Abstract
cGMP-dependent protein kinases are key intracellular transducers of cell signaling. We identified a novel dominant mutation in the C. elegans egl-4 cGMP-dependent protein kinase (PKG) and show that this mutation causes increased normal gene activity although it is associated with a reduced EGL-4 protein level. Prior phenotypic analyses of this gain-of-function mutant demonstrated a reduced longevity and a reduced feeding behavior when the animals were left unperturbed. We characterize several additional phenotypes caused by increased gene activity of egl-4. These phenotypes include a small body size, reduced locomotion in the presence of food, a pale intestine, increased intestinal fat storage, and a decreased propensity to form dauer larvae. The multiple phenotypes of egl-4 dominant mutants are consistent with an instructive signaling role of PKG to control many aspects of animal physiology. This is among the first reported gain-of-function mutations in this enzyme of central physiological importance. In a genetic screen we have identified extragenic suppressors of this gain-of-function mutant. Thus, this mutant promises to be a useful tool for identifying downstream targets of PKG.
CYCLIC GMP (cGMP) is an important second messenger that regulates diverse cellular processes. The levels of cGMP are controlled by activities of a family of several soluble and membrane-bound guanylate cyclases (Wedel and Garbers 2001) and by the opposing activities of phosphodiestereases (Rybalkin et al. 2003). In contrast to this rich regulation of cGMP levels, known effectors of this second messenger are few. One of these key effectors is the cGMP-dependent protein kinase (PKG). In mammals, there are three main isozymes of PKG, PKGIα, PKG1β, and PKGII, which are encoded by two different genes. PKGIα and PKG1β differ only in their N terminus, a protein domain that is thought to promote dimerization, provide autoinhibition to the catalytic domain, and influence substrate specificity by conferring subcellular localization to the enzyme (Pfeifer et al. 1999). Biochemical and cell culture studies of PKG have identified several potential targets, both direct and indirect for PKG (Pfeifer et al. 1999). However, the physiological relevance of many of these targets has yet to be demonstrated.
The study of the role of PKG in metazoan physiology has been greatly advanced with the use of invertebrate animal models. Levels of PKG activity in Drosophila determine the feeding strategy of fly larvae and adults in natural isolates (Osborne et al. 1997). Higher levels of PKG result in the rover phenotype, where the animal travels far from the food source, whereas low levels result in the sitter phenotype, where the animal remains close to the food source. Loss-of-function mutations in the Caenorhabditis elegans egl-4 locus, which encodes a PKG with greater similarity to mammalian PKG1 than to PKGII (Hirose et al. 2003), has many physiological consequences including a large body size (Daniels et al. 2000; Fujiwara et al. 2002; Hirose et al. 2003), impaired sensory adaptation (L'Etoile et al. 2002), increased locomotion in the presence of food (Fujiwara et al. 2002), defective regulation of egg laying (Trent et al. 1983; Daniels et al. 2000), and an increased propensity for dauer development (Daniels et al. 2000). Although direct targets of PKG have not been identified in invertebrates, genetic studies suggest that the EGL-4 PKG may act through different signaling pathways to control different physiological processes.
To date, most studies of PKG physiology have relied on analysis of the consequences of reduced gene function using either dominant negative constructs in cell culture or gene inactivation in mice, Drosophila, and C. elegans. Such studies would be greatly aided by a mutation that increases gene activity. In this study, we report the molecular and genetic characterization of a novel mutation in the C. elegans egl-4 gene that confers increased gene activity. We use this mutant to study the physiological consequences of activation of PKG signaling in worms.
MATERIALS AND METHODS
General:
The wild-type strain used was variety Bristol, strain N2 (Brenner 1974). Worms were maintained on the surface of NGM agar media. Food source was OP50 (Brenner 1974) and cultivation temperature was 15° for strains with an increased propensity to form dauers and 20° for all other strains.
Strains used:
Strains used were MT1074 egl-4(n479) IV (Trent et al. 1983), DA521 ad450sd IV (Avery 1993), DA1113 eat-2(ad1113) II (Raizen et al. 1995), DR40 daf-1(m40) IV (Riddle 1977), CB1364 daf-4(e1364) III (Riddle 1977), CB1372 daf-7(e1372) III (Riddle 1977), CB1393 daf-8(e1393) I (Riddle et al. 1981), JT195 daf-11(sa195) V (Vowels and Thomas 1994), DR77 daf-14(m77) IV (Riddle et al. 1981), JT6130 daf-21(p678) V (Vowels and Thomas 1994), and JK2958 nT1 [qIs51] (IV; V)/dpy-11(e224) unc-42(e270) V (Siegfried et al. 2004).
Isolation of egl-4 loss-of-function mutations in cis to ad450sd and of extragenic suppressors of ad450sd:
We screened the progeny of 1875 daughters of EMS-mutagenized ad450sd hermaphrodites for worms that were larger or darker than ad450sd worms. We then assessed identified suppressors for linkage to chromosome IV and ad450sd by crossing the suppressor strain with nT1[qIS51]/+ males and then examining nongreen progeny of nT1[qIS51]/sup. If linked, then all nongreen hermaphrodites would be suppressed whereas if unlinked or weakly linked, then some of the nongreen hermaphrodites would not be suppressed. Linked suppressors were then tested for complementation of the egl-4 loss-of-function allele n479 by examining the nongreen non-Dpy progeny of ad450 sup/nT1[qIS51] males mated with n479; dpy-11 hermaphrodites. Using this approach, we identified four loss-of-function egl-4 alleles and three extragenic suppressors.
Genetic cis–trans test:
Males of genotype nT1[qIs51]/+ were crossed by ad450cs80 doubly mutant worms and by ad450sd singly mutant worms. Cross-progeny hermaphrodites of genotype nT1[qIs51]/ad450cs80 and nT11[qIs51]/ad450, respectively, were identified as green worms. To make ad450/ad450cs80 heterozygote animals, ad450sd homozygous males were mated with ad450cs80 hermaphrodites under conditions that favored efficient mating (Hodgkin 1983) and cross-progeny were identified as those that were not egg-laying defective.
Construction of double-mutant strains:
To construct the ad450sd double mutants with the various Daf-c mutants, we made the assumption that ad450sd would not completely suppress the dauer constitutive phenotype of these mutants. From the double-heterozygous daf/+; ad450sd/ nT1[qIs51], we isolated nongreen dauer worms at 25°. We then recovered the double mutant at 15°. In the case of daf-1 mutants, whose Daf-c phenotype is maternally rescued, we used the dark intestine and egg-laying-defective phenotypes to first identify daf-1 homozygotes and then identified ad450sd homozygotes in this background on the basis of their small appearance and segregation of 100% small progeny.
To construct the eat-2(ad1113) II; egl-4(ad45sd) IV double mutant, we picked several cross-progeny individually from a cross between eat-2/+; nT1[qIs51]/+ males and ad450sd hermaphrodites. We identified eat-2 homozygous worms among the progeny of these worms on the basis of a pharyngeal pumping rate <60/min (Raizen et al. 1995). Among these eat-2 homozygous worms, animals that did not carry the nT1 translocation were also homozygous for ad450sd.
Molecular biology and sequence analysis:
First-strand cDNA was reverse transcribed from total RNA isolated from ad450sd mutants using a GIBCO (Grand Island, NY) BRL kit. The EGL-4A coding region was PCR amplified from this cDNA and subjected directly to sequencing. The sequence of the mutated residue in ad450sd was confirmed by sequencing two separate cDNA PCR reactions as well as a genomic PCR fragment spanning the residue. For sequencing of ad450cs80 double mutants, we were unable to obtain a PCR product when using this mutant's cDNA as template so we sequenced all 10 exons and intron–exon boundaries using PCR products amplified from genomic DNA.
Analysis of sequence chromatogram primary data was done using Sequencher 4.2 (Gene Codes, Ann Arbor, MI). The alignment of PKG and PKA proteins shown in Figure 1 was performed using MacVector version 7.2 (Accelrys) and the alignment with the Escherichia coli catabolite gene activator protein (CAP) was taken from Weber et al. (1989).
Figure 1.
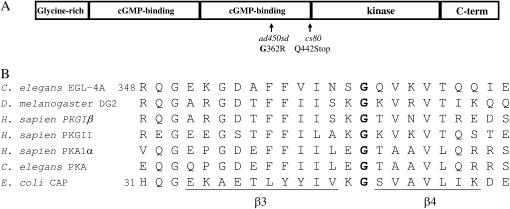
A conserved glycine in PKG is mutated in ad450sd mutants. (A) Domain structure of EGL-4A with the location of ad450sd and cs80, the two mutations discussed in the text, marked. (B) Alignment of the amino acid surrounding the mutated glycine. Included in the alignment are the protein sequence of EGL-4A from C. elegans (Stansberry et al. 2001), dg2 from D. melanogaster (Kalderon and Rubin 1989), type I cGMP-dependent protein kinase from humans (Orstavik et al. 1997), type II cGMP-dependent protein kinase from humans (Orstavik et al. 1996), the catalytic subnit of cAMP-dependent protein kinase from humans and from C. elegans (Gross et al. 1990), and the catabolite activator protein (CAP) from E. coli (Aiba et al. 1982). The amino acids that form the β3 and β4 β-sheets, defined by the crystal structure (Weber and Steitz 1987), are underlined below the sequence of CAP.
Protein level measurement:
Total worm protein was prepared from mixed-stage populations of well-fed worms by sonication followed by centrifugation to remove worm debris. Total protein level was estimated using the Bradford method (Bradford 1976) and 20 μg of total protein was loaded in each lane on the gel. Proteins were separated using SDS–PAGE and transferred to nitrocellulose membranes. Primary antibodies used to probe the blots were the anti-EGL-4 polyclonal antibody described by Hirose et al. (2003) and a commercial anti-tubulin antibody [DM1A from Sigma (St. Louis)]. The EGL-4 antibody detects a doublet protein band at 89 kDa, which is absent in egl-4 null mutants (Hirose et al. 2003) (and data not shown). Detection and quantification of the protein was performed using the Li-Cor Odyssey infrared protein detection system (Li-Cor Biosciences, Lincoln, NE).
Microscopy and photography:
A Spot Insight B/W digital camera (Diagnostic Instruments) mounted on a Zeiss Stemi 2000 stereomicroscope was used to capture the images. Images were stored with 8-bit grayscale resolution. Illumination of the worms was provided by a Fostec DCRIII direct current light source. We found that the use of a direct current light source was far superior to that of an alternating current light source in providing consistent specimen illumination and allowing us to compare grayscale measurements quantitatively.
Body length measurements:
Body length measurements were made on adult worms 2 days (Table 6) or 1 day (all other length data) after the L4 larval stage. Digital images of these adult worms were then subjected to analysis using IPP software (Media Cybernetics, Silver Springs, MD). The spine of the worm was traced using short line segments and the sum of the lengths of these lines was then calculated. The tail of hermaphroditic worms, which forms a gradual taper that is often difficult to perceive, was not included in the measurement. By contrast, male tails, which are short and triangular and therefore easier to measure, were included in length measurements. We attribute the higher coefficient of variation (CV) in measurements from hermaphrodites (CV of seven hermaphrodites was 4.3% whereas CV of seven males was 3.6%) to the imprecision of identifying the back end of hermaphroditic worms.
TABLE 6.
Phenotypic analysis of ad450sd suppressors
| Genotype | Length: mean ± SD (N) | Tracking: mean ± SD (N) | Darkness: mean ± SD (N) |
|---|---|---|---|
| +/ + | 1.00 ± 0.04 (11) | 1.00 ± 0.25 (5) | 1.00 ± 0.09 |
| ad450sd | 0.78 ± 0.02 (15) | 0.11 ± 0.04 (6) | 0.75 ± 0.20 |
| cs82; ad450sd | 0.79 ± 0.05 (19)a | 0.15 ± 0.06 (6)a | 1.08 ± 0.12 |
| ad450sd; cs83 | 0.87 ± 0.07 (13)b | 0.19 ± 0.06 (6)c | 0.89 ± 0.09d |
| ad450sd; cs84 | 0.90 ± 0.02 (11)e | 0.33 ± 0.21 (6)f | 0.90 ± 0.06d |
Measurements shown are relative to wild-type measurements. Two-tailed t-tests were performed to determine statistical significance.
Not significantly different from ad450sd, P > 0.1.
Different from ad450sd, P = 0.001.
Different from ad450sd, P = 0.02.
Different from ad450sd, P < 0.01.
Different from ad450sd, P < 0.000001.
Different from ad450sd, P = 0.05.
Tracking behavior:
The tracking assay was performed essentially as described by Fujiwara et al. (2002). Briefly, a late L4 hermaphrodite was placed in the center on an agar surface of an NGM plate of diameter 10 cm that had a confluent lawn of OP50 bacteria. Seventeen ± 0.5 hr later, the worm was either removed from the plate or immobilized by transferring the plate to 4°. Tracks formed by the worm were analyzed by placing a transparency of a grid that was composed of squares with side dimension of 0.5 cm under the agar plate and counting the number of squares that contained worm tracks. Because of day-to-day variability in the results of the tracking assay, comparison between genotypes was made by performing the test on the same day for all genotypes of interest.
Measurements of intestinal darkness:
To provide a quantitative measure of worm darkness, we photographed hermaphrodites 2 days after reaching the adult stage. All pictures were taken on the same day, on the same agar plate, using the same magnification and luminance, and with the same digital camera exposure time. Analysis of these images was then performed in ImagePro Plus (Media Cybernetics, Silver Springs, MD). The grayscale value of all pixels within a 30–50 × 30–50 μm rectangular area of interest in a body region anterior to the uterus that contained the intestine was averaged for six to eight worms. To include only the intestine in the measurement and to avoid including shadows formed by the worm's body, which blended with the worm and were often darker than the intestine, the borders of the area of interest were kept within the worm's intestine.
With the exception of daf-8 mutants, all mutants with an increased propensity to form dauers were cultivated at 15° prior to measurements of intestinal darkness. daf-8 mutants, whose dark intestine phenotype appears to be temperature sensitive (Vowels and Thomas 1992), were cultivated at 25° and analyzed 1 day after reaching the adult stage.
Nile red staining:
The Nile red staining procedure was as described by Ashrafi et al. (2003). Nile red (Molecular Probes, Eugene, OR) was added to the agar to a final concentration of 0.05 μg/ml. Eggs and newly hatched worms were placed on these plates seeded with bacteria. The worms were immobilized on a thin agar pad with sodium azide and observed using a Rhodamine filter with a Zeiss Axioskop upright microscope equipped with epifluorescence within 24 hr after reaching the adult stage. Photography of different genotypes was performed on the same day under identical camera exposure times and light conditions.
Dauer formation assay:
Five to 10 adult worms were allowed to lay eggs on NGM plates seeded with a dense lawn of OP50 bacteria for 6 hr at 25 ± 0.2° and then removed. The number of eggs that were laid on a single plate varied from 40 to 200. To minimize the effect of small temperature and humidity differences within the incubator on dauer formation, we placed plates containing worms of pairs of genotype we were comparing in alternate positions in vertical stacks.
The plates were examined 44–54 hr later for dauers and nondauer worms. Worms were removed following counting. By counting all eggs that were laid on a plate, we found that the number of worms that were not accounted for as either dauers or nondauers was <2% of total eggs laid. For many genotypes, e.g., daf-11, daf-21, and daf-14, we systematically observed a smaller percentage of dauers than reported previously (Thomas et al. 1993). Since dauer formation has been shown to be exquisitely sensitive to environmental conditions (Ailion and Thomas 2000), we suspect that either slightly different temperatures or different food or worm density account for our different results.
RESULTS
ad450sd is a gain-of-function allele of egl-4:
The semidominant mutant ad450sd, previously named eat-7, was isolated in a genetic screen for feeding-defective mutants on the basis of its small body size, pale appearance, and reduced locomotion and feeding when left unperturbed (Avery 1993). ad450sd has a genetic map position that is consistent with the genetic map position of the cGMP-dependent protein kinase egl-4, a gene that was previously defined by recessive mutations that cause a large body size and increased locomotion (Trent et al. 1983; Fujiwara et al. 2002; Hirose et al. 2003). Several opposite phenotypes of ad450sd mutants in comparison to egl-4 loss-of-function mutants, summarized in Table 1 and described in more detail below, suggested to us that ad450sd is a gain-of-function mutation in egl-4.
TABLE 1.
Genetic and phenotypic evidence suggesting that ad450sd is a gain-of-function egl-4 allele
| ad450sd | egl-4 (lof) | |
|---|---|---|
| Genetics | Semidominanta | Recessiveb |
| Genetic map location | Between ced-2 and lin-1a | Between ced-2 and lin-1b |
| Longevity | Decreasedc | Increasedd |
| Retained eggs | Decreasede | Increasedbef |
| Body size | Decreasede | Increaseddefg |
| Locomotion in the presence of food | Decreasede | Increasedg |
| Intestinal darkness | Decreasede | Increaseddef |
| Nile red staining | Increasede | Decreasede |
| Dauer formation | Decreasede | Increasedf |
lof, loss of function.
This work.
We sequenced exons and intron–exon boundaries of the egl-4 gene in ad450sd mutants and identified a single G to A transition at position 1084 of the EGL4A cDNA (Figure 1A). This mutation is predicted to change glycine 362 in the C-terminal cGMP-binding domain into an arginine. This glycine is conserved in all cGMP-dependent protein kinases, in the regulatory subunits of cAMP-dependent protein kinases, and in the cAMP-binding domain of the E. coli CAP as shown by alignment of the cyclic nucleotide binding domains of these three protein types (Figure 1B). The absolute conservation of glycine 362 suggests that the G362R mutation would have major consequences to enzyme function (see discussion).
To confirm that the G362R mutation causes the ad450sd dominant phenotypes on body size and tracking behavior (additional phenotypic details are below), we performed a genetic cis–trans test (Hengartner et al. 1992; Levin and Horvitz 1993; Lee et al. 1997), using the following logic. If the G362R mutation is responsible for the ad450sd dominant phenotypes, then a null mutation of egl-4 on the same chromosome as ad450sd, the cis configuration, should occlude the dominant phenotypes of ad450sd. The same egl-4 null mutation, however, should not occlude the effect of the ad450sd dominant mutation when present on the opposite chromosome, in the trans configuration. In contrast, if egl-4 and the gene mutated to cause the ad450sd phenotypes were separate but linked genes, then the effect of an egl-4 null mutation in the cis and trans configuration would be the same.
Following chemical mutagenesis, we searched for loss-of-function mutations in egl-4 on the same chromosome as the G362R mutation by screening for suppressors of the small and pale phenotypes of ad450sd (see materials and methods). We identified four loss-of-function egl-4 alleles at a rate of 1/937 mutagenized haploid genomes, a rate that is consistent with the isolation of loss-of-function mutants (Brenner 1974). These four mutants formed an allelic series with respect to the severity of body size and tracking abnormality with egl-4(cs80) being the most severe (Table 2). The body size and tracking phenotypes of cs80 were not significantly different from those of the previously characterized egl-4 null allele n479 (Daniels et al. 2000) (Table 2), suggesting that cs80 too is a null egl-4 allele. We sequenced the egl-4 exons and intron–exon boundaries in cs80 mutants and identified a C → T transition that is predicted to result in a mutation of a glutamine to a premature stop codon at amino acid 442 of the EGL-4A protein. Although there are several alternative splice forms of EGL-4 (Stansberry et al. 2001; Fujiwara et al. 2002; Hirose et al. 2003), Q442 is in exon eight, the same exon in which the G362R mutation is located, and therefore the G362R mutation could not be expressed by alternative splicing. Q442STOP is predicted to truncate the protein before the kinase domain (Figure 1A) and would therefore have no kinase activity.
TABLE 2.
Body length and tracking phenotypes of egl-4 loss-of-function alleles generated on an ad450sd chromosome
| egl-4 allele | Body length in micrometers: mean ± SD (N) | Tracking:a mean ± SD (N) |
|---|---|---|
| + | 1029 ± 83 (5) | 112 ± 25 (5) |
| ad450sd | 873 ± 45 (15) | 13 ± 6 (6) |
| ad450sd cs78 | 1122 ± 53 (20) | 105 ± 38 (7) |
| ad450sd cs79 | 1119 ± 62 (21) | 151 ± 50 (7) |
| ad450sd cs81 | 1096 ± 64 (11) | 143 ± 30 (7) |
| ad450sd cs80b | 1222 ± 66 (24) | 164 ± 69 (7) |
| N479 | 1237 ± 43 (15) | 188 ± 28 (6) |
In this and subsequent tables, tracking refers to the number of 0.5-cm squares entered by the worm in a 17-hr period (see materials and methods).
Body length and tracking values were not significantly different from those of the egl-4 null allele n479 (P > 0.1, two-tailed t-test). The data shown for ad450sd homozygous worms are the same data shown in Table 3. SD, standard deviation; N, number tested.
The cs80 egl-4 null mutation completely suppressed the dominant effect of ad450sd on body length and on locomotion when present in the cis configuration but not when present in the trans configuration (compare rows 3 and 5 in Table 3). We conclude that the phenotypes of ad450sd mutants are caused by the G362R mutation in egl-4.
TABLE 3.
cis–trans test demonstrating that ad450sd is allelic to egl-4(null)
| Relevant genotypea | Body length in micrometers: mean ± SD (N) | Tracking: mean ± SD (N) |
|---|---|---|
| +/+ | 1029 ± 83 (5) | 112 ± 25 (5) |
| ad450sd/+ | 935 ± 28 (6) | 25 ± 11 (5) |
| ad450 cs80/+ | 1025 ± 43 (5)b | 110 ± 21 (7)b |
| ad450sd/ad450sd | 873 ± 45 (15) | 13 ± 6 (6) |
| ad450sd/ad450cs80 | 951 ± 44 (13)c | 43 ± 16 (8)c |
| ad450sd/n479 | ND | 36 ± 12 (7)c |
ND, not done; SD, standard deviation; N, number tested.
In the first three rows, the worm was heterozygous for nT1[qIs51].
Significantly different from ad450sd/+, P < 0.01.
Not significantly different from ad450sd/+, P > 0.1.
Although egl-4(ad450cs80) in the trans configuration did not significantly suppress either the small body size or the reduced locomotion phenotype of ad450sd/+, there was a trend for partial suppression for both phenotypes (compare rows 5 and 6 to row 2 in Table 3). The trend for partial suppression of ad450sd dominant phenotype by the egl-4 null allele in the trans configuration is consistent with the interpretation that ad450sd behaves as a genetic hypermorph. That is, it causes an increase in normal egl-4 gene function (Park and Horvitz 1986). To provide additional evidence to support this interpretation, we tested the effect of the egl-4 null mutant n479 in trans to ad450sd on male body size. We chose to perform this analysis in males rather than in hermaphrodites because length measurements in males showed lower variance and therefore would be more likely to detect a small effect on size (see materials and methods). Indeed, we found a small but statistically significant suppression of the dominant small body size effect of ad450sd by the egl-4 null allele n479 (Table 4). We conclude that ad450sd is a hypermorphic allele of egl-4.
TABLE 4.
Male body length measurements show that ad450sd behaves as a genetic hypermorph
| Genotype | Body length in micrometers: mean ± SD |
|---|---|
| n479/n479 | 965 ± 72 |
| +/n479 | 924 ± 46 |
| +/+ | 891 ± 49 |
| ad450sd/n479 | 856 ± 21ab |
| ad450sd/+ | 788 ± 41c |
| ad450sd/ad450sd | 721 ± 28 |
For each genotype, shown are the average length and standard deviation (SD) of 8–16 males within 24 hr after reaching the adult stage.
Significantly different from ad450sd/+ at P < 0.001.
Significantly different from +/+ at P < 0.01.
Significantly different from ad450sd/ad450sd at P < 0.001.
EGL-4 protein level is reduced in ad450sd mutants:
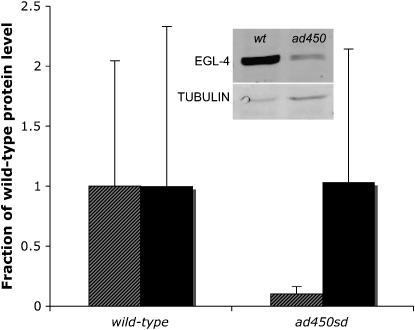
One potential explanation for the gain-of-function phenotype caused by the G362R mutation is an increase in EGL-4 protein abundance. To test for this possibility, we performed quantitative Western blot analysis of EGL-4 in the total protein pool isolated from wild-type and ad450sd mutant worms. Surprisingly, we observed decreased rather than increased EGL-4 protein level in ad450sd mutants (Figure 2). We therefore eliminate one potential explanation for the gain-of-function phenotype. We consider explanations for this reduction of protein level as well as other models to explain the gain-of-function phenotype in the discussion.
Figure 2.
EGL-4 protein level is reduced in egl-4(ad450sd) mutants. Shown are the average and standard deviation measurements relative to the wild-type average level of four biological replicates from wild-type and ad450sd mutant worms. EGL-4 protein level (shaded bars) in ad450sd mutants is reduced compared to that in wild-type worms, P = 0.029 (Wilcoxon's rank sum test). Tubulin protein level (solid bars) by contrast was unchanged, P = 0.34. Shown in the inset is an example showing the staining intensity of EGL-4 and tubulin in wild-type and ad450sd worms.
Phenotypic analysis of egl-4(ad450sd):
ad450sd was previously noted to have decreased longevity (Lakowski and Hekimi 1998), a phenotype opposite to that of egl-4 loss-of-function mutants, which have increased longevity (Hirose et al. 2003). The decreased longevity of ad450sd is particularly striking in light of the fact that virtually all other mutants identified as feeding defective have an increased longevity (Lakowski and Hekimi 1998). We found that in contrast to egl-4 loss-of-function mutants, which have increased retention of eggs in the uterus (35 ± 1 eggs, N = 14) as reported previously (Daniels et al. 2000), egl-4(ad450sd) mutants had a reduced number of retained eggs (8 ± 1 eggs, N = 16) in comparison to wild-type worms (13 ± 1 eggs, N = 13). We analyzed several additional phenotypes, previously noted to be abnormal in egl-4 loss-of-function mutants. These phenotypes are summarized in Table 1 and are detailed in the text below.
Body size:
Body size of ad450sd mutants is decreased whereas body size of egl-4 (lof) mutants is increased (Tables 2–4). Although ad450sd mutants show reduced pharyngeal pumping rates, i.e., feeding, when unperturbed (Avery 1993), the small size of ad450sd mutants is unlikely to be explained solely by caloric restriction. Animals that are doubly mutant for ad450sd and for eat-2, a gene required for the normal fast rate of pharyngeal pumping, are significantly smaller than either single mutant despite no significant change in the feeding rate of the double mutants compared to the eat-2 single mutants (data not shown). eat-2(ad1113); ad450sd double mutants were significantly smaller at 627 ± 56 μm (N = 6) than eat-2(ad1116) (848 ± 63 μm, N = 12) and ad450sd (851 ± 55 μm, N = 8) single mutants (P < 0.0001). Previous studies using loss-of-function mutants suggested that egl-4 affects body size by negatively regulating the activity of the TGF-β ligand dbl-1 (Hirose et al. 2003). Therefore, there appear to be at least two signaling pathways for controlling body size, one that involves TGF-β signaling and the other that is controlled by caloric intake but whose signaling components have yet to be identified.
Locomotion:
ad450sd mutants form fewer tracks on a bacterial lawn, whereas egl-4 loss-of-function mutants form more tracks than wild-type worms, as noted previously (Fujiwara et al. 2002). To demonstrate the reduced locomotion behavior of ad450sd mutants, we measured the tracks formed by single animals left unperturbed for several hours (see materials and methods). Whereas wild-type animals move substantial distances from the point of origin and therefore make tracks that cover close to one-third of the agar surface, ad450sd animals stay close to the center of the agar surface, where they were placed (Tables 2 and 3).
This reduced tracking behavior is not explained by an inability of the animals to move well. Mechanical stimulation of the animals' tails resulted in brisk forward locomotion with a mean number of anterior body bends in 20 sec (15.2 ± 1.4, N = 6) not significantly different from that of wild-type animals (16.3 ± 1.5, N = 5, P > 0.1). Furthermore, introducing a mutation in eat-2 to reduce feeding rates and restrict caloric intake of ad450sd resulted in tracks that were not significantly different from those formed by wild-type animals or by the eat-2 single mutant. eat-2(ad1113); egl-4(ad450sd) double mutants entered 83 ± 38 0.5-cm squares whereas wild-type animals and eat-2 single mutants entered 68 ± 29 and 87 ± 27 squares, respectively, in 17 hr. Finally, ad450sd males appear as active as wild-type males and have normal mating efficiencies (data not shown). Therefore, we conclude that ad450sd mutants have reduced locomotion only in the absence of sufficient motivation to move.
Reduced tracking despite normal ability to move was previously described for mutants with impaired sensory function (Fujiwara et al. 2002). Many of these mutants have structurally defective sensory cilia as shown by defective uptake of lipophilic dyes (Starich et al. 1995). In contrast to these chemosensory mutants, sensory cilia stain normally in ad450sd mutants (data not shown). Furthermore, chemotaxis to the volatile odorant diacetyl is normal in ad450sd (data not shown). The reduced tracking behavior of ad450sd mutants is therefore not the result of grossly abnormal chemosensation.
The reduced locomotion of ad450sd mutants is also not explained by signaling changes in the TGF-β pathway that partially mediate the dauer formation and intestinal darkness phenotypes of ad450sd mutants (see below). daf-7(e1372); egl-4(ad450sd) and daf-8(e1393); egl-4(ad45sd) double mutants formed the same number of tracks as ad450sd single mutants (Table 5). Therefore, the signaling pathway that mediates reduced locomotion by egl-4 remains unknown.
TABLE 5.
Genetic interaction between egl-4(ad450sd) and dauer constitutive mutants
| Genotype | % dauers at 25° (N) | Intestinal darkness | Body length in micrometers: mean ± SD | Tracking: mean ± SD (N) |
|---|---|---|---|---|
| + | 0 (450) | Normal | 1029 ± 83 | 88 ± 39 (7) |
| egl-4(ad450sd) | 0 (792) | Pale | 873 ± 45 | 23 ± 13 (5) |
| egl-4(n479) | 0 (466) | Dark | 1237 ± 43 | 188 ± 28 (6) |
| daf-1(m40) | 99 (512) | Dark | ND | ND |
| daf-1(m40) egl-4(ad450sd) | 78 (389)a | Dark | ND | ND |
| daf-4(e1364) | 100 (337) | Dark | ND | ND |
| daf-4(e1364); egl-4(ad450sd) | 99 (401) | Dark | ND | ND |
| daf-7(e1372) | 99 (741) | Dark | 986 ± 73 | 77 ± 38 (6) |
| daf-7(e1372); egl-4(ad450sd) | 100 (280) | Dark | 830 ± 47 | 15 ± 11 (6) |
| daf-8(e1393) | 59 (393) | Dark | 1002 ± 101 | 86 ± 30 (6) |
| daf-8(e1393); egl-4(ad450sd) | 77 (513)a | Dark | 850 ± 61 | 12 ± 4 (6) |
| daf-14(m77) | 82 (335) | Dark | ND | ND |
| egl-4(ad450sd) daf-14(m77) | 45 (541)a | Dark | ND | ND |
| daf-11(sa195) | 85 (493) | Normal | ND | ND |
| egl-4(ad450sd); daf-11(sa195) | 36 (385)a | Pale | ND | ND |
| daf-21(p673) | 45 (251) | Normal | ND | ND |
| egl-4(ad450sd); daf-21(p673) | 16 (413)a | Pale | ND | ND |
Body length measurements are the average of 5–20 worms. Tracking data for egl-4(n479) are the same as those shown in Table 2. ND, not done.
Significantly different from respective daf single mutant at P < 0.0001, Fisher's exact test.
Intestinal darkness and fat storage:
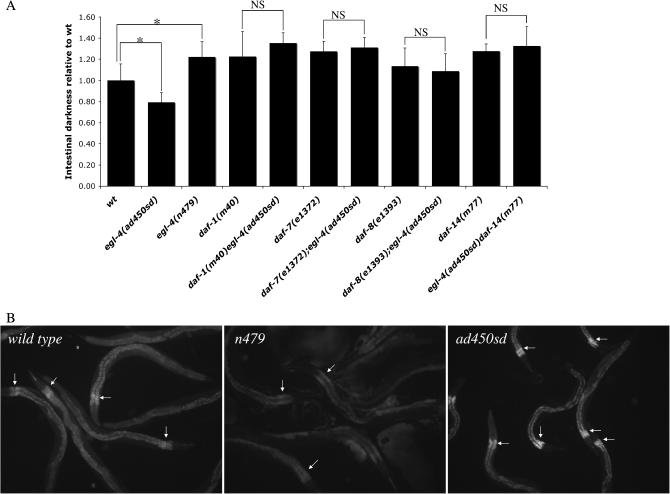
Intestinal darkness under light microscopy appears decreased in ad450sd mutants while it appears increased in egl-4 recessive mutants, as reported previously (Daniels et al. 2000; Hirose et al. 2003). We were able to quantify these differences using digital video imaging methods (Figure 2).
In addition to egl-4 loss-of-function mutants, other mutants with the dark intestine phenotype include those with mutations in genes that mediate TGF-β signaling. These include the TGF-β ligand daf-7, the TGF-β receptor daf-1, and the TGF-β intracellular signaling components daf-8 and daf-14 (Patterson and Padgett 2000). To determine if the pale intestine phenotype of ad450sd mutants requires TGF-β signaling, we constructed strains that were doubly mutant for ad450sd and each of these TGF-β signaling mutants. The intestinal darkness of each of these double mutants was not significantly different from that of the respective TGF-β signaling single mutant (Figure 2). Therefore, egl-4 acts upstream or in parallel to TGF-β signaling to promote a pale intestine.
The C. elegans intestine has been shown previously to be a storage depot for macromolecules, including fat (Ashrafi et al. 2003; McKay et al. 2003). To test whether or not a difference in fat storage could partially explain the difference in intestinal darkness of egl-4 loss- and gain-of-function mutants, we examined Nile red staining in these mutants. Nile red is a vital fluorescent dye that binds to fatty acids and increased Nile red fluorescence has been shown to correlate with increased intestinal fat storage in C. elegans (Ashrafi et al. 2003). We found that ad450sd mutants have increased intestinal Nile red staining, and that the egl-4 recessive mutants have decreased staining (Figure 3). This difference is most prominent in the anterior part of the intestine, immediately posterior to the pharyngeal-intestinal valve. Therefore, the egl-4 gene product promotes intestinal fat storage, particularly in the anterior intestine.
Figure 3.
The effect of egl-4 activity on intestinal darkness and fat storage. (A) The average intestinal grayscale value of six to eight worms subtracted from a maximum grayscale value of 256 and divided by this average value calculated for wild-type worms. A higher value means a darker intestine. Error bars represent standard deviations. To assess for statistical significance of the effect of the n479 and ad450sd mutations on intestinal darkness, six pairwise one-way ANOVAs were performed as marked above the bars. An asterisk indicates statistical significance at P < 0.05 and NS indicates not significant. (B) Shown are six to eight adult worms viewed under a Rhodamine fluorescence filter after cultivation on agar plates containing Nile red. Arrows point to the anterior part of the intestines, where staining appears brightest. Three different observers who were blinded to genotype identified ad450sd as having the brightest staining and n479 as having the faintest staining among these three genotypes.
Dauer formation:
In addition to the dark intestine phenotype, egl-4 loss-of-function mutants share a deregulated dauer formation phenotype with TGF-β pathway mutants. When grown in conditions of higher temperatures, crowding, and reduced food, C. elegans development can proceed through an alternative long-lived third larval stage, called the dauer larva (Riddle and Albert 1997). At least three signaling pathways have been identified that control the dauer formation decision, including a cGMP sensory transduction pathway, a TGF-β signaling pathway, and an insulin-signaling pathway (Riddle and Albert 1997).
egl-4 loss-of-function mutants have previously been noted to be hypersensitive to dauer pheromone and to have a high rate of dauer formation at 27°, a temperature in which wild-type worms only rarely form dauers (Daniels et al. 2000). Genetic epistasis experiments suggested that the egl-4 loss-of-function Daf-c phenotype is explained by the action of the gene in the TGF-β signaling pathway. Consistent with this placement, we found that ad450sd partially suppresses the Daf-c phenotype of daf-11 and daf-21 and does not suppress the Daf-c phenotypes of daf-7 and daf-4 (Table 5). Unlike mutants with defective cilia structure, which completely suppress dauer formation of daf-11 and daf-21 mutants (Vowels and Thomas 1992), the suppression by ad450sd is partial, a result expected for a gene that functions in parallel to daf-11 signaling (Thomas et al. 1993).
Although these results taken alone would support a simple single action of egl-4 in the TGF-β branch of the dauer formation pathway upstream of daf-7, the effect of egl-4(ad450sd) on the Daf-c phenotype of mutants in the other TGF-β signaling genes suggests a more complicated model. We noted a partial suppression of the Daf-c phenotype of daf-1 and daf-14 mutants and a small but significant enhancement of the daf-8 mutants' Daf-c phenotype (Table 5). The partial suppression of daf-1 and daf-14 suggests that egl-4 acts at least partially in parallel to TGF-β signaling in regulating the dauer formation decision. Also supporting a site of action that is parallel to TGF-β signaling is the observation by Daniels et al. (2000) that egl-4 loss-of-function mutations enhance the dauer constitutive phenotypes of daf-7 and daf-14 mutants. Our observed slight enhancement of daf-8 suggests that egl-4 may have dauer-promoting activity in addition to dauer-inhibiting activity, as has been described for other genes that, like egl-4 (Fujiwara et al. 2002), are expressed in multiple sensory neurons (Vowels and Thomas 1992; Coburn et al. 1998).
Suppressors of egl-4(ad450sd):
Genetic suppressor screens of gain-of-function mutants are a powerful way to identify components of signaling pathways in C. elegans (Huang and Sternberg 1995). In the same screen used to identify egl-4 loss-of-function mutations on the same chromosome as ad450sd, we also identified three mutants unlinked to egl-4, cs82, cs83, and cs84, which suppress some or all of the ad450sd mutant phenotypes. As expected on the basis of our double-mutant analysis with Daf-c mutants described above, we found one new allele of daf-8, cs82, which suppresses the pale intestine phenotype of egl-4(ad450sd). Our assignment of cs82 as a daf-8 allele is based on a similar Daf-c and social feeding phenotype, a consistent map position, a failure to complement daf-8 for the Daf-c phenotype, and molecular sequencing data that identified a two-nucleotide deletion in the second exon of the DAF-8 gene R05D11.1 (Riddle and Albert 1997).
cs83 and cs84 partially suppress the small body, reduced locomotion, and pale intestine phenotypes of egl-4(ad450sd) (Table 6). Therefore, suppressor screens of ad450sd promise to be a useful tool in understanding PKG signaling.
DISCUSSION
We have characterized a novel gain-of-function mutation that increases normal activity of the C. elegans cGMP-dependent protein kinase gene egl-4. We use this mutation to demonstrate a signaling role for egl-4 in several physiological processes, including the control of body size, intestinal storage of fat, dauer formation, and locomotion in the presence of food. While for many of these phenotypes a role for egl-4 has been previously described on the basis of analysis of mutants with reduced gene function, analysis of ad450sd, a mutant with increased gene function, allows one to conclude that egl-4 is not simply permissive for these physiological processes but rather plays an instructive signaling role.
Structure and function of cGMP-dependent protein kinases:
Glycine 362 is conserved in all PKG enzymes as well as in the regulatory subunits of PKA. On the basis of amino acid alignment with CAP, whose tertiary structure is known (McKay and Steitz 1981), a three-dimensional structure has been proposed for the cGMP-binding domain of protein kinase G (Weber et al. 1989). By analogy to CAP, the structure of the cGMP-binding domain would consist of eight hydrogen-bonded β-strands that form a compact β-roll structure (Weber and Steitz 1987). cGMP binds in a pocket formed by these β-strands and one α-helix (Weber et al. 1989). At the vertex of this pocket sits glycine 362 (glycine 45 in the CAP structure), as a connector between strand β3 and strand β4 (Figure 1B). This glycine would not be expected to bind cyclic nucleotide monophosphate directly, but must be critical for maintaining the overall tertiary structure of the cyclic nucleotide-binding domain.
We do not yet know the biochemical or cell biological consequences of the G362R mutation and to our knowledge the effect of this particular mutation has never been tested in vitro. The possibility that the gain-of-function phenotype of the G362R mutation is caused by increased steady-state levels of the protein is excluded by our protein level analysis (Figure 2). We in fact observed the opposite result, i.e., of reduced EGL-4 protein level in the ad450sd mutants in comparison to that in wild-type worms. The reduction in protein level might be accounted for by reduced protein folding efficiency of the EGL-4 G362R protein, by reduced stability of the folded protein, or by a negative feedback regulation of EGL-4 protein levels by egl-4 gene activity. Such a negative feedback may occur, for example, if autophosphorylation of the EGL-4 protein marks it for degradation. Autophosphorylation is known to occur in the case of mammalian PKG (Aitken et al. 1984).
Another explanation for the gain-of-function phenotype caused by the G362R mutation is that this mutation causes increased kinase activity of the enzyme. This could occur, for example, by causing enzyme activation during basal conditions, in the absence of cGMP elevation. Autoinhibition of PKG enzymatic activity is thought to occur via interaction between the catalytic domain and an inhibitory domain localized near the N terminus of the protein (Yuasa et al. 2000a). The change from glycine, a small nonpolar amino acid, to arginine, a large charged amino acid, may disrupt the integrity of the β-roll structure and lead to easier access for cGMP binding and therefore faster association kinetics. Alternatively, this mutation may cause such a drastic change to the tertiary structure of the cyclic nucleotide-binding domain with the result that the N-terminal inhibitory domain can no longer effectively inhibit the kinase domain. In the absence of autoinhibition, the enzyme would be active even in the absence of cGMP. Unlike a constitutively active mutant that lacks the whole N-terminal portion of the protein and contains only the catalytic domain (Browning et al. 2001), the G362R PKG mutant should retain substrate specificity by virtue its normal N terminus.
A final possible explanation of this gain-of-function activity is a change in the subcellular distribution of the enzyme. PKG has been shown previously to undergo nuclear translocation in some cell types (Gudi et al. 1997; L'Etoile and Bargmann 2000) and to localize to other discrete cellular compartments in others (Wyatt et al. 1991; Pryzwansky et al. 1995; Surks et al. 1999). One possibility is that nuclear translocation or another subcellular targeting event is altered in G362R mutants. Distinguishing among these possibilities will require future biochemical and cell biological experiments. Regardless of the mechanism, the genetic evidence we describe here provides compelling evidence that the effect of G362R is to increase normal PKG gene activity. Since G362R is a mutation that would not be expected a priori to necessarily cause increased gene activity, our finding underscores the importance of an unbiased genetic approach for the identification of novel mutants.
The use of the ad450sd mutation to identify downstream targets of PKG:
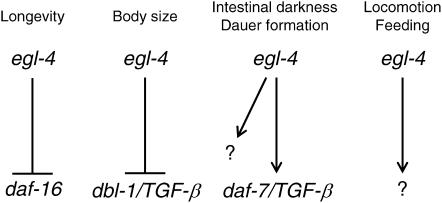
On the basis of analysis of egl-4 loss-of-function and gain-of-function mutant phenotypes, one can conclude that PKG signaling controls multiple physiological processes. The signaling pathway that mediates many of these phenotypes appears distinct (Figure 4). egl-4 promotes reduced longevity via an insulin signaling pathway (Hirose et al. 2003), promotes nondauer development and a pale intestine partially via a TGF-β signaling pathway (Trent et al. 1983; Daniels et al. 2000) (this work), promotes a smaller body size through a different TGF-β signaling pathway (Fujiwara et al. 2002; Hirose et al. 2003), and promotes sensory adaptation through a cGMP-gated cation channel (L'Etoile and Bargmann 2000). The signaling pathway through which egl-4 promotes reduced locomotion on food is not yet known (Figure 4).
Figure 4.
Distinct signaling pathways mediate egl-4 phenotypes. egl-4 promotes a reduced longevity by negatively regulating the activity of the insulin signaling transcription factor daf-16 (Hirose et al. 2003), promotes a reduced body size by negatively regulating the activity of the dbl-1 TGF-β (Fujiwara et al. 2002; Hirose et al. 2003), promotes a pale intestine and nondauer development by positively regulating the daf-7 TGF-β, as well as by another unidentified pathway (Trent et al. 1983; Daniels et al. 2000) (this work), and promotes a reduced locomotion and feeding rate in the presence of food by an as yet unidentified signaling pathway.
Previous approaches for the identification of signaling elements downstream of PKG in mammalian cells have made use of yeast two-hybrid screens and in vitro binding and phosphorylation methods (Butt et al. 1994; Vo et al. 1998; Surks et al. 1999; Yuasa et al. 2000b,c). Verification of the relevance of these targets to mammalian physiology requires in vivo inactivation experiments, which can be costly and difficult to interpret if gene knockouts have pleiotropic effects.
This G362R gain-of-function mutant offers the opportunity to identify downstream signaling targets of egl-4. Indeed, we have demonstrated that extragenic suppressors can be isolated that suppress some or all of the ad450sd phenotypes. Future molecular characterization of the genes affected in these and other suppressors may shed light on PKG signaling in an unbiased hypothesis-independent fashion.
Similarity between the ad450sd phenotype and lethargus behavior:
The phenotype that led to the isolation of ad450sd mutants is the reduced pharyngeal pumping in the presence of abundant food. This mutant was subsequently noted to stop moving when left unperturbed. These two behaviors, pumping cessation and reduced locomotion despite unimpaired ability to move and pump, are reminiscent of behaviors normally exhibited by worms during lethargus behavior. Lethargus is a period that occurs before each of the four molts that separate the larval stages and fourth larval stage and the adult stage (Singh and Sulston 1978). Little is known about the regulation of behavior during lethargus. In current work we are exploring whether or not egl-4 plays a role in the control of lethargus behavior.
Acknowledgments
We are grateful to Yasumi Ohshima for the gift of the EGL-4 antibody. We thank Young You and Julia George-Raizen for comments on this manuscript. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health (NIH) National Center for Research Resources. We acknowledge financial support from the NIH grants K08 NS048914 (D.M.R.), R01 HL60287 (A.I.P.), and R01 GM58540 (M.S.) and from the National Alliance for Research on Schizophrenia and Depression (D.M.R.).
References
- Aiba, H., S. Fujimoto and N. Ozaki, 1982. Molecular cloning and nucleotide sequencing of the gene for E. coli cAMP receptor protein. Nucleic Acids Res. 10: 1345–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ailion, M., and J. H. Thomas, 2000. Dauer formation induced by high temperatures in Caenorhabditis elegans. Genetics 156: 1047–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitken, A., B. A. Hemmings and F. Hoffman, 1984. Identification of the residues on cyclic GMP-dependent protein kinase that are autophosphorylated in the presence of cyclic AMP and cyclic GMP. Biochim. Biophys. Acta 790: 219–225. [DOI] [PubMed] [Google Scholar]
- Ashrafi, K., F. Y. Chang, J. L. Watts, A. G. Fraser, R. S. Kamath et al., 2003. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 421: 220–221. [DOI] [PubMed] [Google Scholar]
- Avery, L., 1993. The genetics of feeding in Caenorhabditis elegans. Genetics 133: 897–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford, M. M., 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254. [DOI] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning, D. D., M. McShane, C. Marty and R. D. Ye, 2001. Functional analysis of type 1 alpha cGMP-dependent protein kinase using green fluorescent fusion proteins. J. Biol. Chem. 276: 13039–13048. [DOI] [PubMed] [Google Scholar]
- Butt, E., K. Abel, M. Krieger, D. Palm, V. Hoppe et al., 1994. cAMP- and cGMP-dependent protein kinase phosphorylation sites of the focal adhesion vasodilator-stimulated phosphoprotein (VASP) in vitro and in intact human platelets. J. Biol. Chem. 269: 14509–14517. [PubMed] [Google Scholar]
- Coburn, C. M., I. Mori, Y. Ohshima and C. I. Bargmann, 1998. A cyclic nucleotide-gated channel inhibits sensory axon outgrowth in larval and adult Caenorhabditis elegans: a distinct pathway for maintenance of sensory axon structure. Development 125: 249–258. [DOI] [PubMed] [Google Scholar]
- Daniels, S. A., M. Ailion, J. H. Thomas and P. Sengupta, 2000. egl-4 acts through a transforming growth factor-β/SMAD pathway in Caenorhabditis elegans to regulate multiple neuronal circuits in response to sensory cues. Genetics 156: 123–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara, M., P. Sengupta and S. L. McIntire, 2002. Regulation of body size and behavioral state of C. elegans by sensory perception and the EGL-4 cGMP-dependent protein kinase. Neuron 36: 1091–1102. [DOI] [PubMed] [Google Scholar]
- Gross, R. E., S. Bagchi, X. Lu and C. S. Rubin, 1990. Cloning, characterization, and expression of the gene for the catalytic subunit of cAMP-dependent protein kinase in Caenorhabditis elegans. Identification of highly conserved and unique isoforms generated by alternative splicing. J. Biol. Chem. 265: 6896–6907. [PubMed] [Google Scholar]
- Gudi, T., H. M. Lohmann and R. B. Pilz, 1997. Regulation of gene expression by cyclic GMP dependent protein kinase requires nuclear translocation of the kinase: identification of a nuclear localization signal. Mol. Cell. Biol. 17: 5244–5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengartner, M. O., R. E. Ellis and H. R. Horvitz, 1992. Caenorhabditis elegans gene ced-9 protects cells from programmed cell death. Nature 356: 494–499. [DOI] [PubMed] [Google Scholar]
- Hirose, T., Y. Nakano, Y. Nagamatsu, T. Misumi, H. Ohta et al., 2003. Cyclic GMP-dependent protein kinase EGL-4 controls body size and lifespan in C. elegans. Development 130: 1089–1099. [DOI] [PubMed] [Google Scholar]
- Hodgkin, J., 1983. Male phenotypes and male mating in Caenorhabditis elegans. Genetics 103: 43–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, L. S., and P. W. Sternberg, 1995. Genetic dissection of developmental pathways. Methods Cell Biol. 48: 97–122. [DOI] [PubMed] [Google Scholar]
- Kalderon, D., and G. M. Rubin, 1989. cGMP-dependent protein kinase genes in Drosophila. J. Biol. Chem. 164: 10738–10748. [PubMed] [Google Scholar]
- Lakowski, B., and S. Hekimi, 1998. The genetics of caloric restriction in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 95: 13091–13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, R. Y., L. Lobel, M. Hengartner, H. R. Horvitz and L. Avery, 1997. Mutations in the alpha1 subunit of an L-type voltage-activated Ca2+ channel cause myotonia in Caenorhabditis elegans. EMBO J. 16: 6066–6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Etoile, N. D., and C. I. Bargmann, 2000. Olfaction and odor discrimination are mediated by the C. elegans guanylyl cyclase ODR-1. Neuron 25: 575–586. [DOI] [PubMed] [Google Scholar]
- L'Etoile, N. D., C. M. Coburn, J. Eastham, A. Kistler, G. Gallegos et al., 2002. The cyclic GMP-dependent protein kinase EGL-4 regulates olfactory adaptation in C. elegans. Neuron 36: 1079–1089. [DOI] [PubMed] [Google Scholar]
- Levin, J. Z., and H. R. Horvitz, 1993. Three new classes of mutations in the Caenorhabditis elegans muscle gene sup-9. Genetics 135: 53–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay, D. B., and T. A. Steitz, 1981. Structure of catabolite gene activator protein at 2.9 Å resolution suggests binding to left-handed B-DNA. Nature 290: 744–749. [DOI] [PubMed] [Google Scholar]
- McKay, R. M., J. P. McKay, L. Avery and J. M. Graff, 2003. C. elegans: a model for exploring the genetics of fat storage. Dev. Cell 4: 131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orstavik, S., R. Solberg, K. Tasken, M. Nordahl, M. R. Altherr et al., 1996. Molecular cloning, cDNA structure, and chromosomal localization of the human type II cGMP-dependent protein kinase. Biochem. Biophys. Res. Commun. 220: 759. [DOI] [PubMed] [Google Scholar]
- Orstavik, S., V. Natarajan, K. Tasken, T. Jahnsen and M. Sandberg, 1997. Characterization of the human gene encoding the type I alpha and type I beta cGMP-dependent protein kinase (PRKG1). Genomics 42: 311–318. [DOI] [PubMed] [Google Scholar]
- Osborne, K. A., A. Robichon, E. Burgess, S. Butland, R. A. Shaw et al., 1997. Natural behavior polymorphism due to a cGMP-dependent protein kinase of Drosophila. Science 277: 834–836. [DOI] [PubMed] [Google Scholar]
- Park, E. C., and H. R. Horvitz, 1986. Mutations with dominant effects on the behavior and morphology of the nematode Caenorhabditis elegans. Genetics 113: 821–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, G. I., and R. W. Padgett, 2000. TGF-β related pathways: roles in Caenorhabditis elegans development. Trends Genet. 16: 27–33. [DOI] [PubMed] [Google Scholar]
- Pfeifer, A., P. Ruth, W. Dostmann, M. Sausbier, P. Klatt et al., 1999. Structure and function of cGMP-dependent protein kinases. Rev. Physiol. Biochem. Pharmacol. 135: 105–149. [DOI] [PubMed] [Google Scholar]
- Pryzwansky, K. B., S. Kidao, T. A. Wyatt, W. Reed and T. M. Lincoln, 1995. Localization of cyclic GMP-dependent protein kinase in human mononuclear phagocytes. J. Leukocyte Biol. 57: 670–678. [DOI] [PubMed] [Google Scholar]
- Raizen, D. M., R. Y. Lee and L. Avery, 1995. Interacting genes required for pharyngeal excitation by motor neuron MC in Caenorhabditis elegans. Genetics 141: 1365–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle, D. L., 1977. A genetic pathway for dauer larvae formation in C. elegans. Stadler Genet. Symp. 9: 101–120. [Google Scholar]
- Riddle, D. L., and P. S. Albert, 1997. Genetic and environmental regulation of dauer larva development, pp. 739–768 in C. elegans II, edited by D. L. Riddle, T. Blumenthal, B. J. Meyer and J. R. Priess. Cold Spring Harbor Laboratory Press, Plainview, NY. [PubMed]
- Riddle, D. L., M. M. Swanson and P. S. Albert, 1981. Interacting genes in nematode dauer larvae formation. Nature 290: 668–671. [DOI] [PubMed] [Google Scholar]
- Rybalkin, S. D., C. Yan, K. E. Bornfeldt and J. A. Beavo, 2003. Cyclic GMP phosphodiesterases and regulation of smooth muscle function. Circ. Res. 93: 280–291. [DOI] [PubMed] [Google Scholar]
- Siegfried, K. R., A. R. Kidd, M. A. Chesney and J. Kimble, 2004. The sys-1 and sys-3 genes cooperate with Wnt signaling to establish the proximal-distal axis of the Caenorhabditis elegans gonad. Genetics 166: 171–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, R. N., and J. E. Sulston, 1978. Some observations on moulting in Caenorhabditis elegans. Nematologica 24: 63–71. [Google Scholar]
- Stansberry, J., E. J. Baude, M. K. Taylor, P. J. Chen, S. W. Jin et al., 2001. A cGMP-dependent protein kinase is implicated in wild-type motility in C. elegans. J. Neurochem. 76: 1177–1187. [DOI] [PubMed] [Google Scholar]
- Starich, T. A., R. K. Herman, C. K. Kari, W. H. Yeh, W. S. Schackwitz et al., 1995. Mutations affecting the chemosensory neurons of Caenorhabditis elegans. Genetics 139: 171–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surks, H. K., N. Mochizuki, Y. Kasai, S. P. Georgescu, K. M. Tang et al., 1999. Regulation of myosin phosphatase by a specific interaction with cGMP- dependent protein kinase Ialpha. Science 286: 1583–1587. [DOI] [PubMed] [Google Scholar]
- Thomas, J. H., D. A. Birnby and J. J. Vowels, 1993. Evidence for parallel processing of sensory information controlling dauer formation in Caenorhabditis elegans. Genetics 134: 1105–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trent, C., N. Tsuing and H. R. Horvitz, 1983. Egg-laying defective mutants of the nematode Caenorhabditis elegans. Genetics 104: 619–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo, N. K., J. M. Gettemy and V. M. Coghlan, 1998. Identification of cGMP-dependent protein kinase anchoring proteins (GKAPs). Biochem. Biophys. Res. Commun. 246: 831–835. [DOI] [PubMed] [Google Scholar]
- Vowels, J. J., and J. H. Thomas, 1992. Genetic analysis of chemosensory control of dauer formation in Caenorhabditis elegans. Genetics 130: 105–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vowels, J. J., and J. H. Thomas, 1994. Multiple chemosensory defects in daf-11 and daf-21 mutants of Caenorhabditis elegans. Genetics 138: 303–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, I. T., and T. A. Steitz, 1987. Structure of a complex of catabolite gene activator protein and cyclic AMP refined at 2.5 Å resolution. J. Mol. Biol. 198: 311–326. [DOI] [PubMed] [Google Scholar]
- Weber, I. T., J. B. Shabb and J. D. Corbin, 1989. Predicted structures of the cGMP binding domains of the cGMP-dependent protein kinases: a key alanine/threonine difference in evolutionary divergence of cAMP and cGMP binding sites. Biochemistry 28: 6122–6127. [DOI] [PubMed] [Google Scholar]
- Wedel, B., and D. Garbers, 2001. The guanylyl cyclase family at Y2K. Annu. Rev. Physiol. 63: 215–233. [DOI] [PubMed] [Google Scholar]
- Wyatt, T. A., T. M. Lincoln and K. B. Pryzwansky, 1991. Vimentin is transiently co-localized with and phosphorylated by cyclic GMP-dependent protein kinase in formyl-peptide-stimulated neutrophils. J. Biol. Chem. 266: 21274–21280. [PubMed] [Google Scholar]
- Yuasa, K., H. Michibata, K. Omori and N. Yanaka, 2000. a Identification of a conserved residue responsible for the autoinhibition of cGMP-dependent protein kinase Ialpha and beta. FEBS Lett. 46: 175–188. [DOI] [PubMed] [Google Scholar]
- Yuasa, K., K. Omori and N. Yanaka, 2000. b Binding and phosphorylation of a novel male germ cell-specific cGMP-dependent protein kinase-anchoring protein by cGMP-dependent protein kinase Ialpha. J. Biol. Chem. 275: 4897–4905. [DOI] [PubMed] [Google Scholar]
- Yuasa, K., K. Omori and N. Yanaka, 2000. c Specific domain of cGMP-dependent protein kinase Ibeta but not Ialpha functions as a transcriptional activator in yeast. IUBMB Life 49: 17–22. [DOI] [PubMed] [Google Scholar]