicating that ethylene and jasmonic acid-induced responses are not required for edr1 resistance. The EDR1 gene was isolated by positional cloning and was found to encode a putative MAP kinase kinase kinase similar to CTR1, a negative regulator of ethylene responses in Arabidopsis. Taken together, these data suggest that EDR1 functions at the top of a MAP kinase cascade that negatively regulates SA-inducible defense responses. Putative orthologs of EDR1 are present in monocots such as rice and barley, indicating that EDR1 may regulate defense responses in a wide range of crop species.
Plants defend themselves against infectious diseases by using both preformed and induced defenses. The latter comprise a complex suite of physiological changes, including a form of programmed cell death called the hypersensitive resistance response (HR) (1). In an effort to identify plant genes that regulate defense responses, we screened for Arabidopsis mutants that displayed enhanced resistance to normally virulent pathogens. The edr1 mutant was identified in a screen for mutants that had become resistant to the bacterium Pseudomonas syringae and was subsequently shown to be resistant to Erysiphe cichoracearum (powdery mildew) (2). Significantly, edr1 mutant plants do not display constitutive expression of the defense gene PR-1, indicating that resistance is not caused by constitutive activation of systemic acquired resistance-associated defenses (3).
Although known defense responses are not constitutively expressed in edr1 plants, several defense responses are induced by E. cichoracearum more rapidly in edr1 plants than in wild-type Arabidopsis variety Col-0 (2). These include deposition of autofluorescent compounds and callose [a β-(1→3) glucan] in mesophyll cell walls, accumulation of PR-1 mRNA, and mesophyll cell death. In wild-type Col-0 plants infected with E. cichoracearum, these defenses are induced more slowly, and very little cell death is observed (2, 4). Because the edr1 mutation is recessive, the EDR1 gene appears to function as a negative regulator. Because these defenses are not expressed in edr1 plants in the absence of pathogens, however, there must be pathogen-associated signals required to induce these defenses. In the absence of EDR1 function, even presumably weak signals from virulent Erysiphe strains are sufficient to induce strong responses.
Because the edr1 mutant appears phenotypically normal in the absence of pathogens but displays enhanced resistance, understanding the molecular basis of this resistance may lead to significant commercial applications. We have therefore assessed the contribution of known plant defense pathways to edr1-mediated resistance and have identified the EDR1 gene. Here we report that EDR1 encodes a putative mitogen-activated protein kinase kinase kinase (MAPKKK) and show that edr1-mediated resistance depends on salicylic acid (SA)-inducible defenses.
Materials and Methods
Construction of Double Mutants.
Double mutants were created by standard genetic crosses. In all cases, edr1 was derived from the Col-0 genotype of Arabidopsis. The eds1–1 and pad4–2 mutations were in a Landsberg erecta genotype, the nim1–1 mutation in a Ws-0 genotype, and the ein2–1 mutation in a Col-0 genotype. For all but ein2–1, double mutants could be identified by PCR-based molecular marker screening of F2 progeny. To identify edr1/ein2–1 double mutants, F2 plants were inoculated with E. cichoracearum, and plants displaying resistance were selected. We then used PCR to amplify the EIN2 gene and identified plants that were homozygous for the ein2–1 mutation by direct sequencing of PCR products. All double mutants were verified to contain the edr1mutation by PCR amplification of EDR1 followed by direct sequencing.
Disease Assays.
Plants were inoculated with E. cichoracearum by transferring conidia (asexual spores) directly from previously infected plants via direct leaf-to-leaf contact. Suppression of the edr1 phenotype was scored by looking for production of visible powder (i.e., abundant conidia) and absence of necrotic lesions 8 days after inoculation.
Chlorophyll Measurements.
Chlorophyll was extracted from Arabidopsis leaves by incubating in 96% ethanol at 50°C for 2 hours. The chlorophyll content of the ethanol extract was then quantified spectrophotometrically, as previously described (5).
Genetic and Physical Mapping of EDR1.
Genetic mapping was accomplished by using a F2 population derived from cross between the edr1 mutant (Columbia genotype, Col-0) and Landsberg erecta (Ler). Genomic DNA was isolated from 239 F2 plants representing 478 meioses and scored with published simple sequence length polymorphism and cleaved amplified polymorphic sequence markers (6, 7). This initial mapping localized edr1 between molecular markers Ateat1 and nga63 (2). We then developed our own molecular markers at intervals between these two markers by amplifying and sequencing 1-kb fragments from both Col-0 and Ler. These analyses identified several additional sequences from this region that were polymorphic between Col-0 edr1 and Ler, which were then mapped relative to edr1 (marker information available on request). Ultimately, edr1 was localized between bacterial artificial chromosome (BAC) end sequence F4014 and an internal sequence from BAC F7G19. This analysis defined a 120-kb region that cosegregated with edr1. This region was fully sequenced and annotated by the Arabidopsis genome project and contained 26 predicted genes. Three of these were selected for amplification and sequencing from the edr1 mutant on the basis of predicted functions that were consistent with the edr1 mutant phenotype (F22013.21, a MAPKKK, F22013.32, a myb-like transcription factor, and F22013.34, a superoxide dismutase). A C→G transversion was identified in gene F22013.21, which produced an early stop codon.
Complementation of the edr1 Mutation.
BAC clone F22013 was partially digested with Sau3A restriction enzyme and ligated to BamHI-digested binary cosmid vector pCLD04541 (8). The ligation mix was packaged by using a Gigapack III Gold packaging extract (Stratagene) and transfected into Escherichia coli strain DH5α. Clones containing EDR1 were identified by colony hybridization by using a PCR-amplified fragment internal to the predicted EDR1 ORF (9). The inserts of these cosmids were then analyzed by restriction enzyme mapping and sequencing of junctions between inserts and vector. Two clones were selected for transformation of Arabidopsis edr1 plants. Clone 1 contained a full-length EDR1 gene, whereas clone 2 was truncated 76 bases 5′ of the EDR1 stop codon but contained all other genes present on clone 1 (predicted genes F22013.19 and F22013.20). Sequence analysis of these two genes amplified from the edr1 mutant revealed no mutations. Both clones were transferred to Agrobacterium tumefaciens strain GV3101 by conjugation. Arabidopsis edr1 plants were transformed by using the floral dip method (10), selecting for kanamycin resistance (50 μg/ml).
DNA and RNA Gel Blot Hybridizations.
DNA and RNA gel blots were hybridized and washed at 55°C (DNA blots) or 65°C (RNA blots) by using Church buffer as described in ref. 11. Total RNA was isolated from Arabidopsis leaf tissue by using TriPure Isolation Reagent (Boehringer Mannheim) following the manufacturer's protocol and poly(A) RNA subsequently purified from the total RNA by using a Qiagen Oligotex midi kit (Qiagen, Chatsworth, CA).
DNA Sequence Analysis of Arabidopsis EDR1.
EDR1 mRNA was amplified by reverse transcription–PCR by using overlapping sets of primers and total RNA as template. During this phase, we were unable to obtain a product by using a primer complementary to the predicted start of the EDR1 ORF. To obtain the 5′ end of the EDR1 cDNA sequence, we used 5′-rapid amplification of cDNA ends (RACE) PCR by using the Marathon 5′-RACE kit from CLONTECH. PCR products were sequenced directly by using an ABI377 automated DNA sequencer and Applied Biosystems BigDye dideoxy terminator reagents (Perkin–Elmer). DNA sequences were assembled by using the sequencher program (Gene Codes, Ann Arbor, MI). Assembly of the full-length EDR1 cDNA sequence revealed that the cDNA sequence predicted from genomic sequence (accession no. AC003981) included an extra exon and intron at the 5′ end that are absent from our experimentally determined cDNA sequence. Our EDR1 protein sequence (accession no. AF305913) thus lacks the first 82 amino acids present in the predicted protein derived from genomic sequence (PIR-T00726).
Isolation of Barley EDR1.
The kinase domain of a putative barley EDR1 gene was amplified by using the degenerate oligonucleotides: 5′-gcIgtIaaIaaIttI(t/c)tIga(t/c)ca(g/a)ga-3′ and 5′-gcIaaIgaIggIc(g/t)IaI(g/a)ttIgg(g/a)tc-3′. Amplification was performed on cDNA from barley variety Ingrid by using the following temperature program: 94°C for 2 minutes followed by 35 cycles of 94°C for 30 sec, 52°C for 30 sec, 72°C for 1 min, followed by 10 min at 72°C. The resulting PCR product was cloned into the vector pGEMT-Easy (Promega) and several independent clones sequenced by using an ABI377 automated sequencer (Applied Biosystems). On the basis of this sequence, two gene-specific primers were designed (5′-TGCGTCGGCTCCGTCATCCAAATATTGT-3′ and 5′-TCTCGTCAATTTGGCAATTAGGGCGATG-3′) for performing 5′ and 3′-RACE PCR by using the Marathon kit (CLONTECH). The 5′ and 3′ RACE PCR products, which overlapped each other, were then cloned into the vector pGEMT-Easy and sequenced.
Results and Discussion
To gain insight into how EDR1 regulates defense responses in Arabidopsis, we assessed the relative contribution of SA, jasmonic acid (JA), and ethylene to edr1-mediated resistance. These three compounds are plant hormones that induce distinct but overlapping sets of defense genes (12, 13). To assess their role in edr1-mediated resistance, we crossed the edr1 mutant to mutants that are blocked at specific steps in the SA, JA, or ethylene response pathways. Double homozygous mutants were then infected with E. cichoracearum and the edr1 mutant phenotype (absence of visible powder and presence of lesions) scored 8 days after infection.
Role of SA, JA, and Ethylene Pathways in edr1-Mediated Resistance.
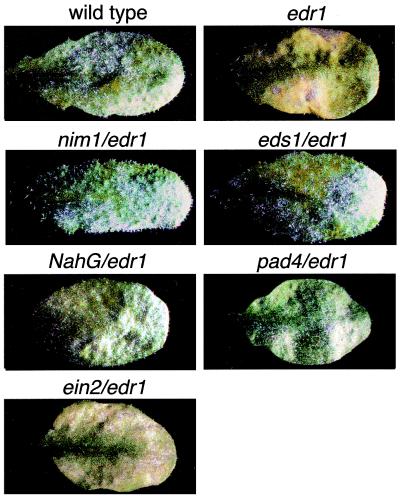
Mutations that block SA-associated defense responses suppressed edr1-mediated resistance (Fig. 1). The dependence on SA-induced responses was verified by mutations in two distinct parts of the SA pathway and by use of a transgene, nahG, that reduces endogenous SA levels (14, 15). The nim1–1 mutation, which blocks a subset of SA-induced responses (16), appeared to completely suppress edr1-mediated resistance, including the cell death response induced by E. cichoracearum. Similarly, the pad4–2 and eds1–2 mutations, which reduce production of SA induced by pathogens (17, 18), also suppressed edr1 (Fig. 1). Finally, the nahG transgene suppressed all edr1-associated phenotypes (Fig. 1).
Figure 1.
Suppression of the edr1 resistance phenotype by mutations that block SA-mediated defense responses. Double mutants containing edr1 and the indicated mutations were constructed by standard genetic crosses. Plants were inoculated with E. cichoracearum and disease phenotypes scored 8 days after inoculation. Single representative leaves were removed from intact plants on the eighth day for photography. In the wild-type and double mutant plants, except for ein2/edr1, abundant asexual sporulation is observed as white patches. Also note the absence of yellow/brown necrotic patches observed in the edr1 mutant leaf.
In contrast to mutations that block SA-mediated defense responses, the ein2 mutation did not suppress edr1-mediated resistance (Fig. 1). Mutations in ein2 block all known ethylene-mediated responses (19) as well as most JA-mediated responses (20–22). Taken together, these data indicate that resistance mediated by the edr1 mutation depends on SA-induced defense responses and is independent of JA- and ethylene-induced defenses.
Molecular Isolation of EDR1.
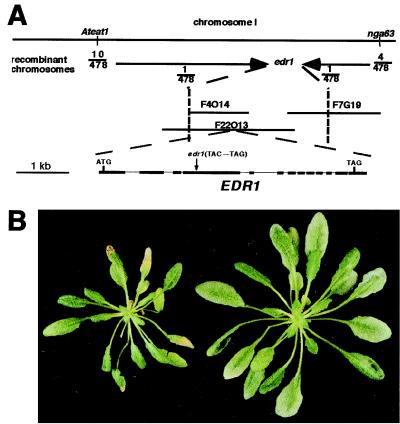
We isolated the EDR1 gene by using a combination of fine-structure genetic mapping and candidate gene analysis. The edr1 mutant was crossed to wild-type Arabidopsis of the Landsberg erecta genotype and the resulting F2 population used for genetic mapping. This work localized edr1 to an approximately 120-kb interval on chromosome 1 (Fig. 2A), which had been fully sequenced as part of the Arabidopsis genome project. Located within this region were several genes that were reasonable candidates to function as negative regulators of defense responses. Three candidate genes (a myb-like transcription factor, a superoxide dismutase, and a MAPKKK) were amplified from the edr1 mutant by using PCR and were sequenced. Analysis of these sequences revealed a C→G transversion in the MAPKKK candidate (F22013.21) that produced an early termination codon (Fig. 2A). No mutations were identified in the other two candidate genes.
Figure 2.
Positional cloning of the EDR1 gene. (A) Genetic and physical map of the region flanking EDR1. The vertical dotted lines indicate the position of the closest markers that defined the edr1 genetic interval. Sequencing of three candidate genes in this interval revealed a C→G transversion in gene F22013.21, which produced an early stop codon. Comparison of the genomic sequence to the cDNA sequence obtained by reverse transcription–PCR revealed the indicated exon (boxes)-intron (lines) structure. (B) Complementation of the edr1 mutation by Agrobacterium-mediated transformation. Shown are sibling plants from the T2 generation that lack the wild-type EDR1 transgene (Left) or that contain the EDR1 transgene (Right). The apparent difference in size is because of necrosis of the larger leaves in the edr1 mutant (resistant) plant.
To confirm that the MAPKKK gene corresponded to EDR1, we transformed two cosmid subclones of BAC F22013 (accession no. AC003981) into the edr1 mutant (see Materials and Methods). Clone 1 contained a full-length EDR1 gene, whereas clone 2 was truncated 76 bases 5′ of the EDR1 stop codon but contained all other genes present on clone 1. Self progeny from kanamycin-resistant transformants were obtained and assayed for resistance to E. cichoracearum. Phenotypically wild-type (i.e., susceptible) plants were observed in 4 of 5 lines transformed with clone 1 (Fig. 2B), and the susceptible phenotype was shown to cosegregate with the T-DNA insert by PCR. No susceptible plants were observed with lines transformed with clone 2, indicating that the MAPKKK gene was responsible for complementation of the edr1 mutant phenotype.
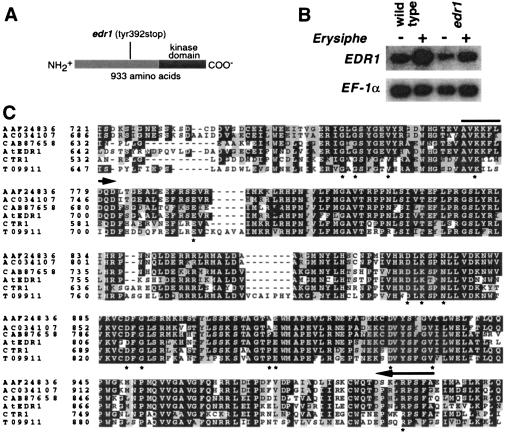
EDR1 encodes a putative MAPKKK consisting of a 276-aa C-terminal kinase domain and a 657-aa N-terminal nonkinase domain (Fig. 3A). The edr1 mutation truncates the predicted EDR1 protein 265 amino acids before the kinase domain. RNA gel blot analysis showed that EDR1 is expressed in uninoculated leaves but is moderately induced (≈5-fold) during infection by E. cichoracearum (Fig. 3B).
Figure 3.
EDR1 encodes a MAPKKK with high similarity to CTR1 and four additional kinases present in Arabidopsis. (A) The predicted EDR1 protein contains 933 amino acids, the carboxyl-terminal third of which contains a kinase domain. (B) RNA gel blot analysis of the EDR1 mRNA. Wild-type and edr1 mutant plants were inoculated with E. cichoracearum (+) or mock inoculated (−) and poly(A) RNA isolated from leaves 3 days after inoculation. The blot was hybridized with an EDR1 cDNA probe corresponding to amino acid residues 180–461. Relative amounts of RNA loaded in each lane were estimated by using an EF-1α cDNA probe. PhosphorImager quantitation of this blot, normalized to the EF-1α signal, revealed that the edr1 message is induced approximately 5-fold by infection with E. cichoracearum. This analysis was repeated three times with similar results. (C) Alignment of the kinase domains of the six known members of the CTR1 kinase family in Arabidopsis. The entire protein for each of the indicated proteins was aligned by using the default parameters of clustalx, a Macintosh version of the clustalw program (47). Only the kinase domain is shown. The full-length alignment can be viewed in supplemental Fig. 6. The alignment file produced by clustalx was formatted by using the boxshade www server (http://www.ch.embnet.org/software/BOX_form.html). White type on a black background indicates a residue that is identical in at least half of the proteins shown, whereas a gray background indicates conservative substitutions as defined by the boxshade 3.21 default parameters. Asterisks indicate residues that are conserved in nearly all protein kinases (48). Arrows indicate regions conserved in the Arabidopsis, tomato, and rice EDR1 orthologs (see supplemental Fig. 6) but divergent in paralogs. Accession nos. for CTR1 and EDR1 are A45178 and AF305913, respectively. Accession no. AC034107 refers to the BAC DNA sequence, from which we derived the indicated protein sequence by using the EDR1 and CTR1 sequences as guides.
EDR1 Belongs to the CTR1 Family of MAPKKKs.
Searches of the GenBank database by using the blastp and tblastn programs (23) uncovered five EDR1 homologs in Arabidopsis with extensive sequence similarity to both the kinase and nonkinase regions (Fig. 3C and Fig. 6, which is published as supplemental data on the PNAS web site, www.pnas.org). The sequence similarity of EDR1 to these five putative kinases was very high in the kinase domain (>65% identity) but much lower in the nonkinase domain (≈25% identity with numerous gaps and insertions). Although the overall similarity among the nonkinase domains is low, there are several blocks of highly conserved sequence (see supplemental Fig. 6), suggesting a conserved function for this domain. No other Arabidopsis proteins displayed large blocks of amino acid sequence similarity to the nonkinase domain, indicating that EDR1 belongs to a distinct subfamily of MAPKKKs consisting of at least six members in Arabidopsis.
Of these six MAPKKKs, only CTR1 has been previously characterized. Loss-of-function mutations in CTR1 confer a constitutive ethylene response phenotype (24); phenotypes include a constitutive triple response (a short fat hypocotyl and an exaggerated apical hook when grown in the dark), constitutive expression of ethylene inducible genes, and severe dwarfing. CTR1 thus functions as a negative regulator of the ethylene-response pathway.
EDR1 Does Not Directly Regulate Ethylene Responses.
The high similarity between the CTR1 and EDR1 protein sequences led us to investigate whether edr1 mutants display any constitutive ethylene responses. Unlike ctr1 mutants, no visible effect on etiolation was observed, and plants were not stunted (data not shown). Furthermore, the ethylene-inducible genes CHIB and HEL were not constitutively expressed and displayed normal ethylene inducibility (data not shown). In addition, edr1 seedlings displayed a normal triple response on exposure to 10 ppm ethylene (data not shown). These data, combined with the lack of suppression by the ein2 mutation, indicate that EDR1 functions in a pathway separate from the ethylene-response pathway.
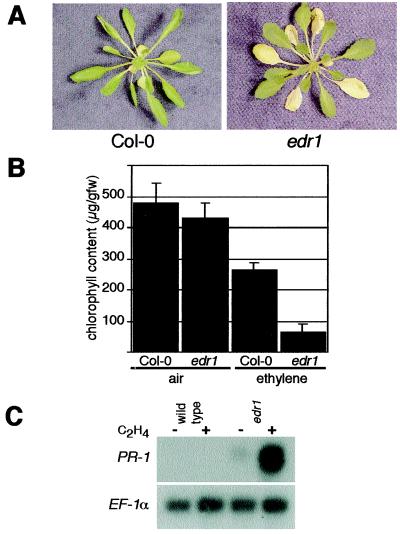
In the course of investigating ethylene responses in edr1 mutants, we observed that edr1 plants displayed an enhanced ethylene-induced senescence response (Fig. 4 A and B). Exposure of 6-week-old wild-type Arabidopsis plants to 100 ppm ethylene induces chlorosis (yellowing) in the oldest 2 leaves starting after 3 days of ethylene exposure. However, in edr1 mutant plants, this yellowing was visible on much younger leaves (Fig. 4A). Quantification of chlorophyll in leaves 3 through 8 (leaf 1 being the oldest true leaf) revealed an approximately 5-fold lower level of chlorophyll in ethylene-treated edr1 plants compared with wild-type Col-0 (Fig. 4B). There was no significant difference between edr1 and wild-type plants not exposed to ethylene.
Figure 4.
Ethylene sensitivity of edr1 mutant plants. (A) Ethylene-induced senescence. Wild-type and edr1 mutant plants were grown in pots for 6 weeks in a growth cabinet set for 9-h days then placed in a sealed chamber containing 100 ppm ethylene. Plants were removed for photography after 72 h. Note the increase in the number of yellow leaves in the ethylene-treated edr1 mutant. (B) Quantification of chlorophyll levels. Leaves 3 through 8 (leaf 1 being the oldest true leaf) were removed from 6-week-old plants after 72 h of exposure to 100 ppm ethylene and chlorophyll levels (microgram/gram fresh weight) measured as described (5). Bars represent the mean and standard deviation of values obtained from six plants. (C) Ethylene-induced expression of PR-1. RNA gel blots were performed with 10 μg of total RNA isolated from rosette leaves (both green and yellow) after 72 h of exposure to ethylene (+) or air (−). Blots were sequentially probed with PR-1 and EF-1α. The latter demonstrates approximately equal RNA loading in all lanes.
The enhanced senescence response of edr1 plants may reflect an overlap between the genetic control of senescence and SA-induced defense responses. The senescence process is regulated in part by SA, as plants containing npr1or pad4 mutations or the nahG transgene display delayed yellowing and reduced necrosis during senescence (25). In addition, several defense related genes are known to be induced during leaf senescence, including the well-characterized defense gene PR-1 (25, 26). Induction of PR-1 by pathogens or during senescence is blocked by the expression of the nahG transgene, and by mutations in NPR1 or PAD4 (25, 27). We used RNA gel-blot analysis to examine expression of PR-1 in ethylene-treated plants. PR-1 was highly expressed in the edr1 mutant plants after 3 days of ethylene exposure, whereas this message is undetectable in wild-type plants (Fig. 4C). This result is consistent with the enhanced senescence phenotype and suggests that both the enhanced disease-resistance phenotype and the enhanced-senescence phenotype may be related to enhanced signaling through the SA pathway.
Identification of EDR1 Orthologs in Crop Species.
Powdery mildew is an economically important disease on many monocot (e.g., wheat and barley) and dicot (e.g., tomato and grape) crops. The enhanced resistance conferred by the edr1 mutation may thus provide a novel means of conferring resistance to this disease, provided that the EDR1 pathway is conserved in crop plants. We therefore searched the available DNA and protein databases for homologs of EDR1. A blastp search of the GenBank nonredundant database identified a predicted protein sequence from tomato (TCTR2, accession no. AJ005077) that has high similarity to the Arabidopsis EDR1 protein over its full length (see supplemental Fig. 6), including 86% identity in the kinase domain. We also identified a partial cDNA sequence from rice in the dbEST database (accession no. D41138) that was 85% identical to the kinase domain of EDR1. We obtained a nearly full-length cDNA sequence for this rice gene by using 5′ and 3′-RACE methods (CLONTECH). Both the rice and tomato genes encode proteins that are more similar to the Arabidopsis EDR1 protein than are any of the 5 Arabidopsis EDR1 paralogs, including CTR1, whose kinase domain is only 65% identical to EDR1 (Fig. 5). This relationship strongly suggests that the rice and tomato genes are orthologs of EDR1 and that this gene family came into existence before the divergence of monocots and eudicots. Consistent with this view is the presence of a second tomato sequence, TCTR1 (accession no. AF096250) that is most similar to the Arabidopsis CTR1 protein (Fig. 5). Furthermore, it indicates that EDR1 has been conserved during angiosperm evolution, presumably because it is performing a conserved function.
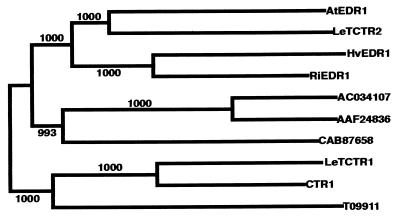
Figure 5.
Conservation of EDR1 among diverse angiosperms. Full-length protein sequences were aligned by using the clustalw program (see supplemental Fig. 6) and a phylogenetic tree derived by using the neighbor-joining method (47). Numbers indicate bootstrap values supporting branch points based on 1,000 replicates. T09911, CAB87658, ACO34107, and AAF24836 are Arabidopsis CTR1/EDR1 homologs of unknown function. Putative EDR1 and CTR1 orthologs are preceded by a two-letter prefix to indicate species of origin: Hv, Hordeum vulgare (barley), Ri, O. sativa (rice), Le, Lycopersicon esculentum (tomato), At, A. thaliana.
Alignment of the rice and tomato EDR1-like sequences with the six Arabidopsis homologs enabled us to identify regions that were conserved among the putative EDR1 orthologs but divergent in the five Arabidopsis paralogs (indicated by arrows in Fig. 3C and supplemental Fig. 6). On the basis of these amino acid sequences, we designed degenerate oligonucleotide primers for amplification of EDR1 orthologs from other plant species (see Materials and Methods). Using these primers, we successfully isolated an EDR1 ortholog from barley that is highly similar to the rice EDR1 ortholog (86% identical over the kinase domain; Fig. 5 and supplemental Fig. 6). It should thus be possible to isolate EDR1 orthologs from most angiosperms.
Relationship of EDR1 to Other MAPKKKs.
EDR1 and CTR1 belong to the Raf subfamily of MAPKKKs, one of three MAPKKK subfamilies found in Arabidopsis. The other two subfamilies have been designated AtMEKK1 and ANPs (28). The AtMEKK1 family has been implicated in mediating touch/cold/drought signal transduction (29, 30), and the ANP family has recently been shown to respond to H2O2 and to negatively regulate auxin responses in protoplast assays (28). The role of these MAPKKKs in intact plants is poorly understood, however, as no mutations in endogenous genes have been described. To date, EDR1 and CTR1 are the only plant MAPKKKs with known mutant phenotypes. The observation that EDR1 and CTR1 function in distinct response pathways suggests that the remaining Raf-like MAPKKKs may function in as yet to be discovered pathways distinct from the ethylene and defense response pathways.
Yeast two-hybrid and in vitro analyses indicate that CTR1 physically interacts with the ethylene receptor protein ETR1 (31). On binding of ethylene to ETR1, the receptor is believed to be inactivated (32), which in turn inactivates CTR1, which then allows the ethylene response pathway to activate. How CTR1 negatively regulates ethylene inducible genes is not known; however, transient expression of the CTR1 kinase domain in maize protoplasts activates endogenous MAP kinases (33), suggesting that CTR1 functions at the top of a classical MAP kinase cascade that is conserved between monocots and eudicots.
A Model for How EDR1 Functions in Disease Resistance.
Because EDR1 and CTR1 belong to the same family of MAPKKKs and because both function as negative regulators, it is tempting to speculate that EDR1 will also interact with an upstream receptor protein and will activate a MAP kinase cascade. By analogy to the CTR1 model, one would predict that a pathogen-associated signal (either induced by or produced by the pathogen) would inactivate this putative receptor, which would lead to inactivation of EDR1, which would then allow activation of SA-mediated defenses by a separate pathway. In essence, before SA-associated defenses are turned on, two signals must be perceived, one to remove the negative regulators (the EDR1 pathway) and one to activate positive regulators (e.g., EDS1/PAD4 pathways). Such a mechanism would reduce the likelihood of inappropriate deployment of defenses and would explain why edr1 plants do not constitutively express defense responses. This mechanism would also explain why eds1 and pad4 mutations suppress edr1 resistance.
The Relationship of EDR1 to Known MAP Kinase Pathways.
Arabidopsis contains at least nine MAP kinases (34), one or more of which could be part of the EDR1 pathway. To date, no mutant phenotypes have been ascribed to any MAP kinase gene of Arabidopsis or of any other plant. However, several plant MAP kinases have been identified that are activated by pathogen infection and by related signal molecules (35–41). For example, the tobacco MAP kinase SIPK is activated by tobacco mosaic virus infection, SA, by several pathogen-derived elicitors and by wounding (42–44). The tobacco WIPK MAP kinase is also activated by all of these stimuli except for SA (44, 45). For reasons described below, however, we do not believe that EDR1 directly regulates either WIPK- or SIPK-like MAP kinases in Arabidopsis.
Orthologs of WIPK and SIPK exist in Arabidopsis and have been designated AtMPK3 and AtMPK6, respectively (41, 46). Recently, Sheen and colleagues used a protoplast assay to show that both of these MAP kinases can be activated by the Arabidopsis MAPKKK ANP1 (28). Furthermore, this ANP1-dependent activation could be triggered by exogenous application of H2O2 but not by cold treatment or the plant hormones auxin and abscisic acid. Because ANP1 belongs to a different subclass of MAPKKKs than EDR1 and CTR1 (28), it is unlikely that EDR1 would also activate AtMPK3 or AtMPK6. This supposition is supported by the observation that AtMPK6 is activated within 2 min of bacterial elicitor application (41), suggesting that it positively regulates primary defense responses, whereas we have shown that EDR1 functions as a negative regulator (2).
Conclusions
We have shown that a loss-of-function mutation in the EDR1 MAPKKK causes enhanced disease resistance to powdery mildew infection of Arabidopsis. The edr1 mutant phenotype indicates that this MAPKKK negatively regulates defense responses. This finding is somewhat unexpected, as there exists a large body of correlative data suggesting that MAP kinase activation is required for induction of plant defenses. We speculate that plant defense responses are regulated at least in part via antagonistic MAP kinase pathways. Because EDR1 and many of the MAP kinases are well conserved in crop plants, it is expected that they function similarly in these species. A clear understanding of how the balance of signaling through these pathways is regulated may provide new approaches to engineering disease resistance in crops.
Supplementary Material
Acknowledgments
We thank P. Schulze-Lefert (the Sainsbury Laboratory) for providing cDNA from rice and barley, the Sainsbury Laboratory for providing research support to R.W.I. during a sabbatical visit, J. Parker (the Sainsbury Laboratory) for providing F2 seed from crosses between edr1 and pad4–1 and between edr1 and eds1–2. J. Danzer, R. Cameron, S. Ludwig, A. Henk, and K. Christiansen for help with genetic mapping and DNA sequencing, and the Arabidopsis Biological Resource Center at Ohio State for providing ein2–1 seed. This work was supported by National Institutes of Health Grant GM46451 and by Novartis Agribusiness Biotechnology Research, Inc.
Abbreviations
- SA
salicylic acid
- HR
hypersensitive resistance response
- JA
jasmonic acid
- MAPKKK
mitogen-activated protein kinase kinase kinase
- RACE
rapid amplification of cDNA ends
- BAC
bacterial artificial chromosome
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The nucleotide and protein sequences reported in this paper have been deposited in the GenBank database (accession nos. AF305913 for the Arabidopsis EDR1 gene, AF305911 for the rice EDR1 gene, and AF305912 for the barley EDR1 gene).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.011405198.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.011405198
References
- 1.Greenberg J T. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:525–545. doi: 10.1146/annurev.arplant.48.1.525. [DOI] [PubMed] [Google Scholar]
- 2.Frye C A, Innes R W. Plant Cell. 1998;10:947–956. doi: 10.1105/tpc.10.6.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ryals J A, Neuenschwander U H, Willits M G, Molina A, Steiner H-Y, Hunt M D. Plant Cell. 1996;8:1809–1819. doi: 10.1105/tpc.8.10.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adam L, Somerville S C. Plant J. 1996;9:341–356. doi: 10.1046/j.1365-313x.1996.09030341.x. [DOI] [PubMed] [Google Scholar]
- 5.Lichtenthaler H K, Wellburn A R. Biochem Soc Trans. 1983;11:591–592. [Google Scholar]
- 6.Konieczny A, Ausubel F M. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- 7.Bell C J, Ecker J R. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- 8.Bancroft I, Love K, Bent E, Sherson S, Lister C, Cobbett C, Goodman H M, Dean C. Weeds World. 1997;4:1–9. [Google Scholar]
- 9.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 10.Clough S J, Bent A F. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 11.Ashfield T, Danzer J R, Held D, Clayton K, Keim P, Saghai Maroof M A, Webb P M, Innes R W. Theor Appl Genet. 1998;96:1013–1021. [Google Scholar]
- 12.Dong X. Curr Opin Plant Biol. 1998;1:316–323. doi: 10.1016/1369-5266(88)80053-0. [DOI] [PubMed] [Google Scholar]
- 13.Schenk P M, Kazan K, Wilson I, Anderson J P, Richmond T, Somerville S C, Manners J M. Proc Natl Acad Sci USA. 2000;97:11655–11660. doi: 10.1073/pnas.97.21.11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaffney T, Friedrich L, Vernooij B, Negrotto D, Ney G, Uknes S, Ward E, Kessmann H, Ryals J. Science. 1993;261:754–756. doi: 10.1126/science.261.5122.754. [DOI] [PubMed] [Google Scholar]
- 15.Lawton K, Weymann K, Friedrich L, Vernooij B, Uknes S, Ryals J. Mol Plant–Microbe Interact. 1995;8:863–870. doi: 10.1094/mpmi-8-0863. [DOI] [PubMed] [Google Scholar]
- 16.Delaney T P, Friedrich L, Ryals J A. Proc Natl Acad Sci USA. 1995;92:6602–6606. doi: 10.1073/pnas.92.14.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Falk A, Feys B J, Frost L N, Jones J D, Daniels M J, Parker J E. Proc Natl Acad Sci USA. 1999;96:3292–3297. doi: 10.1073/pnas.96.6.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou N, Tootle T L, Tsui F, Klessig D F, Glazebrook J. Plant Cell. 1998;10:1021–1030. doi: 10.1105/tpc.10.6.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alonso J M, Hirayama T, Roman G, Nourizadeh S, Ecker J R. Science. 1999;284:2148–2152. doi: 10.1126/science.284.5423.2148. [DOI] [PubMed] [Google Scholar]
- 20.Penninckx I A, Thomma B P, Buchala A, Metraux J P, Broekaert W F. Plant Cell. 1998;10:2103–2113. doi: 10.1105/tpc.10.12.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomma B, Eggermont K, Penninckx I, Mauch-Mani B, Vogelsang R, Cammue B P A, Broekaert W F. Proc Natl Acad Sci USA. 1998;95:15107–15111. doi: 10.1073/pnas.95.25.15107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomma B P H J, Eggermont K, Tierens K F M-J, Broekaert W F. Plant Physiol. 1999;121:1093–1101. doi: 10.1104/pp.121.4.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altschul S F, Gish W, Miller W, E W, M, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 24.Kieber J J, Rothenberg M, Roman G, Feldmann K A, Ecker J R. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- 25.Morris K, MacKerness S A, Page T, John C F, Murphy A M, Carr J P, Buchanan-Wollaston V. Plant J. 2000;23:677–685. doi: 10.1046/j.1365-313x.2000.00836.x. [DOI] [PubMed] [Google Scholar]
- 26.Hanfrey C, Fife M, Buchanan-Wollaston V. Plant Mol Biol. 1996;30:597–609. doi: 10.1007/BF00049334. [DOI] [PubMed] [Google Scholar]
- 27.Delaney T P, Uknes S J, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E, Ryals J. Science. 1994;266:1247–1249. doi: 10.1126/science.266.5188.1247. [DOI] [PubMed] [Google Scholar]
- 28.Kovtun Y, Chiu W L, Tena G, Sheen J. Proc Natl Acad Sci USA. 2000;97:2940–2945. doi: 10.1073/pnas.97.6.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mizoguchi T, Irie K, Hirayama T, Hayashida N, Yamaguchi-Shinozaki K, Matsumoto K, Shinozaki K. Proc Natl Acad Sci USA. 1996;93:765–769. doi: 10.1073/pnas.93.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mizoguchi T, Ichimura K, Yoshida R, Shinozaki K. Results Probl Cell Differ. 2000;27:29–38. doi: 10.1007/978-3-540-49166-8_3. [DOI] [PubMed] [Google Scholar]
- 31.Clark K L, Larsen P B, Wang X, Chang C. Proc Natl Acad Sci USA. 1998;95:5401–5406. doi: 10.1073/pnas.95.9.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hua J, Meyerowitz E M. Cell. 1998;94:261–271. doi: 10.1016/s0092-8674(00)81425-7. [DOI] [PubMed] [Google Scholar]
- 33.Kovtun Y, Chiu W L, Zeng W, Sheen J. Nature (London) 1998;395:716–720. doi: 10.1038/27240. [DOI] [PubMed] [Google Scholar]
- 34.Ligterink W. In: MAP Kinases in Plant Signal Transduction. Hirt H, editor. Heidelberg: Springer; 2000. pp. 11–27. [DOI] [PubMed] [Google Scholar]
- 35.Liu Y, Zhang S, Klessig D F. Mol Plant–Microbe Interact. 2000;13:118–124. doi: 10.1094/MPMI.2000.13.1.118. [DOI] [PubMed] [Google Scholar]
- 36.Romeis T, Piedras P, Zhang S, Klessig D F, Hirt H, Jones J D. Plant Cell. 1999;11:273–287. doi: 10.1105/tpc.11.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schoenbeck M A, Samac D A, Fedorova M, Gregerson R G, Gantt J S, Vance C P. Mol Plant–Microbe Interact. 1999;12:882–893. doi: 10.1094/MPMI.1999.12.10.882. [DOI] [PubMed] [Google Scholar]
- 38.Takezawa D. Plant Mol Biol. 1999;40:921–933. doi: 10.1023/a:1006263607135. [DOI] [PubMed] [Google Scholar]
- 39.Zhang S, Du H, Klessig D F. Plant Cell. 1998;10:435–450. doi: 10.1105/tpc.10.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ligterink W, Kroj T, zur Nieden U, Hirt H, Scheel D. Science. 1997;276:2054–2057. doi: 10.1126/science.276.5321.2054. [DOI] [PubMed] [Google Scholar]
- 41.Nuhse T S, Peck S C, Hirt H, Boller T. J Biol Chem. 2000;275:7521–7526. doi: 10.1074/jbc.275.11.7521. [DOI] [PubMed] [Google Scholar]
- 42.Zhang S, Klessig D F. Plant Cell. 1997;9:809–824. doi: 10.1105/tpc.9.5.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang S, Klessig D F. Proc Natl Acad Sci USA. 1998;95:7225–7230. doi: 10.1073/pnas.95.12.7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumar D, Klessig D F. Mol Plant–Microbe Interact. 2000;13:347–351. doi: 10.1094/MPMI.2000.13.3.347. [DOI] [PubMed] [Google Scholar]
- 45.Seo S, Sano H, Ohashi Y. Plant Cell. 1999;11:289–298. doi: 10.1105/tpc.11.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang S, Klessig D F. In: Map Kinases in Plant Signal Transduction. Hirt H, editor. Heidelberg: Springer; 2000. pp. 65–84. [Google Scholar]
- 47.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hanks S K, Quinn A M. Methods Enzymol. 1991;200:38–62. doi: 10.1016/0076-6879(91)00126-h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.