Abstract
Transferred DNA (T-DNA) transfer from Agrobacterium tumefaciens into eukaryotic cells is the only known example of interkingdom DNA transfer. T-DNA is a single-stranded segment of Agrobacterium's tumor-inducing plasmid that enters the plant cell as a complex with the bacterial virulence proteins VirD2 and VirE2. The VirE2 protein is highly induced on contact of A. tumefaciens with a plant host and has been reported to act in late steps of transfer. One of its previously demonstrated functions is binding to the single-stranded (ss) T-DNA and protecting it from degradation. Recent experiments suggest other functions of the protein. A combination of planar lipid bilayer experiments, vesicle swelling assays, and DNA transport experiments demonstrated that VirE2 can insert itself into artificial membranes and form channels. These channels are voltage gated, anion selective, and single-stranded DNA-specific and can facilitate the efficient transport of single-stranded DNA through membranes. These experiments demonstrate a VirE2 function as a transmembrane DNA transporter, which could have applications in gene delivery systems.
Agrobacterium induces neoplastic growth in many plant species by transporting a single-stranded version of the transferred DNA (T-DNA) into the plant nucleus, where it integrates into the genome (1–3). This transfer process requires the activities of tumor-inducing plasmid-encoded virulence (Vir) proteins. VirA and VirG sense phenolic compounds released from wounded plants and induce expression of the other vir genes, respectively. VirD2, with the help of VirD1, recognizes and cleaves within two 25-bp DNA sequences (called border sequences) that delineate the T-DNA on the tumor-inducing plasmid. On cleavage, VirD2 binds covalently to the 5′ end of the single-stranded T-DNA, forming the T-DNA complex. The VirE2 protein binds to single-stranded DNA (ssDNA) in a sequence-nonspecific manner. Through this activity, it can protect the T-DNA complex from nucleolytic attack (4–6). VirD2 and VirE2 proteins are, together with ss-T-DNA, the only known essential virulence components that must be exported from the bacterial cell. The VirE1 protein, a small VirE2-specific molecular chaperone, stabilizes VirE2, prevents it from self-aggregating, and maintains it in an export-competent state in Agrobacterium (7, 8). Such a need for a chaperone activity is reminiscent of effector proteins of type III secretion systems that also require specific chaperones before transfer to eukaryotic cells (9). The virB operon products as well as the VirD4 protein are thought to assemble into the type IV secretion that spans the inner and outer membranes of Agrobacterium and are required for export of the T-DNA complex and VirE2 to the plant cells (10–12). VirE2 and VirD2 proteins can also be exported from the bacterial cells lacking the VirB apparatus (13). The mechanism driving the T-DNA complex across the plant plasma membrane remains elusive; however, once in the plant cytoplasm, VirD2 pilots the T-DNA complex through the nuclear pore. Finally, the T-DNA integrates into the plant genome with the help of plant enzymes (2, 14).
The ssDNA-binding protein VirE2 is thought to act at a late step of T-DNA transfer, inside the plant cell. Extracellular complementation experiments showed that coinoculation of plants with a combination of Agrobacterium strains that are individually avirulent (one lacking the VirE2 protein and the other lacking the T-DNA) resulted in wild-type levels of transfer (15). Infection of VirE2-expressing tobacco plants with VirE2 mutant bacteria also resulted in full complementation (16). Although the ssDNA-binding activity of VirE2 is essential for DNA transfer, VirE2 may have additional functions that are absolutely required for transfer but are independent of the DNA-binding activity of the protein; transgenic plants expressing the bacterial ssDNA-binding protein RecA do not complement VirE2 mutants of Agrobacterium, although the RecA protein can substitute for VirE2 function in nuclear import of the T-DNA complex (A. Ziemienowicz, personal communication).
Although VirE2 protein was found primarily in the cytoplasmic fraction of induced Agrobacterium (presumably because of the presence of the VirE1 chaperone), a small but significant proportion of VirE2 was also present in the inner membrane fraction, and a low level was detected in both the outer membrane and periplasmic fraction (17). To find out if membrane association of VirE2 has any functional significance, we used biophysical methods. Monolayer and bilayer experiments showed that VirE2 indeed can interact with lipids and can form large channels. These channels are anion selective and specific for ssDNA. Vesicle swelling and transport experiments showed that VirE2 can facilitate efficient diffusion of ssDNA through a lipid bilayer. We incorporate these new data into a model according to which VirE2 is the channel mediating uptake of the T-DNA complex by the plant cell.
Materials and Methods
Cloning of VirE2-His6 in the Expression Vector pET3a.
Plasmid pSW108 (18) contains an XhoI fragment from the tumor-inducing plasmid pTiA6 encompassing the virE operon promoter and the open reading frames of VirE1 and VirE2. A StuI–SmaI fragment from plasmid pSW108 containing the coding region of virE2 was inserted in the HincII site of pUC18, resulting in plasmid pUCSS. An NdeI restriction site was introduced at the level of the first ATG of VirE2 by PCR, by using primer p5 (5′-ATCGTAGCCTGCAGAGTCATATGGATCTTTCTGGCAATGAGAAATCC-3′), primer p6 (5′-GTTTGATAAAAGATCTCTGTGCC-3′), and plasmid pSW108 as a template. The amplified PCR product was cut with PstI and BglII and inserted into pUCSS previously cut with the same restriction enzymes, resulting in plasmid pUCE2. An NdeI–BamHI fragment from plasmid pUCE2 was inserted into plasmid pET3a (19) previously digested with the same enzymes, resulting in plasmid pETE2. An His tag and a PstI site were introduced at the 3′ end of virE2 by PCR, by using the primers 5′-AAGACGTCCTCAGTGATGGTGATGTGATG-3′ and 5′-TATCTGGAATCCTGGGAACG-3′. The PCR was performed at 50°C with Taq polymerase (Qiagen, Chatsworth, CA) for 10 cycles with 300 ng of plasmid and 4 min of extension. The amplified PCR product was cleaved with SacI and PstI and inserted into pETE2 cleaved with the same restriction enzymes, resulting in pETE2-His6.
Purification of the VirE2-His6 Protein.
pETE2-His6 was used to transform Escherichia coli BL21 (DE3) strain (Stratagene). Cells were grown to OD600 0.6, and virE2 expression was induced with 1 mM isopropyl β-d-thiogalactopyranoside for 4 h at 28°C. Cells were harvested, and the recombinant protein was purified under native conditions, by using Ni-nitrilotriacetic acid agarose (Qiagen). Specifically, the pellet was resuspended in lysis buffer (50 mM NaH2PO4, pH 8/300 mM NaCl/20 mM imidazole/1 mg/ml lysozyme), incubated on ice for 30 min, sonicated, and centrifuged at 18,000 × g, and the supernatant was loaded onto a Ni-nitrilotriacetic acid column. The column was washed with 10 volumes of washing buffer (50 mM NaH2PO4, pH 8/300 mM NaCl/50 mM imidazole), and the protein was eluted with elution buffer (50 mM NaH2PO4, pH 8/300 mM NaCl/250 mM imidazole). The fractions containing the protein were pooled and dialyzed overnight against 50 mM NaH2PO4 (pH 8), 300 mM NaCl. The dialyzed protein was centrifuged at 18,000 × g for 20 min; the supernatant was divided into aliquots, flash frozen, and kept at −80°C.
Alternatively, the VirE2 protein was purified under denaturing conditions, by using the Ni-nitrilotriacetic acid purification procedure. The pellet was resuspended in lysis buffer (4 M urea/50 mM NaH2PO4, pH 8/300 mM NaCl/20 mM imidazole), sonicated, and centrifuged at 18,000 × g. The supernatant was loaded onto a Ni-nitrilotriacetic acid column, and a gradient from 4 M urea to 0 M urea in the same buffer over a period of 90 min was run to renature the protein on the column. The subsequent washing, elution, and dialysis steps were similar to the protocol for the purification of native VirE2. The two VirE2 preparations were at least 95% pure and yielded similar experimental results, supporting the argument that all effects observed were indeed VirE2 specific and did not result from the presence of a contaminating activity copurifying with VirE2 in one of the protocols.
Langmuir Trough Experiments.
The procedure used is that of Schwarz and Taylor (20), whereby the VirE2 protein was added with a calibrated microsyringe directly to the subphase composed of a 50 mM NaH2PO4 (pH 8.0), 300 mM NaCl buffer; measurements were started 30 min after injection. The resulting compression isotherm curves of 1-palmitoyl-2-oleoyl-phosphatidylcholine were recorded in the absence or presence of different amounts of purified His-tagged VirE2.
Bilayer Experiments.
Measurements by using black lipid bilayers and current fluctuation assays were performed as described previously (21) in a 10 mM Tris, 1 mM CaCl2, 1 M KCl (pH 7.4) buffer at 20°C. Briefly, VirE2 was reconstituted into black lipid membranes, and ssDNA was added gradually to either side of the membrane. ssDNA entering the channel is expected to lead to a decrease in the current in a concentration-dependent manner.
Assays of Vesicle Swelling and Import of DNA into Vesicles.
Vesicles were prepared according to the method of Luckey and Nikaido (22). Briefly, 2 μmol of soybean phospholipids was dried as a thin film at the bottom of a tube and resuspended by vortex mixing in 600 μl of 10 mM Tris, 150 mM NaCl buffer (pH 7.4) and 5 μg purified VirE2 (or lacking VirE2 in the control sample). Samples were then relaxed for 1 h at 4°C.
For swelling assays, 30 μl of the vesicle suspension was diluted into 600 μl of the same buffer containing or lacking ssDNA, and swelling was followed by measuring OD at 500 nm.
For transport experiments 3 μM 5′ end-labeled [32P]ssDNA was added to 600 μl of the vesicles prepared with 5% negatively charged egg phosphatidyl glycerol to decrease nonspecific DNA binding to lipids. At different time points after the addition of the DNA, 75 μl of the suspension was filtered through a Whatman GF-C filter and washed with 5 ml buffer. The filters were dried for 10 min at 60°C, and the bound radioactive signal was measured by using a scintillation counter.
Results
VirE2 Protein Interacts with Lipids.
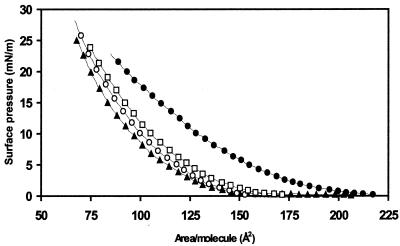
To determine whether VirE2 interacts with lipids, Langmuir trough experiments (20) were performed, and the resulting compression isotherm curves of 1-palmitoyl-2-oleoyl-phosphatidylcholine were recorded in the absence or presence of different amounts of purified His-tagged VirE2 (Fig. 1). The addition of VirE2 to the subphase of the monolayer film induced a significant shift toward larger effective area per lipid molecule in the compression isotherms. This shift can be explained either by insertion of VirE2 into the monolayer or by an absorption process modifying the lipid organization at the air–water interface.
Figure 1.
Compression isotherms of 1-palmitoyl-2-oleoyl-phosphatidylcholine monolayers in the presence of different amounts of VirE2 in a subphase composed of a 50 mM NaH2PO4 (pH 8.0), 300 mM NaCl buffer. The different traces correspond to the 1-palmitoyl-2-oleoyl-phosphatidylcholine compression isotherms recorded with 0 nM (▴), 2 nM (○), 5.9 nM (□), and 15.7 nM (●) of VirE2 in the subphase.
VirE2 Forms Large Voltage-Gated Transmembrane Channels.
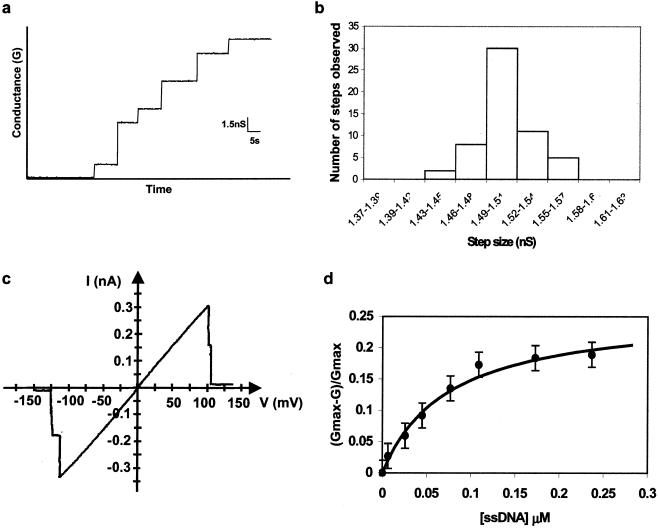
Planar bilayer experiments were performed (21) to test for possible channel formation of inserted VirE2. After formation of a lipid bilayer, purified VirE2 was added to the cis chamber, and an electrical potential difference of 100 mV was applied across the membrane. Discrete conductance steps could be observed, indicating that VirE2 inserted and formed discrete channels in the membrane (Fig. 2a). From the analysis of 56 conductance jumps observed in seven independent experiments, we determined the conductance for a single channel as 1.5 ± 0.15 nS (Fig. 2b). This value is 10 times larger than that measured for the YopB-YopD channel of Yersinia capable of translocating partially unfolded proteins (23). If similar “holes” would also exist in the membrane of plant cells, they may render them very sensitive to external stresses. Planar lipid bilayer and patch-clamp experiments have demonstrated that most membrane channels are not permanently open but can be closed on application of a membrane potential above a certain threshold value (24–26). This phenomenon, known as voltage gating, is also observed with the VirE2 channels. In 100 gating experiments, all channels closed at a transmembrane potential of 120 ± 20 mV (Fig. 2c). Because the plasma membrane potential of plant cells is in the range of 100–250 mV (27, 28), VirE2 channels, if they exist in plant cells, are probably in a closed state in vivo. Regulation of channel opening at the molecular level presents an attractive possibility for the survival of the plant cell.
Figure 2.
Planar lipid bilayer experiments in 10 mM Tris, 1 mM CaCl2, 1 M KCl (pH 7.4) buffer at 20°C. (a) Stepwise increase of conductance on the addition of VirE2 to the bilayer chamber. VirE2 was added to the cis compartment (0.5 μg/ml final concentration), and a 100-mV potential was applied between the electrodes. (b) Statistical distribution of conductance steps normalized to a single step. (c) Current–voltage relationship of two VirE2 channels in a phospholipid bilayer. The applied membrane voltage was changed gradually from −150 mV to +150 mV, by using a voltage ramp. (d) Plot of the relative conductance inhibition (Gmax − G)/Gmax, as a function of the 19-mer ssDNA concentration in the aqueous bulk phase. Gmax corresponds to the conductance measured in the absence of ssDNA, and G is the conductance obtained in the presence of different amounts of ssDNA. The applied voltage was 20 mV.
VirE2 Channels Are Anion-Specific and Interact with ssDNA.
To test for ion selectivity of the VirE2 channel, planar lipid membrane measurements were performed under asymmetrical ionic concentrations (0.1 M and 1 M of KCl in the cis and trans sides of the membrane, respectively). These experiments revealed an anion selectivity of the channels; permeability (PCl−/PK+) calculated according to the Goldman–Hodgkin–Katz equation (29) showed a ratio of 9 of anion over cation influx.
The large diameter of the channel, its anion selectivity, and the possible relevance for the transfer mechanism prompted us to test the channel's ability to interact with ssDNA. A 19-mer DNA oligonucleotide was added to one side of the planar lipid bilayer. The ssDNA was able to partially block the channel in a concentration-dependent manner, decreasing the ion flux across the membrane by approximately 20% (Fig. 2d). In experiments by using a 19-mer oligonucleotide, all channels were saturated at 0.2 μM ssDNA and had a Kd of 0.075 μM. Experiments by using other oligonucleotides (26 nt, 32 nt) or denatured salmon sperm DNA produced similar effects (data not shown), demonstrating that the binding of ssDNA to the channel is neither length nor sequence specific. This ability of the VirE2 pore to interact with ssDNA clearly distinguishes this protein from other channels. For example, PhoE, an anion-selective porin from E. coli (30), did not show any decrease in conductance in the presence of ssDNA (data not shown).
The VirE2 Pore Facilitates Transfer of ssDNA Through the Lipid Bilayer.
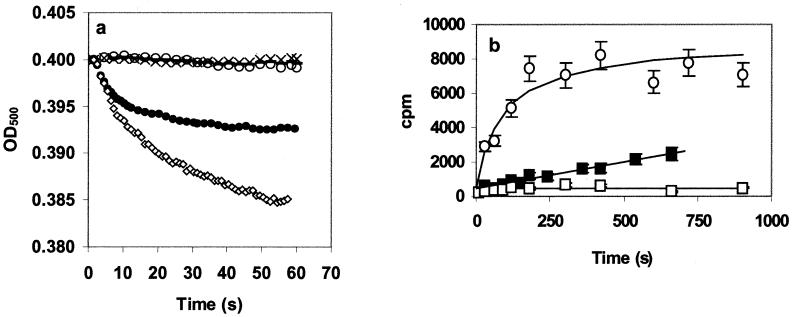
The previous experiments clearly demonstrate an interaction between ssDNA and the VirE2 channel. They do not, however, address the question of whether ssDNA can actually pass through the channel. To investigate this possibility, an osmotic swelling assay was performed (22). In this assay, the rough proteoliposome surface becomes smooth because of the compensatory uptake of water caused by the entry of solute (in this case ssDNA) into the liposome. The swelling of the vesicles results in a decreased light scattering. Vesicles reconstituted in the presence or absence of purified VirE2 protein were added to a ssDNA solution. When VirE2 proteoliposomes were incubated with a 2 μM 19-mer or 8-mer DNA oligonucleotide solution, we observed a decrease in the OD500 indicating, vesicle swelling (Fig. 3a). Swelling rates were faster with shorter oligonucleotides. When liposomes or proteoliposomes were added to plain buffer, no change in the vesicle light scattering was observed. Interestingly, an 18-bp double-stranded DNA could not induce swelling (Fig. 3a). This observation strongly suggests that the observed swelling was indeed induced by a VirE2-facilitated diffusion of ssDNA into the vesicle.
Figure 3.
ssDNA transport experiments. Lipid vesicles were made in 10 mM Tris, 150 mM NaCl (pH 7.4) buffer (22) with and without 5 μg of purified VirE2. (a) Optical density evolution during liposome swelling experiments (22). Thirty microliters of the vesicle suspension was diluted into 600 μl of the same buffer containing or lacking ssDNA, and swelling was followed at 500 nm. VirE2-containing vesicles were diluted in buffer without ssDNA (○); VirE2-containing vesicles were diluted in buffer containing 2 μM 19-mer ssDNA (●), 8-mer ssDNA (◊), or 18-bp double-stranded DNA (−); and liposomes were diluted in buffer containing 2 μM 19-mer (×). (b) [32P]ssDNA transport in proteoliposomes. End-labeled [32P]ssDNA (3 μM) was added to 600 μl of the vesicles. At different time points after the addition of the DNA, 75 μl of the suspension was filtered through a glass microfiber filter and washed with 5 ml buffer. Proteoliposomes were diluted with 19-mer (■) and 8-mer (○), and liposomes were diluted with 19-mer (□). Each curve is the mean of three independent experiments.
To confirm this conclusion, a direct import assay with radiolabeled ssDNA in proteoliposomes was performed. We observed accumulation of the ssDNA only in the vesicles containing VirE2 (Fig. 3b). The combination of vesicle swelling and import experiments demonstrates that the ssDNA enters the vesicles via the VirE2 protein.
Discussion
Several bacteria are known to transport proteins into eukaryotic organisms; examples are bacteria possessing the type III secretion systems (Yersinia spp., Shigella spp., and Xanthomonas spp.; for a review see ref. 9 and the type IV secretion systems (A. tumefaciens, Helicobacter pylori, Legionella pneumophila, Bordetella pertussis, Brucella suis, E. coli, and Rickettsia prowazekii, reviewed in ref. 31). However, apart from Agrobacterium, no other prokaryotic organism is known to (also) transport DNA into the eukaryotic target organism. It is therefore not surprising that Agrobacterium evolved a unique and specialized system to accomplish this task. Data presented above should allow us to better understand one of the steps of T-DNA transfer, the crossing by the T-DNA complex of the plant plasma membrane.
By using biophysical experiments we demonstrated that VirE2 is able to interact with lipids and to form a transmembrane channel with a high conductance value. Vesicle swelling assays showed that the anion-selective VirE2 channel is able to facilitate the transport of ssDNA into liposomes. Swelling rates were faster with shorter oligonucleotides. This observation constitutes an interesting correlation to results obtained with maltooligosaccharides diffusing in maltoporin-containing vesicles where smaller sugars also led to a more pronounced swelling (22). The specificity of the VirE2 channel for ssDNA contrasts with that of another anion-selective channel, PhoE, which did not allow passage of ssDNA through lipid bilayers. The VirE2 channel's specificity for ssDNA is additionally highlighted by the potentially relevant finding that only ssDNA, but not double-stranded DNA, led to vesicle swelling.
The final proof that DNA actually entered the vesicles came from import experiments, in which radiolabeled ssDNA was shown to accumulate inside the vesicles. The VirE2 channel imported a minimum of 80 DNA molecules per added VirE2 molecule, further supporting the actual ssDNA transport. This constitutes a minimal amount of transported DNA molecules because only a fraction of the added VirE2 molecules is expected to have inserted into the vesicle membrane.
If extrapolated to in vivo conditions, a pore allowing passage of ssDNA, a 3.5-Å-diameter molecule, could be deleterious for plant cells. However, VirE2 transgenic plants as well as plants infected with Agrobacterium are viable (16). This potential problem seems to be circumvented by the voltage gating of the VirE2 pore: at 120 mV the pore is closed in vitro. Extrapolated to an in vivo situation, the pore should be closed at the plant-specific membrane potential of 100–250 mV (27, 28).
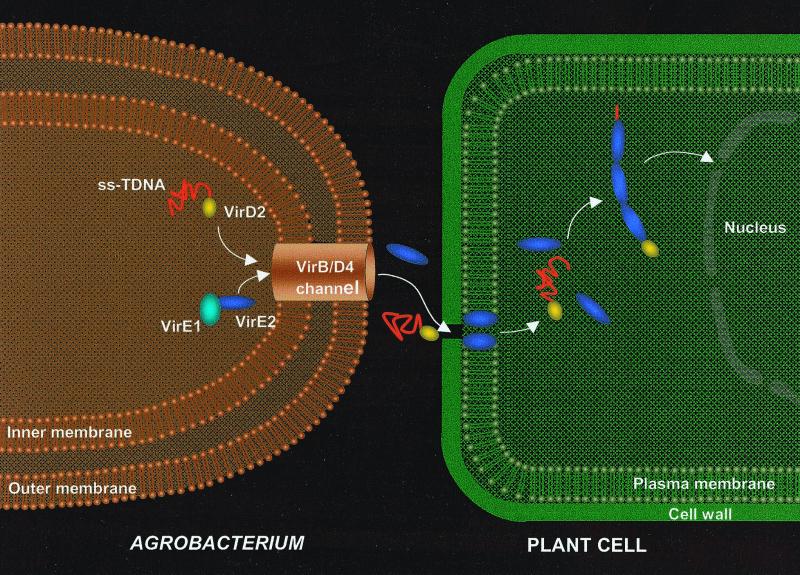
Experimental results reported here invite speculations about the possible biological significance of the findings with respect to T-DNA transfer. The model (Fig. 4) incorporates data from the literature, in combination with biophysical evidence presented above. VirE2 is transported through the VirB-VirD4 channel (10–12) or the alternative route (13) and subsequently inserts into the plant plasma membrane, allowing the transport of the ssDNA–VirD2 complex. VirE2 molecules enter the plant cytoplasm in an unknown way. However, once in the cytoplasm, VirE2 protein molecules coat the DNA, forming a full complex and permitting its transfer to the nucleus (1–3). The proof of the model will require biological validation by in vivo experiments. Nevertheless, it may be helpful in directing research in the field of T-DNA transfer.
Figure 4.
Hypothetical model for T-DNA transfer from the bacteria into the plant cell. In the bacterial cell, the VirE1 chaperone prevents binding of VirE2 to ssDNA, as has been suggested (10–12), and possibly prevents VirE2-dependent channel formation in the bacterial membrane. VirE2 is transported through the VirB-VirD4 channel and subsequently inserts into the plant plasma membrane, allowing the transport of the ssDNA-VirD2 complex. The way in which the VirE2 molecules enter the cytoplasm is unclear. However, once in the cytoplasm, VirE2 protein molecules coat the complex, permitting its transfer to the nucleus (1–3). For simplicity, the pilus and its involvement in T-DNA transfer are omitted from the scheme.
This model also raises a number of intriguing questions. First, Agrobacterium has to prevent VirE2 from inserting into its own membrane if efficient transport to plant membranes is to be guaranteed. VirE1 chaperone is an attractive candidate for this function. In addition to inhibiting VirE2's ssDNA-binding activity (8, 32, 33), VirE1 binding may block the ability of VirE2 to interact with membranes in the bacterial cells.
Once transferred to the plant membrane, a conformational change on transition of VirE2 from the soluble to the membrane-embedded form is expected. Indeed, examples of proteins that can alternate from soluble to membrane-bound have been described previously. α-Hemolysin, for example, is a bacterial toxin of 33 kDa and is water soluble as a monomer, but on oligomerization it inserts into membrane, forming an heptameric transmembrane pore (34).
Another puzzling question concerns the gating properties of the VirE2 pore. The voltage-gating property of the VirE2 pore indicates the possible closing of the pore in plant membranes to maintain the integrity of the plant cell. At the same time, the pore is expected to open on VirD2–T-DNA complex arrival. A variety of factors such as pH, hydrostatic pressure, lipids, or peptides have been shown to modulate the opening/closing of channels (30, 35–37). The factors regulating the opening/closing of the VirE2 pore have yet to be determined. Another question that still remains unsolved is how does a long T-DNA transit the pore in planta? Additional factors are probably required to drive the T-DNA through the pore in vivo. Finally, the mode in which VirE2 ultimately enters the plant cell to perform its cytoplasmic function (1–3) remains to be analyzed.
This is a report of a membrane channel specialized in transporting DNA. The availability of a protein specialized in DNA transport across biological membranes could facilitate the development of cell delivery techniques for DNA or other anionic molecules. These may contribute to the development of ex vivo gene therapy approaches or the optimization of lipid-based drug delivery vehicles. Specifically, the use of VirE2 proteoliposomes may be an elegant alternative for circumventing the problems caused by the respective immunogenicity and toxicity of the viral and cationic/lipid-based delivery systems (38–40).
Acknowledgments
We thank H. Riezman, J. Rosenbusch, J. Hofsteenge, K. D'Hondt, J. Ramsden, and J. Lucht for critical reading of the manuscript. This work was sponsored by Grant 31.036352.92 from the Swiss National Science Foundation. We also acknowledge the financial support of the Novartis Research Foundation to M.D., P.P., and B.H.
Abbreviations
- T-DNA
transferred DNA
- ssDNA
single-stranded DNA
- Vir
virulence
Footnotes
See commentary on page 385.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.011477898.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.011477898
References
- 1.Hansen G, Chilton M D. Curr Top Microbiol Immunol. 1999;240:21–58. doi: 10.1007/978-3-642-60234-4_2. [DOI] [PubMed] [Google Scholar]
- 2.Gelvin S B. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:223–256. doi: 10.1146/annurev.arplant.51.1.223. [DOI] [PubMed] [Google Scholar]
- 3.Rossi L, Tinland B, Hohn B. The Rhizobiaceae. Dordrecht, The Netherlands: Kluwer; 1998. pp. 303–321. [Google Scholar]
- 4.Rossi L, Hohn B, Tinland B. Proc Natl Acad Sci USA. 1996;93:126–130. doi: 10.1073/pnas.93.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sen P, Pazour G J, Anderson D, Das A. J Bacteriol. 1989;171:2573–2580. doi: 10.1128/jb.171.5.2573-2580.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Citovsky V, Wong M L, Zambryski P. Proc Natl Acad Sci USA. 1989;86:1193–1197. doi: 10.1073/pnas.86.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sundberg C, Meek L, Carroll K, Das A, Ream W. J Bacteriol. 1996;178:1207–1212. doi: 10.1128/jb.178.4.1207-1212.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng W, Chen L, Peng W T, Liang X, Sekiguchi S, Gordon M P, Comai L, Nester E W. Mol Microbiol. 1999;31:1795–1807. doi: 10.1046/j.1365-2958.1999.01316.x. [DOI] [PubMed] [Google Scholar]
- 9.Galan J E, Collmer A. Science. 1999;284:1322–1328. doi: 10.1126/science.284.5418.1322. [DOI] [PubMed] [Google Scholar]
- 10.Christie P J. J Bacteriol. 1997;179:3085–3094. doi: 10.1128/jb.179.10.3085-3094.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fullner K J, Lara J C, Nester E W. Science. 1996;273:1107–1109. doi: 10.1126/science.273.5278.1107. [DOI] [PubMed] [Google Scholar]
- 12.Fullner K J. J Bacteriol. 1998;180:430–434. doi: 10.1128/jb.180.2.430-434.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen L, Li C M, Nester E W. Proc Natl Acad Sci USA. 2000;97:7545–7550. doi: 10.1073/pnas.120156997. . (First Published June 13, 2000; 10.1073/pnas.120156997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ziemienowicz A, Tinland B, Bryant J, Gloeckler V, Hohn B. Mol Cell Biol. 2000;20:6317–6322. doi: 10.1128/mcb.20.17.6317-6322.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Otten L, DeGreeve H, J, L, Hain R, J, H J, Schell J. Mol Gen Genet. 1983;175:159–166. [Google Scholar]
- 16.Citovsky V, Zupan J, Warnick D, Zambryski P. Science. 1992;256:1802–1805. doi: 10.1126/science.1615325. [DOI] [PubMed] [Google Scholar]
- 17.Christie P J, Ward J E, Winans S C, Nester E W. J Bacteriol. 1988;170:2659–2667. doi: 10.1128/jb.170.6.2659-2667.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Winans S C, Allenza P, Stachel S E, McBride K E, Nester E W. Nucleic Acids Res. 1987;15:825–837. doi: 10.1093/nar/15.2.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 20.Schwarz G, Taylor S. Langmuir. 1995;180:4341–4349. [Google Scholar]
- 21.Van Gelder P, Dumas F, Rosenbusch J P, Winterhalter M. Eur J Biochem. 2000;267:79–84. doi: 10.1046/j.1432-1327.2000.00960.x. [DOI] [PubMed] [Google Scholar]
- 22.Luckey M, Nikaido H. Proc Natl Acad Sci USA. 1980;77:167–171. doi: 10.1073/pnas.77.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tardy F, Homble F, Neyt C, Wattiez R, Cornelis G R, Ruysschaert J M, Cabiaux V. EMBO J. 1999;18:6793–6799. doi: 10.1093/emboj/18.23.6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bainbridge G, Gokce I, Lakey J H. FEBS Lett. 1998;431:305–308. doi: 10.1016/s0014-5793(98)00761-3. [DOI] [PubMed] [Google Scholar]
- 25.Delcour A H. FEMS Microbiol Lett. 1997;151:115–123. doi: 10.1111/j.1574-6968.1997.tb12558.x. [DOI] [PubMed] [Google Scholar]
- 26.Morgan H, Lonsdale J T, Alder G. Biochim Biophys Acta. 1990;1021:175–181. doi: 10.1016/0005-2736(90)90031-i. [DOI] [PubMed] [Google Scholar]
- 27.Shabala S N, Newman I A. J Membr Biol. 1998;161:45–54. doi: 10.1007/s002329900313. [DOI] [PubMed] [Google Scholar]
- 28.Pouliquin P, Grouzis J, Gibrat R. Biophys J. 1999;76:360–373. doi: 10.1016/s0006-3495(99)77203-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hille B. Ionic Channels of Excitable Membranes. Sunderland, MA: Sinauer; 1980. [Google Scholar]
- 30.Le Dain A C, Hase C C, Tommassen J, Martinac B. EMBO J. 1996;15:3524–3528. [PMC free article] [PubMed] [Google Scholar]
- 31.Covacci A, Telford J L, Del Giudice G, Parsonnet J, Rappuoli R. Science. 1999;284:1328–1333. doi: 10.1126/science.284.5418.1328. [DOI] [PubMed] [Google Scholar]
- 32.Sundberg C D, Ream W. J Bacteriol. 1999;181:6850–6855. doi: 10.1128/jb.181.21.6850-6855.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou X R, Christie P J. J Bacteriol. 1999;181:4342–4352. doi: 10.1128/jb.181.14.4342-4352.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song L, Hobaugh M R, Shustak C, Cheley S, Bayley H, Gouaux J E. Science. 1996;274:1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 35.Todt J C, McGroarty E J. Biochem Biophys Res Commun. 1992;189:1498–1502. doi: 10.1016/0006-291x(92)90244-f. [DOI] [PubMed] [Google Scholar]
- 36.Simon S M, Blobel G. Cell. 1992;69:677–684. doi: 10.1016/0092-8674(92)90231-z. [DOI] [PubMed] [Google Scholar]
- 37.Ishii J, Nakae T. FEBS Lett. 1993;320:251–255. doi: 10.1016/0014-5793(93)80597-n. [DOI] [PubMed] [Google Scholar]
- 38.Crystal R G. Science. 1995;270:404–410. doi: 10.1126/science.270.5235.404. [DOI] [PubMed] [Google Scholar]
- 39.Gao X, Huang L. Biochemistry. 1996;35:1027–1036. doi: 10.1021/bi952436a. [DOI] [PubMed] [Google Scholar]
- 40.Belting M, Petersson P. Biochem J. 1999;342:281–286. [PMC free article] [PubMed] [Google Scholar]