Abstract
Complete resistance (CR) and partial resistance (PR) of rice (Oryza sativa L.) to its bacterial pathogen, Xanthomonas oryzae pv. oryzae (Xoo), was genetically dissected by using 2 mapping populations and 10 Xoo races. Two CR genes, 50 quantitative resistance loci, and 60 digenic interactions were identified, which showed various degrees of race specificity to the Xoo races. The complex epistasis between these loci led us to the discovery of complex genetic networks underlying the rice defensive system to Xoo. The networks consisted of two major components: one representing interactions between alleles at the R loci of rice and alleles at the corresponding avirulence loci of Xoo for CR and the other comprising interactions between quantitative resistance loci in rice and their corresponding aggressiveness loci in Xoo for PR. The race specificity of PR and its strong genetic overlap with CR indicate that PR is essentially “weaker” CR. The genetic networks discovered are expected to maintain a high level of the allelic diversity at avirulent loci in the pathogen by stabilizing selection, which may maintain a high allelic diversity at R loci in the host by the frequency-dependent selection.
Keywords: complete resistance, epistasis, partial resistance, plant–pathogen interaction, rice bacterial blight
The interactions between host plants and their pathogens are characterized with two types of disease resistance, complete resistance (CR) and partial resistance (PR) (1, 2). CR in many plant host–pathogen relationships is hypersensitive, race-specific, and governed by the gene-for-gene system (3, 4). In contrast, PR is quantitative, presumably non-race-specific, and controlled by polygenes (1, 5). Rice (Oryza sativa L.) and its bacterial blight pathogen, Xanthomonas oryzae pv. oryzae (Xoo) are considered a model system to study interactions between host plants and their pathogens. The presence of CR and PR to Xoo in rice has been reported (6, 7). The former is governed by many R genes in a gene-for-gene manner, and the latter is controlled by numerous quantitative resistance loci (QRL) (8). However, the genetic system underlying PR and its relationship with CR in rice remains poorly understood. Here, we report an extensive effort in searching the whole rice genome for R genes for CR and QRL affecting PR in a recombinant inbred line (RIL) population and a doubled haploid line (DHL) population of rice using multiple Xoo races. The results led us to the discovery of complex genetic networks underlying the rice defensive system against Xoo, which shed insights into the coevolution of plants and their pathogens.
Results
Segregation and Race Specificity of the RILs and DHLs in Resistance to Xoo.
The RILs showed two types of segregation in lesion length (LL). When against avirulent Xoo races 1 and 5, the RILs exhibited a bimodal distribution, suggesting involvement of a major R gene(s). When against virulent races 2, 3, 4, and 6, the RILs exhibited an approximately normal distribution with transgressive segregation toward both directions. ANOVA indicated highly significant differences among the genotypes (R2 = 33.9%), the Xoo races (R2 = 23.2%), and the genotype x race interaction (R2 = 38.0%). For virulent races 2, 3, 4, and 6, the genotype x race interaction was more pronounced (R2 = 28.4%), indicating that the PR of rice to Xoo is race-specific.
Similarly, the DHLs exhibited a bimodal distribution against avirulent Xoo races 1, 5, 7, 8, 9, and 10, and an approximately normal distribution against virulent races 2, 3, 4, and 6. Highly significant differences were observed among the genotypes (R2 = 43.7%), among the Xoo races (R2 = 10.2%), and the genotype x race interaction (R2 = 40.1%). Again, the race-specificity of PR to virulent races 2, 3, 4, and 6 was revealed by the highly significant genotype x race interaction in LL (R2 = 21.0%).
Major Genes for CR and PR.
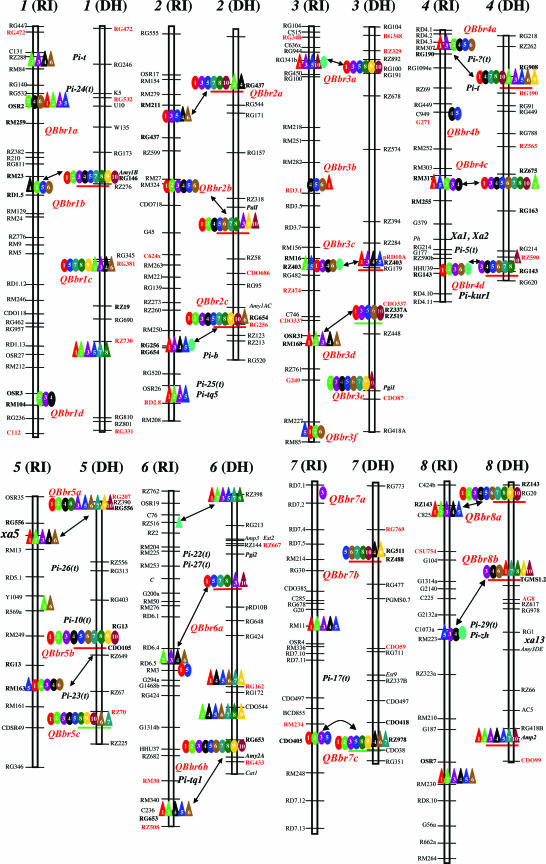
Two major genes for CR were identified. Xa4 mapped to chromosome 11 in both populations (Fig. 1). The indica (cultivars Teqing and IR64) allele at Xa4 had effects of 4.2–6.9 cm for reduced LL against avirulent races and much smaller effects (1.0–3.3 cm) against virulent races 4 and 6, with no detectable effects against virulent races 2, 3, and 9. The Azucena allele at Xa4 acted as a QRL that reduced LL by 0.8–1.5 cm against virulent races 2, 3, and 9 when interacting with three other loci (Table 1, which is published as supporting information on the PNAS web site). A second R gene, Xa25(t) was mapped to chromosome 12 in the DHLs. The Azucena allele at Xa25(t) reduced LL by 4.3 cm against avirulent race 9. The IR64 allele at Xa25(t) acted as a major QRL, with effects ranging from 0.6 to 3.1 cm against virulent races 1–8 (Fig. 1). In the RILs, Xa25(t) was detected as a QRL, and the Teqing allele had effects of 0.7–3.6 cm for reduced LL against all races.
Fig. 1.
Genomic locations of Xa4, Xa25(t), QRL (in red italics), and epistatic loci (triangles) associated with complete and partial resistance to 10 Philippine and 3 Chinese Xoo races detected in the Lemont/Teqing RILs (RI) and IR64/Azucena DHLs (DH). Xa, Pi, and the red-colored markers are major R genes or markers associated with CR to Xoo and rice blast caused by M. grisea detected in the same populations (refs. 12–16 and unpublished data). QRL underlined by red and green lines are closely linked to disease-resistance-gene homologs and defense response genes (16). The double-headed arrows connecting pairwise QRL detected in the RIL and DHL populations indicate cases for which those QRL locate at approximately the same positions based on the common markers in the two populations and the consensus rice linkage map at the Gramene web site (9).
QRL and Their Race Specificity.
In the RILs, 22 QRL were mapped to 12 rice chromosomes (Fig. 1). The Teqing alleles at 14 QRL was associated with resistance, whereas the Lemont alleles at 6 of the QRL resulted in resistance. Alleles from parents at QBbr2a and QBbr4b resulted in resistance, depending on the Xoo races (Table 2 which is published as supporting information on the PNAS web site). On average, each of the QRL was effective against 3.5 races. We detected 31 pairwise interactions between 21 QRL and 17 modifying loci (Fig. 2; see also Table 3, which is published as supporting information on the PNAS web site). On average, each pair of interacting loci was effective against 3.5 races. In 20 cases, the parental type interactions (ITIT/JTJT and ILIL/JLJL) between alleles at two loci resulted in resistance, whereas the recombinant type epistasis (ITIT/JLJL and ILIL/JTJT) conferred resistance in nine cases. In only two cases, either the parental type or recombinant type epistasis resulted in resistance, depending on the Xoo races.
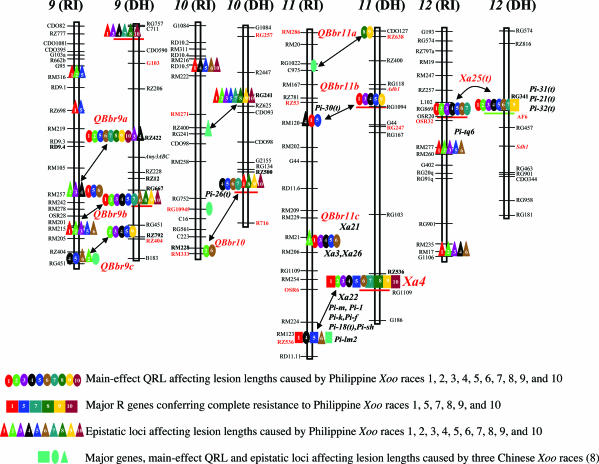
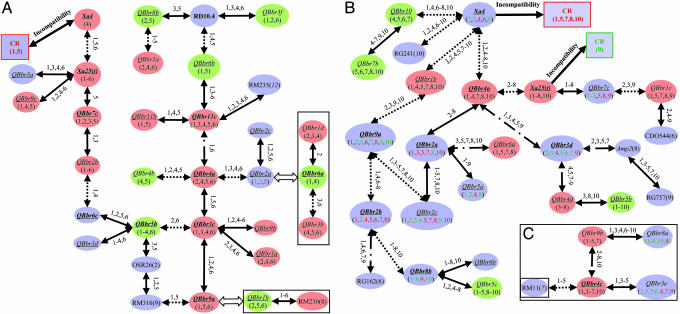
Fig. 2.
Putative genetic networks underlying the defensive system of rice based on the epistatic relationships between R genes and QRL detected. (A) The putative genetic networks underlying CR and PR to six Philippine Xoo races detected in the Lemont/Teqing RILs. (B) The putative genetic networks underlying CR and PR to 10 Philippine Xoo races detected in the IR64/Azucena DHLs. QRL in orange and green represent loci at which the indica (Teqing or IR64) and japonica (Lemont or Azucena) alleles, respectively, resulted in resistance, whereas those in light blue present cases for which resistance resulted from alleles of either parent, depending on Xoo races. Underlined QRL are those detected in both populations and are inferred based on the common markers using the comparative mapping approach (9). The numbers under each QRL represent the Xoo races against which the QRL was effective (Fig. 1). Lines connecting two QRL indicate that significant epistasis was detected between them, and the numbers above the lines are Xoo races against which the epistasis was detected (Tables 3 and 4). Cases for which two interacting loci are connected by solid lines represent the parental type of interaction (the parental genotypes were associated with resistance), those connected by dotted lines represent the recombinant type of interaction (the recombinant genotypes were associated with resistance), and those connected by dotted–dashed lines represent cases for which either the parental or recombinant type interaction was associated with resistance, depending on Xoo races. Rectangular boxes in A and C represent cases for which groups of interacting loci could be included into the networks based on the comparative mapping results in Fig. 1.
In the DHLs, 26 QRL were mapped to 12 rice chromosomes (Fig. 1). The IR64 alleles at 10 QRL were associated with resistance, whereas the Azucena alleles at 6 QRL resulted in resistance. Resistance at the remaining 10 QRL was associated with the IR64 or Azucena alleles, depending on the Xoo races (Table 1). On average, each of the QRL was effective against 6.5 races. A total of 29 highly significant pairwise interactions between 21 QRL and 10 modifying loci were detected in the DHLs (Fig. 2; see also Table 4, which is published as supporting information on the PNAS web site). On average, each pair of interactions was effective against 6.4 Xoo races. Of these, the parental type interactions (IIR64IIR64/JIR64JIR64 and IAZIAZ/JAZJAZ) resulted in resistance in 13 cases, whereas the recombinant type (IIR64IIR64/JAZJAZ and IAZIAZ/JIR64JIR64) epistasis was associated with resistance in 11 cases. In the remaining five cases, either the parental type or recombinant type interactions resulted in resistance, depending on the Xoo races.
The comparative mapping using common markers and the consensus rice linkage map (9) indicated that 28 (54%) of the identified QRL mapped to approximately the same locations across the two mapping populations, including 17 matched QRL and 11 cases where a QRL detected in one population matched a modifying factor in the other (Fig. 1).
Putative Genetic Networks Underlying CR and PR of Rice to Xoo.
Genetically, strong epistasis between two loci would suggest their involvement in the same metabolic pathway. Thus, the epistatic relationships between the QRL allowed us to construct the putative genetic networks underlying the defensive responses of rice to Xoo.
Fig. 2A shows genetic networks detected in the RILs, which involved 27 (66%) of the 41 identified loci and 27 (87%) of the detected interactions. The networks appeared to have two major components. The first component contributed to CR (races 1 and 5) by Xa4. The second component involved 22 QRL and 4 modifying loci that individually and interactively contributed to PR. Five loci (QBbr3c, QBbr4a, QBbr5b, QBbr11c, and QBbr12) appeared to have played important roles in the network by simultaneously interacting with three to five other loci. The remaining four pairs of interacting loci formed three separate groups between QBbr6a and QBbr1d or QBbr3b and between QBbr1b and a modifying factor on chromosome 8, which could be built into the genetic networks indirectly based on the comparative mapping information (Figs. 1 and 2B). The only remaining interaction between QBbr4c and a modifying factor on chromosome 7 could be indirectly integrated into a small group containing five interacting loci (Fig. 2C).
For the DHLs, the epistatic relationships between the QRL resulted in similar genetic networks that involved 25 (69.4%) of the 36 loci and 25 (86.2%) of the interactions detected (Fig. 2B). Similar to Fig. 2A, the qualitative component resulted in CR to races 1, 5, 7, 8, and 10 by Xa4 and to race 9 by Xa25(t). The quantitative component contributed to PR against all Xoo races. Six QRL (QBbr4a, QBbr2b, QBbr2a, QBbr3d, QBbr8a, and QBbr9a) appeared to be important in the networks because of their larger effects and/or interactions with other loci (Fig. 2B). Four of the remaining five interacting loci constituted a small group (Fig. 2C). Only one interaction between two complementary loci (RZ730 and RZ398 on chromosomes 1 and 6) could not be included in the genetic networks based on available information.
Discussion
In this study, we were able to detect the large numbers of QRL and digenic interactions by using multiple differentiating Xoo races. Our threshold for QRL detection was ≈5% at the genome-wide level and the 31 (detected in 103 cases) and 29 (detected in 184 cases) observed significant interactions in the RILs and DHLs represented 0.35% and 0.37%, respectively, of all possible pairwise tests between random marker pairs at logarithm of odds ratio = 3.5 (P = 0.00006), or ≈57 and 63 times as many as expected by chance. Thus, the false positives, if any, should be few in this study.
The most important finding of this study was the discovery of the complex genetic networks that covered 50 (96%) of 52 detected resistance loci and 59 of the 60 detected interactions and uncovered two major genetic components underlying the rice defensive system to Xoo, each of which was apparently controlled by multiple levels of gene-for-gene interactions in the rice–Xoo system and could lead to either CR, PR, or susceptibility. The first component was reflected by interactions between alleles at R loci and alleles at the corresponding avirulence loci in Xoo, leading to CR. When against avirulent races 1, 5, 7, 8, and 10, CR resulted from interactions between the resistance (dominant) alleles at Xa4 in the host and the recessive alleles at avrXa4 in the Xoo races. When against virulent races 2, 3, 4, 6, and 9, the resistance allele from IR64 or Teqing functioned as a QRL. Interestingly, the Azucena allele, Xa4AZ, functioned as QRL against virulent races 2, 3, and 9 epistatically, and the Lemont allele, Xa4L, was associated with susceptibility to all races. At Xa25(t), the Azucena allele resulted in CR against race 9, whereas the IR64 and Teqing alleles acted as QRL conferring PR to all Xoo races, and the Lemont allele was associated with susceptibility to all races. The consistent association of a QRL at either Xa4 or Xa25(t) against virulent races was observed for most dominant R genes in the rice–Xoo relationship (8, 10). The correspondence between major R genes and QRL in genomic locations has been reported in several studies involving rice blast (11–13). Because of the low resolution in QTL mapping, this association between CR genes and QRL for PR could result from differentiated functions of different members in the R gene families (14), from the secondary functions of the same alleles of the R genes that can activate pathways leading to PR, or from both. This uncertainty will be resolved when all rice R genes are cloned in the future.
The second component of the genetic networks was controlled by multiple levels of interactions between QRL in rice and their corresponding aggressiveness loci in Xoo, suggested by various race specificities of the QRL in the networks. Several interesting features of this component were noted. First, all QRL appeared to act in the basal defensive system of rice that inhibited the spread of virulent Xoo races after the onset of disease through a chain reaction of many QRL that acted individually and interactively. Second, there were multiple interrelated and/or independent branches of the networks, suggesting that multiple defensive pathways could be involved. Third, some QRL appeared to be more important. For example, QBbr2b, QBbr5b, and QBbr6b had large and consistent effects against most Xoo races, whereas QBbr4a, QBbr3d, QBbr3c, QBbr11c, and QBbr9b interacted simultaneously with more than three other QRL (Figs. 1 and 2). Those small-effect QRL probably represent the more plastic and superficial components of the basal defensive system. Fourth, there appeared multiple alleles with various functions at many QRL, suggested by the considerable differences among alleles from the four parents in race specificity, epistatic pattern, and magnitude of effects (Tables 1–4). The race specificity of the QRL lends strong support to the notion that there is a gene-for-gene relationship at the polygenic level between rice and Xoo (7). Finally, we noted that 32 (64%) of the detected QRL mapped to approximately the same locations where R genes, defensive candidate genes, or QRL for resistances to Xoo, Magnaporthe grisea (blast), Rhizoctonia solani (sheath blight), and Nilaparvata lugens (brown plant-hopper) were identified in the same populations (Fig. 1) (refs. 12–17 and unpublished data). Thus, the defensive system controlled by the genetic networks may be effective against a wide spectrum of rice pathogens. The race specificity and strong epistasis among different QRL imply that interacting QRL genes would show a coregulated differential expression in response to different Xoo races. For example, based on the strong interaction between Xa25(t) and QBbr7c against virulent races 1–8 resulting in increased resistance (Fig. 2B and Table 4), one can predict a different expression pattern and phenotype of the Xa25(t) gene, depending on the presence or absence of QBbr7c, and vise versa. Thus, the coincidence between the expression pattern of QRL positional candidate genes at multiple loci and their race specificity/epistatic patterns may provide valuable information to identify genes underlying the QRL, which are difficult to clone and characterize using the classical map-based cloning approach.
In conclusion, CR and PR in rice are race-specific, and there is a significant genetic overlap between CR and PR. Thus, PR is essentially “weaker” CR controlled by the complex genetic networks discussed above. The complex genetic networks are expected to maintain a high level of the allelic diversity at avirulence loci in pathogens by stabilizing selection (1, 2), which may maintain a high level of allelic diversity at R loci in the host by the frequency-dependent selection (18). Thus, a full understanding of the genetic networks underlying the plant defensive system requires a genome-wide approach and simultaneous study on a large number of loci involved. In application, a high level and durable resistance can be achieved by pyramiding multiple R genes and QRL from divergent parents by subjecting segregating plant populations to multiple virulent races.
Materials and Methods
Plant Materials and Phenotyping.
Two mapping populations were used in this study. The first population consisted of 292 F13 RILs from a cross between Lemont (japonica) and Teqing (indica) (8), and the second comprised 125 DHLs from a cross between IR64 (indica) and Azucena (japonica) (19).
In the wet season of 1999, the RILs and parents were sown in the seed boxes, and 18-day-old seedlings were transplanted into the screen house at the International Rice Research Institute. Each of the RILs and parents were planted in single row plots with 12 randomly arranged plants per plot, with three replications for each RIL and each parent. All plants were managed under the standard cultural practices. Six Xoo races, PXO611 (race 1), PXO85 (race 2), PXO79 (race 3), PXO71 (race 4), PXO112 (race 5) and PXO99 (race 6) were used for inoculation. Tillers of each plant were divided into two groups, with each group inoculated with two different Xoo races. Four plants in each plot were inoculated at 60 days after transplanting with one of the six Xoo races using the standard leaf clipping method (20). LL on each inoculated leaf was measured 18 days after inoculation. In the wet season of 2001, the DHLs and parents were sown in seed-boxes, and 21-day-old seedlings were transplanted into the screen house. All DHLs and parents were randomly arranged in row plots in three replications, with each DHL represented by 15 plants in one plot. Ten Philippine Xoo races, including races 1–6 (mentioned above) plus PXO145 (race 7), PXO280 (race 8), PXO339 (race 9), and PXO341 (race 10), were used for inoculation. At the booting stage, tillers of three plants in each plot were divided into two groups, with each group inoculated with a single Xoo race using the same method (20). LL was measured 18 days after inoculation on 5 leaves per plant per race in each replication; a total of 45 leaves per DHL were measured for each race.
Genotyping Experiments.
The RILs were assayed with 279 markers, and the resulting map has a total length of 2,001.7 cM, with an average distance of 7.5 cM between adjacent markers (8, 21). For the DHLs, a restriction fragment length polymorphism map was established from an initial population of 135 DHLs, which contains 176 markers and spans a total of 2,003.4 cM, with an average distance of 12.2 cM between markers (19).
Data Analyses.
proc glm (SAS Institute, Cary, NC) (22) was used to test the differences among different genotypes (the parents, RILs, and DHLs), Xoo races, and genotype x race interactions. A computer program based on the mixed linear model approach, qtlmapper 1.0, was used to map QRL and digenic epistasis for resistance using the mean LL and log-transformed LL from three replications and the genotypic data of the RILs and DHLs as input data (23). Data from each race were analyzed separately, and significant QRL and epistatic locus pairs were included in the model to control the background genetic variation. The threshold of logarithm of odds ratio claiming significant QRL was 3.0 for the RILs and 2.7 for the DHLs obtained by using the permutation method (24). The threshold for claiming an epistatic loci pair was logarithm of odds ratio ≥ 3.5 (P = 0.00006).
Supplementary Material
Acknowledgments
We thank D. Mackill and two anonymous reviewers for invaluable comments and suggestions on an early version of the manuscript and B. Hardy for editorial help. This work was supported by Chinese Ministry of Science and Technology Grant 2006CD100201, Chinese Ministry of Agriculture Grant 2004-Z18, and the Rockefeller Foundation.
Abbreviations
- PR
partial resistance
- CR
complete resistance
- Xoo
Xanthomonas oryzae pv. oryzae
- QRL
quantitative resistance loci
- RIL
recombinant inbred line
- DHL
doubled haploid line
- LL
lesion length.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Nelson R. R. J. Environ. Qual. 1972;1:220–227. [Google Scholar]
- 2.Leonard K. J., Czochor R. J. Ann. Rev. Phytopathol. 1980;18:237–258. [Google Scholar]
- 3.Crute I. R. In: Mechanisms of Resistance in Plant Diseases. Fraser R. S. S., editor. Dordrecht, The Netherlands: Nijhoff and Junk; 1986. pp. 80–142. [Google Scholar]
- 4.Hulbert S. H., Webb C. A., Smith S. M., Sun Q. Ann. Rev. Phytopathol. 2001;39:285–312. doi: 10.1146/annurev.phyto.39.1.285. [DOI] [PubMed] [Google Scholar]
- 5.Keen N. T. Annu. Rev. Genet. 1990;24:447–463. doi: 10.1146/annurev.ge.24.120190.002311. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Q., Mew T. W. Plant Dis. 1985;69:896–898. [Google Scholar]
- 7.Parlevliet J. E., Zadoks J. C. Euphytica. 1977;26:5–21. [Google Scholar]
- 8.Li Z., Luo L. J., Mei H. W., Paterson A. H., Zhao X. H., Zhong D. B., Wang D., Wang Y. P., Yu X.Q., Zhu L., et al. Mol. Gen. Genet. 1999;261:58–63. doi: 10.1007/s004380050941. [DOI] [PubMed] [Google Scholar]
- 9.Ware D., Jaiswal P., Ni J., Pan X., Chang K., Clark K., Teytelman L., Schmidt S., Zhao W., Cartinhour S., et al. Nucl. Acids Res. 2002;30:103–105. doi: 10.1093/nar/30.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koch M., Parlevliet J. E. Euphytica. 1991;53:187–193. [Google Scholar]
- 11.Wang G., Mackill D. J., Bonman J. M., McCouch S. R., Champoux M. C. Genetics. 1994;136:1421–1434. doi: 10.1093/genetics/136.4.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tabien R. E., Li Z., Paterson A. H., Marchetti M. A., Park W. D., Stansel J. W., Pinson S. R. M. Theor. Appl. Genet. 2000;101:1215–1225. [Google Scholar]
- 13.Tabien R. E., Li Z., Paterson A. H., Marchetti M. A., Park W. D., Stansel J. W., Pinson S. R. M. Theor. Appl. Genet. 2002;105:313–324. doi: 10.1007/s00122-002-0940-2. [DOI] [PubMed] [Google Scholar]
- 14.Monosi B., Wisser R. J., Pennill L., Hulbert S. H. Theor. Appl. Genet. 2004;109:1434–1447. doi: 10.1007/s00122-004-1758-x. [DOI] [PubMed] [Google Scholar]
- 15.Sallaud C., Lorieux M., Roumen E., Tharreau D., Burruyer R., Svestasrani P., Garsmeur O., Ghesquiere A., Notteghem J. L. Theor. Appl. Genet. 2002;106:794–803. doi: 10.1007/s00122-002-1088-9. [DOI] [PubMed] [Google Scholar]
- 16.Ramalingam J., Vera Cruz C. M., Kukreja K., Chittoor J. M., Wu J. L., Lee S. W., Baraoidan M., George M. L., Cohen M. B., Hulbert S. H., et al. Mol. Plant–Microbe Interact. 2003;16:14–24. doi: 10.1094/MPMI.2003.16.1.14. [DOI] [PubMed] [Google Scholar]
- 17.Vera Cruz C. M., Bai J., Ona I., Leung H., Nelson R. J., Mew T. W., Leach J. Proc. Natl. Acad. Sci. USA. 2000;97:13500–13505. doi: 10.1073/pnas.250271997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brunet J., Mundt C. C. Evolution. 1999;54:406–415. doi: 10.1111/j.0014-3820.2000.tb00043.x. [DOI] [PubMed] [Google Scholar]
- 19.Huang N., Parco A., Mew T., Magpantay G., McCouch S., Guiderdoni E., Xu J., Subudhi P., Angeles E. R., Khush G. S. Mol. Breeding. 1997;3:105–113. [Google Scholar]
- 20.Kauffman H. E., Reddy A. P. K., Hsieh S. P. U., Merca S. D. Plant Dis. Rep. 1973;57:537–541. [Google Scholar]
- 21.Xu J. L., Zhong D. B., Yu S. B., Luo L. J., Khush G. S., Li Z. K. Plant Breeding. 2004;123:43–50. [Google Scholar]
- 22.SAS Institute. SAS Users Guide: Statistics. Cary, NC: SAS Institute; 1996. [Google Scholar]
- 23.Wang D. L., Zhu J., Li Z. K., Paterson A. H. . Theor. Appl. Genet. 1999;99:1255–1264. [Google Scholar]
- 24.Churchill G. A., Doerge R. W. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.