Abstract
The oral pathogen Porphyromonas gingivalis expresses a homolog of the internalin family protein InlJ. Inactivation of inlJ reduced monospecies biofilm formation by P. gingivalis. In contrast, heterotypic P. gingivalis-Streptococcus gordonii biofilm formation was enhanced in the InlJ-deficient mutant. The results indicate a nuanced role for InlJ in regulating biofilm accumulations of P. gingivalis.
The internalin protein family was originally identified in Listeria monocytogenes and is characterized by the presence of an N-terminal leucine-rich repeat (LRR) domain (1). LRRs are involved in protein-protein interactions including ligand-receptor binding (5). Indeed, internalin A (InlA) and InlB of L. monocytogenes are involved in adherence and invasion of the organism (1, 3). Recently, a novel internalin, InlJ, was identified in L. monocytogenes (12). InlJ defines a new subclass family of cysteine-containing LRR proteins. An InlJ mutant of Listeria was found to be significantly attenuated in virulence in mice (12). InlJ type proteins are currently identified in only six bacterial species, one of which is the gram-negative oral anaerobe Porphyromonas gingivalis (12). Expression of InlJ (PG0350) (http://www.lanl.gov) in P. gingivalis was originally detected when the organism was in contact with host epithelial cells (2). However, an InlJ mutant of P. gingivalis did not exhibit an invasion-related phenotype (18), as is also the case for the Listeria InlJ mutant (12). In addition to an intracellular location, a significant component of the P. gingivalis lifestyle is within the complex multispecies biofilm (dental plaque) that develops on tooth surfaces. In this study, we investigated the role of the P. gingivalis InlJ protein in single-species and multispecies biofilm formation by the organism. Interestingly, an InlJ-null mutant exhibited reduced monospecies biofilm development but enhanced heterotypic biofilm formation with Streptococcus gordonii.
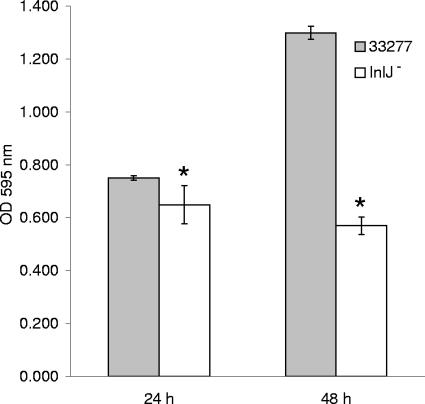
An InlJ-deficient mutant was generated by insertional inactivation in P. gingivalis strain 33277. A central 847-bp fragment of inlJ (PG0350) was amplified by PCR using primers 5′-TCTTCTGCAGGGGACTATGG-3′ and 5′-TTTCCACGTGTTCGGTTGTA-3′ and subcloned into the suicide plasmid pVA3000. The recombinant plasmid was introduced into P. gingivalis 33277 by conjugation as described previously (10). The absence of inlJ transcript was confirmed by reverse transcription-PCR. As the inlJ gene is located between two genes transcribed in the opposite direction, the potential for pleiotropic effects of this mutation are diminished. The adjacent genes are PG0349, a putative hydrolase of the haloacid dehalogenase-like family, and PG0351, a hypothetical protein, neither of which has a documented role in biofilm biogenesis. Homotypic biofilm formation was first tested in the microtiter plate assay described previously by O'Toole and Kolter (8). Parental and mutant strains were suspended in prereduced phosphate-buffered saline, and 5 × 107 cells were incubated at 37°C anaerobically in individual wells of 96-well plates. The resulting biofilms were washed, stained with 1% crystal violet, and destained with 95% ethanol. Absorbance at 595 nm was determined using a Benchmark microplate reader. Figure 1 shows that monospecies biofilm formation was reduced in the InlJ mutant by 13.6% after 24 h and 56.1% after 48 h. For visualization and quantification of biofilm structure, biofilms were generated in a 16-well Culture Well chambered coverglass system, stained with 5 (and 6)-carboxyfluorescein succinimidyl ester (fluorescein isothiocyanate [FITC], 4 μg ml−1; Molecular Probes), and examined by confocal microscopy (Bio-Rad MRC600 confocal scanning laser microscope [Kr/Ar] system with an MS plan ×60 1.4-numerical-aperture objective). Biofilms were observed with the reflected laser light of combined 488-, 546-, and 647-nm wavelengths. The images were analyzed with Image J 1.35c and Adobe Photoshop 6.0 software. MCID-M5 5.1 software was used to determine the total grain area. Biofilm formation by the InlJ mutant was visibly more sparse than that by the parent strain (Fig. 2A), and total accumulation was reduced by 46.2% (Fig. 2B). In addition, the average height across three random x-z sections of the mutant biofilm was reduced by 36.5% compared to the parent strain (Fig. 2C). These experiments demonstrate that InlJ is required for optimal homotypic biofilm formation by P. gingivalis.
FIG. 1.
Microtiter plate monospecies biofilm production by P. gingivalis 33277 and the InlJ mutant at 24 h and 48 h. Asterisks indicate a significant difference (P < 0.05, t test; n = 3) between the mutant and parental strains. OD, optical density.
FIG. 2.
(A) Confocal laser scanning microscopy projections of monospecies biofilm formation by P. gingivalis strains 33277 and the InlJ mutant after 24 h. Magnification, ×40. (B) Total grain area analysis of a 268.6- by 268.6-μm x-y section. (C) Average biofilm height of P. gingivalis accumulation across three random x-z sections. Asterisks indicate a significant difference (P < 0.05, t test; n = 3) between the mutant and parental strains.
On the tooth surfaces, P. gingivalis will be in contact with the diverse species that comprise the plaque biofilm. Thus, to begin to assess the role of InlJ in heterotypic biofilms, mixed S. gordonii-P. gingivalis biofilms were examined. S. gordonii is a common component of dental plaque (11, 13, 14) and is encountered by P. gingivalis upon initial colonization. S. gordonii cells were cultured for 24 h on chambered coverglass and stained with hexidium iodide (15 μg ml−1; Molecular Probes). P. gingivalis cells were stained with FITC as described above and reacted anaerobically with the S. gordonii biofilm for 24 h at 37°C in prereduced phosphate-buffered saline. After washing, accumulations of heterotypic biofilms were observed by confocal microscopy as described above. In contrast to the monospecies biofilm, the InlJ mutant formed more abundant accumulations within the mixed P. gingivalis-S. gordonii biofilm (Fig. 3A). This observation was supported by the total grain analysis of P. gingivalis accumulation (Fig. 3B). In addition, measurement of average biofilm height across three random x-z sections showed higher vertical accretion of the InlJ mutant than the parental strain (Fig. 3C). Hence, in the absence of InlJ, more luxuriant heterotypic biofilms are formed by P. gingivalis.
FIG. 3.
(A) Confocal laser scanning microscopy projections of mixed biofilms of S. gordonii DL1 with P. gingivalis strains 33277 and the InlJ mutant after 24 h. S. gordonii was prestained with hexidium iodide (red), and P. gingivalis was prestained with FITC (green). Magnification, ×40. (B) Total grain area analysis of a 268.6- × 268.6-μm x-y section. (C) Average biofilm height of P. gingivalis accumulation across three random x-z sections. Asterisks indicate a significant difference (P < 0.05, t test; n = 3) between the mutant and wild-type strains.
Biofilm accumulation proceeds through a series of developmental steps involving attachment of bacterial cells to a surface; accumulation by the recruitment of additional cells and proliferation; and, in certain cases, inclusion of additional species. Defined genetic profiles are considered important for distinct phases of biofilm development (7, 15). Regulation of biofilm development can be hypothesized to involve mechanisms that both stimulate an increase in biomass and limit or stabilize accumulation according to environmental constraints. For example, in Pseudomonas aeruginosa, biofilm depth is reduced by the transcription factor RpoS (4, 17). However, RpoS mutants of P. aeruginosa form biofilms of greater depth under flowing conditions (17). RpoS production is regulated at multiple levels, including transcription, translation, and proteolysis, in response to different stress conditions such as nutrient limitation (16). The results of the current study indicate that the InlJ protein of P. gingivalis is exploited to perform roles both in the stimulation of biofilm accumulation on abiotic surfaces and in biofilm control in the more complex and in vivo-relevant situation where other organisms are present. Such a multifunctional role is not inconsistent with the structure and properties of LRR proteins. Internalins of Listeria are involved in adherence, and adhesive activity mediated by InlJ may be important for biofilm initiation on abiotic surfaces. InlJ does possess a signal peptide and is therefore likely to be present on the surface of P. gingivalis. Adherence of P. gingivalis to S. gordonii, however, is mediated through the long and short fimbriae (6, 9) and thus may not require the presence of InlJ. LRRs can also be involved in signal transduction through their capacity to provide a versatile structural framework for the formation of protein-protein interactions. Such a role for InlJ may be important in constraining biofilm growth, possibly to avoid excessive exposure to oxygen in the oral cavity. As a strict anaerobe, P. gingivalis is likely to favor an existence deep within the plaque biofilm. Alternatively, restriction of biofilm development may be important to maintain the integrity of channels that allow nutrient penetration into the biofilm.
Acknowledgments
This work was supported by NIDCR DE12505 and DE11111.
Editor: V. J. DiRita
REFERENCES
- 1.Cabanes, D., P. Dehoux, O. Dussurget, L. Frangeul, and P. Cossart. 2002. Surface proteins and the pathogenic potential of Listeria monocytogenes. Trends Microbiol. 10:238-245. [DOI] [PubMed] [Google Scholar]
- 2.Chen, W., K. E. Laidig, Y. Park, K. Park, J. R. Yates III, R. J. Lamont, and M. Hackett. 2001. Searching the Porphyromonas gingivalis genome with peptide fragmentation mass spectra. Analyst 126:52-57. [DOI] [PubMed] [Google Scholar]
- 3.Cossart, P., J. Pizarro-Cerda, and M. Lecuit. 2003. Invasion of mammalian cells by Listeria monocytogenes: functional mimicry to subvert cellular functions. Trends Cell Biol. 13:23-31. [DOI] [PubMed] [Google Scholar]
- 4.Heydorn, A., B. Ersboll, J. Kato, M. Hentzer, M. R. Parsek, T. Tolker-Nielsen, M. Givskov, and S. Molin. 2002. Statistical analysis of Pseudomonas aeruginosa biofilm development: impact of mutations in genes involved in twitching motility, cell-to-cell signaling, and stationary-phase sigma factor expression. Appl. Environ. Microbiol. 68:2008-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kajava, A. V. 1998. Structural diversity of leucine-rich repeat proteins. J. Mol. Biol. 277:519-527. [DOI] [PubMed] [Google Scholar]
- 6.Lamont, R. J., A. El-Sabaeny, Y. Park, G. S. Cook, J. W. Costerton, and D. R. Demuth. 2002. Role of the Streptococcus gordonii SspB protein in the development of Porphyromonas gingivalis biofilms on streptococcal substrates. Microbiology 148:1627-1636. [DOI] [PubMed] [Google Scholar]
- 7.Moorthy, S., and P. I. Watnick. 2005. Identification of novel stage-specific genetic requirements through whole genome transcription profiling of Vibrio cholerae biofilm development. Mol. Microbiol. 57:1623-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 9.Park, Y., R. Simionato, K. Sekiya, Y. Murakami, D. James, W. Chen, M. Hackett, F. Yoshimura, D. R. Demuth, and R. J. Lamont. 2005. Short fimbriae of Porphyromonas gingivalis and their role in coadhesion with Streptococcus gordonii. Infect. Immun. 73:3983-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park, Y., O. Yilmaz, I. Y. Jung, and R. J. Lamont. 2004. Identification of Porphyromonas gingivalis genes specifically expressed in human gingival epithelial cells by using differential display reverse transcription-PCR. Infect. Immun. 72:3752-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quirynen, M., R. Vogels, M. Pauwels, A. D. Haffajee, S. S. Socransky, N. G. Uzel, and D. van Steenberghe. 2005. Initial subgingival colonization of ‘pristine’ pockets. J. Dent. Res. 84:340-344. [DOI] [PubMed] [Google Scholar]
- 12.Sabet, C., M. Lecuit, D. Cabanes, P. Cossart, and H. Bierne. 2005. LPXTG protein InlJ, a newly identified internalin involved in Listeria monocytogenes virulence. Infect. Immun. 73:6912-6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scannapieco, F. A., L. Solomon, and R. O. Wadenya. 1994. Emergence of human dental plaque and host distribution of amylase-binding streptococci. J. Dent. Res. 73:1627-1635. [DOI] [PubMed] [Google Scholar]
- 14.Socransky, S. S., A. D. Haffajee, M. A. Cugini, C. Smith, and R. L. Kent. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134-144. [DOI] [PubMed] [Google Scholar]
- 15.Stanley, N. R., and B. A. Lazazzera. 2004. Environmental signals and regulatory pathways that influence biofilm formation. Mol. Microbiol. 52:917-924. [DOI] [PubMed] [Google Scholar]
- 16.Venturi, V. 2003. Control of rpoS transcription in Escherichia coli and Pseudomonas: why so different. Mol. Microbiol. 49:1-9. [DOI] [PubMed] [Google Scholar]
- 17.Whiteley, M., M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teitzel, S. Lory, and E. P. Greenberg. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860-864. [DOI] [PubMed] [Google Scholar]
- 18.Zhang, Y., T. Wang, W. Chen, O. Yilmaz, Y. Park, I. Y. Jung, M. Hackett, and R. J. Lamont. 2005. Differential protein expression by Porphyromonas gingivalis in response to secreted epithelial cell components. Proteomics 5:198-211. [DOI] [PMC free article] [PubMed] [Google Scholar]