Abstract
Mutations in the clk-1 gene of the nematode Caenorhabditis elegans result in slowed development, sluggish adult behaviors, and an increased lifespan. CLK-1 is a mitochondrial polypeptide with sequence and functional conservation from human to yeast. Coq7p, the Saccharomyces cerevisiae homologue, is essential for ubiquinone (coenzyme Q or Q) synthesis and therefore respiration. However, based on assays of respiratory function, it has been reported that the primary defect in the C. elegans clk-1 mutants is not in Q biosynthesis. How do the clk-1 mutant worms have essentially normal rates of respiration, when biochemical studies in yeast suggest a Q deficiency? Nematodes are routinely fed Escherichia coli strains containing a rich supply of Q. To study the Q synthesized by C. elegans, we cultured worms on an E. coli mutant that lacks Q and found that clk-1 mutants display early developmental arrest from eggs, or sterility emerging from dauer stage. Provision of Q-replete E. coli rescues these defects. Lipid analysis showed that clk-1 worms lack the nematode Q9 isoform and instead contain a large amount of a metabolite that is slightly more polar than Q9. The clk-1 mutants also have increased levels of Q8, the E. coli isoform, and rhodoquinone-9. These results show that the clk-1 mutations result in Q auxotrophy evident only when Q is removed from the diet, and that the aging and developmental phenotypes previously described are consistent with altered Q levels and distribution.
The nematode Caenorhabditis elegans has been used as a model for genetic studies of longevity, and multiple lifespan extension mutants have been identified that affect various aspects of development (1). One of the genes identified to function in determination of lifespan was clk-1. The clk-1 mutants exhibit a pleiotropic phenotype, characterized by delayed embryonic and postembryonic development, slowed adult behaviors such as swimming, pharyngeal pumping, and defecation, and an extended lifespan (2, 3). The clk-1 mutants also have an increased resistance to stress induced by UV treatment (4). The C. elegans clk-1 gene was characterized and found to be conserved among eukaryotes, including humans and rodents, and was identified as a homologue of the Saccharomyces cerevisiae COQ7 gene (5).
The S. cerevisiae COQ7 gene is required for the biosynthesis of coenzyme Q (ubiquinone or Q) (6). Q is a prenylated benzoquinone lipid that is synthesized in mitochondria, where much of it remains to act in mitochondrial respiration (7), fatty acid β oxidation (8), and uridine synthesis (9, 10). However, Q is also transported to other intracellular membranes, such as the Golgi membranes and lysosomes, and to the plasma membrane, where it functions as an antioxidant (11, 12). As with other yeast coq mutants, the coq7 mutants lack Q, are respiration defective, and hence are incapable of growing on nonfermentable carbon sources (6, 13). The COQ7 homologue from rat (14), human (15), and C. elegans (5) each rescued the yeast coq7 mutant for growth on nonfermentable carbon sources, suggesting a conservation of function from yeast to humans.
Given the functional conservation of yeast, rat, human, and C. elegans CLK-1 (COQ7) polypeptides, it is crucial to know whether changes in the levels of Q may be responsible for the slowed development, behavior, and rate of aging in strains of C. elegans harboring mutant alleles of clk-1 (COQ7). Felkai et al. (16) studied mitochondrial activity by two indirect assays in clk-1 mutants bearing alleles e2519 (E148K), qm30 (590-bp deletion at amino acid 152), or qm51 (maintains intron 2 due to an alteration of codon 93, which leads to an early stop codon) (5). Worms harboring these clk-1 alleles were analyzed for respiratory function by mitochondrial uptake of the fluorescent dye G6-rhodamine in individual worms or by assays of succinate:cytochrome c reductase activity in isolated mitochondria. These assays provided information on the functional state of Q, and both indicated that Q levels in the clk-1 mutants were only slightly diminished compared with wild type. The addition of ubiquinone (Q) to mitochondria isolated from the clk-1 mutants produced only a marginal increase in succinate:cytochrome c reductase activity. Other studies showed that oxygen consumption rates were only slightly lower in the clk-1(e2519) mutant compared with wild type, whereas ATP levels were higher (17). It was concluded that the CLK-1 polypeptide was not essential for either respiration or Q biosynthesis in nematodes. Instead, a model was proposed that involves CLK-1 signaling the status of mitochondrial energy metabolism to the nucleus (18).
Although the indirect assays performed by Felkai et al. (16) indicated that the Q content of the clk-1 mutants was close to normal, the measurements were performed on worms fed the OP50 strain of Escherichia coli, which produces Q8. The phenomenon of dietary supplementation affecting Q levels has been seen in a number of other organisms including yeast (19), mice (20), rats (21, 22), and humans (23). The resultant increases in Q levels upon oral administration have definite phenotypic effects on the model animals. For instance, in rats, Q dietary supplementation markedly attenuated striatal lesions produced by administration of 3-nitropropionic acid, and Q also increased the lifespan of a transgenic mouse model of familial amyotrophic lateral sclerosis (21). Based on these types of studies, Q is now widely used in a variety of clinical therapies and as a nutritional supplement. For example, oral administration of Q is currently being investigated in clinical trials in Huntington's and Parkinson's disease patients (21). To avoid influences by bacterial enzymes or compounds that might confound interpretation of the results, the worms must be cultured on an E. coli mutant that lacks the component under study (24). For these studies on Q biosynthesis in C. elegans, the worms were cultured on Q-less E. coli. The findings reported here show that the clk-1 mutant strains of C. elegans have severe defects in growth and reproduction unless they are provided with a dietary source of Q.
Materials and Methods
Culture Conditions.
Methods for C. elegans were standard (25) except for the E. coli strains GD1 or GD1:pAHG used as an alternate food source. GD1 is a Q-less E. coli strain (ubiG∷Kan, zei∷Tn10dTet), and the vector pAHG contains the ubiG gene in the vector pAH01 and restores Q8 synthesis (26). N2 (Bristol strain) was used as the wild type. The clk-1 alleles used in this study were e2519, qm30, and qm51 (5). The daf-2 alleles used in this study were e1368, e1370, m577, m579, m596, and m41 (27).
Quantification of Q Levels in Bacteria and Worms.
Quantification of Q8 in E. coli strains was performed as described (28). Nematodes from liquid cultures were separated from bacteria and debris via sucrose floatation, and dauer larvae were isolated by using a 1% SDS treatment. The worms were dripped into liquid nitrogen and stored at −80°C until ready for use. The samples were resuspended in 10 ml of H2O and transferred to 50-ml preweighed glass tubes. The animals were collected by centrifugation and the total wet weight of the pellet was determined. Animals were lysed and lipids extracted as described (29), except that Q6 was used as an internal standard (2 μl of 1.688 μg/μl). The lipids were resuspended in 150 μl of 9:1 MeOH/EtOH. Q6, Q8, Q9, rhodoquinone-9 (RQ9), and Q10 were separated by reversed-phase HPLC with a C18 column (Alltech Econosphere 5-um, 4.6 × 250-mm column) and quantitated with an ESA Coulochem II electrochemical detector (E1 −500 mV, E2 +350 mV). An ESA precolumn electrode in an oxidizing mode (E +450 mV) was used to convert all hydroquinone forms into quinones. This was necessary because the signal for reduced menaquinone, a prenylated napthaquinone found in bacteria, was found to interfere with the Q9 signal. Because this procedure converts all quinones to the oxidized form, no special techniques were used to maintain reduced hydroquinones. The electrode potentials listed are slightly different from those previously reported (29); they have been adjusted to decrease sensitivity to oxidized menaquinone. All quinones were quantitated directly from the electrochemical detection (ECD) results using external standards of Q6, Q9, and RQ9 [extinction coefficients: E275MQ6, 14,900 M−1⋅cm−1; E275MQ9, 14,700 M−1⋅cm−1 (30); E283MRQ9, 10,262 M−1⋅cm−1 (31)]. Q6 and Q9 standards were purchased from Sigma.
Isolation and Identification of a RQ9 Standard.
To obtain a RQ9 standard, a known amount of frozen Ascaris suum tissue was homogenized in a Virtis 23 homogenizer with 3:2 hexane/2-propanol (18:1 ml solvent/g tissue wt) for 1 min (32). The homogenate was placed in 50-ml glass conical tubes and shaken at 200 rpm in the dark at 4°C overnight. The tubes were centrifuged (2,000 rpm, 10 min), and the organic layer was removed, placed into fresh tubes, and dried under N2. The pellet was re-extracted with 7:2 hexane/2-propanol (18:1 ml solvent/g tissue wt) using shaking at 4°C, 200 rpm in the dark for 1 h and centrifugation (2,000 rpm, 10 min). The organic phases were pooled and dried. The extract was resuspended in 5 ml of hexane, transferred to a 10-ml tube, and backwashed with dH2O, and the resulting pink solvent was dried under N2 and resuspended in a small amount of hexane. This extract was purified via the above HPLC/ECD system by collecting the pink fraction that eluted at approximately 18.3 min, corresponding to the only ECD- and UV-detectable peak. This compound was verified as RQ9 using mass spectrometry. The predicted molecular weight of the RQ9 product was calculated and compared with that determined by electrospray ionization mass spectrometry (ESI-MS, Perkin-Elmer Sciex, API III triple quadropole mass spectrometer fitted with an Ion Spray source). Negative ion spectra were produced by injection (10–20 μl/injection) of the samples diluted in methanol/ethanol/formic acid (90:10:0.1, vol/vol/%) into a stream of the same solvent (minus formic acid) entering the ion source (10 μl/min). Instrument conditions were 0.3 Da step size, 6.50 s/scan, m/z 50–1,000 scan range, and orifice at 70 V. The parent ion was detected at 780.8 (RQ9 protonated at the amino group), and the daughter ion showed a mass of 182.0 (quinone head group). These ions were comparable to an RQ10 standard (parent ion at 849.8, daughter ion at 182.0).
Results
To determine whether the clk-1 worms are relying on dietary Q for their survival, eggs were isolated from N2 and clk-1 (e2519, qm30, and qm51) gravid adults and were transferred to NGM plates containing bacterial lawns of either OP50, the standard Q-replete E. coli strain, or GD1, a Q-deficient (Q-less) strain of E. coli (26). Each of the nematode strains matured and reproduced on OP50, with the mutants showing a slight developmental lag as described (2). When fed Q-replete OP50 E. coli, the clk-1 mutants developing from eggs had smaller brood sizes than N2 (Table 1), as observed previously with clk-1 (e2519 and qm30) (2, 5). Whereas wild-type worms thrived on the Q-less GD1 E. coli strain, all of the clk-1 mutants arrested in the L2 larval stage and did not produce any progeny due to immaturity (Table 1). This growth arrest on the Q-less food source shows complete penetrance and appears to be a unique property of the clk-1 mutants and not a general property for long-lived C. elegans strains, as no arrest was seen with daf-2 (e1368, e1370, m41, m577, m579, or m596) (data not shown). When the clk-1 developmentally arrested mutant worms were transferred to either OP50 (data not shown) or to a GD1 strain in which Q production had been restored, GD1:pAHG (26), the mutant worms resumed development to adulthood and proceeded to lay viable eggs (Table 1). The worms developing on GD1:pAHG have an equivalent, if not greater, number of progeny as compared with their siblings on OP50. However, in comparing these GD1:pAHG grown clk-1 mutants with their N2 counterparts, a significant decrease in brood size is still evident, similar to the decrease seen on OP50. Thus, the brood size phenotype is affected by diet but does not exhibit complete rescue to wild-type levels, regardless of the food source provided. Taken together, the results indicate that clk-1 mutants require a dietary source of Q, suggesting that clk-1 mutations cause a defect in Q biosynthesis that results in a conditionally lethal phenotype.
Table 1.
Self-brood size (number of progeny per parent) of wild-type and mutant worms on different food sources
| C. elegans genotype | E. coli food source | Q | Progeny per parent on food source
|
|
|---|---|---|---|---|
| From egg | From dauer | |||
| N2 | OP50 | + | 210 ± 45 (26) | ND |
| GD1:pAHG | + | 242 ± 47 (21)* | 236 ± 45 (11) | |
| GD1 | − | 204 ± 40 (11) | 251 ± 40 (4) | |
| clk-1(e2519) | OP50 | + | 150 ± 38 (15)† | ND |
| GD1:pAHG | + | 179 ± 34 (11)† | 174 ± 35 (9)† | |
| GD1 | − | 0 (44)*† | 0 (10)† | |
| clk-1(qm30) | OP50 | + | 141 ± 45 (11)† | ND |
| GD1:pAHG | + | 163 ± 42 (12)† | 163 ± 35 (12)† | |
| GD1 | − | 0 (15)*† | 0 (11)† | |
| clk-1(qm51) | OP50 | + | 79 ± 23 (15)† | ND |
| GD1:pAHG | + | 133 ± 55 (17)*† | 181 ± 34 (14)† | |
| GD1 | − | 0 (52)*† | 0 (12)† | |
Numbers in parentheses indicate the sample sizes.
The sample is significantly different from its respective siblings on OP50; P values = 0.05 according to the 2-tailed Student's t test.
The sample is significantly different from the N2 sample on the same food source; P values = 0.01. ND, not done. The worms often flee from the GD1 food and migrate off the plate, perhaps in search of a more nutritious food source. Both types of Q-replete E. coli strains contain similar amounts of quinone; OP50 contains 117.9 ± 2.7 pmol of Q8 per mg wet wt, whereas GD1:pAHG produces 138.2 ± 3.4 pmol of Q8 per mg wet wt.
To bypass the L2 larval arrest, worms in an alternate third-stage larval form, dauer larvae, were isolated from media containing OP50. SDS-treated dauer larvae were transferred to plates with either Q-less or Q-replete bacteria. N2 wild-type dauer larvae fed GD-1 recovered and grew into fertile adults but showed a 1-day delay in egg laying. The clk-1 mutant dauer larvae fed Q-less GD1 all reach adulthood but failed to produce progeny (Table 1) and appeared quite sickly. As with animals isolated as eggs, the clk-1 mutant dauer larvae developed into fertile adults when grown on GD1:pAHG, the Q-replete strain. These brood sizes are quite similar to those of mutants isolated as eggs on GD1:pAHG (Table 1). The sterility phenotype was not observed when clk-1 mutants were fed OP50 throughout development until late in the L4 larval stage. Transfer of these late L4 larvae to the GD1 plates yielded adults that were capable of laying eggs (data not shown). These late larvae would have completed most of development, including the gonad, while being fed the Q-replete bacteria. Hence, there are two points in development where dietary Q is required by the clk-1 mutants: (i) growth beyond the L2 larval stage as mutants develop from eggs, and (ii) growth to reproductively competent adults as mutants develop from dauer larvae.
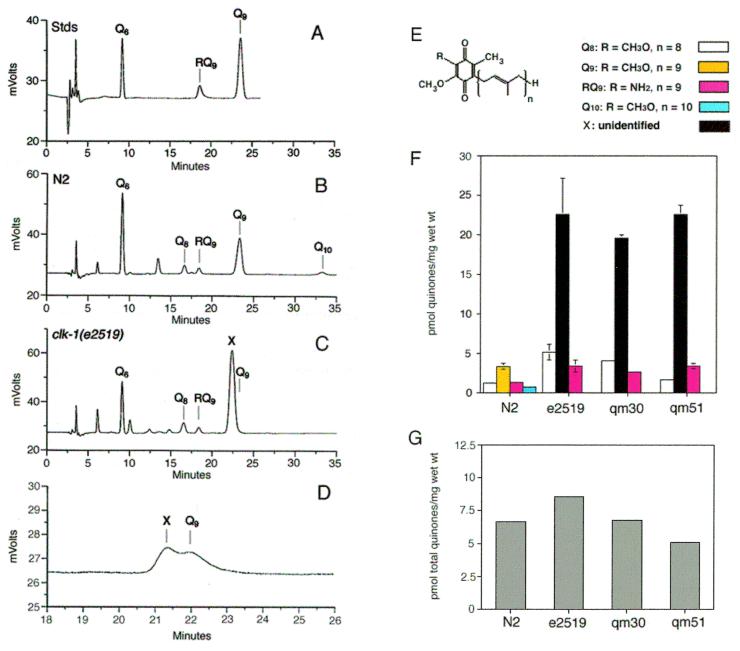
The phenotypes described above depend on the status of dietary Q and suggest the clk-1 mutants have a defect in Q biosynthesis. To quantify directly the amount of Q in the clk-1 mutant worms as compared with wild type, it was important to first obtain a distinct life stage of each genotype. However, growth rates of clk-1 mutants fed OP50 E. coli are known to be quite variable (2). To enable collection of tightly synchronized animals, N2 and clk-1 dauer larvae were isolated from large liquid cultures containing OP50. These standard culture conditions produce the phenotype that has previously defined the clk-1 locus. The dauer larvae were collected, and the total lipid extract was analyzed for Q via an HPLC/ECD system. This system was calibrated to detect nanogram amounts of the following quinones: Q6, used as an internal standard; Q9 and RQ9, the quinones previously identified in C. elegans (refs. 33 and 34 and Fig. 1A); Q8, the E. coli isoform; and Q10. The designation of the quinone compounds in Fig. 1 is based on coelution of redox-active compounds in the sample with quinone standards. Total lipid extracts from N2 (Fig. 1B) and clk-1 mutant dauer larvae (Fig. 1C) contained small amounts of RQ9 (1–3 pmol/mg wet wt) and small amounts of the Q8 E. coli isoform (1–5 pmol/mg wet wt). The predominant Q isoform in the N2 wild-type strain is Q9 (Fig. 1B). In the clk-1(e2519) mutant a peak coeluting with the Q9 standard is not detected; instead, there is a strikingly large quantity of a metabolite eluting 0.5 min earlier than Q9 (Fig. 1C). This metabolite was also observed in clk-1(qm30) and clk-1(qm51) dauer larvae (data not shown). The elution position of this metabolite (referred to as compound X) indicates it is slightly more polar than Q9. When compound X and Q9 fractions are collected, mixed together, and then coinjected onto the reverse-phase HPLC system, they elute separately, indicating that compound X and Q9 are distinct (Fig. 1D).
Figure 1.
Analysis of Q content in N2 and clk-1 dauer larvae. HPLC linked with an ECD was used to analyze Q levels in wild-type and clk-1 mutant worms. (A) Quinone standards: Q6, used as an internal standard; Q9 and RQ9, the quinones previously identified in C. elegans (31, 32). The elution positions of Q8 and Q10 standards are indicated in B and C. The amounts of Q and RQ are determined by analyzing the areas under the peaks and comparing them against standards run the same day. The quantities of Q8, the E. coli isoform, and Q10 were determined by comparing the areas under their peaks and that of the Q6 standard and calculating the molar amounts. Chromatograms are shown of lipid extracts of N2 (B) or clk-1(e2519) (C) nematodes cultured on E. coli OP50. All assignments are based on coelution with a known quinone standard. An unknown peak appears in the clk-1 mutants and is designated “X.” (D) Material corresponding to Q9 and compound X was collected from N2 and clk-1 mutant extracts, respectively, mixed together, and coinjected on the reverse-phase HPLC. (E) Line drawings are presented of Q and RQ. The quinone isoforms that were identified and quantified were Q8 (white bars), Q9 (yellow bars), RQ9 (magenta bars), and Q10 (cyan bars). The respective amounts of the unidentified compound X (black bars) were calculated as compared with the Q9 standard. (F) The quinone content was determined by analyzing the chromatograms in B and C, and similar chromatograms of the clk-1(qm30) and clk-1(qm51) mutant lipid extracts. Error bars represent standard deviation from three injections. Data shown are representative of three separate preparations of lipid extracts from different cultures. (G) Stacked bar graph depicting the sum total level of Q8, Q9, RQ9, and Q10 for N2 and the clk-1 mutants.
Levels of quinone isoforms were quantified from HPLC/ECD chromatograms of each of the three clk-1 mutants and compared with N2 (see Fig. 1F). A very small amount of redox-active material that coelutes with a Q10 standard was identified in the N2 worms (0.8 pmol/mg wet wt; Fig. 1F, cyan bars). Comparison of the peak area of compound X to the Q9 standard indicates that the clk-1 mutants accumulate approximately 5- to 7-fold more of compound X as compared with the amount of Q9 in N2 (Fig. 1F, black bars). However, this method provides only a rough approximation of the amount, as we have not yet determined the structure of compound X. Although compound X is clearly the dominant species in the clk-1 mutant extracts, there is significantly more RQ9 and Q8 in each of the clk-1 mutant dauer larvae as compared with N2 (Fig. 1 E and F, magenta and white bars, respectively). In fact, the amount of Q8 and RQ9 in each of the clk-1 mutant extracts roughly approximates the level of all of the quinones (Q8, Q9, RQ9, and Q10) present in N2 (Fig. 1G). It is possible that the increase in Q8 and RQ9 is compensatory and functions to partially offset the lack of Q9, allowing growth, albeit slow.
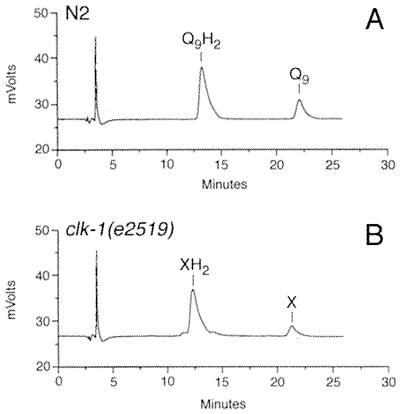
To determine whether compound X displays a quinone-like shift in elution position, the peak corresponding to compound X was collected and again analyzed by HPLC/ECD with the precolumn electrode in a reducing mode (instead of the oxidizing mode as shown in Fig. 1). The elution position of compound X shifts to about 12.5 min (Fig. 2B). Reduction of Q9 results in a similar shift in elution position to about 13 min (Fig. 2A). In summary, the data in Figs. 1 and 2 suggest that the clk-1 mutants are defective in Q biosynthesis and instead produce compound X, which is slightly more polar than Q9, and has quinone-like redox and chromatographic properties.
Figure 2.
Compound X and Q9 show similar chromatographic behavior under reducing conditions. HPLC/ECD was performed as described in Fig. 1 except that the potential of the precolumn electrode was set to a reducing mode (E −550 mV). Chromatograms are shown when the material corresponding to either the Q9 peak in an N2 lipid extract (A) or to compound X in a clk-1(e2519) extract (B) is collected and then injected onto the HPLC system with the precolumn electrode in a reducing mode.
Discussion
The data presented here demonstrate that the clk-1 mutants are defective in Q biosynthesis; the clk-1 mutant dauer larvae lack Q9 and accumulate an unidentified quinone-like metabolite (termed compound X) that is slightly more polar than Q9. Without a dietary source of Q, the C. elegans clk-1 mutants arrest in the L2 larval stage when developing from eggs or are sterile when developing from dauer larvae. Our results provide an explanation for the observation that clk-1 mutants are unable to grow on axenic media (17), as it is another type of Q-less growth medium. Provision of Q-replete E. coli rescues the growth on either axenic media (17) or on GD1 and also restores fertility in clk-1 mutants developing from dauer larvae (Table 1). The studies presented here show that the dietary Q8 contributed by E. coli effectively masks a profound Q auxotrophy that is evident when Q is removed from the diet.
Can the finding that the CLK-1 polypeptide is required for Q biosynthesis in C. elegans be reconciled with the recessive maternal effect inheritance pattern of the Clk-1 phenotype? The Clk-1 phenotype is observed only in clk-1 mutant progeny produced by homozygous mutant mothers (2). Because clk-1(qm51) is likely to be a null allele, the argument has been made that a complete loss of a critical biosynthetic pathway is not compatible with a maternal effect gene (5, 16). However, our experiments indicate that the pattern of maternal effect inheritance of the Clk-1 phenotype may result from an environmental interaction (dietary Q) with genotype and, in fact, is due in part to the product of a metabolic pathway (Q) being placed in the eggs. We postulate that heterozygous mothers can deposit Q9 into their eggs, including those eggs that contain homozygous clk-1(e2519)/clk-1(e2519) mutant individuals. Thus, eggs of heterozygous mothers would have reserves of Q9 in the mitochondria. Normal amounts of Q appear to be in excess of respiratory needs for animal cells in culture (35), so the maternal Q9 could be sufficient for early larval development. For the lifespan extension to show maternal effect, the Q9 should be stable, as appears to be the case (33). Homozygous mutant progeny also can take up exogenous Q8 once they hatch to increase their total Q levels and possibly induce RQ9 biosynthesis.
Can other aspects of the published Clk-1 phenotype be explained in terms of what is known about the CLK-1 polypeptide and its function in Q biosynthesis? Certainly the mitochondrial localization of the CLK-1 polypeptide in C. elegans (16) is consistent with the mitochondrial localization of Q biosynthesis in yeast (19, 36, 37). The ubiquitous expression of CLK-1 throughout the worm is also consistent with the finding that most tissues in rats and humans are competent for Q biosynthesis (38). RNAs corresponding to rat and human homologs of clk-1 (COQ7) and COQ3 are present in a wide variety of tissue types (15, 39, 40). Although the clk-1 mutations produced only a mild effect on respiration and oxygen consumption in C. elegans (16, 17), it seems likely that these effects will be much more dramatic when the animals are cultured on Q-less food. We speculate that both the Clk-1 pleiotropic phenotype (slowed development and adult behaviors) and the variability of timing among individuals may also be a reflection of partial rescue by dietary Q. In fact, the data in Fig. 1 show that the clk-1 dauer larvae accumulate increased amounts of Q8 relative to N2 wild type. This rescue is due to fairly complex phenomena, as dietary Q must be absorbed, distributed, and assimilated throughout the multicellular animal and delivered to the respiratory complexes in the inner mitochondrial membrane. Individual animals and tissues could easily differ in the successful execution of each of these steps and thereby differ in the extent to which Q function is repaired. The mechanisms of uptake and distribution of dietary Q are not understood, and in fact the intracellular distribution of dietary Q differs substantially from Q synthesized endogenously (41). Our studies suggest that C. elegans provides an important model system for the analysis of exogenous Q uptake and metabolism in higher eukaryotes.
Because exogenous Q does not afford complete repair of mitochondrial function and the mitochondria in older animals have a tendency to lose membrane potential (16), the maintenance of NAD+/NADH redox balance is probably a major challenge for the clk-1 mutants. The growth of respiratory-deficient cells in culture is limited by the oxidation of NADH; ρ° cells (lacking mitochondrial DNA) rely on glycolysis for ATP generation and depend on the presence of pyruvate in the growth media for reoxidation of excess NADH (42). The addition of either ferricyanide or exogenous Q to growth media can replace the requirement of ρ° cells for pyruvate (43–45). The exogenous Q is known to function in a trans-plasma membrane electron transport system, where intracellular NADH is oxidized and electrons are transferred across the plasma membrane to a wide variety of extracellular acceptors, including ferricyanide (46, 47). In fact, this trans-plasma membrane electron transport system is up-regulated in ρ° cells (43), and there is a concomitant increase in the level of Q at the plasma membrane (48). Respiratory-deficient yeast cells (atp2 and cor1 mutants) also have increased levels of Q in the plasma membrane (49). Based on these studies in yeast and mammalian cells, it seems likely that the rescue of the clk-1 mutant growth arrest by dietary Q may be acting, at least in part, at the level of the plasma membrane electron transport system as a means of restoring NAD+/NADH redox balance.
The accumulation of RQ9 in the clk-1 mutant dauer larvae would be expected to provide another pathway for the oxidation of NADH. C. elegans dauer larvae have relatively high levels of enzymes that carry out the first two committed steps of malate dismutation, phosphoenolpyruvate-carboxykinase (to form oxaloacetate from pyruvate), and cytoplasmic malate dehydrogenase (50). A highly conserved feature of malate dismutation is the use of fumarate as an electron sink instead of oxygen. In parasitic helminths such as A. suum, RQ9 functions in anaerobic respiratory metabolism as a low potential electron carrier between complex I (NADH-quinone oxidoreductase) and fumarate reductase (33, 51). Hence, the overall reduction of malate to succinate in malate dismutation oxidizes NADH to maintain redox balance.
We hypothesize that the altered state of mitochondrial Q, coupled with an increase of Q at the plasma membrane, could account for the clk-1 mutant phenotype, including slowed development and behaviors, increased stress resistance, and longer lifespan. According to this model increased stress resistance may result from the plasma membrane Q functioning either directly as an antioxidant or indirectly as a coantioxidant that reduces alpha-tocopheroxyl and ascorbyl radicals (49, 52, 53). The longer lifespan of the clk-1 Q-fed worms could result from this increase in stress resistance. Alternatively, the longer lifespan may result from lower levels of mitochondrial Q. It is intriguing that overexpression of wild-type CLK-1 (but not mutant CLK-1) shortens lifespan in C. elegans (16). Expression of wild-type COQ7 on a multicopy plasmid in yeast results in higher levels of Q6 (D. Fernandez-Ayala, C. Santos-Ocana and C.F.C., unpublished observations). It is tempting to speculate that the level of Q in the C. elegans CLK-1 overproducing strains also may be higher than wild type and induce the high rate of living attributed to these animals. Significantly, generation of reactive oxygen species by mitochondria, which have been implicated in the aging process, may depend more on the type of Q isoform present than on the total level of Q (54, 55). In summary, the potential interplay between mitochondrial and plasma membrane Q could have a critical role in energy production, growth control, cell defense, and longevity.
Acknowledgments
We thank all of the members of the Clarke lab for their support. We thank Drs. Albert Courey, Edith Gralla, Sabeeha Merchant, Joan Valentine, Paul Sternberg, and Charles West for their insightful comments and critiques. We especially thank Dr. Beth Marbois (University of California, Los Angeles) and the UCLA Mass Spectrometry Facility for their help in identifying RQ9. The clk-1 (e2519, qm30, and qm51) strains, along with N2, were graciously provided by Dr. S. Hekimi (McGill University, Quebec). The A. suum muscle was kindly provided by Dr. R. Komuniecki (University of Toledo, Ohio). This work was supported by National Institutes of Health Grant GM45952, National Institute on Aging Grant AG18085, and the Siegel Life Project/UCLA Center on Aging.
Abbreviations
- Q
coenzyme Q or ubiquinone
- RQ
rhodoquinone
- ECD
electrochemical detection
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.021337498.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.021337498
References
- 1.Vanfleteren J R, Braeckman B P. Neurobiol Aging. 1999;20:487–502. doi: 10.1016/s0197-4580(99)00087-1. [DOI] [PubMed] [Google Scholar]
- 2.Wong A, Boutis P, Hekimi S. Genetics. 1995;139:1247–1259. doi: 10.1093/genetics/139.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lakowski B, Hekimi S. Science. 1996;272:1010–1013. doi: 10.1126/science.272.5264.1010. [DOI] [PubMed] [Google Scholar]
- 4.Murakami S, Johnson T E. Genetics. 1996;143:1207–1218. doi: 10.1093/genetics/143.3.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ewbank J J, Barnes T M, Lakowski B, Lussier M, Bussey H, Hekimi S. Science. 1997;275:980–983. doi: 10.1126/science.275.5302.980. [DOI] [PubMed] [Google Scholar]
- 6.Marbois B N, Clarke C F. J Biol Chem. 1996;271:2995–3004. doi: 10.1074/jbc.271.6.2995. [DOI] [PubMed] [Google Scholar]
- 7.Trumpower B. J Bioenerg Biomembr. 1981;13:1–24. doi: 10.1007/BF00744743. [DOI] [PubMed] [Google Scholar]
- 8.Frerman F E. Biochem Soc Trans. 1988;16:416–418. doi: 10.1042/bst0160416. [DOI] [PubMed] [Google Scholar]
- 9.Nagy M, Lacroute F, Thomas D. Proc Natl Acad Sci USA. 1992;89:8966–8970. doi: 10.1073/pnas.89.19.8966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones M E. Annu Rev Biochem. 1980;49:253–279. doi: 10.1146/annurev.bi.49.070180.001345. [DOI] [PubMed] [Google Scholar]
- 11.Ernster L, Dallner G. Biochim Biophys Acta. 1995;1271:195–204. doi: 10.1016/0925-4439(95)00028-3. [DOI] [PubMed] [Google Scholar]
- 12.Villalba J M, Crane F L, Navas P. In: Plasma Membrane Redox Systems and Their Role in Biological Stress and Disease. Asard H, Berczi A, Caubergs R J, editors. Dordrecht, The Netherlands: Kluwer; 1998. pp. 247–265. [Google Scholar]
- 13.Tzagoloff A, Dieckmann C L. Microbiol Rev. 1990;54:211–225. doi: 10.1128/mr.54.3.211-225.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jonassen T, Marbois B N, Kim L, Chin A, Xia Y-R, Lusis A J, Clarke C F. Arch Biochem Biophys. 1996;330:285–289. doi: 10.1006/abbi.1996.0255. [DOI] [PubMed] [Google Scholar]
- 15.Vajo Z, King L M, Jonassen T, Wilkin D J, Ho N, Munnich A, Clarke C F, Francomano C A. Mamm Genome. 1999;10:1000–1004. doi: 10.1007/s003359901147. [DOI] [PubMed] [Google Scholar]
- 16.Felkai S, Ewbank J J, Lemieux J, Labbe J C, Brown G G, Hekimi S. EMBO J. 1999;18:1783–1792. doi: 10.1093/emboj/18.7.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Braeckman B P, Houthoofd K, DeVreese A, Vanfletern J R. Curr Biol. 1999;9:493–496. doi: 10.1016/s0960-9822(99)80216-4. [DOI] [PubMed] [Google Scholar]
- 18.Branicky R, Benard C, Hekimi S. BioEssays. 2000;22:48–56. doi: 10.1002/(SICI)1521-1878(200001)22:1<48::AID-BIES9>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 19.Jonassen T, Proft M, Randez-Gil F, Schultz J R, Marbois B N, Entian K D, Clarke C F. J Biol Chem. 1998;273:3351–3357. doi: 10.1074/jbc.273.6.3351. [DOI] [PubMed] [Google Scholar]
- 20.Lass A, Forster M J, Sohal R S. Free Radical Biol Med. 1999;26:1375–1382. doi: 10.1016/s0891-5849(98)00330-x. [DOI] [PubMed] [Google Scholar]
- 21.Matthews R T, Yang L, Browne S, Baik M, Beal M F. Proc Natl Acad Sci USA. 1998;95:8892–8897. doi: 10.1073/pnas.95.15.8892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lonnrot K, Holm P, Lagerstedt A, Huhtala H, Alho H. Biochem Mol Biol Int. 1998;44:727–737. doi: 10.1080/15216549800201772. [DOI] [PubMed] [Google Scholar]
- 23.Mohr D, Bowry V W, Stocker R. Biochim Biophys Acta. 1992;1126:247–254. doi: 10.1016/0005-2760(92)90237-p. [DOI] [PubMed] [Google Scholar]
- 24.Larsen P L. Proc Natl Acad Sci USA. 1993;90:8905–8909. doi: 10.1073/pnas.90.19.8905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sulston J E, Hodgkin J. In: Methods in Nematode Caenorhadbitis elegans. Wood W B, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. pp. 587–606. [Google Scholar]
- 26.Hsu A, Poon W W, Shepherd J A, Myles D C, Clarke C F. Biochemistry. 1996;35:9797–9806. doi: 10.1021/bi9602932. [DOI] [PubMed] [Google Scholar]
- 27.Gems D, Sutton A J, Sundermeyer M L, Albert P S, King K V, Edgley M L, Larsen P L, Riddle D L. Genetics. 1998;150:129–155. doi: 10.1093/genetics/150.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poon W W, Davis D E, Ha H T, Jonassen T, Rather P N, Clarke C F. J Bacteriol. 2000;182:5139–5146. doi: 10.1128/jb.182.18.5139-5146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jonassen T, Clarke C F. J Biol Chem. 2000;275:12381–12387. doi: 10.1074/jbc.275.17.12381. [DOI] [PubMed] [Google Scholar]
- 30.Hatefi Y. Adv Enzymol. 1963;25:275–328. doi: 10.1002/9780470122709.ch5. [DOI] [PubMed] [Google Scholar]
- 31.Moore H W, Folkers K. J Am Chem Soc. 1966;88:567–570. doi: 10.1021/ja00955a031. [DOI] [PubMed] [Google Scholar]
- 32.Radin N S. Methods Enzymol. 1981;72:5–7. doi: 10.1016/s0076-6879(81)72003-2. [DOI] [PubMed] [Google Scholar]
- 33.Van Hellemond J J, Luijten M, Flesch F M, Gaasenbeek C P H, Teilens A G M. Mol Biochem Parasitol. 1996;82:217–226. doi: 10.1016/0166-6851(96)02738-7. [DOI] [PubMed] [Google Scholar]
- 34.Takamiya S, Matsui T, Taka H, Murayama K, Matsuda M, Aoki T. Arch Biochem Biophys. 1999;371:284–290. doi: 10.1006/abbi.1999.1465. [DOI] [PubMed] [Google Scholar]
- 35.Maltese W A, Aprille J R. J Biol Chem. 1985;260:11524–11529. [PubMed] [Google Scholar]
- 36.Poon W W, Barkovich R J, Hsu A Y, Frankel A, Lee P T, Shepherd J N, Myles D C, Clarke C F. J Biol Chem. 1999;274:21665–21672. doi: 10.1074/jbc.274.31.21665. [DOI] [PubMed] [Google Scholar]
- 37.Jonassen T, Clarke C F. In: Coenzyme Q: From Molecular Mechanisms to Nutrition and Health. Kagan V E, Quinn P J, editors. Boca Raton, FL: CRC; 2000. pp. 185–208. [Google Scholar]
- 38.Elmberger P G, Kalen A, Appelkvist E L, Dallner G. Eur J Biochem. 1987;168:1–11. doi: 10.1111/j.1432-1033.1987.tb13379.x. [DOI] [PubMed] [Google Scholar]
- 39.Marbois B N, Xia Y-R, Lusis A J, Clarke C F. Arch Biochem Biophys. 1994;313:83–88. doi: 10.1006/abbi.1994.1362. [DOI] [PubMed] [Google Scholar]
- 40.Asaumi S, Kuroyanagi H, Seki N, Shirasawa T. Genomics. 1999;58:293–301. doi: 10.1006/geno.1999.5838. [DOI] [PubMed] [Google Scholar]
- 41.Turunen M, Appelkvist E L, Sindelar P, Dallner G. J Nutr. 1999;129:2113–2118. doi: 10.1093/jn/129.12.2113. [DOI] [PubMed] [Google Scholar]
- 42.King M P, Attardi G. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- 43.Larm J A, Vaillant F, Linnane A W, Lawen A. J Biol Chem. 1994;269:30097–30100. [PubMed] [Google Scholar]
- 44.Lawen A, Martinus R D, McMullen G L, Nagley P, Vaillant F, Wolvetang E J, Linnane A W. Mol Aspects Med. 1994;15:S13–S27. doi: 10.1016/0098-2997(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 45.Martinus R D, Linnane A W, Nagley P. Biochem Mol Biol Int. 1993;31:997–1005. [PubMed] [Google Scholar]
- 46.Sun I L, Sun E E, Crane F L, Morre D J, Lindgren A, Low H. Proc Natl Acad Sci USA. 1992;89:11126–11130. doi: 10.1073/pnas.89.23.11126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Villalba J M, Navarro F, Cordoba F, Serrano A, Arroyo A, Crane F L, Navas P. Proc Natl Acad Sci USA. 1995;92:4887–4891. doi: 10.1073/pnas.92.11.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gomez-Diaz C, Villalba J M, Perez-Vicente R, Crane F L, Navas P. Biochem Biophys Res Commun. 1997;234:79–81. doi: 10.1006/bbrc.1997.6582. [DOI] [PubMed] [Google Scholar]
- 49.Santos-Ocana C, Cordoba F, Crane F L, Clarke C F, Navas P. J Biol Chem. 1998;273:8099–8105. doi: 10.1074/jbc.273.14.8099. [DOI] [PubMed] [Google Scholar]
- 50.O'Riordan V B, Burnell A M. Comp Biochem Physiol. 1989;92:233–238. [Google Scholar]
- 51.Van Hellemond J J, Klockiewicz M, Gaasenbeek C P H, Roos M H, Tielens A G M. J Biol Chem. 1995;270:31065–31070. doi: 10.1074/jbc.270.52.31065. [DOI] [PubMed] [Google Scholar]
- 52.Santos-Ocana C, Villalba J M, Cordoba F, Padilla S, Crane F L, Clarke C F, Navas P. J Bioenerg Biomembr. 1998;30:465–475. doi: 10.1023/a:1020542230308. [DOI] [PubMed] [Google Scholar]
- 53.Kagan V E, Nohl H, Quinn P J. In: Handbook of Antioxidants. Cadenas E, Packer L, editors. New York: Dekker; 1996. pp. 157–201. [Google Scholar]
- 54.Lass A, Agarwal S, Sohal R S. J Biol Chem. 1997;272:19199–19204. doi: 10.1074/jbc.272.31.19199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lass A, Sohal R S. FASEB J. 2000;14:87–94. doi: 10.1096/fasebj.14.1.87. [DOI] [PubMed] [Google Scholar]