Abstract
Pseudomonas aeruginosa is a critical colonizer of the respiratory tract in cystic fibrosis. The chronic infections with this microorganism contribute to excessive inflammation and progressive lung damage in cystic fibrosis patients. The full repertoire of Pseudomonas products that promote inflammation in the cystic fibrosis lung is not known. Here we show that P. aeruginosa DNA released from the bacterium, but not human DNA from epithelial cells or Escherichia coli DNA, displays proinflammatory properties and induces human respiratory epithelial cells to secrete interleukin-8 (IL-8), a key chemokine causing excessive neutrophil infiltration in the cystic fibrosis lung. IL-8 secretion was not due to an increase in NF-κB- or activator protein-1-dependent IL-8 promoter transcription, but instead depended on p38 and Erk mitogen-activated protein kinases. No secretion of IL-8 was observed using conventional Toll-like receptor 9 ligands (CpG oligonucleotides), although it could be demonstrated that parts of the Toll-like receptor 9-signaling pathway were functional, since class B and C CpG oligonucleotide ligands stimulated production of RANTES chemokine. The IL-8 secretion in response to P. aeruginosa DNA was decreased by treatments that inhibit acidification of intracellular organelles, using chloroquine, a pH-neutralizing compound, or bafilomycin A1, an inhibitor of vacuolar H+-ATPase. These data indicate that DNA released from P. aeruginosa during chronic infections may significantly contribute to the proinflammatory processes in cystic fibrosis. Our findings also show that treatments with drugs diminishing organellar acidification may reduce the inflammatory response in cystic fibrosis.
Cystic fibrosis (CF) is the most common autosomal recessive disease in Caucasians and is caused by mutations in the gene encoding the cystic fibrosis transmembrane conductance regulator (CFTR) (57). This complex disease is characterized by chronic bacterial infection and inflammation of the airways, pancreatic exocrine insufficiency, gastrointestinal involvement, infertility in males, and inefficient salt absorption from sweat fluids (57). The most common cause of death in CF is respiratory failure as a result of progressive pulmonary decline (47) due to chronic infections and inflammation (24). It has not been resolved whether the excessive inflammation is simply a response to persistent infection (6) or whether the primary defect in CFTR directly contributes to the intrinsically higher excessive proinflammatory response (36) by CF respiratory epithelial cells (41). Regardless of the exact etiology, the chronic lung inflammation remains largely responsible for the life-shortening effects of CF. A hallmark of CF airway inflammation is accumulation of interleukin-8 (IL-8) acting as a chemokine, attracting polymorphonuclear cells/neutrophils, which in turn contribute to tissue destruction (8, 36, 38, 45).
The predominant lung pathogen in CF is Pseudomonas aeruginosa, cultured in specimens from more than 80% of CF patients aged 26 years or older (24). Many P. aeruginosa products, including lipoproteins, flagella, exopolysaccharide alginate, pili, lipopolysaccharide (LPS), and toxins (1, 14, 17, 20, 27), belong in the general group of pathogen-associated molecular patterns (PAMPs). PAMPs reflect conserved microbial molecular features and are recognized by host cells via pattern recognition receptors (PRRs), which discriminate between self and pathogen products (34, 48). Toll-like receptors (TLRs) and nucleotide-binding oligomerization domain (Nod) proteins are two classes of PRRs involved in innate immune detection (4). Signaling from these receptors can result in activation of mitogen-activated protein kinases (MAPKs), NF-κB, or type I interferon pathways (4, 48). Pseudomonas challenge of human tracheobronchial epithelial cell cultures causes MAPK activation, besides NF-κB, and is associated with IL-8 secretion (65). MAPK activation by PAMPs has been demonstrated with various ligands, including bacterial lipopeptides, LPS, flagellin, and CpG DNA (2, 3, 23, 29, 58, 67, 69). There are different patterns of MAPK activation, depending upon the stimulus and cell type, with p38 being the most common MAPK activated upon engagement of TLRs.
Signals from various PRRs are essential for innate immunity against P. aeruginosa (61). For example, TLR4 plays a protective role against cytotoxic P. aeruginosa (19). Although P. aeruginosa LPS is a relatively weak TLR4 agonist (18), LPS modifications seem to modulate its potency (27). Pseudomonas flagella activate airway epithelia via TLR5 or through TLR2 in cooperation with aGM1 (1). Pseudomonas ExoS stimulates both TLR2 and TLR4 (17), while alginate rich in mannuronate may too act via TRL2 and TLR4 (21). However, when innate immunity clearance fails, a continuing stimulation of PRRs can become a double-edged sword. We have recently shown that upon conversion to the mucoid, alginate-overproducing phenotype typically encountered in CF, P. aeruginosa upregulates a large number of lipoproteins, termed lipotoxins (20), which further augment proinflammatory proteins. A P. aeruginosa lipotoxin has also been shown to play a role in the allergic response via prolonged stimulation of the TLR2 and TLR4 signaling pathways (55). Interestingly, in the absence of normal recognition of P. aeruginosa pili by TLR2, there is a strong hyperreactive response modulated by TLR4 (42), attesting to the contribution of PAMPs and PRRs to unproductive and potentially damaging inflammatory responses.
PRRs seem to be equally expressed in CF and non-CF respiratory epithelial cells, and their responses appear mainly indistinguishable (25, 46), with the exception of aGM1-augmented effects on TLR2 signaling in CF cells (1, 46, 62). In general, it is possible that continuous stimulation with bacterial products, instead of helping clear the microbes, stokes the runaway inflammation in CF. Although a number of candidate P. aeruginosa PAMPs have been identified (1, 17, 20, 27, 55, 62), the full repertoire remains to be assessed, and it is not known at present whether even-more-potent stimulators of inflammation are generated by P. aeruginosa. Due to a large number of bacterial cells generally found in the CF lung, it is certain that the DNA released from the dying bacteria may act in a way similar to CpG oligonucleotides. The CpG oligonucleotides have been used to study TLR9 signaling, as they mimic the absence of CpG methylated motifs in bacterial chromosomal DNA (30, 40). In this work we explored the possibility that P. aeruginosa DNA, due to its high GC content (66.6%) and hence overabundance of unmethylated CpG motifs, may act as a proinflammatory P. aeruginosa product. We tested whether P. aeruginosa DNA stimulated IL-8 secretion from human airway epithelial cells and investigated the signaling pathways controlling these processes.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Nonmucoid P. aeruginosa PAO1 was from a stock originally received from B. Holloway. Mucoid P. aeruginosa PA578I (Alg+; mucA22) has the most common mucA mutation found in mucoid CF isolates (9). Bacteria were grown in L broth.
Cell culture.
The bronchial epithelial cell lines IB3-1 (ΔF508/W1282X heterozygote) and S9 (created by correcting IB3-1 cells via introduction of a functional CFTR) (16) were grown on LHC-8 medium (Bio-fluids, Rockville, MD) supplemented with 10% fetal bovine serum and antibiotics. Primary normal human bronchial epithelial (NHBE) cells were obtained from a commercial source (Cambrex Bio Science, Baltimore, MD) without identifiers (exempt status from the Institutional Review Board and NIH). NHBE cells were cultured in serum-free bronchial epithelial growth medium with BEGM Single Quots supplements (Cambrex Bio Science). CuFi1 cells, derived from a CF patient, were developed by J. Zabner (70) and were cultured in collagen (Sigma, St. Louis, MO)-coated flasks in serum-free bronchial epithelial growth medium supplemented with BEGM Single Quots (Cambrex Bio Science). All cells were grown at 37°C in 5% CO2. Submerged cultures of respiratory epithelial cells have been used as reported routinely in studies addressing immunological responses. All control samples contained carrier solvent added in equivalent amounts to the treatment specimens.
Genomic DNA preparations and CpG oligonucleotides.
DNAs were prepared using a Wizard genomic DNA purification kit (Promega). All DNAs were suspended in DNase-free, LPS-free distilled water. DNA preparations were denatured before stimulation by heating at 95°C for 5 min, followed by rapid cooling on ice. For some experiments, DNA was methylated with 2 U/μg DNA of CpG methylase M.SssI (New England Biolabs) for 4 h at 37°C. When indicated, DNA was digested with DNase I (Novagen) in 20 mM Tris-HCl, pH 7.5, containing 2 mM MgCl2 and 1 U DNase I/μg DNA for 1 h at 37°C, and DNase I was inactivated by heating for 5 min at 95°C. Human DNA was prepared from respiratory epithelial cells. CpG oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA): CpG A D19, ggTGCATCGATGCAGggggG (63); CpG B 2006, tcgtcgttttgtcgttttgtcgtT (28); and CpG C 2395, tcgtcgttttcggcgcgcgccG (64). (Lowercase letters indicate phosphorothioate linkages; capital letters indicate phosphodiester linkage 3′ of the base.)
Transfections and luciferase reporter assays.
For monitoring transient NF-κB, pAP-1, or pIL-8 activation, the IB3-1 cell line was seeded at 2 × 104 cells per well in a 96-well plate, and 18 to 22 h later cells were transfected using Effectene transfection reagent (QIAGEN) with 48 ng of an NF-κB-responsive luciferase reporter construct (pBVIII-luc, containing three tandem repeats of two NF-κB sites; from G. Nunez, University of Michigan Medical School), 16 ng of a β-galactosidase construct (pEF1-Bos; generously provided by G. Nunez, University of Michigan Medical School) for normalization, and 36 ng of pCDNA3, or 60 ng of pAP-1-Luc plasmid (Stratagene), 20 ng of a β-galactosidase construct for normalization, and 20 ng of pCDNA3, or 75 ng of pIL-8:luc reporter plasmid (containing the 5′-flanking region of the IL-8 gene from −133 to −50; from N. Mukaida, Kanazawa University, Ishikawa, Japan), and 25 ng of a β-galactosidase construct for normalization. The positive control for AP-1-transfected cells was cotransfection with the pFC-MEKK plasmid (Stratagene), transfecting with 60 ng of pAP-1-Luc plasmid, 20 ng of a β-galactosidase construct, 12 ng of pCDNA3 plasmid, and 8 ng of pFC-MEKK plasmid. Transfection mixtures were placed on the cells for 6 h, after which the transfection mixture was removed and replaced with complete growth medium. At 24 h following transfection, cells were incubated with growth medium, serum free, for 21 h and then cells were incubated with stimulus in serum-free medium for 2, 4, 6, 8, or 24 h. Luciferase activity was assayed using the luciferase reagent (Promega), and β-galactosidase activity was assayed using the Galacto-Star luminescence system (Tropix, Bedford, MA). Transfection efficiency was controlled by standardizing luciferase activity to constitutive β-galactosidase production. Stimuli were human tumor necrosis factor alpha (TNF-α) at 20 ng/ml (Sigma Chemical Co.) or DNA at 25 μg/ml.
Ratiometric fluorescence microscopy, pH measurements, and endosomal pH manipulation.
Cellubrevin-pHluorin (pH-sensitive green fluorescent protein [GFP]) was from J. Rothman (43). Cells were transfected with 1 μg/ml DNA using Lipofectin for 6 h at 37°C, 5% CO2. Ratiometric fluorescence microscopy was carried out using an Olympus IX-70 microscope and Olympix KAF1400 charge-coupled-device camera (LSR). The ratio of emission at 508 nm upon excitation at 410 nm versus 470 nm and calibration curves were generated as previously described (52, 53). For pH normalization, cells were grown in the presence of 0.1 mM chloroquine, as previously described (52, 53).
NF-κB nuclear translocation assay and confocal microscopy.
Respiratory cells were seeded in a 24-well plate and 18 to 22 h later incubated in serum-free culture medium for an additional 21 h. Cells were stimulated for the indicated times with medium alone (control), 20 ng/ml TNF-α, or 25 μg/ml PAO1 DNA. Cells were washed three times with phosphate-buffered saline (PBS), fixed with 2% paraformaldehyde for 20 min, washed three times with PBS, permeabilized with 0.2% saponin for 5 min, washed three times with PBS, blocked for 30 min, and incubated with monoclonal antibodies against p65 NF-κB (Santa Cruz) overnight. Slides were washed three times with PBS, incubated with a secondary anti-mouse fluorescein isothiocyanate-conjugated antibody, washed, incubated with 300 nM 4′,6′-diamidino-2-phenylindole (DAPI; Molecular Probes) for 5 min, and finally washed and mounted in mounting medium and examined by confocal microscopy using a Zeiss 510 META microscope. Overlaps between DAPI and NF-κB stains were scored as positive nuclear translocation events.
IL-8 and RANTES assays.
To analyze IL-8 protein expression, 2 × 104 cells/well (IB3-1, S9, or NHBE cells) or 1.5 × 104 cells/well (CuFi1) were seeded in 96-well plates, and 18 to 22 h later cells were incubated with serum-free growth medium for 21 h (IB3-1 and S9 cells) and then cultured in the presence and absence of 20 ng/ml human TNF-α (Sigma Chemical Co.), 10 ng/ml P. aeruginosa LPS (Sigma Chemical Co.), 5 to 30 μg/ml DNA, or 3 μM CpG oligonucleotides for the specific indicated time. CuFi1 and NHBE primary cells were stimulated 18 to 22 h after seeding them, using the growth medium with supplements. After culture, IL-8 was measured in supernatants using an enzyme-linked immunosorbent assay (ELISA) kit for IL-8 (R&D Systems, Minneapolis, MN). ELISAs were performed according to the manufacturer's protocol. When indicated, preincubation (1 h at 37°C) of DNAs or LPS with 10 μg/ml polymyxin B (Sigma Chemical Co.) was used to abrogate the effect of LPS. When indicated, 15 μg/ml cycloheximide was added to cells 2 h before stimulation. When indicated, the MAPK inhibitors were added to cells 2 h (SB203580 and PD98059 [Calbiochem]) or 30 min (SP600125 [A.G. Scientific, Inc., California]) before stimulation at the indicated concentrations. When indicated, 20 μM chloroquine (Sigma Chemical Co.) or 30 and 300 nM bafilomycin A1 (Calbiochem) was added to the cells 30 min before stimulation. RANTES was assayed using culture supernatants from 2 × 104 cells/well seeded in a 96-well plate for 18 to 22 h. Adherent cells were incubated in serum-free growth medium for 21 h and then cultured in the presence and absence of 20 ng/ml human TNF-α, 25 μg/ml PAO1 DNA, or 3 μM oligonucleotides for 24, 48, or 72 h. RANTES was measured in supernatants using a human RANTES/CCL5 ELISA kit (Biosource International, Camarillo, CA).
Statistical analysis.
Experiments were repeated 3 (Fig. 1A and B, 2A, 3A and C, 5, 6, 7A and 8A and B), 6 (Fig. 1C, 3B, and 7B), 9 (Fig. 2B), or 10 times (Fig. 8C and D). All the statistical analyses were performed using Fisher's protected least significant difference post hoc test analysis of variance (ANOVA; SuperANOVA version 1.11; Abacus Concepts, Inc., Berkeley, CA). Data are reported as means ± standard errors. A P value of <0.05 was considered significant relative to the untreated control.
FIG. 1.
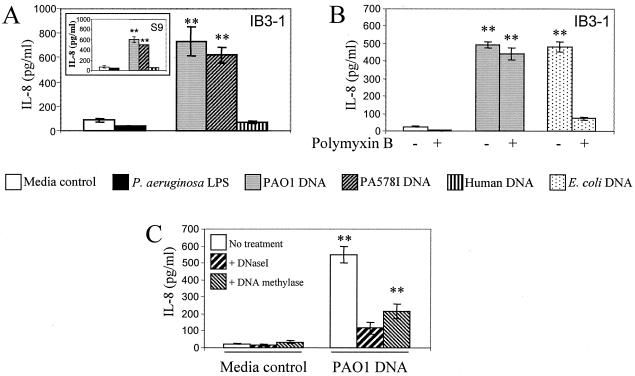
P. aeruginosa DNA stimulates IL-8 secretion in human respiratory epithelial cell lines. A. IB3-1 cells derived from a CF patient were incubated for 24 h with medium alone, 10 ng/ml P. aeruginosa LPS, 25 μg/ml of P. aeruginosa PAO1 DNA, P. aeruginosa PA578I DNA, or DNA from human cells (C38 cell line). Inset, IL-8 secretion by S9 cells (IB3-1 corrected by stable transfection with full-size functional CFTR). B. IB3-1 cells were incubated with medium control, 25 μg/ml P. aeruginosa PAO1 DNA, or E. coli DNA for 24 h, with or without polymyxin B (10 μg/ml). C. IB3-1 cells were incubated with 25 μg/ml P. aeruginosa PAO1 DNA pretreated with DNase I for 1 h at 37°C or with CpG methylase (M.SssI) for 4 h at 37°C. Values represent means and standard errors. **, P < 0.01 (ANOVA).
FIG. 2.
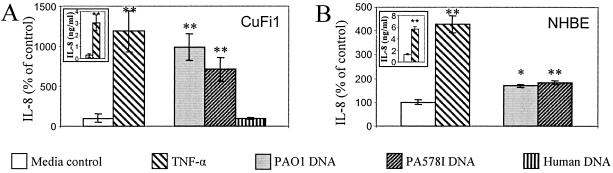
P. aeruginosa DNA stimulates IL-8 secretion in primary human airway epithelial cells. A. CuFi1 cells (derived from a CF patient) were incubated for 24 h with medium alone, 20 ng/ml TNF-α, 25 μg/ml of DNA from P. aeruginosa PAO1, P. aeruginosa PA578I, or human cells (S9). B. Primary NHBE cells. Values represent percent relative to control. Insets, absolute IL-8 values, in nanograms per milliliter. *, P < 0.05; **, P < 0.01 (ANOVA).
FIG. 3.
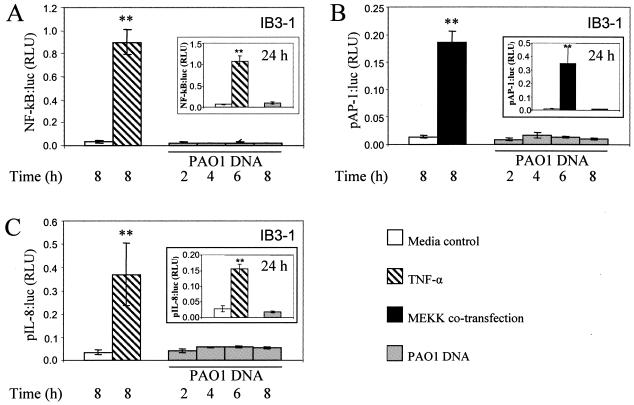
P. aeruginosa DNA does not activate the IL-8 promoter in airway epithelial cells. IB3-1 cells were cotransfected with β-galactosidase and NF-κB:luciferase (A), pAP-1:luciferase (B), or pIL-8:luciferase (C) as reporters. As a positive control for pAP-1:luciferase, cells were cotransfected with the MEKK gene (B). Cells were incubated for 8 or 24 h (insets) with medium alone, with 20 ng/ml TNF-α, or for 2, 4, 6, 8, or 24 h (insets) with 25 μg/ml P. aeruginosa PAO1 DNA. Luciferase activities were normalized to β-galactosidase. **, P < 0.01 (ANOVA).
FIG. 5.
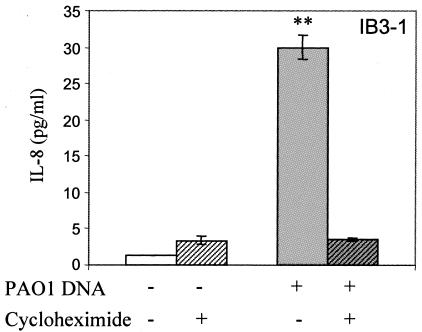
IL-8 secretion in response to P. aeruginosa DNA is protein synthesis dependent. IB3-1 cells were incubated with medium (open bars) or with 25 μg/ml P. aeruginosa PAO1 DNA (gray bars) for 2 h, without or with (striped bars) 15 μg/ml cycloheximide. Cells were preincubated for 2 h with cycloheximide before stimulation. **, P < 0.01 (ANOVA).
FIG. 6.
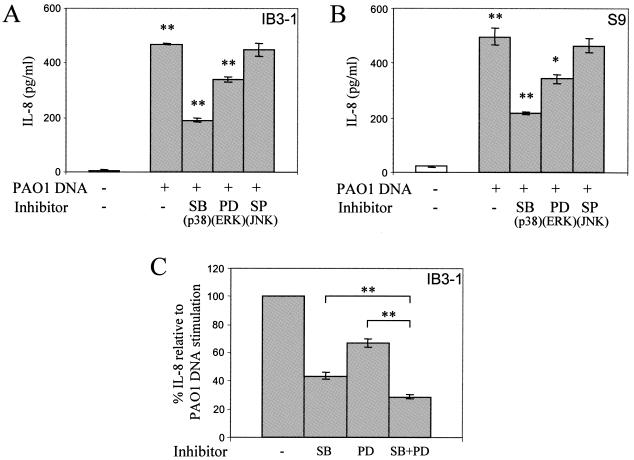
Upregulation of IL-8 secretion by P. aeruginosa DNA in airway epithelial cells depends on p38 and ERK MAPK signaling. IB3-1 (A and C) and S9 (B) cells were incubated with medium alone (open bars) or with 25 μg/ml P. aeruginosa PAO1 DNA (gray bars) for 24 h in the absence or presence of 10 μM SB203580 (p38 inhibitor), 20 μM PD98059 (Erk signaling inhibitor), or 5 μM SP600125 (JNK inhibitor). Cells were preincubated for 2 h before the stimulation with SB203580 and PD98059 and for 30 min with SP600125. **, P < 0.01 (ANOVA); lines indicate compared samples.
FIG. 7.
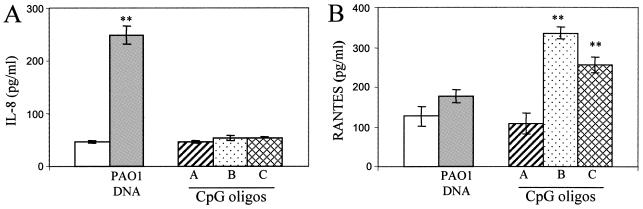
Human respiratory epithelial cells are differentially responsive to CpG oligonucleotides and P. aeruginosa DNA, as reflected in RANTES versus IL-8 secretion profiles. IB3-1 cells were incubated for 24 h (IL-8) or 72 h (RANTES) in medium (open bars), 25 μg/ml PAO1 DNA (striped bars), or 3 μM CpG oligonucleotides (patterned bars). Class A CpG oligonucleotide (ggTGCATCGATGCAGggggG), class B CpG oligonucleotide (tcgtcgttttgtcgtcgttttgtcgtT), and class C CpG oligonucleotide (tcgtcgttttcggcgcgcgccG) (lowercase letters indicate phosphorothioate linkages; uppercase letters indicate a phosphodiester linkage 3′ of the base) have been previously described (28, 63, 64).
FIG. 8.
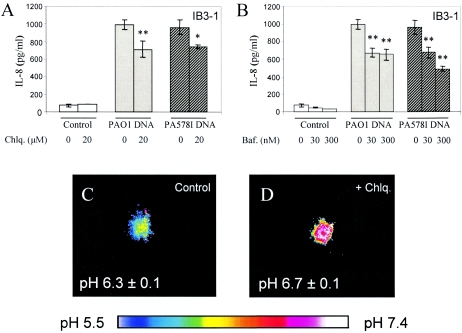
IL-8 secretion by airway epithelial cells and organellar acidification. IB3-1 cells were incubated for 24 h with medium alone (open bars), 25 μg/ml of PAO1 P. aeruginosa DNA (gray bars), or PA578I P. aeruginosa DNA (hatched bars) in the absence or presence of 20 μM chloroquine (A) or in the absence or presence of 30 or 300 nM bafilomycin A1 (B). When chloroquine or bafilomycin A1 was present, cells were preincubated with inhibitors for 30 min prior to stimulation. (C and D) Correction of endosomal hyperacidification by chloroquine treatment in IB3-1 cells transfected with cellubrevin-pHluorin (pH-sensitive GFP) used as an organelle lumenal pH probe (see Materials and Methods). The color-coded look-up table corresponds to pH values in panels C and D. Ratiometric analysis was carried out as previously described (52, 53). n = 10; P = 0.0488 (ANOVA).
RESULTS
Pseudomonas aeruginosa DNA stimulates IL-8 secretion in CF respiratory epithelial cells.
Total DNA was prepared from P. aeruginosa and tested for effects on IL-8 secretion by lung epithelial cells. IB3-1 bronchial epithelial cells, derived from a CF patient carrying CFTR mutations ΔF508 and W1282X, were incubated with PAO1 DNA. This resulted in stimulated secretion of IL-8 relative to resting cells (Fig. 1A). Human DNA, extracted from respiratory epithelial cells, used at the same concentration did not stimulate IL-8 secretion (Fig. 1A). To rule out the possibility that it was the LPS contamination in the DNA preparation that stimulated IL-8 secretion, we incubated IB3-1 cells with P. aeruginosa LPS for the same amount of time. P. aeruginosa LPS did not stimulate cells to secrete IL-8 (Fig. 1A). This was further supported by experiments in which polymyxin B was added to P. aeruginosa DNA, to block any contaminating LPS effects. As shown in Fig. 1B, the ability of P. aeruginosa DNA to induce IL-8 secretion in IB3-1 cells was not affected by coincubation with polymyxin B. However, polymyxin B decreased IL-8 secretion in response to Escherichia coli DNA (low GC content) to control levels (Fig. 1B). With increasing amounts of P. aeruginosa DNA, from 5 to 30 μg/ml, IL-8 secretion was stimulated in a dose-dependent manner (data not shown). Since stimulation with 25 μg/ml gave maximal induction, this was used as the standard concentration in all subsequent experiments.
We next tested whether genetically matched CF and CFTR-corrected cells had different responses to P. aeruginosa DNA. S9 cells, which are IB3-1 CFTR-corrected cells (16), responded at levels similar to the parental CF cells (IB3-1) upon stimulation with P. aeruginosa DNA (Fig. 1A, inset). TNF-α also elicited similar amounts of IL-8 in IB3-1 and S9 cells (4,500 ± 254 versus 4,566 ± 512 pg/ml of IL-8). S9 cells, like IB3-1 cells, did not respond to human DNA or to P. aeruginosa LPS (Fig. 1A, inset).
To show that DNA was responsible for our observations, P. aeruginosa DNA preparations were pretreated with DNase I prior to evaluating IL-8 secretion in IB3-1 cells. Treatment with DNase I abrogated the stimulatory effect of the P. aeruginosa DNA preparation (Fig. 1C). As the known receptors for bacterial DNA (e.g., TLR9) specifically recognize nonmethylated CpG motifs in DNA (30), we pretreated DNA with CpG methylase to convert the CpG motifs in P. aeruginosa DNA to their methylated form. Figure 1C shows that CpG methylation decreased the response to P. aeruginosa DNA.
To examine whether P. aeruginosa DNA stimulates IL-8 secretion in respiratory epithelial cells in general, we tested additional primary cells and cell lines. CuFi1, a cell line derived from a patient homozygous for CFTR ΔF508 (70), also responded to P. aeruginosa DNA by increasing IL-8 secretion (Fig. 2A). The response to P. aeruginosa DNA preparations was comparable to the response to TNF-α. Just like IB3-1 cells, CuFi1 did not respond to human DNA (Fig. 2A).
We next tested the response in primary, NHBE cells. NHBE cells showed increased IL-8 secretion after P. aeruginosa DNA stimulation, albeit to a lesser extent than IB3-1 and CuFi1 cells, but responded effectively to TNF-α stimulation (Fig. 2B).
DNA preparations from mucoid or nonmucoid P. aeruginosa display similar IL-8 stimulatory potential.
We next compared DNA preparations from mucoid alginate-overproducing P. aeruginosa (PA578I) and its nonmucoid ancestral strain (PAO1), since conversion to mucoidy is a well-recognized pathogenic determinant of P. aeruginosa expressed in CF and associated with disease deterioration (24). DNA preparations from mucoid and nonmucoid P. aeruginosa elicited similar IL-8 levels in all cells tested (Fig. 1 and 2).
P. aeruginosa DNA engages a signaling pathway independent of NF-κB activation.
The conventional mechanism of IL-8 increase upon stimulation with CpG DNA is activation of an NF-κB pathway following TLR9 engagement (5, 32). To test whether NF-κB was involved in stimulation of IL-8 production in response to P. aeruginosa DNA, IB3-1 cells were transfected with an NF-κB-luciferase reporter and NF-κB-dependent promoter activation was monitored in response to Pseudomonas DNA. Unexpectedly, NF-κB was not activated by P. aeruginosa DNA at 2, 4, 6, 8, or 24 h, while a typical strong NF-κB response was observed upon activation with TNF-α (Fig. 3A). NF-κB activation was not detected, although increased levels of IL-8 were detected in the supernatants (data not shown). As AP-1 is another important transcriptional activator of the IL-8 promoter, we tested whether an AP-1 induction pathway was responsible for IL-8 production/secretion. IB3-1 cells were transfected with a pAP-1-luciferase reporter plasmid and stimulated with P. aeruginosa DNA. As is the case with NF-κB, P. aeruginosa DNA did not activate pAP-1 at 2, 4, 6, 8, or 24 h (Fig. 3B). This contrasted with the positive control, consisting of cells cotransfected with MEKK, which showed a strong AP-1:luc activation (Fig. 3B).
Finally, we tested the entire previously characterized IL-8 promoter, containing all known regulatory sites that control IL-8 transcription (including NF-κB and AP-1 sites) (32). IB3-1 cells were transfected with the IL-8 promoter-luciferase reporter plasmid and then stimulated with P. aeruginosa DNA for 2, 4, 6, 8, or 24 h. As with individually tested NF-κB and AP-1 probes, we did not detect transcriptional activation of the IL-8 promoter upon stimulation with P. aeruginosa DNA, albeit the promoter responded to TNF-α as expected (Fig. 3C).
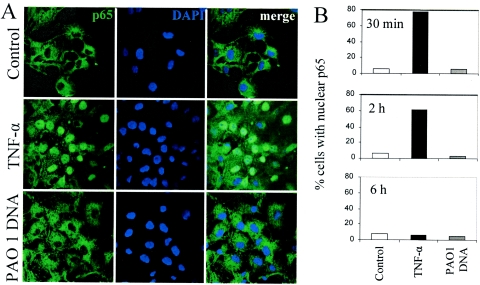
NF-κB activation could not be detected by a variety of assays (not shown). This included absence of p65 translocation to the nucleus upon DNA stimulation, in contrast to the positive control with TNF-α (Fig. 4). The p65 polypeptide is a member of the Rel/NF-κB family of proteins and is considered to be a strong activator when present in dimeric complexes that constitute active, DNA-binding forms of NF-κB (60).
FIG. 4.
Nuclear translocation of p65 NF-κB in human respiratory epithelial cells in response to stimulation with TNF-α but not to P. aeruginosa DNA. IB3-1 cells in serum-free medium were incubated for different times in control medium or medium with 20 ng/ml TNF-α or 25 μg/ml PAO1 DNA. Cells were fixed and subjected to immunofluorescence analysis with antibodies against the p65 subunit of NF-κB and counterstained with the nuclear stain DAPI. (A) Confocal immunofluorescence microcopy images with cells incubated for 30 min. (B) Quantification of p65 nuclear translocation in cells incubated for 30 min, 2 h, and 6 h. Sample sizes and numbers of nuclei counted: for the 30-min stimulation, n = 139 (control), n = 136 (TNF-α), and n = 118 (PAO1 DNA); for the 2-h stimulation, n = 127 (control), n = 115 (TNF-α), and n = 148 (PAO1 DNA); for the 6-h stimulation, n = 135 (control), n = 113 (TNF-α), and n = 173 (PAO1 DNA).
P. aeruginosa DNA activates IL-8 in a protein synthesis-dependent manner.
We next tested whether the increased IL-8 secretion was dependent on protein synthesis. For this purpose, we stimulated IB3-1 cells with P. aeruginosa DNA in the presence or absence of 15 μg/ml cycloheximide, which acts as a protein synthesis inhibitor. We selected 2 h as the time to monitor IL-8 levels, due to the previously described cycloheximide superinduction phenomenon, manifested as a paradoxical increase in IL-8 production upon treatment with this protein synthesis inhibitor, which occurs by mRNA stabilization and/or enhancing transcription (data not shown) (7, 56). IL-8 secretion after 2 h of incubation in the presence of P. aeruginosa DNA was reduced to the basal level in the presence of cycloheximide (Fig. 5). A small enhancement in the control sample is a reflection of the superinduction phenomenon, which was more pronounced at later times (data not shown).
P. aeruginosa DNA stimulation of IL-8 production depends on MAPK signaling.
Secretion of a number of cytokines (including IL-8) can be stimulated by MAPK via posttranscriptional regulatory mechanisms (22, 35, 69). We tested whether MAPK signaling was involved in the IB3-1 cell response to P. aeruginosa DNA by using specific inhibitors for the three classical MAPK pathways: p38, ERK, and JNK. Figure 6A shows that SB203580, a specific inhibitor of p38, decreased IL-8 secretion stimulated by P. aeruginosa DNA in IB3-1 cells. Also, PD98059, a specific ERK inhibitor, decreased DNA-stimulated IL-8 secretion. In contrast, inhibition of JNK with SP600125 did not affect P. aeruginosa DNA stimulation of IL-8 secretion. A similar pattern of inhibition was observed in S9 cells (Fig. 6B). The actions of p38 and ERK inhibitors were partially additive (Fig. 6C).
Differential induction of chemokine secretion in human respiratory epithelial cells in response to CpG oligonucleotides and P. aeruginosa DNA.
We next attempted to clarify whether TLR9 responses are functional in human respiratory epithelial cells by using the well-defined CpG oligonucleotide ligands for TLR9. Applying three different classes of CpG oligonucleotides, we found that they did not activate NF-κB (data not shown) and they also did not stimulate IL-8 secretion (Fig. 7A). Since it has been recently described (33) that TLR7 and TLR9 can induce type I interferons (IFN-α/β) via an IRF-dependent pathway that can moreover interfere with NF-κB activation, we analyzed secretion of RANTES as a type I IFN-stimulated chemokine (12). Secretion of RANTES was detected in IB3-1 cells stimulated for 72 h with 3 μM CpG oligonucleotides (classes B and C, but not A) (Fig. 7B). RANTES secretion was not stimulated by CpG ligands at 24 h, and it was only mildly stimulated at 48 h (data not shown), as expected from the type I IFN response. Conversely, PAO1 DNA did not stimulate RANTES secretion in IB3-1 cells at 24, 48 (data not shown), or 72 h (Fig. 7B), but it did stimulate IL-8 secretion (Fig. 7A), as discussed above.
IL-8 secretion in response to P. aeruginosa DNA stimulation is affected by organellar acidification.
Bacterial DNA recognition by host cells depends on endosomal acidification (26, 29, 40). We tested whether the response to P. aeruginosa DNA can be affected by modulating endosomal acidification. Chloroquine, a weak base which diminishes CpG signaling by neutralizing endosomal pH (26, 29, 66-68), and bafilomycin A1, an inhibitor of the vacuolar H+ pump, were used to treat cells during stimulation with P. aeruginosa DNA. The data shown in Fig. 8A and B indicate that IL-8 secretion by IB3-1 cells in response to P. aeruginosa DNA was decreased when organellar acidification was blocked. Using pH-sensitive GFP targeted to endosomal compartments (43, 53), we examined the pH of endosomes in IB3-1 cells treated with chloroquine and determined that the endosomal pH was increased from 6.3 ± 0.1 to 6.7 ± 0.1 (Fig. 8C and D).
DISCUSSION
Inflammation in the CF lung is a neutrophil-dominated event. The respiratory epithelium is believed to play an important role, since CFTR is primarily expressed in epithelial cells (57). The epithelium may regulate neutrophil recruitment via production of IL-8 (41), although intrinsic abnormalities in IL-8 secretion by CF epithelia have been questioned by others (6). IL-8 is a potent activator and chemoattractant of neutrophils and is secreted by epithelial cells in response to a variety of stimuli (32). Here we report that DNA released from P. aeruginosa acts as a potent stimulator of IL-8 production by human respiratory epithelial cells.
The IL-8 response in human respiratory epithelial cells in our experiments is most likely specific for P. aeruginosa DNA. (i) DNase I treatment decreased stimulation to the basal level. (ii) P. aeruginosa LPS was not a stimulus contaminating DNA preparations, because polymyxin B did not block P. aeruginosa DNA stimulation. We also observed that LPS purified from P. aeruginosa did not elicit IL-8 secretion in airway epithelial cell lines. (iii) It seems highly unlikely that the response observed in our studies was due to contamination with lipopeptides (20), flagellin (10, 14), or pilin (14), because all of these PAMPs activate NF-κB and P. aeruginosa DNA did not do so in our experiments. (iv) Alginate as a contaminant could not be a contributory factor because we did not see any difference between DNA preparations from mucoid or nonmucoid Pseudomonas strains. Moreover, a previous report showed no IL-8 secretion by alginate-stimulated airway epithelial cells (10). (v) Pseudomonas autoinducer could not be present as a contaminant in the concentration necessary to induce IL-8 secretion (more than 10 μM), because PAO1 cultures in stationary phase contain ∼5 μM of Pseudomonas autoinducer (14, 50). (vi) Pyocyanin is not a likely contaminant inducing IL-8 secretion, since pyocyanin increases IL-8 secretion by inducing IL-8 transcription (13) and we did not detect transcriptional activation of the IL-8 promoter.
The concentration of P. aeruginosa DNA that elicited the maximum IL-8 secretion in 24 h was 25 μg/ml. This corresponded approximately to a bacterial density of 2.5 × 108 CFU/ml, a bacterial load that can be found in CF patients (51). Although the total DNA concentration in bronchoalveolar lavage fluids in CF patients is 17.6 ± 11.2 μg/ml (mean ± standard deviation) (54), most likely including human neutrophil and bacterial DNA (37), the local concentration of bacterial DNA in contact with epithelial cells may be higher due to bacterial adhesion or biofilm formation. Significantly, DNase I treatment decreased P. aeruginosa DNA-dependent stimulation of IL-8 secretion in our studies. These results support the benefits of treatments in CF with agents that degrade DNA. For example, Dornase alpha, a recombinant human DNase, has been shown to improve lung function and reduce the number of pulmonary exacerbations in patients with CF (31, 49). Dornase alpha effects have been attributed to facilitated expectoration upon reduction of mucus viscosity due to degradation of DNA released from dead neutrophils (49, 54). In the present report, we show that degradation of P. aeruginosa DNA may be a component of the beneficial action of Dornase alpha by decreasing its contribution to proinflammatory action (49, 59).
In vitro CpG methylation of P. aeruginosa DNA diminished its ability to stimulate IL-8 secretion in our experiments. These observations indicate the possibility that the TLR9 signaling pathway may be involved. To examine TLR9 functional signaling in IB3-1 cells, we stimulated airway epithelial cells with several CpG oligonucleotides (classes A and B) at the most commonly used concentration, 1 μM, but we could not see any NF-κB activation or IL-8 secretion upon 24 h of stimulation. Greene et al. showed a low-level increase of IL-8 secretion after 48 to 72 h of stimulation with high concentrations (15 μM) of CpG oligonucleotide (25). Nevertheless, in concurrence with our findings, Greene and colleagues reported that IL-8 secretion was stimulated by CpG oligonucleotides in a CF tracheal epithelial cell line in a process independent of NF-κB activation (25). In keeping with these and our observations, a CpG-dependent increase of IL-8 secretion in human colonic epithelial cells has been shown to be NF-κB independent and MAPK dependent (3).
Our results with a lack of activation of the IL-8 promoter in the luciferase assay and P. aeruginosa DNA stimulation of IL-8 secretion by airway epithelial cells in a p38- and ERK-dependent manner are best explained by posttranslational mechanisms. The MAPK p38 is known as an mRNA stabilizer of many cytokine transcripts containing adenylate- and uridylate-rich elements in the 3′ untranslated region, including IL-8 transcript (22, 35). The MAPK p38 can also participate in derepression of translation via these elements (39, 69). ERK has been involved in transport from the nucleus to the cytoplasm of cytokine mRNA (15). According to our results, it seems possible that IL-8 secretion is being stimulated by P. aeruginosa DNA in airway epithelial cells by inducing posttranscriptional mechanisms akin to the ones listed above. Furthermore, exocytosis induction of the preformed IL-8 protein, via p38 and ERK pathways, may also be taking place (11, 44).
P. aeruginosa DNA stimulation was partially sensitive to chloroquine and bafilomycin A1, compounds that block endosomal acidification essential in TLR9 signaling (26, 29, 40, 66-68). It has been demonstrated that organelles are hyperacidified in CF respiratory epithelial cells, including endosomal compartments (52, 53). Taking this into account, along with the fact that endosomal acidification is requisite to CpG signaling via TLR9 (26, 29, 40) and that airway epithelial cells express a variety of TLRs, including TLR9 (3, 25, 29, 46, 51), we anticipated that CF cells might respond more than non-CF cells to the GC-rich P. aeruginosa DNA. However, this turned out not to be the case. Nevertheless, IL-8 secretion stimulated by P. aeruginosa DNA was affected by blockers of endosomal acidification.
Collectively, our experiments show that human respiratory epithelial cells respond to P. aeruginosa DNA by secreting IL-8. Our findings suggest that P. aeruginosa DNA-stimulated secretion of IL-8 by human respiratory epithelial cells represents a protein synthesis-dependent process, independent of NF-κB activation and transcriptional activation of IL-8. In spite of the ability of human respiratory epithelial cells to respond to typical CpG TLR9 ligands, the pattern of RANTES induction by CpG oligonucleotides versus IL-8 by P. aeruginosa DNA strongly suggests activation of two separate pathways: (i) type I interferon pathways in the former process and (ii) p38 and ERK pathways in the latter. Significantly, the signaling involved in P. aeruginosa DNA-elicited IL-8 secretion depends on endosomal acidification, based on a partial inhibitory effect of chloroquine and pharmacological blockers of vacuolar H+-ATPase. Our findings suggest that elimination of bacterial DNA by DNase treatment and use of lysosomotropic drugs may help decrease inflammation in CF.
Acknowledgments
This work was supported by NIH grant AI31193 and in part by grant AI050825 from NIH and a grant from Philip Morris USA Inc. and Philip Morris International.
Editor: J. D. Clements
REFERENCES
- 1.Adamo, R., S. Sokol, G. Soong, M. I. Gomez, and A. Prince. 2004. Pseudomonas aeruginosa flagella activate airway epithelial cells through asialo GM1 and toll-like receptor 2 as well as toll-like receptor 5. Am. J. Respir. Cell Mol. Biol. 30:627-634. [DOI] [PubMed] [Google Scholar]
- 2.Agrawal, S., A. Agrawal, B. Doughty, A. Gerwitz, J. Blenis, T. Van Dyke, and B. Pulendran. 2003. Cutting edge: different Toll-like receptor agonists instruct dendritic cells to induce distinct Th responses via differential modulation of extracellular signal-regulated kinase-mitogen-activated protein kinase and c-Fos. J. Immunol. 171:4984-4989. [DOI] [PubMed] [Google Scholar]
- 3.Akhtar, M., J. L. Watson, A. Nazli, and D. M. McKay. 2003. Bacterial DNA evokes epithelial IL-8 production by a MAPK-dependent, NF-κB-independent pathway. FASEB J. 17:1319-1321. [DOI] [PubMed] [Google Scholar]
- 4.Athman, R., and D. Philpott. 2004. Innate immunity via Toll-like receptors and Nod proteins. Curr. Opin. Microbiol. 7:25-32. [DOI] [PubMed] [Google Scholar]
- 5.Bauer, S., C. J. Kirschning, H. Hacker, V. Redecke, S. Hausmann, S. Akira, H. Wagner, and G. B. Lipford. 2001. Human TLR9 confers responsiveness to bacterial DNA via species-specific CpG motif recognition. Proc. Natl. Acad. Sci. USA 98:9237-9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becker, M. N., M. S. Sauer, M. S. Muhlebach, A. J. Hirsh, Q. Wu, M. W. Verghese, and S. H. Randell. 2004. Cytokine secretion by cystic fibrosis airway epithelial cells. Am. J. Respir. Crit. Care Med. 169:645-653. [DOI] [PubMed] [Google Scholar]
- 7.Bischoff, D. S., J. H. Zhu, N. S. Makhijani, and D. T. Yamaguchi. 2005. KC chemokine expression by TGF-beta in C3H10T1/2 cells induced towards osteoblasts. Biochem. Biophys. Res. Commun. 326:364-370. [DOI] [PubMed] [Google Scholar]
- 8.Bonfield, T. L., J. R. Panuska, M. W. Konstan, K. A. Hilliard, J. B. Hilliard, H. Ghnaim, and M. Berger. 1995. Inflammatory cytokines in cystic fibrosis lungs. Am. J. Respir. Crit. Care Med. 152:2111-2118. [DOI] [PubMed] [Google Scholar]
- 9.Boucher, J. C., H. Yu, M. H. Mudd, and V. Deretic. 1997. Mucoid Pseudomonas aeruginosa in cystic fibrosis: characterization of muc mutations in clinical isolates and analysis of clearance in a mouse model of respiratory infection. Infect. Immun. 65:3838-3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cobb, L. M., J. C. Mychaleckyj, D. J. Wozniak, and Y. S. Lopez-Boado. 2004. Pseudomonas aeruginosa flagellin and alginate elicit very distinct gene expression patterns in airway epithelial cells: implications for cystic fibrosis disease. J. Immunol. 173:5659-5670. [DOI] [PubMed] [Google Scholar]
- 11.Coxon, P. Y., M. J. Rane, S. Uriarte, D. W. Powell, S. Singh, W. Butt, Q. Chen, and K. R. McLeish. 2003. MAPK-activated protein kinase-2 participates in p38 MAPK-dependent and ERK-dependent functions in human neutrophils. Cell Signal. 15:993-1001. [DOI] [PubMed] [Google Scholar]
- 12.Cremer, I., J. Ghysdael, and V. Vieillard. 2002. A non-classical ISRE/ISGF3 pathway mediates induction of RANTES gene transcription by type I IFNs. FEBS Lett. 511:41-45. [DOI] [PubMed] [Google Scholar]
- 13.Denning, G. M., L. A. Wollenweber, M. A. Railsback, C. D. Cox, L. L. Stoll, and B. E. Britigan. 1998. Pseudomonas pyocyanin increases interleukin-8 expression by human airway epithelial cells. Infect. Immun. 66:5777-5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiMango, E., H. J. Zar, R. Bryan, and A. Prince. 1995. Diverse Pseudomonas aeruginosa gene products stimulate respiratory epithelial cells to produce interleukin-8. J. Clin. Investig. 96:2204-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dumitru, C. D., J. D. Ceci, C. Tsatsanis, D. Kontoyiannis, K. Stamatakis, J. H. Lin, C. Patriotis, N. A. Jenkins, N. G. Copeland, G. Kollias, and P. N. Tsichlis. 2000. TNF-alpha induction by LPS is regulated posttranscriptionally via a Tpl2/ERK-dependent pathway. Cell 103:1071-1083. [DOI] [PubMed] [Google Scholar]
- 16.Egan, M., T. Flotte, S. Afione, R. Solow, P. L. Zeitlin, B. J. Carter, and W. B. Guggino. 1992. Defective regulation of outwardly rectifying Cl- channels by protein kinase A corrected by insertion of CFTR. Nature 358:581-584. [DOI] [PubMed] [Google Scholar]
- 17.Epelman, S., D. Stack, C. Bell, E. Wong, G. G. Neely, S. Krutzik, K. Miyake, P. Kubes, L. D. Zbytnuik, L. L. Ma, X. Xie, D. E. Woods, and C. H. Mody. 2004. Different domains of Pseudomonas aeruginosa exoenzyme S activate distinct TLRs. J. Immunol. 173:2031-2040. [DOI] [PubMed] [Google Scholar]
- 18.Erridge, C., A. Pridmore, A. Eley, J. Stewart, and I. R. Poxton. 2004. Lipopolysaccharides of Bacteroides fragilis, Chlamydia trachomatis and Pseudomonas aeruginosa signal via toll-like receptor 2. J. Med. Microbiol. 53:735-740. [DOI] [PubMed] [Google Scholar]
- 19.Faure, K., T. Sawa, T. Ajayi, J. Fujimoto, K. Moriyama, N. Shime, and J. P. Wiener-Kronish. 2004. TLR4 signaling is essential for survival in acute lung injury induced by virulent Pseudomonas aeruginosa secreting type III secretory toxins. Respir. Res. 5:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Firoved, A. M., W. Ornatowski, and V. Deretic. 2004. Microarray analysis reveals induction of lipoprotein genes in mucoid Pseudomonas aeruginosa: implications for inflammation in cystic fibrosis. Infect. Immun. 72:5012-5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flo, T. H., L. Ryan, E. Latz, O. Takeuchi, B. G. Monks, E. Lien, O. Halaas, S. Akira, G. Skjak-Braek, D. T. Golenbock, and T. Espevik. 2002. Involvement of toll-like receptor (TLR) 2 and TLR4 in cell activation by mannuronic acid polymers. J. Biol. Chem. 277:35489-35495. [DOI] [PubMed] [Google Scholar]
- 22.Frevel, M. A., T. Bakheet, A. M. Silva, J. G. Hissong, K. S. Khabar, and B. R. Williams. 2003. p38 mitogen-activated protein kinase-dependent and -independent signaling of mRNA stability of AU-rich element-containing transcripts. Mol. Cell. Biol. 23:425-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goral, J., and E. J. Kovacs. 2005. In vivo ethanol exposure down-regulates TLR2-, TLR4-, and TLR9-mediated macrophage inflammatory response by limiting p38 and ERK1/2 activation. J. Immunol. 174:456-463. [DOI] [PubMed] [Google Scholar]
- 24.Govan, J. R., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greene, C. M., T. P. Carroll, S. G. Smith, C. C. Taggart, J. Devaney, S. Griffin, S. J. O'Neill, and N. G. McElvaney. 2005. TLR-induced inflammation in cystic fibrosis and non-cystic fibrosis airway epithelial cells. J. Immunol. 174:1638-1646. [DOI] [PubMed] [Google Scholar]
- 26.Hacker, H., H. Mischak, T. Miethke, S. Liptay, R. Schmid, T. Sparwasser, K. Heeg, G. B. Lipford, and H. Wagner. 1998. CpG-DNA-specific activation of antigen-presenting cells requires stress kinase activity and is preceded by non-specific endocytosis and endosomal maturation. EMBO J. 17:6230-6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hajjar, A. M., R. K. Ernst, J. H. Tsai, C. B. Wilson, and S. I. Miller. 2002. Human Toll-like receptor 4 recognizes host-specific LPS modifications. Nat. Immunol. 3:354-359. [DOI] [PubMed] [Google Scholar]
- 28.Hartmann, G., R. D. Weeratna, Z. K. Ballas, P. Payette, S. Blackwell, I. Suparto, W. L. Rasmussen, M. Waldschmidt, D. Sajuthi, R. H. Purcell, H. L. Davis, and A. M. Krieg. 2000. Delineation of a CpG phosphorothioate oligodeoxynucleotide for activating primate immune responses in vitro and in vivo. J. Immunol. 164:1617-1624. [DOI] [PubMed] [Google Scholar]
- 29.Heil, F., P. Ahmad-Nejad, H. Hemmi, H. Hochrein, F. Ampenberger, T. Gellert, H. Dietrich, G. Lipford, K. Takeda, S. Akira, H. Wagner, and S. Bauer. 2003. The Toll-like receptor 7 (TLR7)-specific stimulus loxoribine uncovers a strong relationship within the TLR7, 8 and 9 subfamily. Eur. J. Immunol. 33:2987-2997. [DOI] [PubMed] [Google Scholar]
- 30.Hemmi, H., O. Takeuchi, T. Kawai, T. Kaisho, S. Sato, H. Sanjo, M. Matsumoto, K. Hoshino, H. Wagner, K. Takeda, and S. Akira. 2000. A Toll-like receptor recognizes bacterial DNA. Nature 408:740-745. [DOI] [PubMed] [Google Scholar]
- 31.Hodson, M. E., S. McKenzie, H. K. Harms, C. Koch, G. Mastella, J. Navarro, and B. Strandvik. 2003. Dornase alfa in the treatment of cystic fibrosis in Europe: a report from the Epidemiologic Registry of Cystic Fibrosis. Pediatr. Pulmonol. 36:427-432. [DOI] [PubMed] [Google Scholar]
- 32.Hoffmann, E., O. Dittrich-Breiholz, H. Holtmann, and M. Kracht. 2002. Multiple control of interleukin-8 gene expression. J. Leukoc. Biol. 72:847-855. [PubMed] [Google Scholar]
- 33.Honda, K., H. Yanai, T. Mizutani, H. Negishi, N. Shimada, N. Suzuki, Y. Ohba, A. Takaoka, W. C. Yeh, and T. Taniguchi. 2004. Role of a transductional-transcriptional processor complex involving MyD88 and IRF-7 in Toll-like receptor signaling. Proc. Natl. Acad. Sci. USA 101:15416-15421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Janeway, C. A., Jr., and R. Medzhitov. 2002. Innate immune recognition. Annu. Rev. Immunol. 20:197-216. [DOI] [PubMed] [Google Scholar]
- 35.Jijon, H. B., W. J. Panenka, K. L. Madsen, and H. G. Parsons. 2002. MAP kinases contribute to IL-8 secretion by intestinal epithelial cells via a posttranscriptional mechanism. Am. J. Physiol. Cell Physiol. 283:C31-C41. [DOI] [PubMed] [Google Scholar]
- 36.Khan, T. Z., J. S. Wagener, T. Bost, J. Martinez, F. J. Accurso, and D. W. Riches. 1995. Early pulmonary inflammation in infants with cystic fibrosis. Am. J. Respir. Crit. Care Med. 151:1075-1082. [DOI] [PubMed] [Google Scholar]
- 37.Kirchner, K. K., J. S. Wagener, T. Z. Khan, S. C. Copenhaver, and F. J. Accurso. 1996. Increased DNA levels in bronchoalveolar lavage fluid obtained from infants with cystic fibrosis. Am. J. Respir. Crit. Care Med. 154:1426-1429. [DOI] [PubMed] [Google Scholar]
- 38.Konstan, M. W., K. A. Hilliard, T. M. Norvell, and M. Berger. 1994. Bronchoalveolar lavage findings in cystic fibrosis patients with stable, clinically mild lung disease suggest ongoing infection and inflammation. Am. J. Respir. Crit. Care Med. 150:448-454. [DOI] [PubMed] [Google Scholar]
- 39.Kontoyiannis, D., M. Pasparakis, T. T. Pizarro, F. Cominelli, and G. Kollias. 1999. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity 10:387-398. [DOI] [PubMed] [Google Scholar]
- 40.Krieg, A. M. 2002. CpG motifs in bacterial DNA and their immune effects. Annu. Rev. Immunol. 20:709-760. [DOI] [PubMed] [Google Scholar]
- 41.Kube, D., U. Sontich, D. Fletcher, and P. B. Davis. 2001. Proinflammatory cytokine responses to P. aeruginosa infection in human airway epithelial cell lines. Am. J. Physiol. Lung Cell Mol. Physiol. 280:L493-L502. [DOI] [PubMed] [Google Scholar]
- 42.Lorenz, E., D. C. Chemotti, K. Vandal, and P. A. Tessier. 2004. Toll-like receptor 2 represses non-pilus adhesin-induced signaling in acute infections with the Pseudomonas aeruginosa pilA mutant. Infect. Immun. 72:4561-4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miesenbock, G., D. A. De Angelis, and J. E. Rothman. 1998. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 394:192-195. [DOI] [PubMed] [Google Scholar]
- 44.Milella, M., A. Gismondi, P. Roncaioli, L. Bisogno, G. Palmieri, L. Frati, M. G. Cifone, and A. Santoni. 1997. CD16 cross-linking induces both secretory and extracellular signal-regulated kinase (ERK)-dependent cytosolic phospholipase A2 (PLA2) activity in human natural killer cells: involvement of ERK, but not PLA2, in CD16-triggered granule exocytosis. J. Immunol. 158:3148-3154. [PubMed] [Google Scholar]
- 45.Muhlebach, M. S., W. Reed, and T. L. Noah. 2004. Quantitative cytokine gene expression in CF airway. Pediatr. Pulmonol. 37:393-399. [DOI] [PubMed] [Google Scholar]
- 46.Muir, A., G. Soong, S. Sokol, B. Reddy, M. I. Gomez, A. Van Heeckeren, and A. Prince. 2004. Toll-like receptors in normal and cystic fibrosis airway epithelial cells. Am. J. Respir. Cell Mol. Biol. 30:777-783. [DOI] [PubMed] [Google Scholar]
- 47.Orenstein, D. M., G. B. Winnie, and H. Altman. 2002. Cystic fibrosis: a 2002 update. J. Pediatr. 140:156-164. [DOI] [PubMed] [Google Scholar]
- 48.Pasare, C., and R. Medzhitov. 2005. Toll-like receptors: linking innate and adaptive immunity. Adv. Exp. Med. Biol. 560:11-18. [DOI] [PubMed] [Google Scholar]
- 49.Paul, K., E. Rietschel, M. Ballmann, M. Griese, D. Worlitzsch, J. Shute, C. Chen, T. Schink, G. Doring, S. van Koningsbruggen, U. Wahn, and F. Ratjen. 2004. Effect of treatment with dornase alpha on airway inflammation in patients with cystic fibrosis. Am. J. Respir. Crit. Care Med. 169:719-725. [DOI] [PubMed] [Google Scholar]
- 50.Pearson, J. P., L. Passador, B. H. Iglewski, and E. P. Greenberg. 1995. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 92:1490-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Platz, J., C. Beisswenger, A. Dalpke, R. Koczulla, O. Pinkenburg, C. Vogelmeier, and R. Bals. 2004. Microbial DNA induces a host defense reaction of human respiratory epithelial cells. J. Immunol. 173:1219-1223. [DOI] [PubMed] [Google Scholar]
- 52.Poschet, J. F., J. C. Boucher, L. Tatterson, J. Skidmore, R. W. Van Dyke, and V. Deretic. 2001. Molecular basis for defective glycosylation and Pseudomonas pathogenesis in cystic fibrosis lung. Proc. Natl. Acad. Sci. USA 98:13972-13977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Poschet, J. F., J. Skidmore, J. C. Boucher, A. M. Firoved, R. W. Van Dyke, and V. Deretic. 2002. Hyperacidification of cellubrevin endocytic compartments and defective endosomal recycling in cystic fibrosis respiratory epithelial cells. J. Biol. Chem. 277:13959-13965. [DOI] [PubMed] [Google Scholar]
- 54.Ratjen, F., K. Paul, S. van Koningsbruggen, S. Breitenstein, E. Rietschel, and W. Nikolaizik. 2005. DNA concentrations in BAL fluid of cystic fibrosis patients with early lung disease: influence of treatment with dornase alpha. Pediatr. Pulmonol. 39:1-4. [DOI] [PubMed] [Google Scholar]
- 55.Revets, H., G. Pynaert, J. Grooten, and P. De Baetselier. 2005. Lipoprotein I, a TLR2/4 ligand modulates Th2-driven allergic immune responses. J. Immunol. 174:1097-1103. [DOI] [PubMed] [Google Scholar]
- 56.Roger, T., T. Out, N. Mukaida, K. Matsushima, H. Jansen, and R. Lutter. 1998. Enhanced AP-1 and NF-κB activities and stability of interleukin 8 (IL-8) transcripts are implicated in IL-8 mRNA superinduction in lung epithelial H292 cells. Biochem. J. 330:429-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rowe, S. M., S. Miller, and E. J. Sorscher. 2005. Cystic fibrosis. N. Engl. J. Med. 352:1992-2001. [DOI] [PubMed] [Google Scholar]
- 58.Schroder, N. W., D. Pfeil, B. Opitz, K. S. Michelsen, J. Amberger, U. Zahringer, U. B. Gobel, and R. R. Schumann. 2001. Activation of mitogen-activated protein kinases p42/44, p38, and stress-activated protein kinases in myelo-monocytic cells by Treponema lipoteichoic acid. J. Biol. Chem. 276:9713-9719. [DOI] [PubMed] [Google Scholar]
- 59.Shah, P. L., S. F. Scott, R. A. Knight, and M. E. Hodson. 1996. The effects of recombinant human DNase on neutrophil elastase activity and interleukin-8 levels in the sputum of patients with cystic fibrosis. Eur. Respir. J. 9:531-534. [DOI] [PubMed] [Google Scholar]
- 60.Siebenlist, U., G. Franzoso, and K. Brown. 1994. Structure, regulation and function of NF-kappa B. Annu. Rev. Cell Biol. 10:405-455. [DOI] [PubMed] [Google Scholar]
- 61.Skerrett, S. J., H. D. Liggitt, A. M. Hajjar, and C. B. Wilson. 2004. Cutting edge: myeloid differentiation factor 88 is essential for pulmonary host defense against Pseudomonas aeruginosa but not Staphylococcus aureus. J. Immunol. 172:3377-3381. [DOI] [PubMed] [Google Scholar]
- 62.Soong, G., B. Reddy, S. Sokol, R. Adamo, and A. Prince. 2004. TLR2 is mobilized into an apical lipid raft receptor complex to signal infection in airway epithelial cells. J. Clin. Investig. 113:1482-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Verthelyi, D., K. J. Ishii, M. Gursel, F. Takeshita, and D. M. Klinman. 2001. Human peripheral blood cells differentially recognize and respond to two distinct CPG motifs. J. Immunol. 166:2372-2377. [DOI] [PubMed] [Google Scholar]
- 64.Vollmer, J., R. Weeratna, P. Payette, M. Jurk, C. Schetter, M. Laucht, T. Wader, S. Tluk, M. Liu, H. L. Davis, and A. M. Krieg. 2004. Characterization of three CpG oligodeoxynucleotide classes with distinct immunostimulatory activities. Eur. J. Immunol. 34:251-262. [DOI] [PubMed] [Google Scholar]
- 65.Wu, Q., Z. Lu, M. W. Verghese, and S. H. Randell. 2005. Airway epithelial cell tolerance to Pseudomonas aeruginosa. Respir. Res. 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yeo, S. J., D. Gravis, J. G. Yoon, and A. K. Yi. 2003. Myeloid differentiation factor 88-dependent transcriptional regulation of cyclooxygenase-2 expression by CpG DNA: role of NF-κB and p38. J. Biol. Chem. 278:22563-22573. [DOI] [PubMed] [Google Scholar]
- 67.Yi, A. K., and A. M. Krieg. 1998. Rapid induction of mitogen-activated protein kinases by immune stimulatory CpG DNA. J. Immunol. 161:4493-4497. [PubMed] [Google Scholar]
- 68.Yi, A. K., R. Tuetken, T. Redford, M. Waldschmidt, J. Kirsch, and A. M. Krieg. 1998. CpG motifs in bacterial DNA activate leukocytes through the pH-dependent generation of reactive oxygen species. J. Immunol. 160:4755-4761. [PubMed] [Google Scholar]
- 69.Yu, Y., H. Zeng, S. Lyons, A. Carlson, D. Merlin, A. S. Neish, and A. T. Gewirtz. 2003. TLR5-mediated activation of p38 MAPK regulates epithelial IL-8 expression via posttranscriptional mechanism. Am. J. Physiol. Gastrointest. Liver Physiol. 285:G282-G290. [DOI] [PubMed] [Google Scholar]
- 70.Zabner, J., P. Karp, M. Seiler, S. L. Phillips, C. J. Mitchell, M. Saavedra, M. Welsh, and A. J. Klingelhutz. 2003. Development of cystic fibrosis and noncystic fibrosis airway cell lines. Am. J. Physiol. Lung Cell Mol. Physiol. 284:L844-L854. [DOI] [PubMed] [Google Scholar]