Abstract
The Saccharomyces cerevisiae silencing protein Sir2 is the founding member of a universally conserved family of proteins that have been shown to possess NAD-dependent histone deacetylation and ADP-ribosylation activities. Here we show that histone deacetylation by Sir2 is coupled to cleavage of the high-energy bond that links the ADP-ribose moiety of NAD to nicotinamide. Analysis of the NAD cleavage products revealed the presence of nicotinamide, ADP-ribose, and a third product that appeared to be related to ADP-ribose. With the use of label transfer experiments, we show that the acetyl group in the histone substrate is transferred to this NAD breakdown product during deacetylation, forming a product that we conclude to be O-acetyl-ADP-ribose. Detection of this species strongly argues for obligate coupling of histone deacetylation to NAD breakdown by Sir2. We propose reaction mechanisms that could account for this coupling via acetyl-ADP-ribose formation. The unprecedented coupling of amide bond cleavage to cleavage of a high-energy bond raises the possibility that NAD breakdown by Sir2 plays an important role in silencing that is independent of its requirement for deacetylation.
Reversible acetylation of lysine residues in the conserved tails of histones, and in other proteins, is closely associated with changes in chromatin structure during activation and repression of transcription. A number of transcriptional repressors have been shown to possess histone deacetylase activity (reviewed in refs. 1 and 2). Among these is the Sir2 protein (3–5), a structural component of silent chromatin that is required for gene silencing and extension of the life span in budding yeast (6–14). Sir2 is a unique deacetylase for two reasons. First, Sir2 appears to be a universally conserved protein, with homologues in all three kingdoms of life (15, 16). Second, the deacetylase activity of Sir2 depends on NAD (3–5).
In addition to deacetylating histones, Sir2-like proteins can perform a weak ADP-ribosylation reaction in which they transfer the ADP-ribose moiety of NAD to protein substrates (3, 17, 18). Moreover, Sir2 is capable of forming labeled NAD from labeled nicotinamide and unlabeled NAD (4), a further indication that it can catalyze breakage of the C-N bond between the ADP-ribose and nicotinamide moieties of NAD. Interestingly, this exchange reaction was shown to depend on the presence of acetylated histone substrates. However, the efficiency of this reaction, as well as its relationship to deacetylation, is unclear. Mutational analyses aimed at discerning the importance of the enzymatic activities of Sir2 for its in vivo function have suggested that some form of Sir2 enzymatic activity is required for silencing (3, 18). However, none of the mutants tested to date have completely separated the two activities.
The apparent dependence of each Sir2 activity on the substrate for the other is puzzling, given that numerous enzymes catalyze one reaction or the other without any need of cofactors. The amide hydrolysis reaction associated with deacetylation has been particularly well characterized, because it is similar to the hydrolysis of peptide bonds catalyzed by proteases. No cofactor-dependent proteases have been described to date. In addition, the prevalent Rpd3 class of histone deacetylases catalyzes deacetylation in a cofactor-independent manner via a mechanism that has been proposed to be similar to that of metalloproteases (19). Similarly, ADP-ribosyltransferases and NAD-glycohydrolases, which catalyze cleavage of the N-glycosidic bond linking nicotinamide to ADP-ribose, do not require cofactors (20, 21). Of central importance to understanding how Sir2 contributes to silencing is deciphering how and why Sir2 unites these seemingly disparate activities.
By using a variety of separation methods, we have examined the reaction products formed by Sir2 in in vitro reactions containing NAD and acetylated histone substrates. We find that histone deacetylation by Sir2 is accompanied by the efficient breakdown of NAD. Surprisingly, we find that the acetyl group on the substrate is transferred to an ADP-ribose-related NAD cleavage product, indicating a direct relationship between the two activities. Based on its chemical properties and its mode of formation, we propose that this novel compound is acetyl-ADP-ribose. Consistent with the observed coupling of deacetylation and NAD cleavage, point mutations in the conserved core domain of Sir2 fail to separate the two activities. These findings indicate that NAD is directly involved in histone deacetylation and provide an example of an amide hydrolysis reaction that requires a nucleotide coenzyme.
Materials and Methods
Reaction Conditions.
All reactions contained buffer R [50 mM Tris (pH 8.0)/100 mM NaCl/1 mM DTT] and 0.1–1 μM glutathione S-transferase (GST) or GST-Sir2 fusion protein. Incubations were at 25°C for the indicated times. Reactions at pH 10 used 50 mM glycine/KOH, pH 10.0, instead of Tris.
TLC.
Reactions for TLC contained 5 μCi [α-32P]NAD (NEN; 800 Ci/mmol) with or without 100 μM cold NAD, or 0.05 μCi [carbonyl-14C]NAD (Amersham Pharmacia; 51 mCi/mmol).Histone substrates were either calf thymus histones (3 μg) or histone H4 N-terminal peptides (500 μM unless otherwise indicated). Histone H4 N-terminal peptides corresponded to amino acids 1–19 of histone H4, with or without an acetyl group at one of the four lysines (positions 5, 8, 12, and 16) and were synthesized and HPLC-purified at the Tufts University Core Facility. All peptides were supplied as lyophilized powder and dissolved in H2O. NADase was purchased from Sigma.
Reactions were carried out in 10-μl volumes and then diluted 100-fold with buffer R. Two microliters of the dilution was spotted on a TLC plate (Whatman; LHPKDF Silica 60 A). Plates were developed in 80% ethanol/20% 2.5 M ammonium acetate, dried, and exposed to a Bio-Rad PhosphorImager screen for 5–30 h. Images were processed with the use of quantity one software. [14C]NAD reactions were not diluted before TLC. Migration positions of unlabeled NAD, ADP-ribose, and nicotinamide standards (all from Sigma) were determined by UV shadowing.
Calculation of NAD Cleaved:Peptide Deacetylated.
Throughout this work, the given peptide concentrations refer to molarities calculated from the predicted mass of the peptide. The peptides used in this study were purified by high-performance liquid chromatography (HPLC) in a buffer system containing trifluoroacetic acid (TFA) and then lyophilized. TFA is therefore likely to contribute to the mass of the lyophilized material. We estimate that each peptide molecule is associated with up to seven molecules of TFA in solution (corresponding to the seven positive charges in the peptide). Thus the effective mass of the peptide is equal to its predicted mass plus 7 times the mass of TFA. In reactions containing 500 μM peptide as calculated from the predicted mass, the effective peptide concentration is 370 μM. The ratios of moles of NAD cleaved to moles of peptide deacetylated in the reactions described in Fig. 3 are 0.63 ± 0.16 from the predicted concentration and 0.85 ± 0.18 from the effective concentration (n = 5).
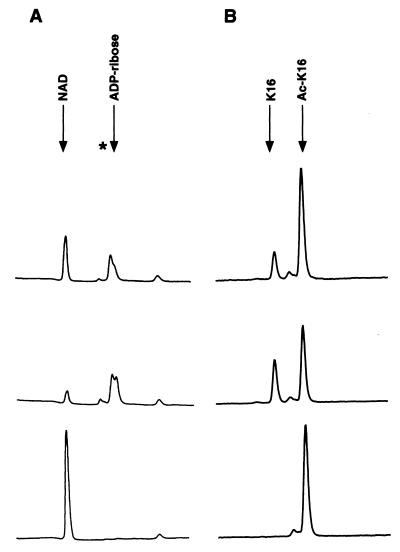
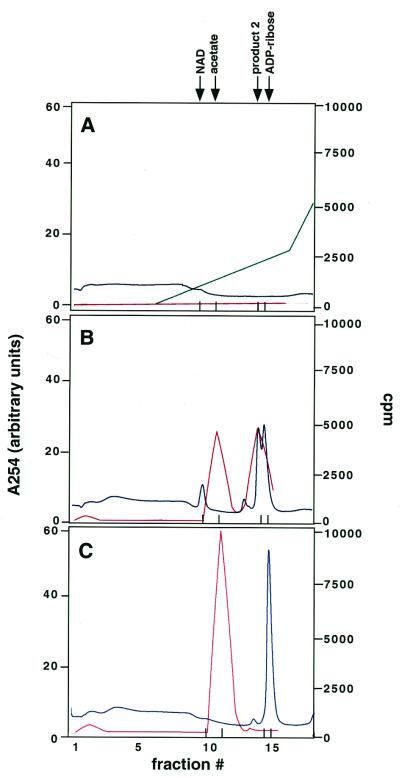
Figure 3.
Coupling of NAD breakdown and histone deacetylation. Reactions containing wild-type Sir2 were incubated with or without 100 μM NAD and 500 μM AcK-16 peptide and fractionated either by ion-exchange chromatography (A) or by HPLC (B). Shown are chromatograms of reactions stopped after incubation for 1 h (Top) or 6 h (Middle). The bottom panel in A shows a control reaction in which the Ac-K16 peptide was replaced by an unacetylated peptide. The bottom panel in B shows a control reaction to which no NAD was added. Arrows indicate elution times of various standards. The asterisk indicates the elution time of the product 2 peak.
ADP-Ribosylation and Deacetylation Assays.
ADP-ribosylation was assayed as previously described (18). For acetate release assays, recombinant yeast histone H3 produced in Escherichia coli (provided by A. Farrell, Harvard Medical School, Boston) was first acetylated in vitro. Acetylation reactions were in buffer R with 10 μCi [3H]acetyl-CoA (ICN; 12 Ci/mmol), 1 μM 6× His-Esa1, and 10–50 μM substrate for 4 h at 30°C. Labeled histones were precipitated by adding trichloroacetic acid to a final concentration of 20% and incubating on ice for 30 min. Precipitates were collected by centrifugation, washed with 10% trichloroacetic acid, and dissolved in 100 mM Tris (pH 8.0). Approximately 2 μg of labeled histone was then added to a 10-μl reaction with or without 100 μM NAD. Acetate release was quantitated by adding 20 μl H2O and 7.5 μl of 0.1 M HCl/0.16 M acetic acid, followed by extraction with 100 μl ethyl acetate. The ethyl acetate phase was removed and counted in 2 ml scintillation fluid (Research Products International, Mt. Prospect, IL).
Ion-Exchange Chromatography.
Large-scale reactions contained 100 μM NAD and 500 μM Ac-K16 peptide and were carried out in 100-μl volumes. Fractionation was carried out on a 1-ml prepacked MonoQ column (Amersham Pharmacia), with the use of an AKTA fast performance liquid chromatography (FPLC) apparatus (Amersham Pharmacia). Ninety microliters of the reaction was diluted in 900 μl H2O and injected onto the column. The column was eluted in a gradient of 0–500 mM NH4HCO3 in H2O, and 1-ml fractions were collected. UV absorbance of each fraction was measured at 254 nm. The 3H-acetylated histone H3 included in some reactions was acetylated in vitro as described above. 3H-containing fractions were counted by diluting 0.5 ml of each fraction into 4.5 ml of scintillation fluid. [2-14C]Acetic acid (54 mCi/mmol) was from NEN. Data were compiled with the use of unicorn software.
High-Performance Liquid Chromatography.
HPLC was carried out on a C18 column (Vydac, Hesperia, CA; 218TP54) with the use of a Hewlett–Packard Series II 1090 apparatus. Ten microliters of a 100-μl reaction were diluted in 90 μl of 0.08% TFA and injected onto the column. Elution used a gradient of 0–80% acetonitrile in H2O with 0.1% TFA included throughout. UV absorbance was measured at 214 nm. Data were compiled with the use of hpchemstation software.
Plasmid Construction and Protein Purification.
Plasmids pDM111a and pDM360, encoding GST-Sir2 and GST-H364Y, respectively, have been described (18). Plasmids pJT23 and pJT25, encoding GST-G262A and GST-G270A, respectively, were constructed by site-directed mutagenesis of pDM111a with the use of overlap PCR (22) and sequenced at the Harvard Medical School Biopolymers Facility. Primer sequences are available on request.
GST-fusion proteins were expressed in E. coli DH5α and purified as described (18). 6xHis-Esa1 was expressed from plasmid pRSET-Esa1 (a gift from D. Allis, University of Virginia, Charlottesville, VA) and purified as described (23).
Results
The NADase Activity of Sir2 Is Stimulated by Acetyl-Lysine.
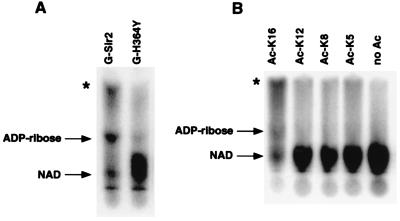
We had previously shown that wild-type Sir2 could mediate the transfer of label from [32P]NAD to itself and histones in vitro (18). We noticed that free [32P]NAD was also modified by Sir2 in these reactions. To characterize the NAD modification activity further, we incubated purified GST-Sir2 with [α-32P]NAD and calf thymus histones and analyzed the reaction products by TLC and autoradiography (Fig. 1A). In the presence of wild-type Sir2, NAD was converted into two distinct phosphate-containing products. One of these products comigrated with ADP-ribose, as judged by comparison with migration of standards. The second product (marked by * in Fig. 1A) is referred to here as product 2. Quantitation of signal intensities by PhosphorImager analysis showed that nearly 70% of the free NAD was modified in these reactions. In contrast, ≈1% of the NAD was covalently attached to protein under the same conditions (data not shown). A mutant form of Sir2 (G-H364Y) that is defective for silencing in vivo and ADP-ribosylation in vitro was also unable to modify free [32P]NAD, suggesting that these activities of Sir2 may be linked (Fig. 1A).
Figure 1.
Acetyl-lysine-dependent NAD breakdown by Sir2: autoradiographs of TLC plates used to separate labeled reaction products. Arrows indicate the migration positions of cold standards. Asterisks denote product 2. (A) Stimulation of NAD breakdown by calf thymus histones. Reactions contained 5 μCi [32P]NAD (1 μM final concentration) and either wild-type Sir2 (G-Sir2) or the H364Y point mutant (G-H364Y). Incubations were for 6 h at 25°C. (B) Stimulation of NAD breakdown by histone H4 N-terminal peptides. Reactions contained 5 μCi [32P]NAD, 100 μM cold NAD, 500 μM peptide acetylated at the indicated lysine, and wild-type Sir2. Incubations were for 6 h at 25°C.
The discovery of deacetylase activity in Sir2 (3–5) led us to ask whether the NAD breakdown activity we observed was related to deacetylation. We repeated the experiments shown in Fig. 1A with the use of recombinant yeast histones produced in E. coli and observed no modification of NAD. We also found that Sir2 failed to transfer label from [32P]NAD onto these histones (J.C.T. and D.M., unpublished observations). These results suggested that breakdown of NAD by Sir2 required the presence of acetylated substrates. To investigate this possibility in more detail, Sir2 was incubated with chemically synthesized peptides corresponding to the N terminus of histone H4 that were either unacetylated or contained a single acetyl-lysine residue at position 5, 8, 12, or 16. As shown in Fig. 1B, the unacetylated peptide failed to stimulate the NAD breakdown activity. However, a peptide acetylated at lysine 16 potently stimulated NAD breakdown. Peptides acetylated at other positions were relatively ineffective. Interestingly, the Ac-K16 H4 peptide was previously shown to be the favored substrate for deacetylation by Sir2 as well (3), providing additional support for the possibility that NAD breakdown and deacetylation are related.
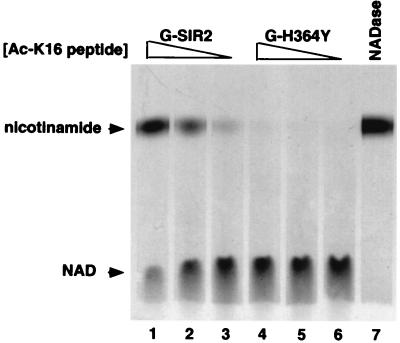
Because previous reports indicated that Sir2 could act on the nicotinamide ribose C-N bond in NAD, the appearance of a phosphate-containing product distinct from ADP-ribose in Sir2 reactions was surprising. To gain some insight into the chemical nature of product 2, we incubated GST-Sir2 with the Ac-K16 peptide described above and NAD that was labeled at the amide carbonyl carbon of the nicotinamide group ([carbonyl-14C]NAD; see Fig. 6 B and C). Use of this form of NAD allowed us to assess whether product 2 was generated as a result of cleavage of the C-N bond linking ADP-ribose to nicotinamide or some other modification that left the C-N bond intact. We detected only two labeled products in these reactions, and comparison of their migration with that of unlabeled standards indicated that they corresponded to NAD and nicotinamide (Fig. 2 and data not shown). The upper band in Sir2 reactions also comigrated with the labeled product formed by NADase (compare Fig. 2, lanes 1–3 with lane 7), an enzyme known to generate ADP-ribose and nicotinamide from NAD. Formation of free nicotinamide by Sir2 required the acetylated substrate in a dose-dependent manner and was not observed in reactions that contained the H364Y mutant form of Sir2 (Fig. 2). These results indicated that all products of NAD breakdown must result from breakage of the glycosidic bond that links nicotinamide to the C1" position of ribose. Product 2 must therefore be an ADP-ribose derivative.
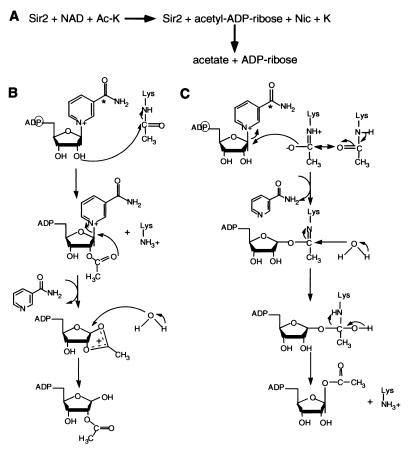
Figure 6.
Proposed mechanisms for NAD-dependent histone deacetylation by Sir2. (A) Summary of overall reaction scheme. (B) Model 1, nucleophilic attack of the 2" hydroxyl of NAD on the N-acetyl carbonyl group (see text for details). (C) Model 2, nucleophilic attack of the nicotinamide-ribose C-N bond by the isoamide form of the N-acetyl carbonyl group (see text for details). The positions of the radioactive atoms in the labeled versions of NAD used in this study are indicated by a circle (for the α-phosphate) and an asterisk (for the amide carbonyl carbon).
Figure 2.
NAD breakdown activity of Sir2 involves cleavage of the C-N bond between the nicotinamide and ribose moieties of NAD. Wild-type Sir2 (G-Sir2) or the H364Y point mutant (G-H364Y) was incubated with 1000 μM (Lanes 1 and 4), 100 μM (Lanes 2 and 5), or 10 μM (Lanes 3 and 6) Ac-K16 peptide and 0.05 μCi of [14C]NAD (the final concentration was 100 μM). In Lane 7, 0.05 units of NADase was incubated with 0.05 μCi of [14C]NAD. Incubations were for 6 h at 25°C. The reactions were subjected to TLC and autoradiographed, as shown. Arrows indicate the migration positions of cold standards.
Quantitation of Sir2 Reaction Product Formation.
If NAD breakdown were coupled to deacetylation, we would expect a consistent relationship between the amounts of NAD breakdown products and deacetylation products formed in the reaction. To test this possibility, we used a system that allowed the analysis of both deacetylation and NAD breakdown in the same reaction. Large-scale Sir2 reactions containing 1 μM GST-Sir2, 100 μM NAD, and 500 μM Ac-K16 peptide were incubated for 1 h or 6 h at 25°C. Portions of each reaction were then fractionated either by ion-exchange chromatography (to follow nucleotide products) or by reverse-phase high-performance liquid chromatography (HPLC) (to follow peptides). Three major peaks were present on ion-exchange chromatograms (Fig. 3A Top and Middle). Two of these peaks coelute with NAD and ADP-ribose, as judged by comparison with elution profiles of standards. The third peak (marked by an asterisk) significantly overlapped the ADP-ribose peak, and its abundance relative to the ADP-ribose peak argued that it represented product 2 observed by TLC. Consistent with the TLC experiments in Figs. 1 and 2, the reaction depended on the presence of acetyl-lysine, because a reaction with an unacetylated peptide showed only a single peak corresponding to NAD (see Fig. 3A Bottom).
HPLC chromatograms of the same reactions revealed two major peaks corresponding to the deacetylated and acetylated forms of the Ac-K16 peptide, as judged by comparison to the elution of standards (Fig. 3B). A control reaction that did not contain NAD gave only a single peak at the position of the acetylated peptide. By calculating the areas under the various peaks generated by the two fractionation procedures, we found that the ratio of moles of NAD modified to moles of peptide deacetylated in our reactions was 0.85 ± 0.18 (see also Materials and Methods). We observed the same reaction products at similar relative amounts under a variety of reaction conditions. These included NAD and peptide concentrations, each ranging from 100 to 500 μM, enzyme concentrations from 0.1 to 1 μM, and incubations times ranging from 10 min to 6 h (data not shown). These data supported a stoichiometric relationship between NAD breakdown and histone deacetylation by Sir2.
Acetyl Transfer from Substrate to ADP-Ribose.
We considered two models to account for the coupling of histone deacetylation and NAD breakdown by Sir2. In one model, the substrates of one activity would act as allosteric effectors of the other activity. For example, association of an acetylated substrate with Sir2 might trigger NAD hydrolysis, which in turn would activate the deacetylase activity of Sir2 via a conformational switch. In this case, the chemistry of catalysis of one reaction would be completely independent of that of the other. Alternatively, we thought it possible that the reactions were mechanistically coupled, such that the chemistry of catalysis of one reaction would be linked to that of the other.
We thought it possible that mechanistic coupling of the two reactions would involve transfer of the acetyl group from the acetylated substrate to an NAD breakdown product. Tpt1, a yeast enzyme involved in tRNA splicing, is known to catalyze an analogous phosphotransfer reaction involving NAD (24, 25). A similar mechanism could also result in the formation of product 2. If product 2 is produced as a result of acetyl transfer from histones to ADP-ribose, we would expect that deacetylation of 3H-acetylated histones would result in the elution of label at the position of product 2 during chromatography. We tested this possibility by following the fate of [3H]acetyl groups on labeled histone substrates in our reactions. Reactions were set up as in Fig. 3, except that ≈30,000 cpm of 3H-labeled, acetylated histone H3 was included in the reactions. Fractionation by ion-exchange chromatography showed that most of the NAD in the reactions was converted to product 2 and ADP-ribose (Fig. 4B, blue trace), as observed in experiments presented in Fig. 3. Assay of radioactivity in these column fractions revealed the presence of two peaks (Fig. 4B, red trace). The peak at fraction 11 eluted at the same position as free acetate, as determined by running ≈500 cpm of [14C]acetate on the column under the same conditions. Strikingly, the peak at fraction 14 coeluted precisely with product 2. Coelution of labeled acetate and a phosphate-containing product of NAD breakdown strongly argued that product 2 is an acetyl-containing ADP-ribose derivative. Because the labeled histone substrates remained bound to the column under the elution conditions used, no peaks of radioactivity were observed in the absence of NAD (Fig. 4A). Similar results were obtained in experiments that used the H364Y mutant form of Sir2 (data not shown), indicating that Sir2 activity was required to generate the labeled species observed. We obtained similar results with the use of a 3H-labeled H4 N-terminal peptide instead of histone H3, and with a range of incubation times from 1 to 6 h (data not shown).
Figure 4.
Sir2 transfers the acetyl group in the substrate to a breakdown product of NAD. Shown are ion-exchange chromatograms depicting fractionation of reactions that contained Sir2, 500 μM Ac-K16 peptide, and ≈30,000 cpm of 3H-labeled, acetylated histone H3. Incubation was for 6 h at 25°C. The labeled substrates remained bound to the column under the elution conditions used. Red traces represent cpm, blue traces represent A254. The green trace in A represents the salt gradient from 0 to 500 mM used in these experiments; it was omitted in the two lower panels for simplicity. The standards indicated at the top elute as follows: NAD, fraction 10; acetate, fraction 11; product 2, fraction 14; ADP-ribose, fraction 15 (also indicated by tick marks at the bottom of each panel). (A) No NAD was included in the reaction. (B) NAD (100 μM) was included in the reaction. (C) The reaction was carried out at pH 10 and included 100 μM NAD.
The pH sensitivity of the labeled product also suggested that it contained both acetate and ADP-ribose components (Fig. 4C). In reactions carried out at pH 10, product 2 could not be detected, either by ion-exchange chromatography or by TLC (Fig. 4C and data not shown). In addition, the peak of radioactivity observed at fraction 14 was no longer detectable, and there were corresponding increases in the sizes of the free acetate peak in fraction 11 and the ADP-ribose peak in fraction 15. Alkali treatment of fraction 14 from a reaction carried out at pH 8 also revealed an acetate peak and an ADP-ribose peak on rechromatography (data not shown). This finding demonstrated that elevated pH changed the structure of product 2 rather than Sir2 activity. Overall, these results indicated that an acetyl-containing derivative of ADP-ribose was generated by Sir2 and supported a direct, mechanistic coupling of NAD cleavage and histone deacetylation.
Analysis of Mutant Sir2 Proteins Provides Independent Support for Coupling of Deacetylation to NAD Hydrolysis.
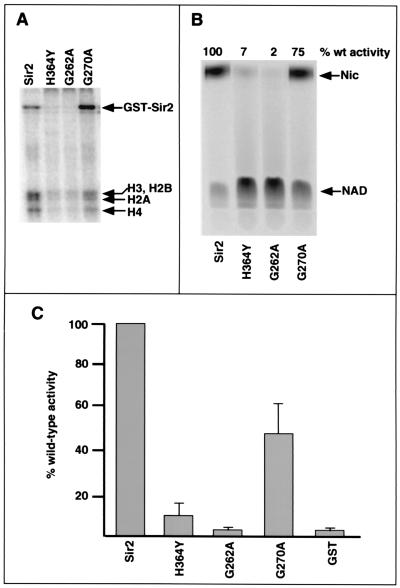
As an independent test of the coupled activity hypothesis, we analyzed the activities of three mutant forms of Sir2 containing single amino acid substitutions at highly conserved positions within the catalytic core domain of the protein. The coupled activity hypothesis predicts that mutants showing a defect in one activity should show a concomitant defect in the other activity. This prediction was borne out for the three Sir2 mutant proteins that were tested. The H364Y and G262A mutants had little or no detectable ADP-ribosyltransferase or NAD breakdown activities (Fig. 5 A and B). Both mutants also showed severe defects in deacetylase activity (Fig. 5C). The G270A mutant retained similar amounts of NAD breakdown and histone deacetylase activities (≈75% and ≈50% of wild type, respectively; see Fig. 5 B and C). This mutant showed a 50% decrease in ADP-ribosylation of histones compared with wild type, although its autolabeling activity was normal (Fig. 5A). The ADP-ribosylation and histone deacetylation activities we found for this mutant are similar to those reported by Imai et al. (3). These data are consistent with a coupled mechanism for NAD breakdown and histone deacetylation.
Figure 5.
Mutations in the conserved core domain of Sir2 fail to separate its activities. (A) ADP-ribosylation assays. Wild-type Sir2 (G-Sir2) or the mutant proteins were assayed as described in ref. 18. (B) NAD breakdown assays. Reactions contained either the wild-type protein or one of the mutants, 500 μM Ac-K16 peptide, and 0.05 μCi [14C]NAD (100 μM final concentration). Products were separated by TLC after 6-h incubations at 25°C. Signals were visualized and quantitated by PhosphorImager analysis, as shown. (C) Histone deacetylase assays. Reactions contained either wild-type Sir2 or one of the mutants, ≈8,000 cpm of 3H-acetylated, recombinant yeast histone H3, and 100 μM NAD. Incubations were for 1–6 h at 25°C. Release of acetate from the labeled substrate was quantitated as described in Materials and Methods. (Error bars denote standard deviations from the mean.)
Discussion
Three lines of evidence in the present study support a direct role for NAD in the catalysis of histone deacetylation by the yeast silencing protein Sir2. First, we observe a stoichiometric relationship between the amount of NAD cleaved and histone substrates deacetylated in Sir2 reactions. Second, Sir2 point mutations have parallel effects on its deacetylation and NAD cleavage activities. (These mutants also show corresponding defects in silencing function in vivo; see refs. 3 and 18.) Finally, and most importantly, the formation of an acetate-containing NAD breakdown product shows that NAD is a direct participant in the deacetylation reaction.
Our results strongly suggest that the mechanism of NAD-dependent histone deacetylation by Sir2 involves transfer of the acetyl group from the substrate to an NAD cleavage product that is likely to be acetyl-ADP-ribose (Fig. 6A). We think it likely that detection of ADP-ribose in our reactions reflects nonenzymatic decay of this product, because at very long reaction times (24 h) only ADP-ribose is present (also compare Fig. 3A Top and Middle). An ester linkage between acetate and one of the hydroxyl groups on the nicotinamide would be consistent with the base sensitivity of the labeled adduct we observe. We propose two plausible mechanisms for NAD-dependent deacetylation involving acetyl-ADP-ribose formation.
In the first model (Fig. 6B), Sir2 would catalyze nucleophilic attack of the 2"-hydroxyl of the nicotinamide ribose on the N-acetyl carbonyl group, forming 2"-acetyl-NAD. Intramolecular attack of the carbonyl oxygen on the C1" would lead to formation of an unstable cyclic acetoxycarbonium ion, which would be susceptible to cleavage by water (or a basic amino acid side chain). This mechanism is analogous to that proposed for the formation of ADP-ribose 1"-2" cyclic phosphate by Tpt1 (25). Although attack of the ionic species could occur at either C1" or C2" at this stage, the anomeric carbon would be the more likely target because of sharing of the positive charge by the ring oxygen. The likely product in this reaction scheme is 2"-O-acetyl-ADP-ribose.
In the second model (Fig. 6C), NAD is cleaved by acetalysis. This mechanism proposes a nucleophilic attack on the C1" carbon by the isoamide form of the N-acetyl carbonyl oxygen, releasing nicotinamide. A basic side chain on the enzyme could help stabilize the isoamide by abstracting the proton from the amide nitrogen. Attack of water on the carbonyl carbon would then generate a tetrahedral intermediate. Removal of a proton (possibly by the enzyme) from the hydroxyl group would then release deacetylated lysine and 1"-O-acetyl-ADP-ribose. Participation of an isoamide in an enzyme-catalyzed nucleophilic attack has also been suggested for cleavage of ATP by formylglycinamide ribonucleotide aminotransferase (26).
Can our findings provide an explanation for the previously described ADP-ribosyltransferase activity of Sir2? In the case of the mechanism shown in Fig. 6B, resolution of the acetoxycarbonium ion by a side chain on the enzyme or the substrate would result in a protein-acetyl-ADP-ribose adduct. The mechanism in Fig. 6C could also account for protein labeling, because it involves the formation of a transient substrate-ADP-ribose intermediate. The detection of ADP-ribosylated histones and peptides in reactions containing 32P-labeled NAD and acetylated substrates may therefore reflect a low-frequency capture of such an intermediate (3, 18). Transient ADP-ribose-substrate or AMP-substrate intermediates have also been detected in the case of a 2′-phosphotransferase involved in tRNA splicing (mentioned above) and E. coli DNA ligase, two other enzymes that use NAD as a cofactor for catalysis (25, 27). Nonetheless, it remains possible that some of the ADP-ribosylated species detected in Sir2 reactions are the result of nonenzymatic linkage of the released ADP-ribose or acetyl-ADP-ribose to amino acid side chains.
Whereas some precedent exists for coupling of NAD hydrolysis to catalysis of additional chemical reactions (24, 27), coupling of amide hydrolysis to additional steps has never been observed and raises questions about the possible biological significance of such a reaction mechanism. The energy of hydrolysis of the nicotinamide ribose glycosidic bond of NAD is −8.2 kcal/mol (28, 29). For comparison, the energy of hydrolysis of ATP to ADP is −7.3 kcal/mol (30). The large amount of energy released through NAD cleavage might be required to overcome an unusual kinetic barrier to deacetylation. Alternatively, this energy may be used to couple histone deacetylation to some other process, such as chromatin condensation or spreading of silencing factors along the chromatin fiber. It is also possible that the formation of acetyl-ADP-ribose itself, rather than the energy used in its formation, is critical to Sir2 function. For example, acetyl-ADP-ribose could serve as an effector of other silencing proteins or perhaps as an acetyl group donor for a novel acetyltransferase.
In summary, we have shown that the yeast silencing protein Sir2 has two coupled enzymatic activities: histone deacetylation and cleavage of a high-energy bond in NAD. Detection of an acetyl-containing NAD breakdown product indicates that these two activities are mechanistically linked. The relative contributions of the NADase and deacetylase activities of Sir2 to the mechanism of silencing remain to be determined.
Acknowledgments
We thank David Allis and Tim Richmond for plasmids, Alison Farrell for purified yeast histone H3, and Alexei Kisselev for help with HPLC. We also thank Lew Cantley, Harry Noller, Tim Mitchison, Steve Miller, Tarun Kapoor, and Chris Walsh for helpful discussions; Tim Mitchison, Steve Miller, and Chris Walsh for suggesting reaction mechanisms and reading the manuscript; and an anonymous reviewer for helpful suggestions. This work was supported by a grant from the National Institutes of Health and a Giovanni Armenise–Harvard Foundation fellowship.
Abbreviations
- TFA
trifluoroacetic acid
- GST
glutathione S-transferase
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.031563798.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.031563798
References
- 1.Kuo M H, Allis C D. BioEssays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 2.Grunstein M. Nature (London) 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 3.Imai S, Armstrong C M, Kaeberlein M, Guarente L. Nature (London) 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 4.Landry J, Sutton A, Tafrov S T, Heller R C, Stebbins J, Pillus L, Sternglanz R. Proc Natl Acad Sci USA. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. . (First Published May 16, 2000; 10.1073/pnas.110148297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith J S, Brachmann C B, Celic I, Kenna M A, Muhammad S, Starai V J, Avalos J L, Escalante-Semerena J C, Grubmeyer C, Wolberger C, et al. Proc Natl Acad Sci USA. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rine J, Herskowitz I. Genetics. 1987;116:9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klar A J S, Fogel S, MacLeod K. Genetics. 1979;93:37–50. doi: 10.1093/genetics/93.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aparicio O M, Billington B L, Gottschling D E. Cell. 1991;66:1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- 9.Gottlieb S, Esposito R E. Cell. 1989;56:771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- 10.Bryk M, Banerjee M, Murphy M, Knudsen K E, Garfinkel D J, Curcio M J. Genes Dev. 1997;11:255–269. doi: 10.1101/gad.11.2.255. [DOI] [PubMed] [Google Scholar]
- 11.Smith J S, Boeke J D. Genes Dev. 1997;11:241–254. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- 12.Hecht A, Stahl-Bolsinger S, Grunstein M. Nature (London) 1996;383:92–96. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- 13.Gotta M, Strahl-Bolsinger S, Renauld H, Laroche T, Kennedy B K, Grunstein M, Gasser S M. EMBO J. 1997;16:3243–3255. doi: 10.1093/emboj/16.11.3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaeberlein M, McVey M, Guarente L. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brachmann C B, Sherman J M, Devine S E, Cameron E E, Pillus L, Boeke J D. Genes Dev. 1995;9:2888–2902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- 16.Frye R A. Biochem Biophys Res Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 17.Frye R A. Biochem Biophys Res Commun. 1999;260:273–279. doi: 10.1006/bbrc.1999.0897. [DOI] [PubMed] [Google Scholar]
- 18.Tanny J C, Dowd G J, Huang J, Hilz H, Moazed D. Cell. 1999;99:735–745. doi: 10.1016/s0092-8674(00)81671-2. [DOI] [PubMed] [Google Scholar]
- 19.Finnin M S, Donigian J R, Cohen A, Richon V M, Rifkind R A, Marks P A, Breslow R, Pavletich N P. Nature (London) 1999;401:188–193. doi: 10.1038/43710. [DOI] [PubMed] [Google Scholar]
- 20.Bell C E, Eisenberg D. Biochemistry. 1996;35:1137–1149. doi: 10.1021/bi9520848. [DOI] [PubMed] [Google Scholar]
- 21.Han S, Craig J A, Putnam C D, Carozzi N B, Tainer J A. Nat Struct Biol. 1999;6:932–936. doi: 10.1038/13300. [DOI] [PubMed] [Google Scholar]
- 22.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 23.Smith E R, Eisen A, Gu W, Sattah M, Pannuti A, Zhou J, Cook R G, Lucchesi J C, Allis C D. Proc Natl Acad Sci USA. 1998;95:3561–3565. doi: 10.1073/pnas.95.7.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Culver G M, McCraith S M, Zillmann M, Kierzek R, Michaud N, LaReau R D, Turner D H, Phizicky E M. Science. 1993;261:206–208. doi: 10.1126/science.8392224. [DOI] [PubMed] [Google Scholar]
- 25.Spinelli S L, Kierzek R, Turner D H, Phizicky E M. J Biol Chem. 1999;274:2637–2644. doi: 10.1074/jbc.274.5.2637. [DOI] [PubMed] [Google Scholar]
- 26.Walsh C. Enzymatic Reaction Mechanisms. San Francisco: Freeman; 1979. [Google Scholar]
- 27.Olivera B M, Hall Z W, Anraku Y, Chien J R, Lehman I R. Cold Spring Harbor Symp Quant Biol. 1968;33:27–34. doi: 10.1101/sqb.1968.033.01.008. [DOI] [PubMed] [Google Scholar]
- 28.Rowen J W, Kornberg A. J Biol Chem. 1951;193:497–507. [PubMed] [Google Scholar]
- 29.Zatman L J, Kaplan N O, Colowick S P. J Biol Chem. 1953;200:197–212. [PubMed] [Google Scholar]
- 30.Alberty R A. J Biol Chem. 1968;243:1337–1343. [PubMed] [Google Scholar]