Abstract
Human nuclear RNase P purified from HeLa cells has ATPase activity. This activity is associated with one of the protein subunits of the enzyme, Rpp20. Thus, human nuclear RNase P, which contains several proteins and one essential RNA, has at least one other enzymatic activity in addition to cleavage of phosphoester bonds in RNA. The amino acid sequence of Rpp20 has a signature motif found in an ATPase-containing subunit of a family of protein complexes (ABC transporters) that mediate a variety of trans-membrane traffic, as well as a segment, DIxxN, that resembles the DEAD box motif of many ATPases: together, these might represent an ATPase signature motif.
Keywords: Rpp20, ATPase motifs, ATPase inhibition, tRNA precursor
Human nuclear RNase P, an essential enzyme required for processing of precursor tRNAs, consists of at least eight distinct proteins associated with an essential RNA subunit, H1 RNA (1–4). Only a few of the subunits exhibit strong homology with proteins found in, for example, Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila melanogaster, or Mus musculus (1–4), an indication that the remaining subset of the subunits found in the RNase P holoenzyme may vary in function, as well as structure, across the evolutionary spectrum. Some protein subunits might be essential only for the chemistry of catalysis by RNase P and others might play additional roles in the function of the enzyme. For example, two RNase P subunits, Rpp29 and Rpp38, function in the localization of the holoenzyme complex to the nucleolus (5).
Many aspects of cellular RNA metabolism and processing involve ATP and RNA unwinding activities (refs. 6 and 7, and refs. therein). Recently, an ATP-dependent helicase from Thermus thermophilus that has a segment of amino acid sequence (DEAD box) in common with many ATPases and that shares a structural motif with a bacterial RNase P protein was described (8). Here, we show that RNase P from Homo sapiens has ATPase activity, but RNase P from Escherichia coli does not. We also describe segments of amino sequence in a protein subunit of human RNase P that are similar to the DEAD box sequence and a signature motif found in the ATPases of ABC transporter complexes. The latter are a large family of protein complexes associated with the transport of a variety of compounds across both prokaryotic and eukaryotic membranes.
Materials and Methods
Materials.
Restriction enzymes were obtained from New England Biolabs. Radiochemicals were obtained from Amersham Pharmacia. Oligonucleotides were synthesized at the Keck Facility of Yale University.
Purification of Human RNase P and Rpp20.
Human nuclear RNase P was purified by four steps (DEAE-Sepharose Fast Flow anion-exchange chromatography, velocity sedimentation in a 15–30% glycerol gradient, FPLC MonoQ HR-10/10 column and FPLC Superose 6 gel filtration column; Amersham Pharmacia) as described (1). Protein concentrations were measured by using a Bio-Rad protein assay reagent with BSA as control.
His-tagged, recombinant Rpp20 protein was expressed in E. coli BL21(DE3) at 22°C and purified on a histidine-binding resin (Novagen) column, according to Jarrous et al. (3).
Enzyme Assays.
RNase P holoenzyme activities were assayed in 1× PA buffer (50 mM Tris⋅HCl, pH 7.5/10 mM MgCl2/100 mM NH4Cl) at 37°C. M1 RNA was assayed in M1 buffer (1× PA with 100 mM MgCl2 and 4% polyethylene glycol). The substrate RNAs, yeast suppressor tRNASer precursor (pSupS1), and E. coli precursor tRNATyr (pTyr) were transcribed in vitro (9) in the presence of [α-32P]GTP, purified on a 7 M urea/5% polyacrylamide gel, and were used at a final concentration of 100 nM (2,000 cpm per reaction).
ATPase activity was detected by the hydrolysis of ATP to ADP and phosphate in a reaction mixture of 1× ATPase buffer (50 mM Tris⋅HCl, pH 8.0/5 mM MgCl2/1 mM DTT), 0.1 mM ATP, and 0.1 μCi of either [α-32P]ATP or [γ-32P]ATP. ATP stock solutions were adjusted to pH 7.5 prior to use. The reaction mixture (5 or 10 μl) was incubated at 37°C for 30 min and stopped by addition of 1 μl of 0.4 M EDTA and placed on ice. An aliquot of the reaction mixture (0.5 μl) was spotted on a polyethyleneimine TLC plate and dried. The TLC plate was developed by using a solution that contained 1 M LiCl and 1 M acetic acid and exposed in a phosphorimager cassette (Fuji) after drying.
RNA helicase activity was assayed in 20 mM Tris⋅HCl (pH 7.5), 1 mM or 5 mM MgCl2, 1 mM DTT at room temperature or at 37°C. The substrate was a duplex of two RNA oligonucleotides (GAAAACGCGGCUUAA and UUAAGUCGCACCA) that were labeled with polynucleotide kinase at their 5′ termini with 32P. Reaction mixtures were analyzed on a 20% native polyacrylamide gel with 20 mM Hepes⋅KOH, pH 7.5, and 1 mM MgCl2 as running buffer.
Site-Directed Mutagenesis.
Mutagenesis of the Rpp20 gene was performed with the “megaprimer” PCR technique (10), and DNA sequences were confirmed by sequencing with T7 Sequenase (Amersham Pharmacia).
Immunoprecipitation of Active Holoenzyme.
Sera (40 μl) from immunized rabbits were mixed with 5 mg of washed protein A-Sepharose CL-4B beads (Amersham Pharmacia) in NET-2 buffer (50 mM Tris⋅HCl, pH 7.5/150 mM NaCl/0.05% Nonidet P-40) (11) by nutating overnight at 4°C. Beads were washed four times with NET-2. Then, 20 μl of RNase P from a MonoQ column or a similar volume of recombinant Rpp20 purified from E. coli was added to the beads. Immunoprecipitations were performed in 250 μl during overnight nutation at 4°C. Beads were collected by a short centrifugation (7,000 rpm for 3 min), the supernatant fluid was removed, and the beads were washed two times with NET-2 and then three times with 1× ATPase buffer. Supernatant fractions and beads were each assayed for enzymatic activity.
Results
Nuclear RNase P Purified from HeLa Cells Has ATPase Activity.
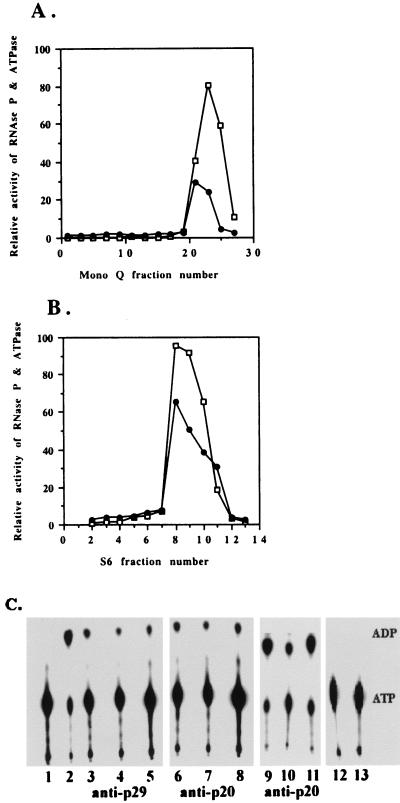
Human nuclear RNase P, purified as described in Materials and Methods, was incubated with [α-32P]ATP. The enzyme cleaves ATP to form [α-32P]ADP and phosphate (Fig. 1C, lane 2). That phosphate was one of the products of the reaction was shown in separate experiments with [γ-32P]ATP. The ATPase activity follows the RNase P activity during purification (Fig. 1 A and B). On the other hand, E. coli RNase P (holoenzyme or M1 RNA alone) did not exhibit any ATPase activity (data not shown).
Figure 1.
Nuclear RNase P purified from HeLa cells has ATPase activity. The RNase P (□) and ATPase (●) activities of the fractions from a Mono Q FPLC column (A) and an S6 column (B) were assayed as described in Materials and Methods. (C) Immunoprecipitation of ATPase activities by polyclonal rabbit antibodies against recombinant Rpp29 and Rpp20 protein subunits of human RNase P. The procedure used is described in Materials and Methods. Lane 1, Incubation of ATP alone. Lane 2, Addition of RNase P from an S6 column. Lanes 3–5, Immunoprecipitation of human RNase P with Rpp29 antibody and (lanes 6–8) with Rpp20 antibody. Lanes 9–11, Immunoprecipitation of Rpp20 with Rpp20 antibody. Lane 12, Rpp29 antibody with no addition of RNase P. Lane 13, Rpp20 antibody with no addition of RNase P. Lanes 3, 6, and 9, aliquots from initial supernatants. Lanes 4, 7, and 10, aliquots from supernatants after overnight immunoprecipitation. Lanes 5, 8, and 11, aliquots from the pellets. The sum of ATPase activity recovered after immunoprecipitation is somewhat greater than that found in the initial supernatant fraction both because of the higher efficiency of the ATPase activity in the undiluted 1× ATPase buffer used to assay the immunoprecipitated fractions and the fact that even rabbit preimmune control serum enhances ATPase activity.
To investigate whether the ATPase activity was associated nonspecifically with other proteins in the same fraction that contained RNase P activity, immunoprecipitation of the enzyme was performed. As previously reported, anti-Rpp29 and anti-Rpp20 antibodies from rabbit can immunoprecipitate the corresponding protein subunits, Rpp29 and Rpp20, of human RNase P and the holoenzyme, itself. These two antibodies can also efficiently bring down more than 65% of ATPase activity from the supernatants of the immunoprecipitation mixture (Fig. 1C), indicating that the ATPase and RNase P activities are very likely in the same complex.
The ATPase activities of both Rpp20 (see below) and RNase P are independent of added RNAs. Adding H1 RNA, M1 RNA, or precursor tRNA to ATPase reaction mixtures does not increase the activity of Rpp20. Incubation of the RNase P with 0.01 μg/ml RNase A for 30 min did not lead to a significant decrease of the ATPase activity (data not shown). However, the H1 RNA in the enzyme was degraded, an indication that the ATPase activity of the holoenzyme can be decoupled from its RNase P activity.
The optimal temperature for RNase P activity in vitro, 50°C, was unchanged in the presence of ATP (data not shown).
Rpp20 Has Amino Acid Sequence Homology with Other ATPases.
To identify which of the human RNase P subunits might be an ATPase, BLAST (National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov) and FASTA (European Bioinformatics Institute of European Molecular Biology Laboratory, http://www.ebi.ac.uk) sequence comparison programs were used to compare all of the protein sequences in these databases with all of the RNase P protein subunits characterized to date. We found that only Rpp20 has significant homology with a putative ATP binding cassette in an ABC transporter ATPase from Arabidopsis thaliana: a C-terminal fragment of that ATPase share more than 25% identity and 30% similarity with Rpp20 (Fig. 2B). We also found by visual inspection that a segment of the Rpp20 sequence shows weak homology, DIxxN (conserved bases: uppercase; semiconserved: lowercase; nonconserved: x), with the ATPase motif of the Upf1p subfamily (of as yet undefined general function) of RNA helicases that contain a DEAD box, but not with the RNA interaction and helicase motifs (Fig. 2A; refs. 6 and 7). Furthermore, the Walker ATPase motif A (GxxxxGKT; ref. 12), which is found in ABC transporter ATPases and all DEAD box ATPases, is absent from the sequence of Rpp20.
Figure 2.
Rpp20 has strong homology with ABC transporter ATPases (in particular a putative ABC transporter ATPase from A. thaliana, see below), and weak homology with a Upf1p subfamily of the DEAD-box ATP-dependent RNA helicase superfamily. (A) Schematic representation of the common motifs of Rpp20, the Upf1p subfamily, and ABC transporters. The alignment of sequence conservation of ABC transporter ATPase super family is described in ref. 12, and that of the Upf1p family is reviewed in refs. 6 and 7. Fully conserved residues are shown in uppercase, semiconserved in lowercase, and nonconserved are designated as x. (B) Sequence alignment of Rpp20 and the C-terminal fragment (starting at residue 163) of the ABC transporter ATPase from A. thaliana. Identical residues are marked by an asterisk; residues of high similarity by a colon; and residues of low similarity by a period. Gaps are shown as dashed lines.
Rpp20 Has ATPase Activity.
Recombinant Rpp20 expressed in and purified from E. coli can hydrolyze ATP to ADP and phosphate. That this activity is the result of Rpp20 rather than a contaminating ATPase from E. coli is demonstrated by the fact that preparations of an Rpp20 mutant, G59S, and Rpp30 (another subunit of RNase P), obtained by an identical purification scheme, do not display any ATPase activity. Furthermore, antibodies against recombinant Rpp20 immunoprecipitate the ATPase activity (Fig. 1C: these antibodies do not precipitate either the ATPase activity or the protein, itself, with 100% efficiency; refs. 3 and 4). The kcat of Rpp20 is about 65 nmol product/min per mg, which is within the range of 10 to 1,650 nmol/min per mg reported for the isolated ATPase domain activity of the ABC transporters (13). However, we could not detect any helicase activity associated with Rpp20 (data not shown), consistent with the fact that there is no RNA helicase motif in the Rpp20 amino acid sequence.
To show further that the signature motif and the putative DIxxN box of Rpp20 play key roles in its ATPase activity, we made several mutations in these motifs. Only one of the mutants, N40A, could not be expressed in E. coli. Several other mutant proteins were purified and their ATPase activities were measured. Mutants N40Q and D36A show activity close to the wild type, but mutant D36E exhibits only 7% of wild-type activity. Mutant G59S in the signature motif had no activity, as mentioned above, whereas others (D57A, G59A, R61A) still retained considerable activity (Table 1).
Table 1.
The effect of mutations in Rpp20 on its ATPase activity
| Enzyme | Relative activity, % |
|---|---|
| Wild type | 100 |
| Mutants in the DIxxN box | |
| D36E | 7 |
| D36A | 110 |
| N40Q | 25 |
| Mutants in the ABC signature motif | |
| D57A | 94 |
| G59A | 89 |
| G59S | 0 |
| R61A | 44 |
The activities of Rpp20 and its mutant derivatives are shown as percentages of the wild-type activity, which is about 65 nmol/min per mg. Each value represents the average of two independent determinations with a variation about 10%.
Enzymatic Properties of ATPases Associated with Rpp20 and Human RNase P.
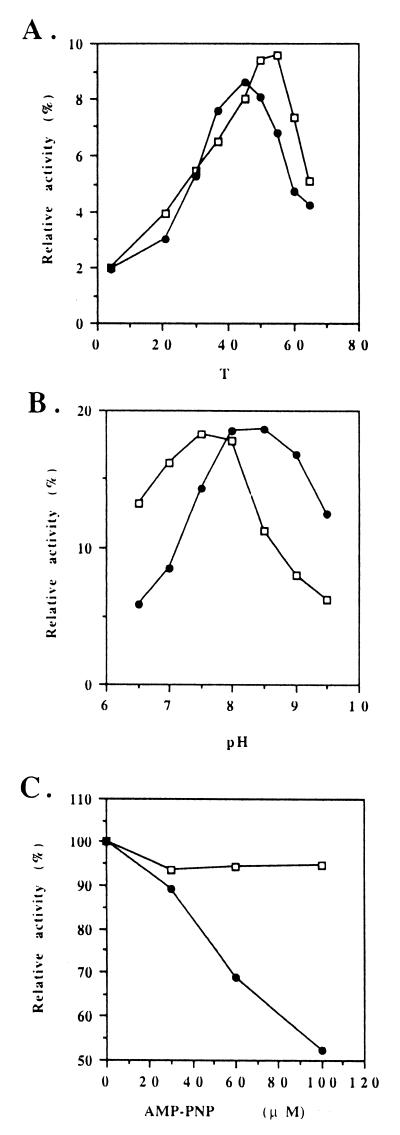
The ATPase activities of Rpp20 and the RNase P holoenzyme did not have precisely the same optimal temperature or pH (Fig. 3 A and B). They also responded differently to the inhibitor AMP-PNP (5′-adenylylimidodiphosphate) (Fig. 3C).
Figure 3.
Enzymatic properties of Rpp20 (□) and human RNase P (●). Final concentration of enzyme was 7.5 μg/ml for Rpp20 and 40 μg protein/ml for human RNase P (MonoQ fraction). (A) Optimal temperature for ATPase activity. (B) Optimal pH for ATPase activity. The 1× ATPase buffer was replaced by 10 mM Bistris propane⋅HCl, 5 mM MgCl2, and 1 mM DTT. (C) Inhibition of ATPase activity by AMP-PNP.
AMP-PNP inhibits the ATPase and RNase P activities of human RNase P, but it cannot inhibit the ATPase activity of Rpp20 (Fig. 3C). This is similar to the effect of the inhibitor on HisQMP2 (periplasmic histidine permease: membrane bound complex), in which the catalytic subunit HisP is not inhibited by vanadate, whereas the ATPase activity of the intact complex is inhibited by this compound (14). Vanadate at 100 μM did not inhibit the ATPase activity of either Rpp20 or the RNase P holoenzyme complex. The holoenzyme ATPase activity was inhibited by about 40% at 1 mM vanadate but the Rpp20 ATPase activity was not.
Both human RNase P and Rpp20 have GTPase activities (data not shown). The GTPase activities (kcat of 42 nmol/min per mg for Rpp20 and 0.78 nmol/min per mg for holoenzyme) are about 60–70% of those for the Rpp20 ATPase, a similar ratio to those found in many other ATPases.
The Km values of the holoenzyme and Rpp20 are very close to each other. The Km value of Rpp20 is 265 μM, whereas that of human RNase P is 280 μM. The kcat of Rpp20 is about 65 nmol/min per mg, whereas that of the human holoenzyme is about 1.1 nmol/min per mg protein. We note, however, that on the basis of protein mass (assuming all subunits are present in single copy), the Rpp20 content of the holoenzyme preparations we use is at most around 7%. If the holoenzyme is approximately 50% pure, which is likely as judged by SDS-gel analysis, then the kcat values for Rpp20 and the holoenzyme are very similar.
Discussion
The ATPase Activity of the Rpp20 Subunit of Human RNase P.
We report that human RNase P has an ATPase activity and that one of the protein subunits of RNase P, Rpp20, is an ATPase. The enzymatic properties of the ATPase of Rpp20 and that of the human RNase P are not absolutely identical. The holoenzyme is somewhat more sensitive to temperature and an ATP analog inhibitor than the isolated protein. The local environment of Rpp20 and its catalytic center will be affected by contacts with other protein subunits. Differences in thermal stability and the shape of the ATP binding pocket (and, therefore, its accessibility to AMP-PNP) will clearly be sensitive to these contacts.
E. coli RNase P has no ATPase activity. Previous reports (15, 16) show that during precursor tRNA cleavage, some unwinding of the substrate occurs. By analogy, a similar unwinding of substrate must occur during cleavage by eukaryotic (human) RNase P. However, we have not yet been able to demonstrate directly any helicase activity associated with either E. coli or human RNase P with the substrate we used. Several RNA helicases in eukaryotic cells, however, do have ATP-dependent unwinding activities (6, 7).
The ATPase Signature Motif and the DIxxN Box.
A comparison of the sequences of several proteins that have ATPase activity with that of Rpp20 has revealed the common presence of certain motifs. The signature motif of ABC transporter ATPases is the most conserved element in the amino acid sequences of this enzyme family (12). In the case of Rpp20, the mutations, D57A and G59A in the segment of the protein that resembles the ABC transporter ATPase signature motif, 56LDGGAR, exhibited lowered ATPase activity, and G59S inactivated the activity completely. These results indicate that the motif may play an important role in ATP binding and or catalysis.
The importance of G59 in Rpp20 can also be inferred from its similar positioning in the ATPase signature motif of ABC transporter ATPases to G795 of Rad50 (12). Both of these residues are found in the fourth position of this motif. G795 binds the γ-phosphate of ATP directly as seen in the crystal structure of Rad50 (12). G59 of Rpp20 may be have a similar function to G759 although we have not yet observed such binding.
Previous reports showed that Rad50 and HisP form dimers in solution and in crystals (12, 17). Although we did not observe cooperativity in ATP hydrolysis by Rpp20 (data not shown), we note that 5–10% of recombinant Rpp20 appears in dimeric form on SDS/PAGE (data not shown).
No experimental data have been provided for the function of the DIxxQ box for the Upf1p family of DEAD-box RNA helicases. For other DEAD box proteins, such as eIF-4A, some residues in the DEAD box appear essential for ATP hydrolysis to ADP and Pi whereas others have no phenotype (18). The mutation, D36E, in the DIxxN box of Rpp20 decreases ATPase activity >90%. The mutation N40Q resulted in reduction of activity by >75%. We propose, therefore, that in Rpp20, the ABC transporter signature motif and the DIxxN box are both critical for ATP binding and catalysis.
Rpp20, An Add-On Component of RNase P?
Human RNase P cleaves precursor tRNA efficiently, even in the absence of ATP. Rpp20 does not interact with H1 RNA, the RNA subunit of human RNase P, directly according to results of a yeast three hybrid analysis (T. Jiang and S.A., unpublished observations), an indication that Rpp20 is not involved in the catalytic center if that center is built around H1 RNA. Accordingly, Rpp20 might be an add-on component to a primitive eukaryotic RNase P. A simple hypothesis might suggest that Rpp20 is a physical and functional link to another RNase P-related intracellular function, In fact, a small, heat shock protein (Hsp27) has been shown to interact with Rpp20 both in a yeast two-hybrid genetic assay and by direct biochemical experimentation. Hsp27 increases RNase P activity significantly in vitro after heat shock (T. Jiang and S.A., unpublished observations). Although the exact mechanism of Hsp27 function in relation to human nuclear RNase P and its ATPase activity is not known, it is clear that Rpp20 can act as a functional bridge between at least one other nuclear protein, Hsp27, and the catalytic activity of RNase P.
Acknowledgments
We thank our colleagues, especially Drs. C. Guerrier-Takada and T. Jiang, for helpful discussions. This work was supported by grants from the Human Frontiers of Science Program and the National Institutes of Health (GM19422; to S.A.).
Abbreviation
- AMP-PNP
5′-adenylylimidodiphosphate
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.021555498.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.021555498
References
- 1.Eder P, Kekuda R, Stolc V, Altman S. Proc Natl Acad Sci USA. 1997;94:1101–1106. doi: 10.1073/pnas.94.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lygerou Z, Pluk H, van Venrooj W J, Seraphin B. EMBO J. 1996;15:5936–5948. [PMC free article] [PubMed] [Google Scholar]
- 3.Jarrous N, Eder P S, Guerrier-Takada C, Hoog C, Altman S. RNA. 1998;4:407–417. [PMC free article] [PubMed] [Google Scholar]
- 4.Jarrous N, Eder P S, Wesolowski D, Altman S. RNA. 1999;5:153–157. doi: 10.1017/s135583829800185x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jarrous N, Wolenski J S, Wesolowski D, Lee C, Altman S. J Cell Biol. 1999;146:559–571. doi: 10.1083/jcb.146.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de la Cruz J, Kressler D, Linder P. Trends Biochem Sci. 1999;24:192–198. doi: 10.1016/s0968-0004(99)01376-6. [DOI] [PubMed] [Google Scholar]
- 7.Linder P, Daugeron M C. Nat Struct Biol. 2000;7:97–99. doi: 10.1038/72464. [DOI] [PubMed] [Google Scholar]
- 8.Morlang S, Weglohner W, Franceschi F. J Mol Biol. 1999;294:795–805. doi: 10.1006/jmbi.1999.3282. [DOI] [PubMed] [Google Scholar]
- 9.Gopalan V, Baxevanis A D, Lansman D, Altman S. J Mol Biol. 1999;264:5098–5103. [Google Scholar]
- 10.Sarkar G, Sommer S S. BioTechniques. 1990;8:404–407. [PubMed] [Google Scholar]
- 11.Gold H A, Craft J, Hardin J A, Bartkiewicz M, Altman S. Proc Natl Acad Sci USA. 1998;85:5483–5487. doi: 10.1073/pnas.85.15.5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hopfner K-P, Darcher A, Sin D S, Craig L, Arthur L, Carney J P, Tainer J A. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 13.Holland I B, Blight M A. J Mol Biol. 1999;293:381–399. doi: 10.1006/jmbi.1999.2993. [DOI] [PubMed] [Google Scholar]
- 14.Nikaido K, Liu P-Q, Ames G F-L. J Biol Chem. 1997;272:27745–27752. doi: 10.1074/jbc.272.44.27745. [DOI] [PubMed] [Google Scholar]
- 15.Knap A K, Wesolowski D, Altman S. Biochemie. 1990;72:779–790. doi: 10.1016/0300-9084(90)90187-l. [DOI] [PubMed] [Google Scholar]
- 16.Krummel D A P, Altman S. Proc Natl Acad Sci USA. 1999;96:11200–11205. doi: 10.1073/pnas.96.20.11200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hung L-W, Wang I X, Nikaido K, Liu P-Q, Ames G F-L, Kim S-H. Nature (London) 1998;396:703–707. doi: 10.1038/25393. [DOI] [PubMed] [Google Scholar]
- 18.Pause A, Sonenberg N. EMBO J. 1992;11:2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]