Abstract
Human P-glycoprotein (ABCB1) is a primary multidrug transporter located in plasma membranes, that, utilizes the energy of ATP hydrolysis to pump toxic xenobiotics out of cells. P-glycoprotein employs a most unusual molecular mechanism to perform this drug transport function. Here we review our work to elucidate the molecular mechanism of drug transport by P-glycoprotein. High level heterologous expression of human P-glycoprotein, in the yeast Saccharomyces cerevisiae, has facilitated biophysical studies in purified proteoliposome preparations. Development of novel spin-labeled transport substrates has allowed for quantitative and rigorous measurements of drug transport in real time by EPR spectroscopy. We have developed a new drug transport model of P-glycoprotein from the results of mutagenic, quantitative thermodynamic and kinetic studies. This model satisfactorily accounts for most of the unusual kinetic, coupling and physiological features of P-glycoprotein. Additionally, an atomic detail structural model of P-glycoprotein has been devised to place our results within a proper structural context.
Keywords: P-glycoprotein, Multidrug Resistance, Transporter, Energy Coupling, Mechanism, Thermodynamics, Kinetics, EPR, Homology Modeling, Heterologous Expression
Introduction
Human P-glycoprotein (ABCB1, MDR1 gene product) is a member of the ATP binding cassette (ABC) transporter superfamily (Hyde et al., 1990). ABC proteins are one of the largest protein families and are involved in transporting a wide variety of biological compounds across the membrane (Holland and Blight, 1999). P-glycoprotein (P-gp) is a single, glycosylated polypeptide of about 170-180 kDa found in plasma membranes of cells. It consists of two homologous halves. Each half contains six Transmembrane helices (TMs), a nucleotide binding domain, and intervening Intra Cellular Domain structures (ICDs). P-glycoprotein transports various and structurally unrelated compounds to the outside of cells using the energy of ATP hydrolysis (Senior et al., 1995b). ATPase activity of this enzyme is stimulated several fold by transported drugs. Most of the transport substrates are hydrophobic and many have positive charge together with characteristic hydrogen bond acceptor patterns (Seelig, 1998; Ecker et al., 1999). Because of their hydrophobicity, drugs partition into lipid bilayers or bind tightly to proteins and chromosomes and have low free concentrations in the cytosol. In the “hydrophobic vacuum cleaner model”, P-glycoprotein takes its substrates from the inner leaflet of the plasma membrane and transports them out to the extracellular bulk water phase (Gottesman and Pastan, 1993). Unique features of P-glycoprotein are its very broad substrate specificity and basal ATPase activity in the absence of transport substrates (Omote and Al-Shawi, 2002). Human P-glycoprotein plays an important role in absorption, distribution, metabolism, excretion and toxicity of pharmacologically relevant drugs (Ambudkar et al., 1999; Leslie et al., 2005). The significant medical importance of this transporter is exemplified by P-glycoprotein mediated multidrug resistance that compromises cancer chemotherapy regimens (Polgar and Bates, 2005). Yet the actual mechanism of drug transport is still not well understood. Here we review our recent work and methods that we have employed to further understand the unusual and very interesting drug transport mechanism of P-glycoprotein and to establish the physiological basis of the unique features of its transport cycle.
Expression, purification and mutagenesis of human P-glycoprotein in yeast
To obtain large amounts of human P-glycoprotein (P-gp), we developed a plasmid-based, high-yield expression system in the yeast Saccharomyces cerevisiae. Expression was controlled either under the strong constitutive PMA1 promoter or the tightly regulated and inducible GAL1 promoter. An important finding was the utilization of glycerol as a chemical chaperone to enhance the expression level and to increase P-gp stability in the plasma membrane (Figler et al., 2000). P-glycoprotein was expressed with an N-terminal poly-histidine tag to facilitate purification by metal ion affinity chromatography. When purified to homogeneity, the yeast-expressed human P-gp, was fully functional and had the highest specific activity reported to date. Purified P-gp could then be easily reconstituted into defined-lipid proteoliposomes that were fully competent for drug transport. Furthermore, the orientation of P-glycoprotein in the liposomes could be changed by controlled reconstitution conditions, such that all nucleotide-binding sites could be made to face the external medium (Al-Shawi et al., 2003). Successes in employing our plasmids and methodologies in high-yield expression of other plasma membrane proteins have been published (Tu and Weissman, 2002; Lee and Altenberg, 2003a; Lee and Altenberg, 2003b; Moiseenkova et al., 2003; Zeng et al., 2004; De Rivoyre et al., 2005; Pisani et al., 2005). The recent expression of human MRP1 (ABCC1 gene product) in yeast (Lee and Altenberg, 2003a; Lee and Altenberg, 2003b) has proven the utility of this system in expressing other mammalian ABC transporters. These authors also confirmed our stated advantages of S. cerevisiae over P. pastoris expression (Figler et al., 2000) namely; expression in the plasma membrane, ease of use of plasmid-borne system, rapid generation of mutant forms, no drug selections required for expression, and simplicity and direct scalability in readily available laboratory equipment.
Development of novel EPR drug transport assays
This was necessary because rigorous quantitative drug transport assays in the field of P-glycoprotein research were lacking. The transport assay problem arises from the fact that most of the transport substrates of P-gp are highly hydrophobic, partition into lipid membranes and can flip-flop or diffuse across the two lipid leaflets of the bilayer. Thus, in a closed system, such as proteoliposomes in a test tube, it was difficult to generate transmembrane drug gradients due to these recycling events. Furthermore the hydrophobicity and “stickiness” of the drugs led to binding to the sides of vessels and filters causing low signal to noise ratios. The classical assays employing radioactive drugs (Sharom et al., 1993; Sharom, 1997) or fluorescent drugs and analogs (Shapiro and Ling, 1995; Shapiro et al., 1997; Shapiro and Ling, 1997a; Shapiro and Ling, 1998; Shapiro et al., 1999) suffer from these problems and the results must be considered qualitative. To overcome these obstacles we synthesized a set of novel spin-labeled transport substrates of P-glycoprotein, including spin-labeled derivatives of verapamil (SL-verapamil) (Omote and Al-Shawi, 2002), rhodamine-123 (SL-R123), tetramethylrhodamine (SL-TMR) and tetramethylrosamine (SL-MTMRos) encompassing both the “H-type” and “R-type” transport drug classes (Shapiro and Ling, 1997b; Shapiro et al., 1999). The most useful and extensively utilized transport substrate was spin-labeled verapamil as described below.
Verapamil is an excellent transport substrate of P-gp (Yusa and Tsuruo, 1989) but can easily and rapidly flip-flop across the lipid leaflets on deprotonation in the physiological pH range. An introduced nitroxide radical and a fixed positive charge were the important features of SL-verapamil. This fixed positive charge was introduced to inhibit flip-flop of this molecule and the nitroxide enabled real time EPR measurements of drug transport (Omote and Al-Shawi, 2002). Binding of an amphipathic molecule to liposomes changes its mobility and accessibility. EPR spectroscopy is a particularly good tool to quantitatively monitor changes in mobility and accessibility (Flewelling and Hubbell, 1986; Victor and Cafiso, 2001).
P-glycoprotein takes up substrates from the cytoplasmic leaflet of the plasma membrane and transports them to the aqueous medium surrounding the cell (Gottesman and Pastan, 1993). Partitioning of drugs into the lipid bilayer is a requirement for transport and is a major determinant of apparent Km of drug transport and activation (KmD; (Romsicki and Sharom, 1999; Seelig and Landwojtowicz, 2000)). SL-verapamil was found to be 120-fold more hydrophilic than verapamil (Omote and Al-Shawi, 2002). SL-verapamil was an excellent P-gp substrate that activated the ATPase activity by 5-fold. Apparent KmD and Ki (inhibition constant for high concentrations of drugs) values were 4.3 and 206 μM respectively. These values were significantly lower than those of verapamil (62 and 640 μM respectively) indicating SL-verapamil was a substrate with higher affinity than verapamil. The apparent KmD and Ki and KmA (KmATP = 0.8 mM) values for SL-verapamil measured by transport assays described below were identical to the values obtained in classical ATPase activity assays. A very important finding was that KmD values were directly referable to high-affinity, cytoplasmic-facing, drug-loading “ON-sites” whereas Ki values were directly referable to low-affinity, extracellular-facing, drug-releasing“OFF-sites” (Omote and Al-Shawi, 2002).
In aqueous solution the three sharp resonance peaks of a highly mobile nitroxide are seen (Fig 1A; supernatant spectrum). These peaks are broadened and their intensities are reduced on addition of high concentrations of lipid as SL-verapamil partitions into the lipid. There are two components of such spectra that can be easily resolved mathematically: a low mobility component due to SL-verapamil partitioning into the membrane and a high mobility aqueous component. On addition of lipids, the aqueous EPR signal changes very rapidly. On dilution, the SL-verapamil reequilibrated very rapidly with the aqueous phase. This indicated that binding of drugs to the membrane was fast and reversible and not rate-limiting as proposed by others (Shapiro and Ling, 1998). It should be noted that SL-verapamil was designed to be hydrophilic to facilitate transport studies. However, in other applications it might be preferable to have very hydrophobic spin-labeled drugs that have little or no aqueous signal because the majority of the drug partitions into the lipid. Both SL-TMR and SL-MTMRos fulfill these requirements.
Fig. 1.
Transport of spin-labeled verapamil by P-glycoprotein. Panel A. The reaction mixture contained 50 μM of SL-verapamil and 2 μM P-glycoprotein, in unilamellar proteoliposomes with an average hydrodynamic radius of 420 Å, at pH 7.5 and 23 °C. Transport was started in the EPR spectrometer by addition of ATP to the reaction mixture containing a potent ATP regenerating system (Omote and Al-Shawi, 2002). The concentration of SL-verapamil in the aqueous phase is plotted as a function of time. After maximum steady state transport was achieved (120 min), vesicles and supernatant were separated by centrifugation. Vesicles were resuspended in buffer and spectra recorded at the times indicated. Representative EPR spectra are shown for various time points along the transport assay. The post centrifugation vesicular spectrum is expanded to show fine details. The sum of the vesicular and supernatant spectra was identical to the transport assay spectrum at 120 min. Panel B. Schematic representation of the steady state concentrations of SL-verapamil at 120 min. Calculated values were obtained from the deconvoluted steady state spectrum at 120 min of panel A. These results were in absolute agreement with results obtained by analysis of the component spectra of the supernatant and vesicles obtained by centrifugation and SL-verapamil concentration analysis.
A great advantage of EPR spectroscopy is that it is possible to count the number of spins present (by double integration) and to accurately determine the concentration of probe present in a given compartment. Thus, one can quantitate the amounts of free and bound SL-verapamil throughout the course of a transport experiment. Both line-shape analysis and line-height analysis yield identical results. In the transport experiment of Fig. 1, 10 μM SL-verapamil immediately entered the external lipid leaflet. On addition of ATP, under strict regeneration conditions, the external aqueous concentration of SL-verapamil was reduced and the lipid bound component increased until steady state was achieved. On centrifugation the supernatant had a completely aqueous spectrum, whereas the vesicles had a predominantly lipid bound spectrum with an aqueous component that was inaccessible to externally applied, membrane impermeable spin relaxing agents (Fig. 1A). This aqueous component was in the lumen of the vesicles. On dilution of the vesicles there was no release of the SL-verapamil trapped inside the vesicles (Fig. 1A). Since SL-verapamil in the outer lipid leaflet can reequilibrate very quickly with the medium, the membrane bound component was clearly trapped in the inner lipid leaflet and could not flip-flop out. If the ATPase inhibitors vanadate or EDTA were added at any point, transport stopped immediately and the gradient was stable for more than 3 hrs, unless detergent was added. With a classical transport substrate such as verapamil, the gradient decays within seconds due to flip-flop events. Concentration changes as a function of time in the four resolved components could be easily calculated (Fig. 1B). Results of centrifugation separations (Fig. 1A) confirm the accuracy of these calculations (for further details see Omote and Al-Shawi, 2002). From such transport experiments we established that SL-verapamil transport was tightly coupled to ATP hydrolysis and that transport was the rate-limiting step for the overall transport cycle. The SL-verapamil gradients generated were in the range of Ki/KmD values expected of a primary transport system, even when there was a surplus of external transport substrate (Omote and Al-Shawi, 2002).
Steady state thermodynamic analysis of P-glycoprotein drug transport
ATPase activity associated with P-glycoprotein is characterized by three drug-dependent phases; basal (no drug), drug-activated (at moderate drug concentrations) and drug-inhibited (at high drug concentrations). This gives rise to bell-shaped curves of ATPase activities as a logarithmic function of drug concentration (Al-Shawi and Senior, 1993; Urbatsch et al., 1994). Using spin-labeled transport substrates, similar bell-shaped curves for drug transport activity are seen but lacking a basal component (Omote and Al-Shawi, 2002). To establish the coupling relationships between ATPase activity and drug transport activity in P-glycoprotein, we determined the intrinsic thermodynamics of the intrinsic rate-limiting steps at saturating ligand concentrations. To achieve this we measured the Arrhenius activation energies as a function of drug concentration during the ATPase and transport reactions. Representative “R-type” and “H-type” transport drugs were used at various concentrations. The thermodynamic analysis was also carried out on drug transport with the aid of SL-verapamil. For this analysis we determined the apparent kinetic constants KmD, Ki, KmA as a function of temperature for each drug and condition employed. We also employed van't Hoff plots to establish the types of chemical interactions that each drug made with the lipids and drug-binding sites. We calculated that the energy available from the hydrolysis of a single ATP molecule was sufficient to forcibly rehydrate a drug molecule bound to P-gp. Full details can be found in Al-Shawi et al., 2003. We also found that KmD and Kd values (high affinity) for drug binding followed the same temperature profiles and were referable to the high-affinity, drug-loading “ON-sites” whereas Ki values followed independent temperature profiles and were referable to the low-affinity, drug-unloading “OFF-sites”. Thus by van't Hoff analysis it is possible to distinguish between drug binding to the “ON-sites” or the “OFF-sites”.
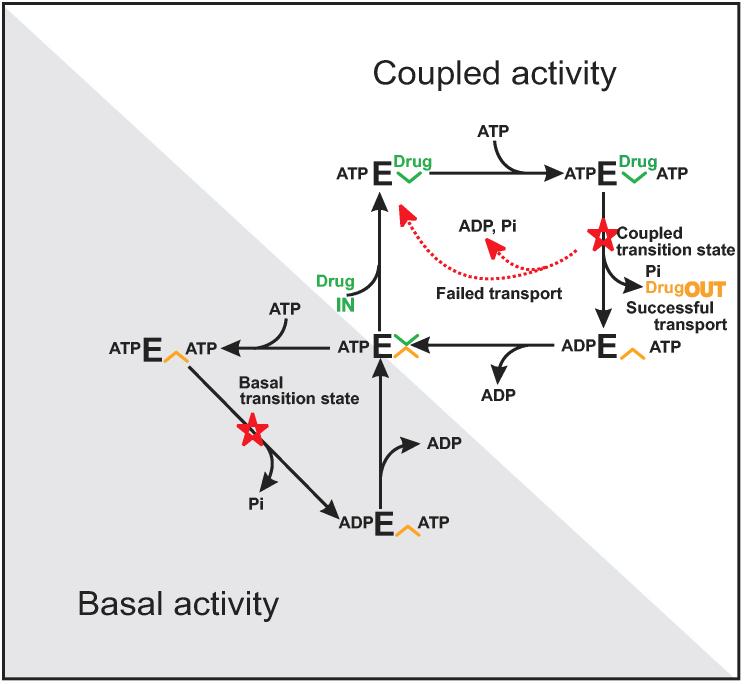
Using linear free energy analyses (Exner, 1973; Nakamoto et al., 1999), two distinct transition states were found that could be assigned to basal ATPase activity and to coupled drug transport activity respectively. The drug inhibition phase was associated with low-affinity drug-unloading “OFF-sites”. Both transition states involved major concerted conformational reorientations of the protein. It was concluded that basal ATPase activity is a mechanistic property of the enzyme and constitutes a separate uncoupled pathway of ATP hydrolysis. No endogenous or exogenous substrate is carried across the transporter in the basal cycle. As the basal ATPase can be rapidly and fully inhibited by the application of high concentrations of transport substrate, it was concluded that P-glycoprotein retains a structure in which the drug binding sites are in a low-affinity drug-unloading conformation. Recently, it has been demonstrated that the two transition states have two distinct physical structures (Rao and Nuti, 2003). These results and others led us to propose a new drug transport model for P-glycoprotein (Fig. 2). In the presence of saturating drugs all activity proceeds by the coupled activity cycle, previously described as the alternating catalytic cycle (Senior et al., 1995a). Transport results obtained with SL-verapmil indicated that this cycle was tightly coupled and that net flux through this pathway ceased when the gradient of transported drug was in the range of the Ki/KmD ratio (Omote and Al-Shawi, 2002). If there is insufficient drug and two ATP molecules bind the empty transporter, P-gp partitions to the uncoupled cycle and hydrolyses ATP without any transport work (Al-Shawi et al., 2003) (Fig. 2; Basal cycle). Numerical simulations of observed ATPase activities and transport activities employing SL-verapamil confirmed this new model. Two-step models for drug pumping, as originally proposed by Sauna and Ambudkar, 2001, see also Refs. (Litman et al., 2003; Davidson and Chen, 2004), do not fit our transport and thermodynamic data as well as the model in Fig. 2 does. Thus our new drug transport model is the only model in the current literature that can accommodate most of the kinetic data available for P-glycoprotein (see Discussion sections of Refs. (Al-Shawi et al., 2003; Omote et al., 2004).
Fig. 2.
Catalytic and drug transport cycles of P-glycoprotein. A partitioning model of P-glycoprotein catalytic and transport cycles is shown. E refers to P-gp species. Inner-leaflet, high-affinity, drug-loading “ON-sites” are shown in green (loading site, [unk]). Extracellular facing, low-affinity, drug-unloading “OFF-sites” are shown in orange (unloading site, [unk]). Red stars show the rate-limiting transition states for the two cycles. The centrally located species ATPE [unk] [unk] indicates a molecule of P-glycoprotein that has one bound ATP. This species is thought to be a mobile carrier form of the protein in which the unloaded high-affinity and unloaded low-affinity drug-binding sites are in equilibrium. Upper right cycle shows the drug-activated, coupled activity; lower left cycle shows the uncoupled basal activity. If there is insufficient drug and two ATP molecules bind, P-gp partitions to the uncoupled cycle and hydrolyses ATP without any transport work. In this cycle the drug sites are in a low-affinity unloading conformation. However, if there is sufficient transport drug present, P-gp partitions to the coupled activity cycle. The coupled cycle is the alternating catalytic cycle previously described (Senior et al., 1995a). Transport drug binds first to a high-affinity loading site followed by ATP to form the ternary complex. After passing through the high-energy transition state, drug is released to the other side of the membrane (successful transport). Different transport drugs lead to different energy levels of the rate-limiting coupling transition state (Al-Shawi et al., 2003; Omote et al., 2004). Due to the instability of the higher-energy transition states, there is a greater probability of nonproductive transition state decay without drug transport for drugs with higher-energy transition states (failed transport). Failed transport is observed when wild-type P-glycoprotein is transporting colchicine or etoposide. The mutation G185V increases the strength of colchicine and etoposide interaction with the transition state and improves the transport of these drugs by reducing the level of failed transport (Omote et al., 2004).
Overall it was found that large conformational changes were associated with the rate-limiting transition state involving an unusually large number of bond-forming and bond-breaking events (Al-Shawi et al., 2003; Omote et al., 2004). For both SL-verapamil transport and associated ATP hydrolysis, the same rate-limiting transition state applied. These were strictly coupled events that must have been mediated by the same global concerted conformational changes. It was also found that each individual drug type made different interactions with the concerted rate-limiting transition state through different specific chemical bonds. Furthermore, it was found that rapidly transported substrates bound tighter to this transition state and required fewer conformational alterations by P-glycoprotein to get to this transition state. As illustrated in Fig. 2 (Coupled cycle), the overall rate-limiting step of P-gp during transport is a concerted carrier reorientation step in which the carried drug is moved from a high-affinity drug-loading “ON-site” to a low-affinity drug-unloading “OFF-site”.
Regulation of ATPase activity between the two catalytic cycles makes P-glycoprotein an energy inefficient transporter. We believe that this unusual transport mechanism (Fig. 2) is absolutely required for P-glycoprotein's physiological function as a very broad drug specificity exporter. A large component of the drug selectivity of P-gp arises from partitioning of hydrophobic transport drugs into the inner leaflet followed by protein binding. The partitioning between these two phases (membrane inner-leaflet and high-affinity drug-loading sites) is nearly isoenergetic (Seelig et al., 2000). Thus the interaction of the drug with P-glycoprotein is weak relative to the membrane. This may be a strict requirement for low drug selectivity by P-gp to maximize the chemical entities that can be transported. Given the high toxicity of P-glycoprotein's natural xenobiotic substrates, it has evolved to maximize successful drug transport. We postulate that the central ATPE [unk] [unk] species of Fig. 2 (located on both the coupled and uncoupled cycles) is a mobile carrier form of the protein. In this species the high and low affinity drug-binding sites are in equilibrium and this species is the only species that is competent to bind drugs for transport. Thus its concentration must be maximized. If P-glycoprotein binds 2 ATP molecules in the presence of high ATP and low drug concentrations it becomes kinetically incompetent for transport. Under such conditions the dissociation of ATP is slow. P- glycoprotein has evolved the uncoupled ATPase cycle (Fig. 2, shaded region) to return quickly to the transport competent form ATPE [unk] [unk] and to maximize its concentration. In other words, Pgp has evolved a parallel process that is always faster than a serial process that is rate limited by the slowest step. Clearly P-glycoprotein pays the price of this cellular protection vigilance with lowered overall coupling efficiency thereby, sacrificing efficiency to achieve higher drug clearance rates (Al-Shawi et al., 2003; Omote et al., 2004).
Improved coupling by the P-glycoprotein mutation G185V
The human colchicine-resistance P-glycoprotein mutation G185V has been extensively studied using non-purified natural systems (Choi et al., 1988; Kioka et al., 1989; Safa et al., 1990; Stein et al., 1994; Rao, 1995; Muller et al., 1996; Ramachandra et al., 1996; Ruth et al., 2001). However the data and interpretations generated by these studies were conflicting and ambiguous. Nevertheless, these reports clearly showed that the improved resistance to colchicine and etoposide afforded by G185V P-glycoprotein was highly correlated with the colchicine transport mechanism and as such was of great interest in understanding drug transport. Thermodynamic analyses of purified G185 mutant forms, reconstituted into transport competent proteoliposomes, indicated that G185V enzyme had specifically modified thermodynamic properties of colchicine or etoposide-dependent activities, whereas the thermodynamic properties of transport of other drugs such as verapamil, SL-verapamil or valinomycin remained unchanged in the coupled cycle. To improve the rate of colchicine or etoposide transport, the G185V transporter lowered the uncoupled basal activity as well as lowering the activation energy of the transport rate-limiting step. The high transition state energy of wild-type P-gp, when transporting colchicine or etoposide, increases the probability of nonproductive degradation of the transition state without transport (failed transport, Fig. 2 Coupled cycle). G185V P-gp transports etoposide or colchicine in an energetically more efficient way with decreased enthalpic and entropic components of the activation energy. EPR analysis of spin-labeled G185C enzyme and kinetic parameters of the G185C enzyme indicate that position 185 is surrounded by other residues, and is volume sensitive. These results and structural modeling suggest that residue 185 is a pivotal point in transmitting conformational changes between the catalytic sites and the colchicine drug-binding domain. This is mediated by ICD1 (Intra Cellular Domain 1) (Omote et al., 2004). Replacement of a highly flexible glycine with a bulky low-flexibility valine alters this communication and results in more efficient colchicine and etoposide transport. Another important conclusion was that drug binding energy was not used to facilitate transport. Full details can be found in Omote et al., 2004.
Structural modeling
To place our results, such as those discussed above for G185V P-glycoprotein, in a proper structural context, we performed homology modeling followed by extensive energy minimization to generate a stable atomic-detail structural-model of P-glycoprotein (Omote et al., 2004). To construct this model we used the highly homologous crystal structure coordinates of two ABC transporters; V. cholera lipid A transporter (MsbA, (Chang, 2003)) and S. typhimurium histidine permease (HisP, (Hung et al., 1998)). Recent structural models agree with the available biochemical data and there is a clear convergence between them (Shilling et al., 2003; Stenham et al., 2003; Omote et al., 2004). Thus our P-gp structural model is a consensus structural model that is expected to be highly representative of the true structure of P-gp. Full details of the modeling procedure can be found in Omote et al., 2004. The coordinates generated are available by request. P-glycoprotein was modeled in a closed active conformation. This model had no atom clashes and generated a Ramachandran plot (Ramachandran and Sasisekharan, 1968) in which 98.7% of the backbone angles were in allowed regions. A distinctive feature of P-glycoprotein and related multidrug resistance ABC transporters are the Intra Cellular Domain helices (ICDs) that connect the Transmembrane helices (TMs) to the nucleotide binding sites (Chang and Roth, 2001). These ICDs provide the logical structural elements for transmitting coupling and conformational information between the drug-binding and nucleotide-binding sites and thus regulate the coupling of the transport and hydrolytic activities of the transporter. A potential drug entry cleft was found between the two halves of the molecule at the level of the membrane interface and ICDs that opens to both sides of the membrane. The putative drug-binding chamber was found between both halves of P-glycoprotein. The order of TMs is 2, 3, 4, 1, 6, 5 for the N-terminal half and 8, 9, 10, 7, 12, 11 for the C-terminal half. Recent cross-linking experiments have verified this order (Loo et al., 2004a; Loo et al., 2004b).
Perspective
A long-standing goal of active transport research is to achieve a molecular understanding of coupling between the catalytic and transport mechanism. As discussed in this review we continue to strive to place the unusual kinetic and thermodynamic features of P-glycoprotein transport cycle within their proper physiological and structural contexts. It is with such understanding that a detailed atomic-level model of the transport processes of this most interesting of transporters can be elucidated. The expectation from this model is that it will allow for rational modulation of P-glycoprotein function by rational drug design and other forms of rational intervention, thus improving cancer treatments, AIDS treatments, oral administration of hydrophobic drugs and treatments of other diseases impacted by this transporter.
Footnotes
Supported by NIH grant GM52502 to M.K.S.
References
- Al-Shawi MK, Polar MK, Omote H, Figler RA. J. Biol. Chem. 2003;278:52629–52640. doi: 10.1074/jbc.M308175200. [DOI] [PubMed] [Google Scholar]
- Al-Shawi MK, Senior AE. J. Biol. Chem. 1993;268:4197–4206. [PubMed] [Google Scholar]
- Ambudkar SV, Dey S, Hrycyna CA, Ramachandra M, Pastan I, Gottesman MM. Annu. Rev. Pharmacol. Toxicol. 1999;39:361–398. doi: 10.1146/annurev.pharmtox.39.1.361. [DOI] [PubMed] [Google Scholar]
- Chang G. J. Mol. Biol. 2003;330:419–430. doi: 10.1016/j.jmb.2003.05.001. [DOI] [PubMed] [Google Scholar]
- Chang G, Roth CB. Science. 2001;293:1793–1800. doi: 10.1126/science.293.5536.1793. [DOI] [PubMed] [Google Scholar]
- Choi KH, Chen CJ, Kriegler M, Roninson IB. Cell. 1988;53:519–529. doi: 10.1016/0092-8674(88)90568-5. [DOI] [PubMed] [Google Scholar]
- Davidson AL, Chen J. Annu. Rev. Biochem. 2004;73:241–268. doi: 10.1146/annurev.biochem.73.011303.073626. [DOI] [PubMed] [Google Scholar]
- De Rivoyre M, Bonino F, Ruel L, Bidet M, Therond P, Mus-Veteau I. FEBS Lett. 2005;579:1529–1533. doi: 10.1016/j.febslet.2005.01.054. [DOI] [PubMed] [Google Scholar]
- Ecker G, Huber M, Schmid D, Chiba P. Mol Pharmacol. 1999;56:791–796. [PubMed] [Google Scholar]
- Exner O. Prog. Phys. Org. Chem. 1973;10:411–482. [Google Scholar]
- Figler RA, Omote H, Nakamoto RK, Al-Shawi MK. Arch. Biochem. Biophys. 2000;376:34–46. doi: 10.1006/abbi.2000.1712. [DOI] [PubMed] [Google Scholar]
- Flewelling RF, Hubbell WL. Biophys. J. 1986;49:531–540. doi: 10.1016/S0006-3495(86)83663-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman MM, Pastan I. Annu. Rev. Biochem. 1993;62:385–427. doi: 10.1146/annurev.bi.62.070193.002125. [DOI] [PubMed] [Google Scholar]
- Holland IB, Blight MA. J. Mol. Biol. 1999;293:381–399. doi: 10.1006/jmbi.1999.2993. [DOI] [PubMed] [Google Scholar]
- Hung LW, Wang IX, Nikaido K, Liu PQ, Ames GF-L, Kim SH. Nature. 1998;396:703–707. doi: 10.1038/25393. [DOI] [PubMed] [Google Scholar]
- Hyde SC, Emsley P, Hartshorn MJ, Mimmack MM, Gileadi U, Pearce SR, Gallagher MP, Gill DR, Hubbard RE, Higgins CF. Nature. 1990;346:362–365. doi: 10.1038/346362a0. [DOI] [PubMed] [Google Scholar]
- Kioka N, Tsubota J, Kakehi Y, Komano T, Gottesman MM, Pastan I, Ueda K. Biochem. Biophys. Res. Commun. 1989;162:224–231. doi: 10.1016/0006-291x(89)91985-2. [DOI] [PubMed] [Google Scholar]
- Lee SH, Altenberg GA. Biochem. Biophys. Res. Commun. 2003a;306:644–649. doi: 10.1016/s0006-291x(03)01029-5. [DOI] [PubMed] [Google Scholar]
- Lee SH, Altenberg GA. Biochem. J. 2003b;370:357–360. doi: 10.1042/BJ20021452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie EM, Deeley RG, Cole SP. Toxicol. Appl. Pharmacol. 2005;204:216–237. doi: 10.1016/j.taap.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Litman T, Skovsgaard T, Stein WD. J. Pharmacol. Exp. Ther. 2003;307:846–853. doi: 10.1124/jpet.103.056960. [DOI] [PubMed] [Google Scholar]
- Loo TW, Bartlett MC, Clarke DM. J. Biol. Chem. 2004a;279:7692–7697. doi: 10.1074/jbc.M311825200. [DOI] [PubMed] [Google Scholar]
- Loo TW, Bartlett MC, Clarke DM. J. Biol. Chem. 2004b;279:18232–18238. doi: 10.1074/jbc.M400229200. [DOI] [PubMed] [Google Scholar]
- Moiseenkova VY, Hellmich HL, Christensen BN. Biochem. Biophys. Res. Commun. 2003;310:196–201. doi: 10.1016/j.bbrc.2003.09.011. [DOI] [PubMed] [Google Scholar]
- Muller M, Bakos E, Welker E, Varadi A, Germann UA, Gottesman MM, Morse BS, Roninson IB, Sarkadi B. J. Biol. Chem. 1996;271:1877–1883. doi: 10.1074/jbc.271.4.1877. [DOI] [PubMed] [Google Scholar]
- Nakamoto RK, Ketchum CJ, Al-Shawi MK. Annu. Rev. Biophys. Biomol. Struct. 1999;28:205–234. doi: 10.1146/annurev.biophys.28.1.205. [DOI] [PubMed] [Google Scholar]
- Omote H, Al-Shawi MK. J. Biol. Chem. 2002;277:45688–45694. doi: 10.1074/jbc.M206479200. [DOI] [PubMed] [Google Scholar]
- Omote H, Figler RA, Polar MK, Al-Shawi MK. Biochemistry. 2004;43:3917–3928. doi: 10.1021/bi035365l. [DOI] [PubMed] [Google Scholar]
- Pisani DF, Rivoyre MD, Ruel L, Bonino F, Bidet M, Dechesne CA, Mus-Veteau I. Biochem. Biophys. Res. Commun. 2005;331:552–556. doi: 10.1016/j.bbrc.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Polgar O, Bates SE. Biochem. Soc. Trans. 2005;33:241–245. doi: 10.1042/BST0330241. [DOI] [PubMed] [Google Scholar]
- Ramachandra M, Ambudkar SV, Gottesman MM, Pastan I, Hrycyna CA. Mol. Biol. Cell. 1996;7:1485–1498. doi: 10.1091/mbc.7.10.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran GN, Sasisekharan V. Adv. Protein. Chem. 1968;23:283–438. doi: 10.1016/s0065-3233(08)60402-7. [DOI] [PubMed] [Google Scholar]
- Rao US. J. Biol. Chem. 1995;270:6686–6690. [PubMed] [Google Scholar]
- Rao US, Nuti SL. J. Biol. Chem. 2003;278:46576–46582. doi: 10.1074/jbc.M308078200. [DOI] [PubMed] [Google Scholar]
- Romsicki Y, Sharom FJ. Biochemistry. 1999;38:6887–6896. doi: 10.1021/bi990064q. [DOI] [PubMed] [Google Scholar]
- Ruth A, Stein WD, Rose E, Roninson IB. Biochemistry. 2001;40:4332–4339. doi: 10.1021/bi001373f. [DOI] [PubMed] [Google Scholar]
- Safa AR, Stern RK, Choi K, Agresti M, Tamai I, Mehta ND, Roninson IB. Proc. Natl. Acad. Sci. USA. 1990;87:7225–7229. doi: 10.1073/pnas.87.18.7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauna ZE, Ambudkar SV. J. of Biol. Chem. 2001;276:11653–11661. doi: 10.1074/jbc.M011294200. [DOI] [PubMed] [Google Scholar]
- Seelig A. Eur. J. Biochem. 1998;251:252–261. doi: 10.1046/j.1432-1327.1998.2510252.x. [DOI] [PubMed] [Google Scholar]
- Seelig A, Blatter XL, Wohnsland F. Int. J. Clin. Pharmacol. Ther. 2000;38:111–121. doi: 10.5414/cpp38111. [DOI] [PubMed] [Google Scholar]
- Seelig A, Landwojtowicz E. Eur. J. Pharm. Sci. 2000;12:31–40. doi: 10.1016/s0928-0987(00)00177-9. [DOI] [PubMed] [Google Scholar]
- Senior AE, Al-Shawi MK, Urbatsch IL. FEBS Lett. 1995a;377:285–289. doi: 10.1016/0014-5793(95)01345-8. [DOI] [PubMed] [Google Scholar]
- Senior AE, Al-Shawi MK, Urbatsch IL. J. Bioenerg. Biomembr. 1995b;27:31–36. doi: 10.1007/BF02110328. [DOI] [PubMed] [Google Scholar]
- Shapiro AB, Corder AB, Ling V. Eur. J. Biochem. 1997;250:115–121. doi: 10.1111/j.1432-1033.1997.00115.x. [DOI] [PubMed] [Google Scholar]
- Shapiro AB, Fox K, Lam P, Ling V. Eur. J. Biochem. 1999;259:841–850. doi: 10.1046/j.1432-1327.1999.00098.x. [DOI] [PubMed] [Google Scholar]
- Shapiro AB, Ling V. J. Biol. Chem. 1995;270:16167–16175. doi: 10.1074/jbc.270.27.16167. [DOI] [PubMed] [Google Scholar]
- Shapiro AB, Ling V. Eur. J. Biochem. 1997a;250:122–129. doi: 10.1111/j.1432-1033.1997.00122.x. [DOI] [PubMed] [Google Scholar]
- Shapiro AB, Ling V. Eur. J. Biochem. 1997b;250:130–137. doi: 10.1111/j.1432-1033.1997.00130.x. [DOI] [PubMed] [Google Scholar]
- Shapiro AB, Ling V. Eur. J. Biochem. 1998;254:181–188. doi: 10.1046/j.1432-1327.1998.2540181.x. [DOI] [PubMed] [Google Scholar]
- Sharom FJ. J. Membr. Biol. 1997;160:161–175. doi: 10.1007/s002329900305. [DOI] [PubMed] [Google Scholar]
- Sharom FJ, Yu X, Doige CA. J. Biol. Chem. 1993;268:24197–24202. [PubMed] [Google Scholar]
- Shilling RA, Balakrishnan L, Shahi S, Venter H, van Veen HW. Int. J. Antimicrob. Agents. 2003;22:200–204. doi: 10.1016/s0924-8579(03)00212-7. [DOI] [PubMed] [Google Scholar]
- Stein WD, Cardarelli C, Pastan I, Gottesman MM. Mol. Pharmacol. 1994;45:763–772. [PubMed] [Google Scholar]
- Stenham DR, Campbell JD, Sansom MS, Higgins CF, Kerr ID, Linton KJ. FASEB J. 2003;17:2287–2289. doi: 10.1096/fj.03-0107fje. [DOI] [PubMed] [Google Scholar]
- Tu BP, Weissman JS. Mol. Cell. 2002;10:983–994. doi: 10.1016/s1097-2765(02)00696-2. [DOI] [PubMed] [Google Scholar]
- Urbatsch IL, Al-Shawi MK, Senior AE. Biochemistry. 1994;33:7069–7076. doi: 10.1021/bi00189a008. [DOI] [PubMed] [Google Scholar]
- Victor KG, Cafiso DS. Biophys. J. 2001;81:2241–2250. doi: 10.1016/S0006-3495(01)75871-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusa K, Tsuruo T. Cancer Research. 1989;49:5002–5006. [PubMed] [Google Scholar]
- Zeng GF, Pypaert M, Slayman CL. J. Biol. Chem. 2004;279:3003–3013. doi: 10.1074/jbc.M309760200. [DOI] [PubMed] [Google Scholar]