Abstract
MalT, the specific activator of the maltose regulon, is the prototype of a family of high-molecular-mass ATP-binding bacterial transcription activators. On binding of its two positive effectors, the inducer maltotriose and ATP, MalT oligomerizes to an active state competent for promoter binding and transcription activation. In addition to its previously known DNA-binding domain, limited proteolysis showed that MalT contains three other domains, the boundaries of which were accurately delimited by N-terminal microsequencing. The N-terminal domain alone binds ATP. Maltotriose binding involves an extended region corresponding to domains 2 and 3, although weak binding to domain 3 alone was also observed. Moreover, maltotriose binding induces a conformational shift involving a movement of both domains 1 and 3 with respect to domain 2, leading to the active form of the protein. Sequence examination of the MalT homologues suggests that these three domains might constitute a signaling module.
A family of bacterial transactivators that display unusual features, the MalT (or LAL) family (1, 2), was recently discovered. Its members differ from all other known activators by their high molecular mass (generally above 90 kDa). They all possess a putative ATP-binding site near their N terminus and share homology with LuxR near their C terminus. They activate operons involved in mainly catabolic functions in the related Gram-positive Streptomyces and Mycobacterium genera. However, some of them, including the best studied protein of the family, the MalT protein of Escherichia coli, are found in Gram-negative bacteria. MalT activates the maltose regulon to allow E. coli to use maltodextrins (starch degradation products) as a carbon and energy source (3, 4).
MalT binds to its target promoters only in the presence of two positive effectors: ATP, which MalT hydrolyzes, and maltotriose, the inducer of the maltose system (5). Maltotriose and ATP act by promoting the formation of a high-order MalT oligomer, a form that most likely favors its cooperative binding to the multiple sites in its target promoters, which in turn allows transcription activation (6). The regulation of MalT activity also involves three negative effector proteins: MalK, the ATPase of the maltodextrin ABC (ATP-binding cassette) transporter (7); MalY, a protein homologous to βC-S lyases (8); and Aes, which is homologous to acylesterases (9). MalK and MalY interact directly with MalT (10, 11), and MalY acts by competing against maltotriose for binding to MalT and preventing the formation of the productive oligomer (11). Hence, the formation of a high-order oligomer constitutes a checkpoint in MalT activity that is under the control of at least four positive or negative signals. This tight regulation is unprecedented among bacterial transactivators.
Little was known about the tertiary structure of the members of the MalT family. Their characteristic high molecular mass suggests that they are multidomain proteins, but only the C-terminal region of MalT that is homologous to LuxR has been isolated (12). This domain, which binds to MalT-binding sites and activates transcription (13), comprises only 95 amino acids, i.e., 10% of the protein. In view of the many signals and interactions involved in the regulation of MalT activity, the identification of the domains comprising MalT constitutes an important step toward understanding the action of each signal, as well as the way different signals are integrated within a unique protein.
In this work, I have determined the domain structure of MalT, purified the isolated domains, and studied their activities with respect to the binding of the positive effectors as well as the conformational changes caused by their binding. The implications of these findings for the MalT family are discussed.
Materials and Methods
Strains and Plasmids.
pop2319 is an MC1000 (14) derivative carrying ΔmalA157 (15), a deletion of the entire malT gene. BL21(DE3) and pLysS have been described (16). pOM151, pOM152, pOM155, pOM156, pOM160, and pOM162 were constructed as follows. Fragments of the malT gene were amplified by PCR, with the use of oligonucleotides DT1G (5′-CGGGGCCATGGGCCTGATTCCGTCAAAACTAA) and DT1D (5′-CGTAGAAGCTTAGGCGCCGCGTGCCGACTTATGG), DT2G (5′-CGGGGCCATGGGCCGCCTGGCGGGAATC) and DT2D (5′-CGTAGAAGCTTAGGCGCCTTCAGCACGGGCTAGC), or DT3G (5′-CGGGGCCATGGGCCATGAAATCAAGGACATCA) and DT3D (5′-CGTAGAAGCTTAGGCGCCATGATGTTGATTGATTTCTC), digested by NcoI and HindIII and inserted between the NcoI and HindIII sites of pARA14 (17), giving rise, respectively, to pOM150, pOM151, and pOM152. The insertion of a blunt-ended His tag encoding linker (sequence of the coding strand: 5′-CATCACCATCACCATCAC) in the EheI site of pOM150 gave pOM155. The insertion of a His tag encoding linker made of two oligonucleotides (HTAG1: 5′-CATGCATCATCATCACCATCA and HTAG2: 5′-CATGTGATGGTGATGATGATG) in the NcoI site of pOM151 gave pOM156. The NcoI–HindIII fragment of pOM155 containing part of the malT sequence was cloned in the corresponding sites of pET28b(+) (Novagen), giving pOM160. pOM162 was constructed by cloning a PCR-amplified fragment of malT (using oligonucleotides DT2G and DT3D), after digestion by NcoI and HindIII, in the corresponding sites of pET28b(+) followed by insertion of the HTAG1-HTAG2 linker in the NcoI site. All cloned PCR fragments were verified by sequencing. All constructs without His tag encoding linker encode a polypeptide with the sequence MG[DTX]GA, where [DTX] designates the sequence of domain DTX (X = 1, 2, 3, or 2–3). The constructs with a His tag encoding linker at the NcoI site or at the EheI site encode a polypeptide with the sequence MHHHHHHMG[DTX]GA or MG[DTX]GHHHHHHA, respectively.
Sequence Comparisons.
The parameters used for Blast (18) searches were database, nonredundant; matrix blosum62; gap weight 11; gap length weight 1; lambda ratio 0.85. clustal x (19) parameters were gap insertion penalty 12; gap extension penalty 2; matrix: blosum30.
Limited Proteolysis and N-Terminal Microsequencing.
Four micrograms of MalT or MalTc26 in 13 μl of a buffer containing 50 mM Tris⋅HCl (pH 7.7), 10% sucrose, 0.3 M KCl, 1 mM Mg acetate, 0.1 mM EDTA, and, when required, 1.2 mM maltotriose, 0.12 mM ATP or 0.58 mM ADP or 0.58 mM 5′-adenylylimidodiphosphate, were preincubated at 20°C for 20 min. Two microliters of proteinase K, trypsin, or subtilisin in the same buffer without maltotriose or adenine nucleotide was added, and the samples were incubated at 25°C for 15 or 30 min. Reactions were arrested by trichloroacetic acid precipitation (5 μl of 60% trichloroacetic acid). Samples were analyzed by SDS/PAGE and staining with Coomassie blue. Samples used for N-terminal microsequencing were prepared in the same way, but scaled up to produce sufficient amounts of the fragments. Microsequencing was carried out by the “Service de Microséquençage” at the Institut Pasteur.
Purification of the MalT Fragments.
DT1H: LB medium (500 ml) (20) containing 25 μg/ml kanamycin sulfate was inoculated with BL21(DE3) (pOM160) at OD 0.1, grown to OD 2 at 37°C, and incubated at 24°C for 15 min. Isopropyl β-d-thiogalactoside (1 mM) was then added, and growth was continued at 24°C for 4 h. Cells were collected, washed, and resuspended in buffer A [sodium phosphate (pH 7.0)/0.5 M KCl/10% sucrose]. MgCl2 (1 mM), 0.1 mM ATP, and 1 mM 4-(2-aminoethyl)benzenesulfonylfluoride (Interchim, Montluçon, France) were added. The cells were disrupted in a French press and centrifuged (30 min at 180,000 × g), and the protein was purified on a 5-ml nickel–nitrilotriacetic acid (Ni-NTA) column (Qiagen, Chatsworth, CA).
HDT2: A 1-liter culture of pop2319(pOM156) in LB medium + 100 μg/ml ticarcillin (SmithKline Beecham) was grown at 37°C, induced at OD 0.5 with 0.5 g/liter arabinose, harvested after 5 h, washed and resuspended in buffer B [50 mM Tris⋅HCl (pH 7.7)/10% sucrose/0.5 M KCl], disrupted in a French press after the addition of 1 mM 4-(2-aminoethyl)benzenesulfonylfluoride, and centrifuged (60 min at 30,000 × g). The protein was purified on a HiTrap chelating affinity column (Amersham Pharmacia) loaded with NiCl2.
DT3: pop2319(pOM152) inoculated at OD 0.15 in 100 ml 2YT medium (20) (100 μg/ml ticarcillin) was grown at 30°C to OD 0.5 and induced with 0.5 g/liter arabinose. Cells were further grown for 15 h, harvested, washed and resuspended in buffer B, disrupted with a French press, and centrifuged (60 min, 25,000 × g). The protein was purified on a 1-ml Ni-NTA column.
HDT2–3: BL21(DE3)(pLysS, pOM162) was inoculated in 1 liter of LB medium (25 μg/ml kanamycin sulfate/10 μg/ml chloramphenicol) at OD 0.1. Cells were grown to OD 1 at 37°C, induced with 1 mM isopropyl β-d-thiogalactoside, transferred at 18°C 1 h 15 min later, and grown for 4 h at 18°C. They were then harvested, washed, and resuspended in buffer B. After the addition of 10 mM 2-mercaptoethanol and 1 mM 4-(2-aminoethyl)benzenesulfonylfluoride, they were disrupted in a French press and centrifuged (30 min, 180,000 × g). The protein was purified on a 5-ml Ni-NTA agarose column followed by chromatography on a POROS HQ/M (Roche Molecular Biochemicals) column.
In all of the metal-chelate chromatography steps, the eluent was imidazole (either a 400 mM step or a 0–400 mM gradient, except for DT3, which was eluted with 20 mM imidazole).
Affinity Chromatography of ATP.
A soluble extract of BL21(DE3)(pOM160) prepared as described above from 20 ml of culture was loaded on a 100 μl Ni-NTA column equilibrated in buffer A + 1 mM MgCl2 and washed with 1 ml buffer B + 1 mM MgCl2 + 80 mM imidazole and 7 ml buffer C [50 mM Tris⋅HCl (pH 7.7)/10% sucrose/0.2 M KCl/1 mM MgCl2]. [γ-32P]ATP (100 μl at 2 mM in buffer C, ≈1 μCi) was added, and the column was spun, allowed to stand for 10 min at room temperature, and rapidly washed with 1 ml buffer C. After this step, 2 × 100 μl buffer C and then 5 × 100 μl of the same buffer containing 400 mM imidazole were passed stepwise through the column by adding the solution, spinning the column, and then letting it stand for 5 min each time. EDTA (25 mM) was added immediately to each fraction. The radioactivity in all fractions and in the starting ATP solution was counted in a Beckman scintillation counter after the addition of 5 ml Ready gel (Beckman). The protein concentration of the fractions was estimated by a Bradford assay performed on 2 μl of each fraction, with BSA as a standard. ATPase activity was measured for 1 μl of each fraction by TLC on polyethyleneimine-cellulose (Schleicher & Schuell), with a solvent containing 1 M formic acid and 0.5 M LiCl, and by counting the radioactivity in the spots in a PhosphorImager. For the control experiment, a soluble extract prepared from 25 ml of pop2319(pOM152) grown as described above was loaded on a column identical to that used for DT1H but equilibrated in buffer B + 1 mM MgCl2 and washed with 5 ml of buffer C. The DT3 column was then treated exactly like the DT1H column.
Fluorimetry.
The fluorescence measurements were performed on a Quantamaster spectrofluorometer (PTI, South Brunswick, NJ) at 20°C in 50 mM Tris⋅HCl (pH 7.7), 0.2 M KCl buffer at 0.83 μM DT3 or 0.96 μM carbonic anhydrase, or at 14°C in the same buffer supplemented with 1 mM MgCl2 and 0.1 mM 5′-adenylylimidodiphosphate at 1.3 μM DT1H. The excitation wavelength was 286 nm. Excitation and emission bandwidths were set to 2 and 5 nm, respectively (DT3), or 1 nm and 5 nm (controls). Maltotriose binding studies were carried out by adding increasing concentrations of maltotriose to 1 ml DT3 (0.83 μM in the same buffer), without exceeding a 5% dilution. Fluorescence emission of the protein excited at 286 nm was recorded at 320 nm for at least 200 s after each maltotriose addition. The average of the first 150 points obtained after equilibrium was reached was used for the plot.
Ammonium Sulfate Precipitation with Radiolabeled Maltotriose.
Thirty microliters of a 7.7 μM HDT2–3, DT3, or BSA solution in 40 mM Hepes-KOH (pH 8.0), 0.23 M KCl, 1 mM DTT containing 0.025 μCi (1 μM) [14C]maltotriose (American Radiolabeled Chemicals, St. Louis), and various concentrations of unlabeled maltotriose were incubated at 22°C for 20 min and then chilled on ice for 2 min. Ammonium sulfate (180 μl at 3.5 M, pH 8.0) was added, and the samples were centrifuged for 30 min, washed with 200 μl 3.5 M ammonium sulfate (pH 8.0), and centrifuged for 5 min. The pellet was redissolved in 500 μl H2O, the undissolved residue was dissolved in 500 μl 1% SDS, and both samples were counted as indicated before. The sum of these two measurements corrected for the background (amount of radioactivity present in the tubes with BSA) was used to calculate the amount of bound maltotriose. Part of the initial mix was also counted to calculate the amount of radioactivity initially present in the samples.
Results
Limited Proteolysis in the Presence of Different Combinations of Ligands.
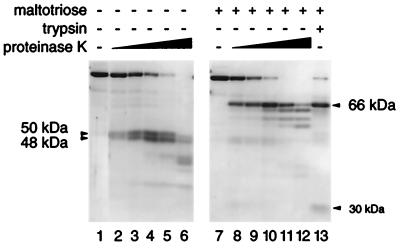
To investigate its domain structure, MalT-ATP was first proteolyzed with increasing concentrations of proteinase K in the presence and absence of the inducer maltotriose. In the presence of maltotriose, a 66-kDa proteolysis-resistant fragment appeared (Fig. 1). A 30-kDa band, which might contain the complement of this fragment, was present at low proteinase K concentrations and in the trypsin digest (see below and Fig. 1, lanes 8 and 13) but was degraded at higher protease concentrations. In the absence of maltotriose, two proteolysis-resistant fragments appeared, with estimated molecular masses of 50 kDa and 48 kDa (Fig. 1). Similar results were obtained with two other proteases, trypsin and subtilisin (Fig. 1, lane 13, and not shown). The results obtained with trypsin and subtilisin support the conclusion that the cleaved regions are well exposed to the solvent and that the resistant fragments correspond to folded regions of the protein. Proteolysis of the 48-kDa fragment sometimes went further (see below) to produce a 45-kDa fragment, probably because of the use of different MalT protein preparations (see Fig. 2B, lanes 2 and 4).
Figure 1.
Proteolysis of MalT by proteinase K or trypsin in the presence of ATP with or without maltotriose. Proteinase K/MalT weight ratios were 1:512, 1:256, 1:128, 1:64, 1:32 (lanes 2–6) or 1:256, 1:128, 1:64, 1:32, 1:16 (lanes 8–12), and the trypsin/MalT weight ratio was 1:50. Mixtures were digested for 30 min. Samples were analyzed by SDS/PAGE (12% acrylamide/bisacrylamide, 75:1). Molecular masses are estimated from the migration of the fragments.
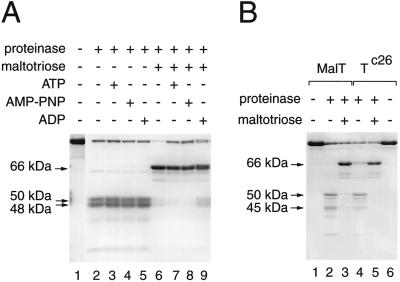
Figure 2.
Influence of ligands and the MalTc26 substitution on the proteolysis of MalT by proteinase K. Mixtures were digested for 15 min. (A) Role of the adenine nucleotide. Proteinase K/MalT ratio: 1:64 (samples with maltotriose) and 1:128 (samples without maltotriose). (B) Proteolysis of the MalTc26 variant by proteinase K. Proteinase K/MalTc26 ratio: 1:64 (samples with maltotriose) and 1:128 (samples without maltotriose).
ATP, ADP, and the nonhydrolyzable ATP analog 5′-adenylylimidodiphosphate exert differential effects on the activation (5) and oligomerization (6) properties of MalT. To study the effect of these nucleotides on the proteolysis pattern, MalT was digested by proteinase K in the presence of all combinations of the ligands maltotriose and ATP, ADP, or 5′-adenylylimidodiphosphate under the same conditions, except that the proteinase K concentration was halved for the samples without maltotriose. As shown in Fig. 2A, the nature or even the presence of the adenine nucleotide had very little influence on the proteolysis pattern. Only ADP seemed to counteract the effect of maltotriose slightly.
Because maltotriose is the only positive effector that markedly alters the proteolysis pattern of MalT, the biochemically well-characterized constitutive variant MalTc26, which is active in the absence of maltotriose (21), was subjected to limited proteolysis. Interestingly, its proteolysis pattern in the absence of maltotriose was a mixture of the patterns obtained for MalT in the absence and in the presence of maltotriose (Fig. 2B). In particular, the 66-kDa band present in the absence of maltotriose in the MalTc26 pattern was shown by N-terminal microsequencing (see below) to be identical to that obtained in the presence of maltotriose in the MalT pattern. This result strongly suggests that the patterns observed in the absence and in the presence of maltotriose are characteristic, respectively, of the inactive and active forms of MalT and that the malTc mutations displace the equilibrium toward the active state. The fact that only a relatively small proportion of the MalTc26 molecules spontaneously adopted the active conformation correlates with the observation that MalTc26 activity is enhanced by maltotriose (21). Indeed, the patterns obtained for MalT and MalTc26 in the presence of maltotriose were identical (Fig. 2B).
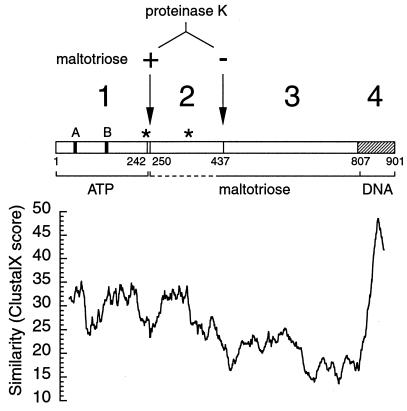
To delineate the resistant regions, the proteinase K fragments were N-terminally microsequenced (see Fig. 3). The sequence of the fragments that appear simultaneously in the absence of maltotriose indicated that they originate at amino acids 1 (48-kDa fragment) and 437 (50-kDa fragment). The predicted sizes of the 1–436 and 437–901 fragments of MalT (49 and 54 kDa, respectively) suggest that the observed fragments are generated by proteolysis of only a small segment (possibly a single peptide bond) immediately upstream from position 437. Microsequencing the 45-kDa fragment obtained (in addition to the 48 kDa) in the absence of maltotriose in certain MalT preparations (see above) showed that the two fragments have the same N terminus. This observation indicates that a small C-terminal region of this fragment is more sensitive to proteolysis than is the rest of the fragment. The 66-kDa band obtained in the presence of maltotriose contained two polypeptides originating at positions 248 and 250. These polypeptides could correspond either to the 248–901 and 250–901 fragments (75 kDa) that would migrate abnormally or to fragments derived from them by a secondary cleavage around position 800. Interestingly, the tryptic 66-kDa band contains two fragments originating at positions 241 and 242, which shows that the protease-sensitive region upstream from position 250 encompasses at least 9 amino acids. Finally, the 66-kDa proteinase K-resistant fragment obtained from MalTc26 in the absence of maltotriose was also shown to begin at positions 248 or 250, which confirms the interpretation of the MalTc26 proteolysis pattern.
Figure 3.
Schematic representation of the MalT protein. (Upper) Solid vertical bars stand for the A and B Walker motifs, stars indicate the location of the two biochemically characterized substitutions [MalTc22 (E359K) and MalTc26 (R242P)] rendering MalT constitutive. Arrows indicate the position of the first amino acid of the proteinase K-resistant fragments obtained in the presence (+) or the absence (−) of maltotriose. The different domains (numbered above) and the 1–2 linker are delineated (small numbers refer to the amino acid sequence of MalT). (Lower) The similarity inside the MalT subfamily (mean clustal x score over a 50-aa window in an alignment including MalT, AcoK from Klebsiella pneumoniae (accession no. U10553), AlkS from Pseudomonas putida (AJ245436), OrfV from Pseudomonas alcaligenes (AF092918), and SC3A7.02c from Streptomyces coelicolor (AL031155) is plotted against the amino acid position (25th of the window) in the MalT sequence and drawn to scale with respect to the upper part of the figure. The mean score obtained by aligning MalT and four random sequences with the amino acid composition of AcoK, AlkS, OrfV, and SC3A7.02c is 16.4.
These results show that there are at least two protease sensitive linker regions in MalT, which leads to the proposition that MalT contains four domains delimited by these two regions and by the 806–807 peptide bond (Fig. 3). To simplify, regions 1–241, 242–436, 437–806, and 242–806 were named DT1, DT2, DT3, and DT2–3, respectively. Moreover, proteolysis patterns in the presence or in the absence of maltotriose arise by completely different proteolytic events, which clearly demonstrates the existence of two conformational states of the protein, one with an exposed 250 region and a buried 437 region, the other with a buried 250 region and an exposed 437 region. The presence of the inducer provides a switch between these two states.
Expression and Purification of the Domains.
To determine whether the three proposed regions constitute structural domains, expression plasmids for DT1H, HDT2 (His-tagged versions of DT1 and DT2, respectively), and DT3 were constructed. The soluble fraction of the three polypeptides was purified by using metal chelate affinity chromatography (DT3 binds a nickel-agarose column, probably because of its two C-terminal histidine residues). DT1H precipitated immediately when eluted from the nickel-agarose column unless ATP was added before this step. This finding suggests that DT1H binds ATP, which fits well with the fact that it contains the Walker boxes of the putative ATP-binding site (2, 22). The fact that all of these polypeptides, which are part of regions resistant to proteinase K under certain conditions, were stable and at least partially soluble indicates that they are structural domains of the protein. Indeed, x-ray diffraction of DT3 crystals showed it to be well structured over its entire length and to be composed of a single globule (C. Steegborn, O.D., and R. Huber, unpublished data).
Domain 1 Binds ATP.
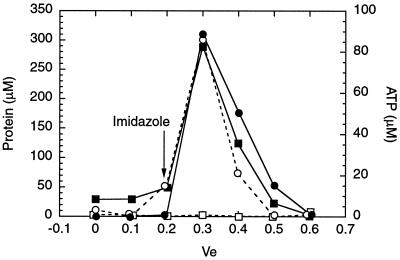
ATP binding, which is required for MalT activity (5), may involve domain 1 (see above). Because of the poor solubility of DT1H in the absence of ATP, ATP binding to immobilized DT1H was examined. [γ-32P]ATP (2 mM) was passed through an Ni-NTA agarose column that had been preloaded with DT1H. The column was then washed, and the bound protein was eluted with imidazole. The amounts of protein and ATP eluted were quantified by a Bradford assay and by counting the radioactivity present in the fractions (Fig. 4, closed symbols). ATP was clearly retained by the DT1H column and coeluted perfectly with the protein, whereas ATP did not bind a column preloaded with the same amount of DT3 (Fig. 4, open symbols). The eluted DT1H was found by SDS/PAGE to be at least 90% pure (not shown). Hence, domain 1 is the ATP-binding domain of MalT, and MalT probably binds ATP through its Walker motifs (Fig. 3).
Figure 4.
Affinity chromatography of ATP on an Ni-NTA DT1H or DT3 column. ●, ■, DT1H column; ○, □, DT3 column; ○, ●, protein concentrations; □, ■, ATP concentrations. The abscissa corresponding to the flow-through obtained at the first spin in the presence of 400 mM imidazole is indicated by the arrow.
TLC analysis of the fractions showed that 24% of the ATP that coeluted with the protein had been hydrolyzed to inorganic phosphate and probably ADP. This finding suggests that the isolated domain 1 also retains the ATPase activity of MalT.
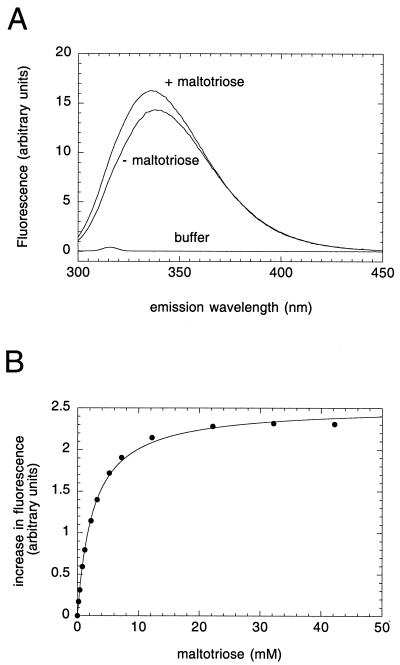
Domain 3 Binds Maltotriose with a Low Affinity.
Comparison of the amino acid sequences of the MalT subfamily revealed very low conservation throughout domain 3 (Fig. 3; see Discussion). An explanation for this fact could be that this domain binds the inducer, which differs from one protein to another. This hypothesis was tested by fluorimetry, by using the intrinsic fluorescence of the tryptophan residues. The addition of 25 mM maltotriose to a solution of DT3 excited at 286 nm (Fig. 5A) increased the emission fluorescence by 15% at the maximum of the spectrum, compared with a 1% decrease for DT1H (not shown; most probably a nonspecific effect, inasmuch as a similar fluorescence decrease was also observed with carbonic anhydrase), and displaced the maximum of the spectrum from 339 to 336 nm. This behavior indicates that maltotriose causes at least one tryptophan to move to a less polar environment. Moreover, prior addition of 25 mM glucose did not elicit any change in the DT3 spectra in the presence and absence of maltotriose (not shown). These data suggest that domain 3 specifically binds maltotriose.
Figure 5.
Fluorescence emission spectra of domain 3 and maltotriose binding. (A) Fluorescence spectra of domain 3 were recorded before and after addition of 25 μl of 1 M maltotriose. (B) Fluorescence study of the binding of maltotriose to DT3. Fluorescence measurements were carried out as described in Materials and Methods.
To measure the affinity of maltotriose for domain 3, the increase in fluorescence emission as a function of the maltotriose concentration was monitored at 320 nm with the use of 0.83 μM DT3, excited at 286 nm. The curve obtained could be fitted by a quadratic equation (Fig. 5B), which is consistent with the binding of maltotriose to a unique site present in domain 3. However, the deduced apparent KD (2.5 mM) is two orders of magnitude higher than that (≈20 μM) found by an indirect method for MalT (11).
Region 2–3 and MalT Bind Maltotriose with Similar Affinities.
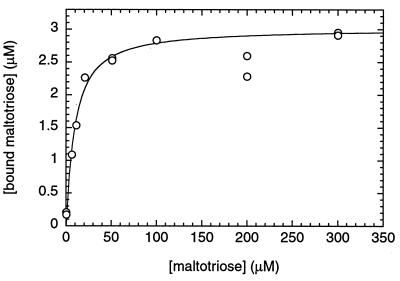
The reason for this discrepancy could be that the binding of maltotriose involves two domains. To test this explanation, the polypeptide comprising domains 2 and 3 was purified as a His-tagged version (HDT2–3). The binding of maltotriose to HDT2–3 (7.7 μM) was monitored by ammonium sulfate precipitation of the protein in the presence of 14C-radiolabeled maltotriose and counting of the radioactivity in the pellet. The values obtained could be fitted to a quadratic equation (Fig. 6), but this time with a 300-fold lower apparent KD (8 μM), in good agreement with the data obtained with MalT, even though only 40% of HDT2–3 sites were occupied by maltotriose. This binding was specific, because 1 mM maltose did not reduce the amount of bound maltotriose at a total maltotriose concentration of 1 μM. The amount of maltotriose precipitated in the presence of 7.7 μM DT3 under the same conditions was indistinguishable from the background, as expected because of its low affinity.
Figure 6.
Maltotriose binding to HDT2–3. Results of ammonium precipitation with 7.7 μM HDT2–3, expressed as concentrations of bound maltotriose, are plotted against the total maltotriose concentration.
Discussion
Maltotriose Activation of MalT Proceeds Through a Conformational Switch.
It is reported here that MalT contains three domains in addition to the small C-terminal DNA-binding domain that shares homology with LuxR. The N-terminal domain binds ATP. The second and the third domains comprise the maltotriose-binding site, although weak but specific binding of maltotriose to domain 3 was also observed. Unexpectedly, whereas the DNA-binding function and part of the activation determinants of MalT are harbored by 10% of the protein, all of the remaining 90% is required to bind the two positive effectors, i.e., for regulation of the activation function.
As suggested by the dramatic effect of ATP on DT1H solubility and the fact that MalT is stabilized by ATP (22) and is mostly in the ATP-bound form in the cell (5), ATP might act as a structural cofactor that ensures correct folding of domain 1. The other cofactor, maltotriose, has a profound effect on the tertiary structure of the whole protein. First, it causes a conformational change within domain 3, because its binding probably results in the burial of a tryptophan residue. Second, the presence of maltotriose simultaneously protects the linker region upstream from position 437 and exposes the linker upstream from position 250 to proteolysis. This finding suggests that maltotriose converts MalT from a form in which the 2–3 linker is solvent exposed and the 1–2 linker is buried to one in which the status of these two linker regions is reversed. The most attractive explanation for these observations is that, by binding to an extended region comprising domains 2 and 3, maltotriose induces a conformational switch resulting in the movement of both domains 3 and 1 with respect to domain 2.
That this conformational change is a step in the pathway of MalT activation is supported by the study of the constitutive variants. The pattern of limited proteolysis of MalTc26 shows that the 1–2 linker is exposed even in the absence of maltotriose, in agreement with a two-state model in which the equilibrium between the active and inactive state can be displaced either completely by maltotriose or incompletely by a malTc mutation. Many malTc mutations (21, 23) map within codons 220–244, i.e., amino acids very close to or within the linker that becomes exposed when MalT binds maltotriose. Furthermore, three of them (including malTc26) introduce a proline, an amino acid that favors the formation of β-turns, in this linker. This observation strongly suggests that these mutations act by forcing the linker to adopt the active conformation that is normally induced by maltotriose or by enhancing its flexibility.
Possible Mechanism for the Conformational Switch.
Altogether, the conformational change involving the 2–3 linker and the substantial increase in the affinity of domain 3 for maltotriose induced by domain 2 suggest an attractive hypothesis. MalT could bind maltotriose according to the “Venus flytrap” mechanism of periplasmic binding proteins (24), i.e., through a rotation of domains 2 and 3, enabling each domain to interact with one side of the maltotriose molecule. It is noteworthy in this respect that periplasmic binding proteins are also signaling proteins, in which the hinge-bending motion enables receptor proteins to differentiate between the loaded and unloaded states (24). However, a second model involving an induced fit between domains 3 and 2 caused by the binding of maltotriose to domain 3 cannot be excluded.
Domain Structure and Cofactor Binding Within the MalT Family.
Sensitive homology searches and motif discovery tools have allowed the identification of 18 homologues of MalT (1, 2), defining the MalT family of activators. Homology between remote members of this family is extremely weak. A Blast search (http://www.ncbi.nlm.nih.gov/BLAST/) starting with the MalT sequence gives only five homologues of MalT: AcoK (Klebsiella pneumoniae), AlkS (Pseudomonas putida), SC3A7.02c (Streptomyces coelicolor), OrfV (Pseudomonas alcaligenes), and PknK (Z83866, Mycobacterium tuberculosis). These proteins constitute a group that we call the MalT subfamily. All of these proteins except PknK are the same length as MalT and have an N-terminal putative ATP-binding site and a C-terminal LuxR-type DNA-binding domain in register with those of MalT. PknK is devoid of the LuxR domain and is consequently ≈130 amino acids shorter at its C terminus, but it possesses an additional ≈330-amino acid, serine/threonine-specific kinase-like region at its N terminus. Examination of the structure of PknK and of those of the more distantly related MalT homologues Rv0386 (AL021931), Rv1358 (Z75555), and Rv2488 (AL021246) of M. tuberculosis, which have both a LuxR DNA-binding domain and ≈200-amino acid N-terminal extensions unrelated to PknK, leads to the following hypothesis. The polypeptide constituted by domains 1, 2, and 3 is a signaling module that can be grafted onto various output domains, which allows the integration of two positive input signals (the presence of the inducer and that of ATP) and possibly negative signals and transfer of the resulting information to the output domain through a conformational change that triggers a high-order oligomerization.
Within the MalT subfamily, the level of sequence similarity is lowest from between 370 and 460 to a position around 820 (in the MalT sequence), a region that roughly corresponds to domain 3, and also around the 241–250 and 437 linker regions (Fig. 3). Thus, the patterns of sequence conservation between MalT and its closest homologues correlate with the results of the limited proteolysis experiments reported here. There is little doubt that the three-dimensional structures of domains 1, 2, and 4 (the DNA-binding domain) are conserved throughout the family. Moreover, even though domain 3 is poorly conserved at the primary sequence level, the conservation of its size and location with respect to the other three domains suggests that its overall fold might be conserved. In conclusion, the MalT family is likely to be the result of divergent rather than convergent evolution, as could have been inferred from the early proposal that only the ATP-binding site and the DNA-binding domain were conserved (2).
The members of the MalT family are exceptionally large: MalT is the largest activator in E. coli, and the five largest proteins from M. tuberculosis belonging to the “regulation” category as defined in the Tuberculist database (http://genolist.pasteur.fr/TubercuList/) are MalT homologues. This work indeed shows that they have one more domain compared with other large bacterial activators, those belonging to the NtrC family and TyrR. These activators share some features with MalT, inasmuch as they also integrate two positive signals and undergo a high-order multimerization (25–27). However, in addition to their absence of sequence homology with MalT, their domain organization is different, because ATP seems to bind to the central domain (28–30). This observation suggests that, despite superficial similarities, their activation mechanism and that of MalT are different.
Acknowledgments
I thank my colleagues E. Richet for many stimulating discussions, V. Schreiber for the gift of MalTc26, D. Deville-Bonne for her advice on fluorescence spectroscopy, A. Chaffotte for a useful suggestion, and J. M. Masson (Institut de Pharmacologie et de Biologie Structurale, Toulouse, France) for the gift of plasmid pARA14. I thank E. Richet, C. Steegborn (Max-Planck-Institut für Biochemie, Martinsried, Germany), and my colleague A. P. Pugsley for critical reading of the manuscript.
Abbreviation
- NTA
nitrilotriacetic acid
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Valdez F, González-Cerón G, Kieser H M, Servín-González L. Microbiology. 1999;145:2365–2374. doi: 10.1099/00221287-145-9-2365. [DOI] [PubMed] [Google Scholar]
- 2.De Schrijver A, De Mot R. Microbiology. 1999;145:1287. doi: 10.1099/13500872-145-6-1287. [DOI] [PubMed] [Google Scholar]
- 3.Boos W, Shuman H. Microbiol Mol Biol Rev. 1998;62:204–229. doi: 10.1128/mmbr.62.1.204-229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwartz M. In: Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1987. pp. 1482–1502. [Google Scholar]
- 5.Richet E, Raibaud O. EMBO J. 1989;8:981–987. doi: 10.1002/j.1460-2075.1989.tb03461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schreiber V, Richet E. J Biol Chem. 1999;274:33220–33226. doi: 10.1074/jbc.274.47.33220. [DOI] [PubMed] [Google Scholar]
- 7.Reyes M, Shuman H A. J Bacteriol. 1988;170:4598–4602. doi: 10.1128/jb.170.10.4598-4602.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reidl J, Boos W. J Bacteriol. 1991;173:4862–4876. doi: 10.1128/jb.173.15.4862-4876.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peist R, Koch A, Bolek P, Sewitz S, Kolbus T, Boos W. J Bacteriol. 1997;179:7679–7686. doi: 10.1128/jb.179.24.7679-7686.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panagiotidis C H, Boos W, Shuman H A. Mol Microbiol. 1998;30:535–546. doi: 10.1046/j.1365-2958.1998.01084.x. [DOI] [PubMed] [Google Scholar]
- 11.Schreiber V, Steegborn C, Clausen T, Boos W, Richet E. Mol Microbiol. 2000;35:765–776. doi: 10.1046/j.1365-2958.2000.01747.x. [DOI] [PubMed] [Google Scholar]
- 12.Vidal-Ingigliardi D, Richet E, Danot O, Raibaud O. J Biol Chem. 1993;268:24527–24530. [PubMed] [Google Scholar]
- 13.Danot O, Vidal-Ingigliardi D, Raibaud O. J Mol Biol. 1996;262:1–11. doi: 10.1006/jmbi.1996.0493. [DOI] [PubMed] [Google Scholar]
- 14.Silhavy T J, Berman M L, Enquist L W. Experiments with Gene Fusions. Plainview, NY: Cold Spring Harbor Lab. Press; 1984. [Google Scholar]
- 15.Hatfield D, Hofnung M, Schwartz M. J Bacteriol. 1969;98:559–567. doi: 10.1128/jb.98.2.559-567.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 17.Cagnon C, Valverde V, Masson J M. Protein Eng. 1991;4:843–847. doi: 10.1093/protein/4.7.843. [DOI] [PubMed] [Google Scholar]
- 18.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 21.Dardonville B, Raibaud O. J Bacteriol. 1990;172:1846–1852. doi: 10.1128/jb.172.4.1846-1852.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richet E, Raibaud O. J Biol Chem. 1987;262:12647–12653. [PubMed] [Google Scholar]
- 23.Notley-McRobb L, Ferenci T. Environ Microbiol. 1999;1:45–52. doi: 10.1046/j.1462-2920.1999.00003.x. [DOI] [PubMed] [Google Scholar]
- 24.Quiocho F A, Ledvina P S. Mol Microbiol. 1996;20:17–25. doi: 10.1111/j.1365-2958.1996.tb02484.x. [DOI] [PubMed] [Google Scholar]
- 25.Wilson T J, Maroudas P, Howlett G J, Davidson B E. J Mol Biol. 1994;238:309–318. doi: 10.1006/jmbi.1994.1294. [DOI] [PubMed] [Google Scholar]
- 26.Pérez-Martín J, de Lorenzo V. Cell. 1996;86:331–339. doi: 10.1016/s0092-8674(00)80104-x. [DOI] [PubMed] [Google Scholar]
- 27.Rippe K, Mucke N, Schulz A. J Mol Biol. 1998;278:915–933. doi: 10.1006/jmbi.1998.1746. [DOI] [PubMed] [Google Scholar]
- 28.Pérez-Martín J, de Lorenzo V. J Mol Biol. 1996;258:575–587. doi: 10.1006/jmbi.1996.0270. [DOI] [PubMed] [Google Scholar]
- 29.Flashner Y, Weiss D S, Keener J, Kustu S. J Mol Biol. 1995;249:700–713. doi: 10.1006/jmbi.1995.0330. [DOI] [PubMed] [Google Scholar]
- 30.Cui J, Somerville R L. J Biol Chem. 1993;268:5040–5047. [PubMed] [Google Scholar]