Abstract
The carboxyl-terminal domain (CTD) of the largest subunit of RNA polymerase II plays an important role in transcription and processing of the nascent transcript by interacting with both transcription and RNA processing factors. We show here that the cleavage/polyadenylation factor IA of Saccharomyces cerevisiae directly contacts CTD. First by affinity chromatography experiments with yeast extracts we demonstrate that the Rna15p, Rna14p, and Pcf11p subunits of this complex are associated with phosphorylated CTD. This interaction is confirmed for Rna15p by yeast two-hybrid analysis. Second, Pcf11p, but not Rna15p, is shown to directly contact phosphorylated CTD based on in vitro binding studies with recombinant proteins. These findings establish a direct interaction of cleavage/polyadenylation factor IA with the CTD. Furthermore, a quantitative analysis of transcription run-on performed on temperature-sensitive mutant strains reveals that the lack of either functional Rna14p or Pcf11p affects transcription termination more severely than the absence of a functional Rna15p. Moreover, these data reinforce the concept that CTD phosphorylation acts as a regulatory mechanism in the maturation of the primary transcript.
In Saccharomyces cerevisiae the largest subunit of RNA polymerase II (pol II) contains a carboxyl-terminal domain (CTD) consisting of 26 tandem repeats of a heptapeptide module with the consensus sequence Tyr-Ser-Pro-Thr-Ser-Pro-Ser (1, 2). Two serines per heptad repeat may undergo reversible phosphorylation during each transcription cycle (3). The phosphorylation status of the CTD is correlated with different stages of the transcription cycle. Thus hypophosphorylated polymerase (IIA) is competent for initiation, whereas hyperphosphorylated polymerase (II0) is associated with transcription elongation. Increasing evidence has been provided in the last few years showing that the CTD acts as a direct physical link between transcription and nascent RNA processing: in mammals, the cleavage-polyadenylation specificity factor (CPSF) and the cleavage stimulation factor (CstF), as well as splicing factors and 5′ capping enzyme, all bind to the CTD (4–9). Furthermore, CPSF and CstF copurify with pol II (4). The concept of a factor recruiting/docking platform has emerged as one likely function of this peculiar polypeptide. Evidence for a more direct role of the CTD in polyadenylation has also been indicated (10).
In S. cerevisiae, the proteins so far known to bind to the CTD are the Mediator complex (11, 12), the Elongator complex (13), proteins involved in 5′ end capping of nascent RNA (8, 14), and the Ess1 peptidylprolyl isomerase (15, 16). To date the interaction of cleavage/polyadenylation factors with the CTD remains largely uncharacterized. In yeast 3′-end processing of pre-mRNA requires cleavage factor IA (CF IA), cleavage factor IB (CF IB), cleavage factor II (CF II), polyadenylation factor I (PFI), poly(A) polymerase, and the poly(A)-binding protein 1 (17–22). CF IA, which is involved in both cleavage and polyadenylation, comprises four polypeptides: Rna15p, Rna14p, Pcf11p, and Clp1p. Biochemical and genetic analyses have shown that these subunits are tightly associated (17, 18, 20, 23). Rna14p and Rna15p have localized sequence similarity with the 77-kDa and 64-kDa subunits of mammalian CstF, respectively (24). Rna15p is the RNA-binding subunit of CF IA, containing a canonical RNA-binding domain with the ribonucleoprotein-2 and ribonucleoprotein-1 motifs, and it has also been reported to bind in vitro to uridine-rich sequences (17, 23, 25). Neither Clp1p (21) nor Pcf11p displays any similarity to known mammalian processing factors. However, Pcf11p does contain a putative CTD-binding domain at the N terminus that is homologous to domains found in a number of polypeptides from various organisms (26–28).
Previous studies in our laboratory have revealed that subunits of CF IA are directly involved in transcription termination: Birse et al. (29) demonstrated that mutations in RNA15, RNA14, and PCF11 affect transcription termination based on transcription run-on analysis. Exactly how these factors affect transcriptional termination has yet to be elucidated.
In this study, we have analyzed the potential interaction of CF IA with the CTD. Affinity chromatography experiments with whole-cell yeast extracts show that CF IA binds specifically to the CTD, as judged by the presence of Rna15p, Rna14p, and Pcf11p in the pellet fraction. The binding is strongly enhanced by the phosphorylation of the CTD. Furthermore, pull-down experiments with purified recombinant proteins demonstrate that Pcf11p is one of the subunits of CF IA that directly contacts the CTD, again exhibiting a much higher affinity for the phosphorylated form. Finally, quantitative transcription run-on analysis highlights the important role of Pcf11p and possibly Rna14p, showing that mutations in PCF11 and RNA14 affect transcription termination more drastically than mutations in RNA15. This observation suggests that Pcf11p and possibly Rna14p may play a crucial role in coupling transcription with pre-mRNA processing by recruiting the whole CF IA complex to the CTD.
Materials and Methods
Strains, Media, and Transformation.
The S. cerevisiae strain used for the preparation of extracts for affinity chromatography experiments was W303-1B, and the reporter host strain used for the two-hybrid analysis was Y187. The temperature-sensitive (ts) mutant strains used for the run-on analysis have been described (18, 23). Escherichia coli DH5α strain was used for both plasmid construction and overproduction of the glutathione S-transferase (GST) fusion protein. E. coli BL21(DE3) was used for recombinant Pcf11p and Rna15p overproduction. Media used to grow yeast strains were yeast extract-peptone containing 2% glucose or selective synthetic complete medium containing 2% glucose. Yeast cells were transformed with the use of LiAc salts.
Construction of pGST-CTD and Purification of the Protein.
DNA fragments encoding all 26 CTD repeats were amplified from genomic DNA. The PCR product was digested and ligated into the overexpression plasmid pGEX-4-T1 (Amersham Pharmacia). The resulting plasmid, pGST-CTD, was transformed into E. coli DH5α. Freshly obtained transformants were grown at 37°C to an OD600 of 0.6; the culture was then induced by the addition of isopropyl-β-d-thiogalactoside at a final concentration of 0.5 mM and transferred to 30°C for 3–16 h. The GST-CTD polypeptide was purified by affinity chromatography on glutathione Sepharose 4B (Amersham Pharmacia) as outlined (30).
Affinity Chromatography.
GST-CTD was phosphorylated for 2 h at 30°C with casein kinase I (New England Biolabs) and 1 mM ATP, with the use of the manufacturer's buffer. Unphosphorylated and phosphorylated GST-CTD and GST proteins were immobilized onto 20 μl of glutathione Sepharose 4B at 2.5 mg/ml by shaking for 1 h at room temperature. The resins were then washed, equilibrated with buffer A (20 mM Hepes⋅KOH, pH 7.6/1 mM EDTA/1 mM DTT/20% glycerol/100 mM potassium acetate/0.03% Nonidet P-40), and then mixed with 50 μl of yeast extract (8 mg/ml) in 200 μl of buffer A containing 150 μg/ml of BSA and protease inhibitors (1 μg/ml of pepstatin A/1 μg/ml of leupeptin/1 mM PMSF) for 1 h at room temperature on a rotating wheel. Yeast extracts were prepared as described (31). The beads were then collected and washed five times with buffer A containing protease inhibitors, resuspended in 30 μl of 2× SDS sample buffer, and boiled for 5 min. Proteins were fractionated by SDS/PAGE, transferred to nitrocellulose membranes, and analyzed by immunoblotting. Detection was performed by chemiluminescence with the enhanced chemiluminescence kit (Amersham Pharmacia). Polyclonal antibodies raised against Rna15p, Rna14p, Clp1p, and Fip1p were a gift from Lionel Minvielle-Sebastia (University of Bordeaux, France). The monoclonal Gal4 DNA-BD antibody was purchased from CLONTECH. For the affinity chromatography experiments with recombinant Pcf11p and Rna15p, 1 μg of each protein was incubated with GST-CTD, phosphorylated GST-CTD, and GST resins, and the above described protocol was used. Both Pcf11p and Rna15p were detected by an antihistidine monoclonal antibody (Qiagen, Chatsworth, CA). The plasmid pGBD-Pcf11 used to transform the pcf11 mutant was constructed by cloning the wild-type PCF11 gene into the vector pAS2–1 (CLONTECH), so that the resulting product was in-frame with the plasmid-encoded Gal4 DNA binding domain.
Yeast Two-Hybrid Analysis.
All of the primers for PCR were based on the coding sequences in the Saccharomyces Genome Database (http://genome-www.stanford.edu/Saccharomyces/). The two-hybrid bait and prey vectors were pAS2–1 and pACT2 (CLONTECH), respectively. The RNA15 gene was amplified by PCR and cloned in pAS2–1. The CTD encoding fragment was cloned into the prey vector pACT2. It was used as a prey rather than a bait, as it had been previously reported to activate transcription per se when fused to the Gal4 DNA binding domain (7). The bait and the prey plasmids were transformed into yeast Y187 and cotransformants selected on synthetic complete dropout medium lacking leucine and tryptophan. β-Galactosidase colony-lift filter assay and liquid assay were performed according to the CLONTECH manual.
Production of Recombinant Pcf11p and Rna15p.
The PCF11 and RNA15 genes were cloned in pET-32b(+) and pET-22b(+) vectors (Novagen), respectively. The His-tagged proteins were overproduced in E. coli BL21(DE3) and purified by Ni2+ affinity chromatography according to the Novagen technical manual. Rna15p was purified directly from the soluble bacterial lysate, and Pcf11p was first solubilized from inclusion bodies and refolded with the Protein refolding kit (Novagen).
Transcription Run-on Analysis.
Transcription run-on experiments were performed as described (29).
Results
The CF IA Complex Contained in Yeast Extracts Binds to the CTD.
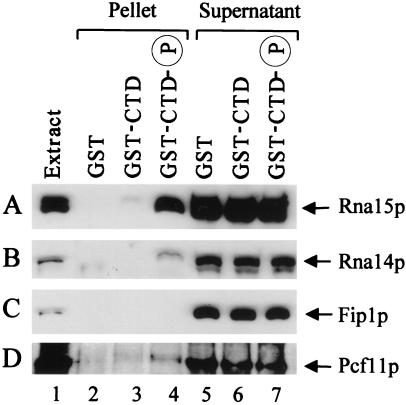
We first investigated the potential interaction of the CF IA complex with the CTD. Affinity chromatography with yeast whole-cell extracts and a GST-CTD column was carried out. A GST-CTD fusion protein and GST control were overproduced in E. coli, purified, and immobilized onto glutathione Sepharose 4B. GST-CTD was further phosphorylated with casein kinase I (New England Biolabs); the phosphorylation of the CTD by this enzyme has previously been shown to be sufficient to recruit capping activities (32). Yeast processing-competent extracts were loaded onto the GST, GST-CTD, and phosphorylated GST-CTD affinity columns, and bound proteins were subsequently eluted with SDS sample buffer. The eluates were then analyzed by SDS/PAGE followed by Western blotting. With this approach we detected a specific interaction of CF IA with the CTD, based on the presence of the Rna15p (Fig. 1A, lanes 3 and 4) and Rna14p (Fig. 1B, lane 4) subunits in the eluates. Both proteins were retained on the CTD resins, but not on the column containing GST alone (Fig. 1 A and B, lane 2). The binding is strongly enhanced by CTD phosphorylation; in the absence of phosphorylation, we invariably observe a barely perceptible association (Fig. 1 A and B, lane 3). The difference in signal intensity for Rna15p and Rna14p in the bound fraction reflects the differential strength of the two antibodies, anti-Rna14p being a much weaker antibody (compare also lane 1 in Fig. 1 A and B). For this reason no Rna14p is detectable in the fraction bound to unphosphorylated CTD (Fig. 1B, lane 3). We also checked for the presence of the Clp1p subunit of CF IA in the eluates, but the low affinity of this antibody precluded the detection of this protein in the pellet fraction. However, given the tight association of the subunits of CF IA (17–23), Clp1p is likely to be recruited to the CTD as part of the CF IA complex. The interaction observed was resistant to treatment with RNase, arguing against the possibility of a RNA bridge (data not shown). As a control for the specificity of CF IA interaction, Fip1p, a component of the polyadenylation factor I, was also tested but did not bind to either unphosphorylated or phosphorylated CTD (Fig. 1C, lanes 3 and 4, respectively), suggesting that polyadenylation factor I (complex) does not interact with the CTD. Antibodies against Pcf11p, the fourth component of CF IA, were unavailable to check for the presence of this subunit in the eluates. However, the possible interaction of this protein with the CTD was investigated by a different approach. The temperature-sensitive strain carrying the pcf11-9 allele (which is defective in both 3′-end cleavage and transcription termination) was transformed with pGBD-Pcf11, a plasmid expressing PCF11 fused to Gal4 DNA binding domain at high levels. Transformants were subjected to a double selection, i.e., selective medium (synthetic complete-minus tryptophan) and temperature (30°C). The GAL4-PCF11 fusion gene complements the pcf11-9 mutation restoring growth at restrictive temperature: transformants harboring pGBD-Pcf11 are capable of growth at 37°C, whereas transformants containing the empty vector (pAS2–1) are not (data not shown). A crude yeast extract was prepared with the use of transformants bearing pGBD-Pcf11 and loaded onto the GST, GST-CTD, and phosphorylated GST-CTD columns. Monoclonal antibodies recognizing the Gal4 DNA binding domain were used to analyze the eluates. An interaction of the Pcf11 fusion protein with phosphorylated CTD was detected (Fig. 1D, lane 4): the signal is not strong, perhaps because the Gal4 DNA binding domain restricts the contact of Pcf11p with the CTD through steric hindrance. However, it has been proved to be reproducible in repeated experiments. Once again, as observed before, in the absence of phosphorylation the signal is barely perceptible (Fig. 1D, lane 3).
Figure 1.
CF IA associates with phosphorylated CTD. A whole-cell extract from strain W303-1B was chromatographed onto GST, GST-CTD, and phosphorylated GST-CTD columns. After binding, the beads were pelleted and washed, and bound proteins were detected by immunoblotting with α-Rna15p (A), α-Rna14p (B), and α-Fip1p (C) antibodies. Lane 1, extract (2% of total input); lanes 2–4, pellet fractions (100%); lanes 5–7, supernatant fractions (8%). (D) Affinity chromatography performed with the use of a crude extract from pcf11-9 mutant transformed with pGBD-Pcf11. The protein was detected by α-Gal4 DNA binding domain antibody. Lane 1, extract (10% of total input); lanes 2–4, pellet fractions (60%); lanes 5–7, supernatant fractions (4%).
Pcf11p Contains a CTD-Binding Domain and Interacts Directly with the CTD.
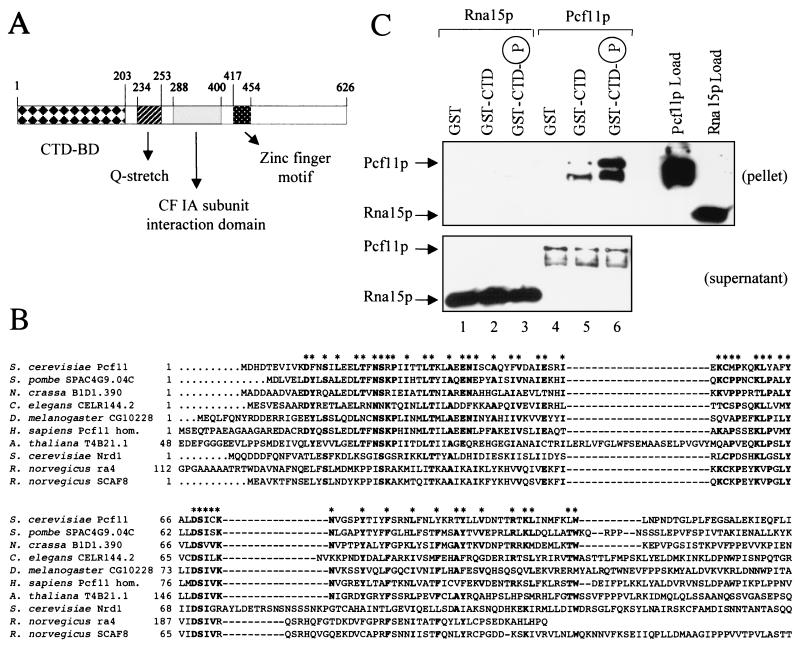
The above experiments used yeast whole-cell extracts to explore the interactions of CF IA with the CTD. These therefore do not discriminate between direct or indirect binding of the individual subunits. If a subunit of CF IA acted as a “bridge” to recruit the whole CF IA complex to the CTD, then Pcf11p would be a likely candidate because it contains a putative CTD-binding domain stretching over the first 203 amino acids of its N terminus (Fig. 2A). Adjacent to this domain is a segment of 20 consecutive glutamine residues (commonly found in transcription factors), followed by the region involved in the interaction with Rna14p and Rna15p (18). The C-terminal domain of the protein exhibits a potential zinc finger motif (amino acids 417–454) followed by an uncharacterized region.
Figure 2.
Pcf11p contains a CTD-binding domain and interacts directly with the CTD. (A) Schematic representation of the domain organization of Pcf11p. The CTD-binding domain is indicated as CTD-BD. (B) Alignment of conserved CTD-binding domains. The N-terminal CTD-binding domain of S. cerevisiae Pcf11p is compared with domains of homologous proteins from S. pombe (SPAC4G9.04C gene product; GenBank accession no. Z69727), Neurospora crassa (B1D1.390 gene product; no. CAB91288), C. elegans (CELR144.2 gene product; no. U23515), Drosophila melanogaster (CG10228 gene product; no. AAF58192), Homo sapiens (Pcf11 homolog; no. AF046935), Arabidopsis thaliana (T4B21.1 gene product; no. AAD03447), S. cerevisiae Nrd1p (no. CAA96158), Rattus norvegicus rA4 (no. U49058), and R. norvegicus SCAF8 (no. U49055). Bold residues indicated with asterisks are identical among at least five of the sequences. In addition to these residues, there are many similarities that are not highlighted. The gaps introduced to maximize the homology are represented by dashed lines. (C) Pcf11p interacts directly with the CTD, whereas Rna15p does not. Recombinant His-tagged Pcf11p and Rna15p were chromatographed on GST, GST-CTD, and phosphorylated GST-CTD resins. The bound fraction was eluted with SDS sample buffer and detected by Western blot with antihistidine monoclonal antibodies. Load (100% of input), pellet (100%), and supernatant (10%) fractions are shown. Lanes 1–3, Rna15p chromatography; lanes 4–6, Pcf11p chromatography. The lower bands detectable in lanes 5 and 6 are degradation products of full-length Pcf11p.
Previous studies on Nrd1 (26) and our own blast searches have shown that Pcf11p shares homology with a number of polypeptides, all possessing a conserved amino-terminal CTD-binding domain (Fig. 2B). Some of those are as yet hypothetical gene products, whereas others have been characterized and shown to bind to the CTD. Nrd1 has been reported to associate with the CTD in a yeast two-hybrid assay (7), and recent genetic evidence supports this finding (27). Likewise, a member of the rat SR (serine–arginine-rich)-like CTD-associated factor family, SCAF8 (previously known as rA8) has been shown to bind to the CTD by both genetic and biochemical experiments (7, 28). Whereas the sequence homology shared by Pcf11p with the above two proteins is confined to the CTD-binding domain, extensive similarity with all of the other hypothetical proteins shown in the alignment is exhibited (Fig. 2B). This similarity had already been noted for the Schizosaccharomyces pombe and Caenorhabditis elegans homologues (26).
These observations prompted us to investigate the potential direct interaction of Pcf11p with the CTD. A recombinant histidine-tagged Pcf11p was overproduced in E. coli and purified from solubilized inclusion bodies. The protein was passed over GST, GST-CTD, and phosphorylated GST-CTD columns. Bound fractions were eluted with SDS sample buffer and monitored by immunoblotting with antihistidine monoclonal antibodies. A specific interaction of Pcf11p with the CTD was detected: the protein binds weakly to unphosphorylated CTD (Fig. 2C, lane 5), but with high affinity to the phosphorylated form (Fig. 2C, lane 6). No association with GST alone (Fig. 2C, lane 4) has been observed. This result shows that Pcf11p interacts directly with the CTD and that the binding is strongly stimulated by CTD phosphorylation.
Recombinant Rna15p was also examined for its association with the CTD. The RNA15 gene was cloned into an overexpression vector, and the histidine-tagged protein was overproduced and purified from E. coli. Rna15p was chromatographed onto GST, GST-CTD, and phosphorylated GST-CTD. In contrast with Pcf11p, Rna15p did not associate at all with either form of CTD and was detected exclusively in the supernatant (not bound) fractions (Fig. 2C, lanes 2 and 3). This result demonstrates that Rna15p does not make direct contact with the CTD, although we cannot exclude the possibility that the recombinant Rna15p lacks posttranslational modifications crucial for establishing contacts with the CTD.
CTD:Rna15p Interaction Detected by Yeast Two-Hybrid Analysis.
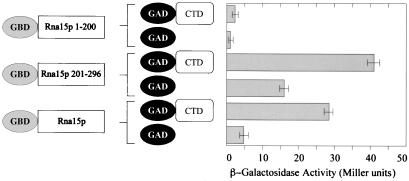
The yeast two-hybrid assay was used to test for in vivo binding of the Rna15p subunit to the CTD. The RNA15 gene was cloned in fusion with the Gal4 DNA binding domain (bait plasmid), and the CTD encoding fragment was cloned in fusion with the Gal4 activation domain (prey plasmid). The bait plasmid was cotransformed into yeast cells together with the prey, and the transformants were assayed for the production of β-galactosidase. We were able to detect an interaction between Rna15p and CTD: the association was detected by filter assay within 2 h of incubation at 30°C. Rna15p has been reported to be able to activate transcription per se in yeast cells (18): we also observed this self-activation of transcription, when the Rna15 bait plasmid was transformed on its own together with the empty prey vector. However, in this case the colonies on the filter developed a pale color only after several hours of incubation at 30°C. To obtain a quantitative measurement of the interaction, we carried out liquid β-galactosidase activity assay in replicates (Fig. 3): a modest, but significant, activity was reproducibly observed, confirming an interaction between Rna15p and the CTD; the background transcription triggered by Rna15p on its own was 5- to 6-fold lower.
Figure 3.
Two-hybrid analysis for Rna15p. Quantitative liquid β-galactosidase assays for the interaction between the CTD and Rna15p (full-length), Rna15p (), and Rna15p () were performed in duplicate for at least two transformants. Mean values are shown in the graph.
To define more precisely the Rna15p region involved in the CTD interaction, we constructed clones comprising either the N-terminal 200 aa or the C-terminal 201–296 aa of Rna15p. The N terminus contains the RNA-binding motif of Rna15p, whereas no function has yet been ascribed to the C terminus. Both regions were fused in-frame with the Gal4 DNA binding domain and assayed in the two-hybrid system for an interaction with the CTD in parallel with the full-length protein. The results obtained by liquid β-galactosidase assay are shown in Fig. 3: the C-terminal domain of Rna15p interacts with the CTD, whereas the N-terminal domain does not. We therefore conclude that the Rna15p N terminus, which contains the RNA binding motif, is dispensable for the CTD interaction, whereas the C terminus plays a role in linking the two proteins together.
This positive result obtained for Rna15p is in apparent conflict with the lack of interaction observed in the pull-down experiments with recombinant Rna15p. A way to reconcile the two findings is the following: the yeast two-hybrid assay does not discriminate between a direct association and an indirect one involving one or more endogenous partners present in yeast cells; therefore Rna15p may contact the CTD through another protein (possibly another subunit of CF IA), and that would explain why the recombinant Rna15p does not bind to the CTD in the affinity chromatography experiment. This explanation would also account for the detection of the Rna15p subunit in the pellet fraction of the affinity chromatography performed with yeast whole-cell extract.
Mutations in PCF11 and RNA14 Affect Transcription Termination More Drastically Than Mutations in RNA15.
Previous studies in our laboratory established the coupling of transcription termination to 3′-end mRNA processing by analyzing strains carrying ts mutations in genes encoding cleavage and polyadenylation factors (29). Transcription run-on analysis was used to measure transcription termination at the 3′ end of the CYC1 gene. The transcription termination profile of the strains carrying mutations in PCF11, RNA14, and RNA15 was found to be severely disrupted with substantial levels of run-on transcripts detected beyond the CYC1 polyadenylation site. In contrast, factors involved in polyadenylation (Fip1p, Pap1p, Yth1p) did not appear to be necessary for termination.
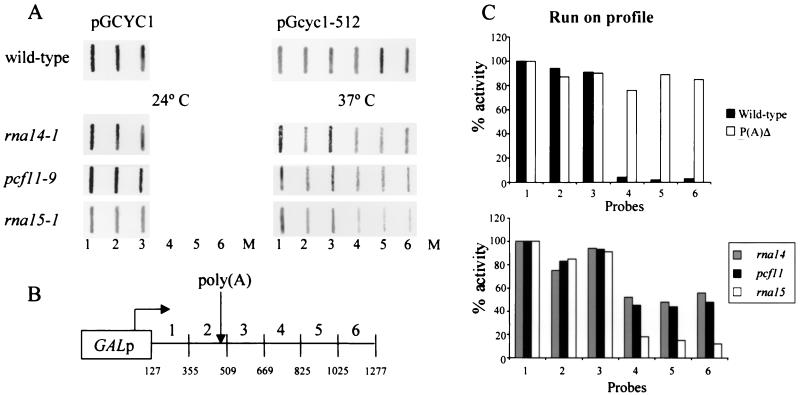
In the light of our CTD binding data, a pattern appeared to emerge: the factors involved in transcription termination were also associated with the CTD of pol II. In contrast, Fip1p, the only polyadenylation factor we tested for CTD interaction, was not involved in termination and was not associated with the CTD. This correlation prompted us to repeat the run-on experiments and to quantify the impact of each cleavage factor (Rna15p, Rna14p, and Pcf11p) on transcription termination. The ts strains carrying rna14–1, pcf11–9, and rna15–1 alleles, as well as the wild-type W303-1B, were transformed with plasmid pGCYC1, in which the CYC1 gene is under the control of the GAL1/10 promoter. As a control, the wild type was also transformed with plasmid pGcyc1-512, which contains a 38-bp deletion in the CYC1 poly(A) site known to disrupt transcription termination (33). The mutant strains were analyzed at permissive (24°C) and restrictive (37°C) temperatures. The distribution of run-on transcripts over the contiguous single-stranded probes 1–6 is shown in Fig. 4A. Consistent with the results obtained by Birse et al. (29), the mutant strains display no defect in permissive conditions, whereas they are severely affected with regard to transcription termination at restrictive temperature with conspicuous run-on transcript detected past the poly(A) site over probes 4, 5, and 6.
Figure 4.
Transcription termination in pcf11 and rna14 mutants is more severely disrupted than in rna15. (A) Run-on analysis performed under permissive conditions (24°C) and after a shift to restrictive temperature (37°C) for 30 min in the strains indicated. The numbers at the base of the panels indicate the probes, and M indicates the M13 probe used as a background hybridization control. (B) Schematic diagram of plasmid pGCYC1 showing the arrangement of M13 probes used in the run-on experiments relative to the CYC1 poly(A) site (position 502). (C) Quantitative analysis of run-on profiles. PhosphorImager quantitation was performed by using imagequant software; the transcription levels of probes 1–6 is normalized according to the U content and corrected for the M13 background signal.
A quantitative analysis of these data revealed that the run-on profile of rna14-1 and pcf11-9 is more drastically disrupted than that observed for rna15-1 (Fig. 4C). These results suggest that Rna14p and Pcf11p have a greater impact on transcription termination than the Rna15p subunit. In view of our CTD binding data, these results strengthen the possibility that Rna14p and Pcf11p may be the two subunits of CF IA that directly contact the CTD to recruit the whole complex to pol II.
Discussion
Over the last decade the CTD of pol II has been the focus of remarkable interest because of its proposed role as a factor docking platform. Increasing evidence has documented its involvement in both transcription regulation and processing of the nascent transcript (reviewed in refs. 34–37).
This study presents biochemical and genetic data demonstrating that CF IA physically interacts with the CTD of pol II in the yeast S. cerevisiae. Affinity chromatography experiments with yeast extracts have shown that this complex binds specifically to phosphorylated CTD based on the presence of Rna15p, Rna14p, and Pcf11p in the eluate fraction. In contrast, Fip1p, a polyadenylation factor component of PFI, does not bind. Pull-down experiments with recombinant Pcf11p, which contains a CTD-binding domain at its N terminus, have demonstrated that this protein interacts directly with the CTD, whereas recombinant Rna15p does not. Once again the binding is greatly enhanced by CTD phosphorylation, consistent with what we observed for CF IA in yeast extracts. Furthermore, a quantitative analysis of transcription run-on profiles of strains carrying mutations in the RNA15, RNA14, and PCF11 genes has revealed that mutant Rna14p and Pcf11p subunits display more pronounced termination defects than defective Rna15p.
The recruitment of pre-mRNA processing factors or complexes to pol II specifically through the phosphorylation of CTD has already been observed in the case of the capping (8, 14) and splicing (6, 7, 38) machinery. Both the yeast capping apparatus and the mammalian capping enzyme bind selectively to phosphorylated CTD. Splicing and splicing-related factors have also been found to be associated with hyperphosphorylated pol II. Interestingly, one such SR-like protein known as SCAF8 has been reported to bind exclusively to phosphorylated CTD (28) and shares homology with Pcf11p in its CTD-binding domain (Fig. 2B). Our observation that recruitment of CF IA to the CTD is strongly stimulated by CTD phosphorylation lends further support to the view that the phosphorylation of the CTD acts as a regulatory mechanism in the processing of the nascent transcript. Changes in the net charge of the CTD affects its conformation, allowing the binding or loading of some factors and the dissociation of others.
In contrast with what we observed in yeast for CF IA, the mammalian CPSF and CstF have been reported to bind equally well to the CTD independently of its phosphorylation status (4); and in a reconstituted system containing all of the mammalian cleavage/polyadenylation factors, both hypophosphorylated and hyperphosphorylated pol II (and likewise unphosphorylated and phosphorylated GST-CTD) activate 3′ cleavage (10). These apparently conflicting observations may reflect an evolutionary divergence between lower and higher eukaryotes in the cleavage/polyadenylation apparatus. Even so, the CTD recruitment of cleavage/polyadenylation factors is clearly conserved throughout eukaryotes. Dantonel et al. (5) demonstrated that the mammalian CPSF is loaded onto the preinitiation transcription complex by TFIID. Whether this observation holds true for the yeast 3′-end forming apparatus remains to be discovered. However, the fact that CF IA subunits exhibit a remarkably higher affinity for the phosphorylated form of CTD versus the unphosphorylated suggests that CF IA may not be recruited at the promoter in the pol IIA preinitiation complex, as is CPSF. Rather, CF IA may associate at a later stage with the elongating pol II0 holoenzyme. Further investigations by protein coimmunoprecipitation and chromatin immunoprecipitation assays could shed light on this aspect.
Using pull-down experiments with purified proteins, we have shown that Pcf11p directly interacts with the CTD, whereas Rna15p is likely to make indirect contacts with it. Pcf11p has a multidomain topology exhibiting interesting features, and the CTD-binding domain at its N terminus is particularly intriguing. In a previous study on the yeast Nrd1 protein (26), Pcf11p was found to be part of a group of polypeptides from different organisms, the common motif of which is a CTD-binding domain. Database searches led us to identify members of this growing family: the N. crassa B1D1.390 protein, the D. melanogaster CG10228 protein, the A. thaliana T4B21.1 protein, and the H. sapiens Pcf11p homologue. The presence of such a CTD-binding domain in disparate organisms argues for an evolutionary conservation of CTD-binding proteins throughout eukaryotes.
Previous investigations in our laboratory had unveiled a connection between cleavage and transcription termination (29): ts mutations in PCF11, RNA15, and RNA14 were found to affect transcription termination, whereas a mutation in FIP1 did not. Remarkably, these data exactly parallel our CTD binding results: Pcf11p, Rna15p, and Rna14p but not Fip1p interact with the CTD. Furthermore, a quantitative analysis has shown that the absence of a functional Pcf11p or Rna14p affects transcription termination more drastically than the lack of a functional Rna15p. pcf11 thermosensitive alleles are synthetically lethal with ts alleles of rna14 (18), and it has recently been shown that overproduction of Pcf11p suppresses rna14-1 mutation in an allele-specific manner (39). Such observations taken together suggest that these two proteins may share an overlapping, interchangeable function. The ability of Pcf11p to directly interact with phosphorylated CTD suggests a possible physiological role for this protein. Pcf11p may in fact bind to the hyperphosphorylated pol II and recruit cleavage factors to the elongating complex, thus coupling transcription and mRNA processing. Rna14p may play an analogous role.
Interestingly, exhaustive two-hybrid screens performed in yeast (40, 41) have highlighted further potential connections between polyadenylation and splicing, and between 3′-end processing and chromatin remodeling: Pcf11 has been found to interact with a Sm-like protein known as Lsm8p that may play a role in allowing conformational rearrangements of the U6 small nuclear ribonucleoprotein in the association–dissociation cycle of spliceosome complexes (42). Last, Pcf11p has been reported to interact with Ada2p, a component of the two nucleosomal histone acetyltransferase complexes SAGA and ADA (41). These additional links once again emphasize how the basic cellular processes of chromatin remodeling, transcription, and pre-mRNA processing are tightly interconnected.
Acknowledgments
We thank Lionel Minvielle-Sebastia and Walter Keller for providing antibodies against Rna15p, Rna14p, Clp1p, and Fip1p; Michaela Sharpe for the gift of vectors pAS2–1 and pACT2 and strain Y187; and Finbarr Hayes for continuous support and advice. We are grateful to Shona Murphy, Agustín Aranda, and Finbarr Hayes for helpful comments on the manuscript. This work was supported by a program grant (032773) from the Wellcome Trust (to N.J.P.).
Abbreviations
- CTD
carboxyl-terminal domain
- pol II
RNA polymerase II
- CF
cleavage factor
- CPSF
cleavage polyadenylation specificity factor
- CstF
cleavage stimulation factor
- PF
polyadenylation factor
- SR
serine–arginine-rich
- SCAF
SR-like CTD-associated factor
- GST
glutathione S-transferase
- ts
temperature-sensitive
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.021545298.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.021545298
References
- 1.Allison L A, Moyle M, Shales M, Ingles C J. Cell. 1985;42:599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- 2.Corden J L, Cadena D L, Ahearn J M, Dahmus M E. Proc Natl Acad Sci USA. 1985;82:7934–7938. doi: 10.1073/pnas.82.23.7934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dahmus M E. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- 4.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson S D, Wickens M, Bentley D L. Nature (London) 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 5.Dantonel J-C, Murthy K G K, Manley J L, Tora L. Nature (London) 1997;389:399–402. doi: 10.1038/38763. [DOI] [PubMed] [Google Scholar]
- 6.Mortillaro M J, Blencowe B J, Wei X, Nakayasu H, Du L, Warren S L, Sharp P A, Berezney R. Proc Natl Acad Sci USA. 1996;93:8253–8257. doi: 10.1073/pnas.93.16.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuryev A, Patturajan M, Litingtung Y, Joshi R V, Gentile C, Gebara M, Corden J L. Proc Natl Acad Sci USA. 1996;93:6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, Bentley D L Amgen EST Program. Genes Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho C K, Sriskanda V, McCracken S, Bentley D, Schwer B, Shuman S. J Biol Chem. 1998;273:9577–9585. doi: 10.1074/jbc.273.16.9577. [DOI] [PubMed] [Google Scholar]
- 10.Hirose Y, Manley J L. Nature (London) 1998;395:93–96. doi: 10.1038/25786. [DOI] [PubMed] [Google Scholar]
- 11.Thompson C M, Koleske A J, Chao D M, Young R. Cell. 1993;73:1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- 12.Myers L C, Gustafsson C M, Bushnell D A, Lui M, Erdjument-Bromage H, Tempst P, Kornberg R D. Genes Dev. 1998;12:45–54. doi: 10.1101/gad.12.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Otero G, Fellows J, Li Y, de Bizemont T, Dirac A M G, Gustafsson C M, Erdjument-Bromage H, Tempst P, Svejstrup J Q. Mol Cell. 1999;3:109–118. doi: 10.1016/s1097-2765(00)80179-3. [DOI] [PubMed] [Google Scholar]
- 14.Cho E J, Takagi T, Moore C R, Buratowski S. Genes Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morris D P, Phatnanai H P, Greenleaf A L. J Biol Chem. 1999;274:31583–31587. doi: 10.1074/jbc.274.44.31583. [DOI] [PubMed] [Google Scholar]
- 16.Wu X, Wilcox C B, Devasahayam G, Hackett R L, Arévalo-Rodríguez M, Cardenas M E, Heitman J, Hanes S D. EMBO J. 2000;19:3727–3738. doi: 10.1093/emboj/19.14.3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kessler M M, Zhao J, Moore C L. J Biol Chem. 1996;271:27167–27175. doi: 10.1074/jbc.271.43.27167. [DOI] [PubMed] [Google Scholar]
- 18.Amrani N, Minet M, Wyers F, Dufour M E, Aggerbeck L P, Lacroute F. Mol Cell Biol. 1997;17:1102–1109. doi: 10.1128/mcb.17.3.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amrani N, Minet M, Le Gouar M, Lacroute F, Wyers F. Mol Cell Biol. 1997;17:3694–3701. doi: 10.1128/mcb.17.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minvielle-Sebastia L, Preker P J, Wiederkehr T, Strahm Y, Keller W. Proc Natl Acad Sci USA. 1997;94:7897–7902. doi: 10.1073/pnas.94.15.7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Preker P J, Ohnacker M, Minvielle-Sebastia L, Keller W. EMBO J. 1997;16:4727–4737. doi: 10.1093/emboj/16.15.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao J, Kessler M M, Moore C L. J Biol Chem. 1997;272:10831–10838. doi: 10.1074/jbc.272.16.10831. [DOI] [PubMed] [Google Scholar]
- 23.Minvielle-Sebastia L, Preker P J, Keller W. Science. 1994;266:1702–1705. doi: 10.1126/science.7992054. [DOI] [PubMed] [Google Scholar]
- 24.Takagaki Y, Manley J L. Nature (London) 1994;372:471–474. doi: 10.1038/372471a0. [DOI] [PubMed] [Google Scholar]
- 25.Takagaki Y, Manley J L. Mol Cell Biol. 1997;17:3907–3914. doi: 10.1128/mcb.17.7.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steinmetz E J, Brow A D. Mol Cell Biol. 1996;16:6993–7003. doi: 10.1128/mcb.16.12.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conrad N K, Wilson S M, Steinmetz E J, Patturajan M, Brow D A, Swanson M S, Corden J L. Genetics. 2000;154:557–571. doi: 10.1093/genetics/154.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patturajan M, Wei X, Berezney R, Corden J L. Mol Cell Biol. 1998;18:2406–2415. doi: 10.1128/mcb.18.4.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Birse C E, Minvielle-Sebastia L, Lee B A, Keller W, Proudfoot N J. Science. 1998;280:298–301. doi: 10.1126/science.280.5361.298. [DOI] [PubMed] [Google Scholar]
- 30.Peterson S R, Dvir A, Anderson C W, Dynan S W. Genes Dev. 1992;6:426–438. doi: 10.1101/gad.6.3.426. [DOI] [PubMed] [Google Scholar]
- 31.Zhao J, Kessler M, Helmling S, O'Connor J P, Moore C. Mol Cell Biol. 1999;19:7733–7740. doi: 10.1128/mcb.19.11.7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez C R, Cho E J, Keogh M C, Moore C L, Greenleaf A L, Buratowski S. Mol Cell Biol. 2000;20:104–112. doi: 10.1128/mcb.20.1.104-112.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zaret K S, Sherman F. Cell. 1982;28:563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]
- 34.Carlson M. Annu Rev Cell Dev Biol. 1997;13:1–23. doi: 10.1146/annurev.cellbio.13.1.1. [DOI] [PubMed] [Google Scholar]
- 35.Neugebauer K M, Roth M B. Genes Dev. 1997;11:3279–3285. doi: 10.1101/gad.11.24.3279. [DOI] [PubMed] [Google Scholar]
- 36.Bentley D. Curr Opin Cell Biol. 1999;11:347–351. doi: 10.1016/S0955-0674(99)80048-9. [DOI] [PubMed] [Google Scholar]
- 37.Proudfoot N J. Trends Biochem Sci. 2000;25:290–293. doi: 10.1016/s0968-0004(00)01591-7. [DOI] [PubMed] [Google Scholar]
- 38.Kim E, Du L, Bregman D B, Warren S L. J Cell Biol. 1997;136:19–28. doi: 10.1083/jcb.136.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rouillard J M, Brendolise C, Lacroute F. Mol Gen Genet. 2000;262:1103–1112. doi: 10.1007/pl00008653. [DOI] [PubMed] [Google Scholar]
- 40.Fromont-Racine M, Rain J-C, Legrain P. Nat Genet. 1997;16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- 41.Uetz P, Giot L, Cagney G, Mansfield T A, Judson R S, Knight J R, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. Nature (London) 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 42.Mayes A E, Verdone L, Legrain P, Beggs J D. EMBO J. 1999;18:4321–4331. doi: 10.1093/emboj/18.15.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]