Abstract
We applied two experiments useful in the study of ligand-regulated DNA binding proteins to AraC, the dimeric regulator of the Escherichia coli l-arabinose operon. In the absence of arabinose, AraC prefers to loop DNA by binding to two half-sites that are separated by 210 base pairs, and in the presence of arabinose it prefers to bind to adjacently located half-sites. The basis for this ligand-regulated shift in binding appears to result from a shift in the rigidity of the system, where rigidity both in AraC protein in the absence of arabinose, and in the DNA are required to generate the free energy differences that produce the binding preferences. Eliminating the dimerization domains and connecting the two DNA binding domains of AraC by a flexible peptide linker should provide a protein whose behavior mimics that of AraC when there is no interaction between its dimerization and DNA binding domains. The resulting protein bound to adjacent half-sites on the DNA, like AraC protein in the presence of arabinose. When the two double-stranded DNA half-sites were connected by 24 bases of single-stranded, flexible DNA, wild-type AraC protein bound to the DNA in the presence and absence of arabinose with equal affinity, showing that AraC modulates its DNA binding affinity in response to arabinose by shifting the relative positions of its DNA binding domains. These results are consistent with the light switch mechanism for the action of AraC, refine the model, and extend the range of experimental tests to which it has been subjected.
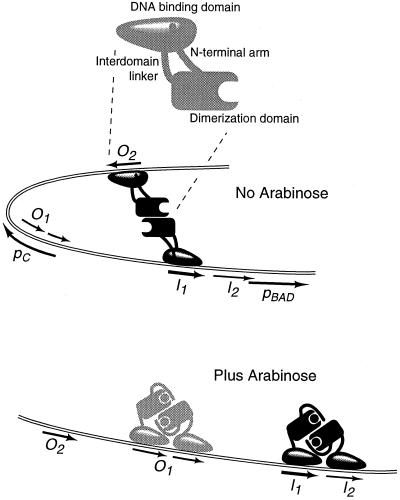
Data obtained from both in vivo and in vitro experiments have shown that, in the absence of arabinose, the dimeric AraC protein prefers binding to the well separated I1 and O2 half-sites and forming a DNA loop (1–4), Fig. 1. On the addition of arabinose, the protein's affinity for the I1 and I2 half-sites increases by about 50-fold, leading the protein to prefer to bind to these adjacent half-sites and induce the pBAD promoter rather than loop and repress pBAD (3, 5–8). The basis for the change in the DNA binding properties appears to result from an arabinose-induced shift of the N-terminal arms from associating with the DNA-binding domains of AraC to associating with the dimerization domains (refs. 4, 9, and 10; Fig. 1). The process involving this shift is called the light switch mechanism because the arms switch expression of the system on and off.
Figure 1.
The regulatory region of the araBAD operon showing the binding sites for AraC, the two promoters (pBAD and pC), and the light switch mechanism for AraC action. AraC bound to the O1 site in the presence of arabinose is shown in gray because occupancy of the site is only partial.
In the absence of arabinose, the arms from the dimerization domains are thought to bind to the DNA-binding domains and rather rigidly hold them apart from one another and in an orientation that favors DNA looping. Whether the arms also affect the DNA binding properties of the individual DNA binding domains has not been addressed before this work. A rigid connection between the domains mediated by the arms is consistent with the fact that reversing the head-to-tail orientation of the asymmetric O2 half-site while retaining its AraC-contacting area on the same face of the DNA eliminates DNA looping (4). Another property that can be attributed to rigidity can be seen in the binding of AraC in the absence of arabinose to two adjacent I1 and I2 half-sites. Whereas such binding normally does not occur in vivo, it can be observed in vitro (3, 6). According to the current view, to bind to adjacent half-sites, part of the DNA binding energy must be used to distort AraC so that the DNA binding domains are correctly positioned adjacent to one another. In the presence of arabinose, however, the DNA-binding domains of AraC are freed from the arms and can easily adopt an orientation compatible with the adjacent DNA half-sites. Thus, the presence of arabinose increases the affinity of AraC for adjacent half-sites, whether they are in direct repeat or inverted repeat orientation (6, 11).
One important input to the formulation of the light switch mechanism was the observation that deleting the N-terminal arms of AraC shifts the protein's preference in the absence of arabinose to that of preferring to bind adjacent half-sites rather than looping (9). Another important input was genetic information indicating an interaction between the N-terminal arm of AraC and the DNA binding domain of AraC (9). Whereas the light switch mechanism seems to be the simplest model compatible with the existing genetic and physiological data, and is consistent with the structure of the dimerization domain in the presence and absence of arabinose (12), the model has not been subjected to extensive biophysical testing. Here, we describe experiments designed to explore this latter area, in particular, the role of rigidity and flexibility of AraC and the DNA in the behavior of AraC in response to arabinose. The experiments also explore in a general way the signals that are sent from the dimerization domains to the DNA binding domains and whether the intrinsic affinity of the DNA binding domains for DNA is modulated. The experimental approaches we used should be applicable to the study of many ligand-regulated DNA binding proteins. In the case of AraC, the results rule out entire classes of general models, but are entirely consistent with the light switch mechanism, and in fact, refine the mechanism.
Materials and Methods
Oligonucleotide Primers.
Oligonucleotide primers, synthesized at the 200-nM scale and purified by PAGE, were purchased from Integrated DNA Technologies (Coralville, IA) or were synthesized on an Applied Biosystems 381A DNA synthesizer and purified by FPLC (13).
Plasmids.
Genes coding for proteins consisting of two AraC DNA binding domains, amino acids 169–292 and amino acids 177–292, connected by flexible linkers of 13 amino acids (-ESLHPPMDEFRGS-) or 19 amino acids (-ESLHPPMDEFVTQDMINGS-), were constructed de novo from ≈60-base synthetic oligonucleotides containing 15-base overhangs and cloned into the plasmid pSE380 (Invitrogen). We used a two-step PCR protocol based on that used to build a synthetic 303-nt-long HIV-2 Rev protein (14). In the design of these oligonucleotides, care was taken to eliminate secondary structure formation, and only oligonucleotides greater than 40% G-C content were considered. Potential ends of oligonucleotides were shifted along the sequence of the gene, and codons in the second AraC DNA binding domain were changed to avoid formation of primer-dimers or regions of internal base pairing of greater than 5 consecutive base pairs, or greater than 3 base pairs at the 3′ ends. A 100-μl mixture containing 100 ng of each oligonucleotide was subjected to seven PCR cycles of 94°C for 1 min, 55°C for 1.5 min, and 72°C for 2 min. Then 100 ng each of two end primers was added to the reaction, and 25 more cycles of PCR with an annealing temperature of 55°C were performed. The final PCR product was purified, digested with NcoI and XbaI, and ligated into the NcoI and XbaI sites of pSE380. The Quikchange procedure (Stratagene) was used to correct the two or three nonsilent point mutations introduced by the PCR steps.
Strains.
The strains used for assay of promoter activity were as follows: RE1 (ΔaraC–leu1022, araB+A+D+, Δlac74, galK−, strr, recA938∷cat; ref. 15); RE5 (ΔaraC–leu1022, Δlac74, galK−, strr, thi1, [λaraI1I2-pBAD-lacZ, ΔO2]; ref. 16); BS1 (F−, Δara–leu1022, [Δlac 74], galK, thi1, strr, [λaraI1I1-lacZ]; ref. 9); and SH321(F−, ΔaraC–leu1022, Δlac74, galK−, strr; ref. 17).
Enzyme Assays.
Cells were grown in liquid minimal salts media with 0.4% glycerol and 0.4% casamino acids to an apparent optical density at A550 nm of between 0.3 and 0.6 in the presence and in the absence of 0.2% arabinose, as noted, and assayed for arabinose isomerase (18), or β-galactosidase (19), as described. The activity of the wild type I1I2-ara pBAD promoter, was measured by using a single-copy chromosomal insertion of I1I2-pBAD fused to lacZ, and β-galactosidase was measured. Repression via looping was measured by using pL, a plasmid containing a modified ara pC fused to lacZ (9). In the absence of DNA looping, pC is active, and β-galactosidase is produced. If AraC protein is present and capable of forming a DNA loop, pC, and therefore β-galactosidase production, is repressed.
Flexible DNA.
A 75-base oligonucleotide with the sequence (with the two I1 half-sites underlined) ACCCTAGCATTTTTATCCATAAGACCTACTGGTACCGTCTCATGCATAGACCCTAGCATTTTTATCCATAAGACC was synthesized and 32P end labeled. A 25-base oligonucleotide with the reversed complimentary I1 sequence (with I1 underlined) GTCTTATGGATAAAAATGCTAGGGT was mixed with the 75-base oligonucleotide in a molar ratio of 20:1 in 10 mM Tris⋅HCl, pH 8.0, 1 mM EDTA, 50 mM KCl, and 5 mM MgCl2. The mixture was held at 95°C for 3 min and gradually cooled to room temperature. This procedure yielded a DNA structure with two double-stranded I1 half-sites connected by 24 bases of single-stranded DNA.
DNA Binding Assay.
The DNA migration retardation assay was performed as described (6). All binding reactions used buffer containing 10 mM Tris⋅HCl, pH 7.4, 1 mM EDTA, 50 or 100 mM KCl as indicated, 1 mM dithioerythritol, 0.05% Nonidet P-40, and 5% glycerol. Arabinose, if present, was at 50 mM. To 200 μl buffer, DNA fragments were added to concentrations of 1–10 nM, and the samples were incubated at 37°C for 10 min. AraC protein was diluted with binding buffer and added to final concentrations of 1–100 nM. Nonspecific competitor calf thymus DNA, 75 ng/ml, and 100× molar excess of nonradioactive specific competitor containing four consecutive I1 half-sites in the same orientation and on the same face of the DNA (I1-I1-I1-I1) (11) were added 20 min later, and 20-μl samples were withdrawn and loaded on the gel at the times indicated.
Results
Flexible Protein.
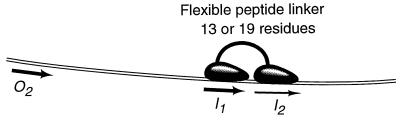
The two DNA binding domains in wild-type AraC protein are connected to the dimerization domains. We eliminated any constraints or interactions between the dimerization domains and the DNA binding domains in AraC by directly connecting two DNA binding domains with a flexible linker. We made two constructs, by using linkers of 13 and 19 aa consisting in part of the natural interdomain linker in AraC and in part the linker region of the yeast mating type repressor protein α2 (20). This linker provides the yeast α2 repressor protein with sufficient flexibility to bind DNA half-sites with a variety of spacings and orientations (21), and is likely to provide an unstructured connection between the AraC DNA binding domains (Fig. 2).
Figure 2.
Two DNA binding domains of AraC connected by a flexible linker.
The presence of long repeated sequences coding for the two DNA binding domains could generate problems with in vitro construction, in DNA sequencing, and in vivo because of intramolecular recombination. Therefore, we used the redundancy of the genetic code to design a new DNA sequence coding for one of the domains that retained the wild-type amino acid sequence. In doing this, we chose frequently used codons when their introduction did not generate the possibility of internal base pairing or stable secondary structures in the DNA. DNA coding for the double DNA binding domain proteins was assembled from oligonucleotides by PCR as described in Materials and Methods and inserted into a plasmid for expression of the proteins.
Both of the double DNA binding domain proteins induced the ara pBAD promoter in the absence of arabinose about as well as wild-type AraC protein induces the promoter in the presence of arabinose (Table 1). The fact that they induced so well indicates that neither looped between araI1 and araO2 to any significant degree. The induction we observe of pBAD requires two DNA binding half-sites and protein with two DNA binding domains, for we observe negligible induction by single DNA binding domain AraC using the araI2 half-site (R.S., unpublished data, and data not shown).
Table 1.
Activities of AraC and double DNA binding domain derivatives
| Protein | Promoter activity, %
|
|
|---|---|---|
| I1I2pBAD | O1−pC-lacZ | |
| No AraC | 2 | 150 |
| WT AraC − arabinose | 3 | 40 |
| WT AraC + arabinose | 100 | 100 |
| 13-amino acid linker | 110 | 140 |
| 19-amino acid linker | 180 | 120 |
Activation abilities of wild-type AraC and double AraC DNA binding domain proteins with linker lengths of 13 and 19 amino acids. Activity at I1I2-pBAD was measured with arabinose isomerase, and activity at O1−pC-lacZ was measured with β-galactosidase. WT, wild-type.
As a further check on possible repression by DNA looping, we also measured the activity of the pC promoter in a plasmid carrying the construct O1−pC-lacZ (Table 1). In this construct, because the O1 binding site for AraC has been inactivated, the pC promoter is repressed only by DNA looping by AraC between the I1 and O2 half-sites. AraC in the absence of arabinose decreases activity from this promoter almost 4-fold (9). In the presence of arabinose, AraC also represses β-galactosidase synthesis in the O1−pC-lacZ construct, but less well. These results are in accord with assay of DNA looping by dimethyl sulfate footprinting (2) as well as with modeling of the regulatory behaviors of pC and pBAD in wild type and in various mutant situations based on thermodynamic measurements (4). As seen in Table 1, neither of the double DNA binding domain proteins shows any significant repression of pC that would indicate the presence of any significant amount of DNA looping. Hence, we can conclude that the double DNA binding proteins prefer to bind to adjacent half-sites.
Flexible DNA.
Are the differences in the DNA binding properties of AraC that are observed in the presence and absence of arabinose a consequence solely of changes in the positioning of the DNA binding domains? If they are, providing a DNA substrate containing binding half-sites that are connected with a flexible linker should allow the half-sites to adjust their positions to accommodate the DNA binding domains' relative positions in the presence and absence of arabinose. As a result, DNA with flexibly connected half-sites should bind to AraC with an affinity nearly independent of arabinose, whereas DNA whose half-sites are connected with normal double-stranded DNA, which is relatively rigid, should display an arabinose dependence. An arabinose dependence will also be seen if the arms modulate the DNA binding affinity of the individual DNA binding domains.
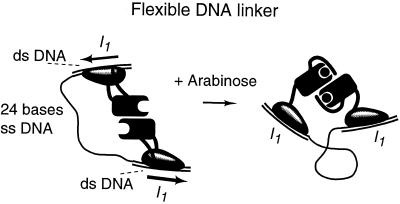
The required flexible DNA can be made by connecting two double-stranded half-sites with single-stranded DNA (Fig. 3). We constructed the DNA from a 75-base single-strand segment and two 25-base single-strand segments hybridized near the 5′ and 3′ ends of the 75-base segment. This yielded two 25-base pair double-stranded regions containing embedded I1 half-sites separated by 24 bases of single-stranded DNA.
Figure 3.
AraC protein in the absence and presence of arabinose binding to two I1 half-sites connected by 24 bases of single-stranded DNA.
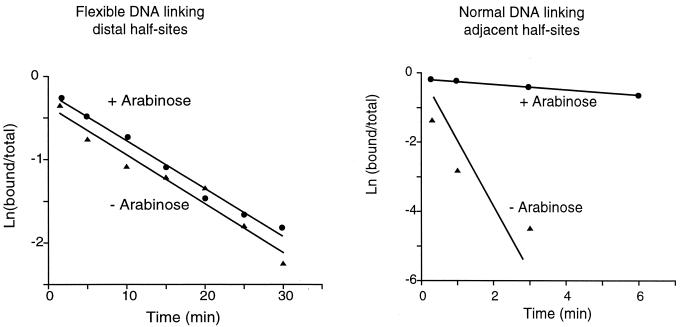
Wild-type AraC shows no significant arabinose dependence in its DNA binding affinity to the special DNA. This independence is shown by the fact that its dissociation rate, as indicated by the slopes of the lines in Fig. 4, from the DNA containing the flexible linker is virtually the same in the presence or absence of arabinose. By contrast, a typically large arabinose response was observed in the control experiment measuring dissociation rate of AraC from the double-stranded DNA containing adjacently located I1 and I2 half-sites.
Figure 4.
Dissociation kinetics of wild-type AraC in the presence and absence of arabinose. (Left) DNA contained two I1 half sites connected by 24 single-stranded nucleotides and buffer contained 100 mM KCl. (Right) DNA contained adjacent I1-I2 half-sites, and buffer contained 50 mM KCl.
Discussion
How do small molecule ligands modulate the DNA binding affinities of DNA binding proteins? One issue relevant to this question concerns the sending of signals as a result of ligand binding. The binding of ligand can result in a signal being sent to the DNA binding domains of such a protein, and a different signal being sent in the absence of ligand. Rather than two different signals being used however, it seems likely that the behavior of many proteins will be well approximated by a signal vs. no signal mode of operation. Thus, one important question in understanding ligand responsive DNA binding proteins is whether there is a “no signal” state, and whether it is with the ligand present or the ligand absent.
Another issue concerns what the signal ultimately does to modulate the DNA binding affinity of the protein. In the case of oligomeric proteins like AraC that contact DNA with more than one domain, two basic modulation mechanisms are possible. First, the affinity of individual domains for DNA can be modulated, and, second, the joint affinity or binding cooperativity between the two units can be modulated. A simple way to accomplish the latter is for the ligand to shift the separation of the protein's DNA binding domains. If they previously were positioned advantageously for simultaneous binding to the two DNA binding half-sites, a shift in their separation or orientation will allow only one at a time to bind the half-sites, and the overall binding affinity of the protein will be much reduced. Of course, utilization of the separation mechanism necessitates that both the DNA and the protein be sufficiently rigid that distorting the complex to allow two mispositioned or misoriented DNA binding domains to bind simultaneously costs a significant amount of free energy.
In our first experimental approach, we removed the dimerization domains from AraC and directly connected the two DNA binding domains with a peptide linker. This construct eliminates the possibility of sending signals from the dimerization domains because they are absent. Hence, the behavior of the resulting protein identifies whether the no signal state is closer to that of free AraC, or AraC bound to arabinose. We found that connecting the DNA binding domains with the linker yielded a protein that behaves like AraC in the presence of arabinose. We therefore conclude that a signal is sent from the dimerization domains of AraC to the DNA binding domains in the absence of arabinose and that, in the presence of arabinose, virtually no signal is sent.
In our second experimental approach, we examined the binding of AraC to a special DNA consisting of the two AraC double-stranded half-sites connected by 24 bases of single-stranded and, hence, highly flexible DNA. If AraC modulates its DNA binding affinity exclusively by changing the relative positioning of its two DNA binding domains in response to arabinose, then it will bind with nearly unchanging affinity to a DNA binding site consisting of two normal half-sites connected by the flexible single-stranded DNA. Such an affinity independence will be observed because the two double-stranded half-site portions of the binding site are free to shift relative positions along with the shifting positions of the protein's DNA binding domains, and there will be no significant energetic difference in binding in the presence or absence of arabinose. In contrast, if AraC modulates its DNA binding affinity by altering the intrinsic affinity of its individual DNA binding domains for DNA, its affinity for the flexible DNA will remain responsive to the presence of ligand. We found that the binding of AraC to the flexible DNA became insensitive to arabinose; hence, AraC modulates its DNA binding almost exclusively by changing the relative positioning of its DNA binding domains.
The first of our experiments shows that a signal is sent from the dimerization domains when arabinose is absent, and the second shows that the signal controls the positioning of the DNA binding domains and that arabinose does not modulate the affinities of the individual DNA binding domains for DNA. Hence, on the basis of these experiments, we can conclude that, in the absence of arabinose, an interaction between the dimerization domains of AraC and the DNA binding domains holds the protein in a state that it prefers to form a loop in the DNA, and that, in the presence of arabinose, the DNA binding domains are free to assume a variety of relative positions and orientations.
Genetic experiments have led to the proposal of an explicit mechanism for AraC response to arabinose (9, 10). It is called the light switch mechanism because the N-terminal arm of AraC is proposed to bind to the DNA binding domain of AraC when the ara system is off, and to bind to the dimerization domain when arabinose is present and the ara system is on. The results of the present experiments are completely compatible with the light switch mechanism and extend the range of experimental tests that have been applied to the mechanism. The present results also refine the light switch mechanism by limiting the role of the N-terminal arms to the positioning or restraining of the DNA binding domains. Before these experiments, the possibility had not been excluded that the binding of the N-terminal arms to the DNA binding domains also alters the domains' affinities for DNA, much like the peptide arm from the yeast mating factor a1 modulates the affinity of α2 for DNA (22).
Surprisingly, knowledge of the structure of the dimerization domain of AraC in the presence and absence of arabinose did not directly indicate the mechanism by which the protein responds to the presence of arabinose. In fact, the crystallographic data led to the proposal that AraC dimerizes by one interface in the absence of arabinose and by a different interface in the presence of arabinose (12). That model was cast in doubt by experiments described earlier where DNA looping is eliminated when the O2 half-site is reversed, and was supplanted by the light switch model on the basis of genetic data (9, 10). The data described in this paper also is not consistent with the alternative interfaces model.
The results we have presented here provide further experimental support and extend the light switch mechanism for AraC function. This mechanism is an example of an arm-domain interaction that generates a ligand response in a regulatory protein. Examples of other proteins that may employ regulated arm-domain interactions are yeast glycogen phosphorylase (23), phenylalanine hydroxylase (24), and the interaction between Ran and the Ran-binding protein (25). Many more examples are known of unregulated arm-domain interactions that join two domains or proteins (26). The rhodopsin-dynein interaction (27), DNA polymerase-Pcn1 interaction (28), and interactions with clathrin and cargo adapters (29), are examples that illustrate the wide diversity of arm-domain interactions. Arm-domain interactions, and regulated arm-domain interactions like those found in AraC may be widespread in nature and perhaps will be found when the structures and mechanisms of action of more multidomain proteins and multiprotein complexes are determined.
Acknowledgments
We thank members of the laboratory for their assistance, and Beverly Wendland and Doug Barrick for comments on the manuscript. This work was supported by National Institutes of Health Grant GM18277.
References
- 1.Dunn T, Hahn S, Ogden S, Schleif R. Proc Natl Acad Sci USA. 1984;81:5017–5020. doi: 10.1073/pnas.81.16.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martin K, Huo L, Schleif R. Proc Natl Acad Sci USA. 1986;83:3654–3658. doi: 10.1073/pnas.83.11.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lobell R B, Schleif R. Science. 1990;250:528–532. doi: 10.1126/science.2237403. [DOI] [PubMed] [Google Scholar]
- 4.Seabold R, Schleif R. J Mol Biol. 1998;278:529–538. doi: 10.1006/jmbi.1998.1713. [DOI] [PubMed] [Google Scholar]
- 5.Greenblatt J, Schleif R. Nat New Biol. 1971;233:166–170. doi: 10.1038/newbio233166a0. [DOI] [PubMed] [Google Scholar]
- 6.Hendrickson W, Schleif R. J Mol Biol. 1984;178:611–628. doi: 10.1016/0022-2836(84)90241-9. [DOI] [PubMed] [Google Scholar]
- 7.Hendrickson W, Schleif R. Proc Natl Acad Sci USA. 1985;82:3129–3133. doi: 10.1073/pnas.82.10.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee N, Francklyn C, Hamilton E. Proc Natl Acad Sci USA. 1987;84:8814–8818. doi: 10.1073/pnas.84.24.8814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saviola B, Seabold R, Schleif R. J Mol Biol. 1998;278:539–548. doi: 10.1006/jmbi.1998.1712. [DOI] [PubMed] [Google Scholar]
- 10.Reed W, Schleif R. J Mol Biol. 1999;294:417–425. doi: 10.1006/jmbi.1999.3224. [DOI] [PubMed] [Google Scholar]
- 11.Carra J H, Schleif R. EMBO J. 1993;12:35–44. doi: 10.1002/j.1460-2075.1993.tb05629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soisson S, MacDougall-Shackleton B, Schleif R, Wolberger C. Science. 1997;276:421–425. doi: 10.1126/science.276.5311.421. [DOI] [PubMed] [Google Scholar]
- 13.Cubellis M V, Marino G, Mayol L, Piccialli G, Sannia G. J Chromatogr. 1985;329:406–414. [Google Scholar]
- 14.Dillon P, Rosen C. Biotechniques. 1990;9:298–300. [PubMed] [Google Scholar]
- 15.Eustance R, Bustos S, Schleif R. J Mol Biol. 1994;242:330–338. doi: 10.1006/jmbi.1994.1584. [DOI] [PubMed] [Google Scholar]
- 16.Eustance R, Schleif R. Proteins. 1996;25:501–505. doi: 10.1002/prot.9. [DOI] [PubMed] [Google Scholar]
- 17.Hahn D, Dunn T, Schleif R. J Mol Biol. 1984;180:61–72. doi: 10.1016/0022-2836(84)90430-3. [DOI] [PubMed] [Google Scholar]
- 18.Schleif R, Wensink P. Practical Methods in Molecular Biology. New York: Springer; 1981. [Google Scholar]
- 19.Miller J. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 20.Astell C, Ahlstrom-Jonasson L, Smith M, Tatchell K, Nasmyth K, Hall B. Cell. 1981;27:15–23. doi: 10.1016/0092-8674(81)90356-1. [DOI] [PubMed] [Google Scholar]
- 21.Sauer R, Smith D, Johnson A. Genes Dev. 1988;2:807–816. doi: 10.1101/gad.2.7.807. [DOI] [PubMed] [Google Scholar]
- 22.Stark M, Escher D, Johnson A. EMBO J. 1999;18:1621–1629. doi: 10.1093/emboj/18.6.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin K, Hwang P, Fletterick R. J Biol Chem. 1995;270:26833–26839. doi: 10.1074/jbc.270.45.26833. [DOI] [PubMed] [Google Scholar]
- 24.Kobe B, Jennings I, House C, Michell B, Goodwill K, Santarsiero B, Stevens R, Cotton R, Kemp B. Nat Struct Biol. 1999;6:442–448. doi: 10.1038/8247. [DOI] [PubMed] [Google Scholar]
- 25.Vetter I, Nowak C, Nishimoto T, Kuhlmann J, Wittinghofer A. Nature (London) 1999;398:39–46. doi: 10.1038/17969. [DOI] [PubMed] [Google Scholar]
- 26.Schleif R. Proteins. 1999;34:1–3. doi: 10.1002/(sici)1097-0134(19990101)34:1<1::aid-prot1>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 27.Tai A, Chuang J, Bode C, Wolfrum U, Sung C. Cell. 1999;97:877–887. doi: 10.1016/s0092-8674(00)80800-4. [DOI] [PubMed] [Google Scholar]
- 28.Reynolds N, Warbrick E, Fantes P, MacNeill S. EMBO J. 2000;19:1108–1118. doi: 10.1093/emboj/19.5.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haar E, Harrison S, Kirchhausen T. Proc Natl Acad Sci USA. 2000;97:1096–1100. doi: 10.1073/pnas.97.3.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]