Abstract
Ribonucleotide reductase (RNR) catalyzes the rate-liming step in de novo deoxyribonucleotide biosynthesis and is essential in DNA replication and repair. Cells have evolved complex mechanisms to modulate RNR activity during normal cell cycle progression and in response to genotoxic stress. A recently characterized mode of RNR regulation is DNA damage-induced RNR subunit redistribution. The RNR holoenzyme consists of a large subunit, R1, and a small subunit, R2. The Saccharomyces cerevisiae R2 is an Rnr2:Rnr4 heterodimer. Rnr2 generates a diferric–tyrosyl radical cofactor required for catalysis; Rnr4 facilitates cofactor assembly and stabilizes the resulting holo-heterodimer. Upon DNA damage, Rnr2 and Rnr4 undergo checkpoint-dependent, nucleus-to-cytoplasm redistribution, resulting in colocalization of R1 and R2. Here we present evidence that Rnr2 and Rnr4 are transported between the nucleus and the cytoplasm as one protein complex. Tagging either Rnr2 or Rnr4 with a nuclear export sequence causes cytoplasmic localization of both proteins. Moreover, mutations at the Rnr2:Rnr4 heterodimer interface can affect the localization of both proteins without disrupting the heterodimeric complex. Finally, the relocalization of Rnr4 appears to involve both active export and blockage of nuclear import. Our findings provide new insights into the mechanism of DNA damage-induced RNR subunit redistribution.
RIBONUCLEOTIDE reductase (RNR) catalyzes the reduction of ribonucleoside diphosphate (NDP) to deoxyribonucleoside diphosphate (dNDP), an essential step in de novo biosynthesis of deoxyribonucleoside triphosphates (dNTPs) (Thelander and Reichard 1979). Class I RNRs were originally identified in Escherichia coli and are conserved from yeast to mammal (Reichard 1993). The prototype class I RNR holoenzyme is composed of two subunits, the large subunit R1 (α, whose oligomeric state is understood incompletely) (Kashlan et al. 2002) and the small subunit R2 (β2) (Jordan and Reichard 1998). R1 contains the catalytic site and two allosteric effector binding sites that control, respectively, substrate specificity and turnover rate of nucleotide reduction. R2 is responsible for generating a diferric–tyrosyl radical [(Fe)2–Y·] cofactor that is required for catalysis (Elledge and Davis 1987; Jordan and Reichard 1998; Stubbe et al. 2001).
The RNR of the budding yeast Saccharomyces cerevisiae has an unusual property: its small subunit is a heterodimer of β and β′, encoded, respectively, by the RNR2 and RNR4 genes (Huang and Elledge 1997; Wang et al. 1997). Both Rnr2 and Rnr4 share extensive sequence homology to all characterized RNR small subunits from other organisms. However, Rnr2 contains all of the six highly conserved amino acids that are absolutely required for iron binding and the subsequent generation of the (Fe)2–Y· cofactor, while Rnr4 has substitutions in three of the six residues (Elledge and Davis 1987; Huang and Elledge 1997). Thus, only Rnr2 is capable of forming the (Fe)2–Y· cofactor required for catalysis and is essential for mitotic growth (Elledge and Davis 1987; Chabes et al. 2000; Ge et al. 2001). RNR4, conversely, is required for viability in some strains under all conditions and in some others only at lower temperature (Huang and Elledge 1997; Wang et al. 1997). Recent studies from several laboratories have established that the active form of the S. cerevisiae R2 is the Rnr2:Rnr4 (ββ′) heterodimer (Chabes et al. 2000; Ge et al. 2001; Perlstein et al. 2005). Immunoprecipitation assays show that the Rnr2:Rnr4 complex can be readily isolated from yeast cell extract (Huang and Elledge 1997; Perlstein et al. 2005). Although Rnr4 cannot form any radical on its own, it is required to facilitate the generation of the (Fe)2–Y· cofactor in Rnr2 and to stabilize the resulting holo-heterodimer both in vitro and in vivo (Perlstein et al. 2005). The recently determined crystal structures of the Rnr2:Rnr4 (ββ′) heterodimer and the homodimers of Rnr2:Rnr2 (β2) and Rnr4:Rnr4 (β′2) have provided structural rationale for why the heterodimer is the preferred complex in vivo (Sommerhalter et al. 2004; Voegtli et al. 2001).
Eukaryotic cells have evolved complex surveillance mechanisms to regulate RNR activity both during normal cell cycle progression and in response to genotoxic stress, to ensure adequate and balanced dNTP pools for high fidelity in DNA replication and repair. Failures in such regulation can lead to cell death, increased mutation rates, and other genomic abnormalities (Desany et al. 1998; Amin et al. 2001; Chabes et al. 2003). In addition to allosteric regulation (Jordan and Reichard 1998), the activity of the S. cerevisiae RNR is regulated by three characterized mechanisms in response to genotoxic stress including DNA damage and stress during DNA replication. All three mechanisms require the function of the DNA damage and replication checkpoint kinases Mec1, Rad53, and Dun1. First, the RNR genes are transcriptionally induced by genotoxic stress. Crt1 is a transcription repressor that normally keeps the RNR2/3/4 expression in check (Zhou and Elledge 1992; Huang et al. 1998). DNA damage induces Mec1/Rad53/Dun1-dependent Crt1 hyperphosphorylation and removal of Crt1 from its target promoters (Huang et al. 1998). Second, DNA damage induces Mec1/Rad53/Dun1-dependent degradation of Sml1, a protein inhibitor of the RNR large subunit (Zhao et al. 1998; Chabes et al. 1999; Zhao and Rothstein 2002). Third, the Mec1/Rad53/Dun1 kinases mediate the colocalization of the large and small subunits of RNR in response to genotoxic stress (Yao et al. 2003).
The S. cerevisiae RNR small subunit is predominantly localized in the nucleus, whereas the large subunit is predominantly localized in the cytoplasm (Yao et al. 2003). When cells encounter DNA-damaging or replication-blocking agents, the RNR small subunits undergo nucleus-to-cytoplasm redistribution and become colocalized with the large subunit, which presumably facilitates formation of the RNR holoenzyme (Yao et al. 2003). In S. pombe, the RNR small subunit Suc22 is also redistributed from the nucleus to the cytoplasm upon DNA damage, while the large subunit Cdc22 remains ubiquitously distributed inside the cell (Liu et al. 2003).
DNA damage-induced RNR redistribution has been reported in organisms other than yeasts. In plant cells the large subunit R1 moves from the cytoplasm to the nucleus after UV irradiation (Lincker et al. 2004). In mammalian cells both the large subunit, hRRM1, and the two small subunits, hRRM2 and p53R2, translocate from the cytoplasm to the nucleus after cells are irradiated with UV (Xue et al. 2003). The translocation of both R1 and R2 from the cytoplasm to the nucleus suggests that dNTPs can be synthesized in the nucleus for DNA repair in mammalian cells. Conversely, the cytoplasmic colocalization of R1 and R2 in both S. cerevisiae and S. pombe upon genotoxic stress suggests that the cytoplasm is the major site of dNTP production in yeast cells (Liu et al. 2003; Yao et al. 2003).
The molecular details of RNR small subunit redistribution in S. cerevisiae are not well understood. In S. pombe the redistribution of the RNR small subunit appears to be mediated via degradation of Spd1 by the Cop9/signalosome (Liu et al. 2003; Bondar et al. 2004; Holmberg et al. 2005). Spd1 was originally identified as an S phase inhibitor (Woollard et al. 1996; Borgne and Nurse 2000). Liu et al. (2003) have demonstrated that Spd1 degradation is required for Suc22 redistribution and proposed that Spd1 is likely to anchor the RNR small subunit in the nucleus. No apparent sequence homolog of Spd1 has been identified in S. cerevisiae. In this study we explore the molecular mechanisms whereby the S. cerevisiae RNR small subunit is imported into the nucleus and redistributed to the cytoplasm upon genotoxic stress. By tagging either Rnr2 or Rnr4 with a nuclear export sequence (NES), we have shown that targeting either one of the two proteins to the cytoplasm results in cytoplasmic localization of the other. Moreover, we have demonstrated that mutations at the Rnr2:Rnr4 heterodimer interface can lead to cytoplasmic accumulation of both proteins without disrupting the heterodimeric complex. Finally, we provide experimental evidence suggesting that the DNA damage-induced redistribution of the RNR small subunit involves both active nuclear export and blockage of nuclear import. Taken together, our results suggest a model in which the heterodimeric RNR small subunit is transported across the nuclear envelope as one protein complex.
MATERIALS AND METHODS
Yeast strains and growth conditions:
All yeast strains in this study (Table 1) were derived from a W303 parental strain, Y300 (MATa can1-100 ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1) (Allen et al. 1994). Growth of yeast strains and genetic manipulations were as described previously (Burke et al. 2000). Hydroxyurea (HU; Sigma) was used at concentrations ranging from 25 to 150 mm, methyl methanesulfonate (MMS; Sigma) at a concentration of 0.025%, and 5-fluoroorotic acid (Sigma) at 1 g/liter.
TABLE 1.
Strains and plasmids
| Genotype | |
|---|---|
| Strains | |
| Y300 | MATacan1-100 ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 |
| MHY20 | as Y300 rnr4∷LEU2 pMH140 |
| MHY43 | as Y300 rnr2∷Tn(TRP1) pNN317 |
| MHY343 | as Y300 rnr2∷PRNR2-FLAG-RNR2-kan |
| MHY346 | as Y300 rnr4∷PRNR4-HA-RNR4-kan |
| MHY444 | as Y300 rnr2∷Tn(TRP1) pMH828 |
| MHY639 | as Y300 rnr2∷Tn(TRP1) pMH813 |
| MHY640 | as Y300 rnr2∷Tn(TRP1) pMH832 |
| MHY638 | as Y300 rnr2∷Tn(TRP1) pMH812 |
| MHY462 | as Y300 rnr4∷LEU2 pMH824 |
| MHY641 | as Y300 rnr4∷LEU2 pMH569 |
| MHY642 | as Y300 rnr4∷LEU2 pMH1307 |
| MHY643 | as Y300 rnr4∷LEU2 pMH1308 |
| Plasmids | |
| pMH569 | pRS413-PRNR4-HA-RNR4 (wild type, KKV at codons 50–52) |
| pMH812 | pRS415-PRNR2-3xMyc-RNR2 (KRA-to-AAA at codons 101–103) |
| pMH813 | pRS415-PRNR2-3xMyc-RNR2 (wild type, KRA at codons 101–103) |
| pMH824 | pRS413-PRNR4-HA-NES-RNR4 |
| pMH826 | pRS314-PRNR2-3xMyc-NES-RNR2 |
| pMH828 | pRS413-PRNR2-3xMyc-NES-RNR2 |
| pMH832 | pRS415-PRNR2-3xMyc-RNR2 (KRAΔ at codons 101–103) |
| pMH1026 | pRS314-PRNR4-FLAG-NES-RNR4 |
| pMH1184 | pRS416-PRNR4-HA-RNR4(1-340)-GFP |
| pMH1206 | pRS416-PRNR2-3xMyc-RNR2(1-393)-GFP |
| pMH1307 | pRS413-PRNR4-HA-RNR4 (KKV-to-AAA at codons 50–52) |
| pMH1308 | pRS413-PRNR4-HA-RNR4 (KKVΔ at codons 50–52) |
| pMH1326 | pRS416-GAL1-RNR4-GFP |
| pMH1470 | pRS416-PRNR3(Xm)-GFP |
Construction of the rnr2∷Tn(TRP1) and rnr4∷LEU2 strains were performed as described previously (Elledge and Davis 1987; Huang and Elledge 1997). Strains carrying various RNR2 and RNR4 alleles on centromeric plasmids in an rnr2Δ and an rnr4Δ background were generated from MHY43 and MHY20, respectively, using a plasmid shuffle strategy (Sikorski and Boeke 1991). MHY343 contains the FLAG-RNR2-kan cassette that is integrated into the chromosomal RNR2 locus under the control of the endogenous RNR2 promoter. The FlagRnr2 has the protein sequence MDYKDDDDKH preceding the Rnr2 sequence. MHY346 contains the HA-RNR4-kan cassette that is integrated into the chromosomal RNR4 locus under the control of the endogenous RNR4 promoter. The HARnr4 has the protein sequence MPYPYDVPDYASLGGH preceding the Rnr4 sequence.
Plasmid constructions:
All plasmids used in this study are listed in Table 1. Plasmids pNN317 and pMH140 were described previously (Elledge and Davis 1987; Huang and Elledge 1997). A 2751-bp NheI–AvrII restriction fragment that contains the RNR4 genomic DNA was subcloned into pRS413 (Sikorski and Hieter 1989) to generate pMH131. A 50-bp DNA sequence encoding the HA epitope followed by a five-residue linker, 5′-GC ATG CCT TAC CCA TAC GAT GTT CCA GAT TAC GCT AGC TTG GGT GGT CAT-3′, was introduced before the RNR4 coding sequence in pMH131 to create an NdeI site at the first ATG by site-directed mutagenesis (Smith 1985) and double-stranded oligonucleotide insertion, yielding pMH569. The rnr4(KKV-to-AAA) (or -3A) and rnr4(KKVΔ) (or -3Δ) mutations were introduced into pMH569 by site-directed mutagenesis to yield pMH1307 and pMH1308, respectively. An NheI site was created immediately 3′ to the HA-encoding sequence in pMH569 by site-directed mutagenesis and a 48-bp double-stranded oligonucleotide encoding for the MAPKK's NES (LQKKLEELELD) (Fukuda et al. 1996) was inserted between the NheI and NdeI sites, yielding pMH824. The HA-NES encoding sequence that is immediately 5′ to the RNR4 ORF in pMH824 is 5′-ATG CCT TAC CCA TAC GAT GTT CCA GAT TAC GCT AGC CTG CAA AAG AAG CTG GAA GAA CTG GAA CTG GAT GGA CAT-3′. The 2.9-kb SacI–ClaI fragment from pMH824 was subcloned into pRS314 (Sikorski and Hieter 1989), and the HA-encoding sequence before the NheI site was replaced by a FLAG-encoding sequence, 5′-ATG CCT GAC TAC AAA GAC GAT GAC GAC AAG-3′, yielding pMH1026. The green fluorescence protein (GFP) fusion constructs pMH1184, pMH1206, and pMH1326 are all based on pRS416 (Sikorski and Hieter 1989) and contain in-frame C-terminal fusions to the GFP (S65T, V163A) variant. pMH1184 contains an RNR4 promoter-driven N-terminally HA-tagged RNR4 (codons 1–340) fused to the GFP. pMH1206 contains an RNR2 promoter-driven N-terminally (Myc)3-tagged RNR2 (codons 1–393) fused to the GFP. pMH1326 contains a GAL1 promoter-driven (Mumberg et al. 1994) RNR4 (codons 1–340) fused to the GFP. The plasmids pMH812, pMH813, and pMH832 are all based on pRS415 (Sikorski and Hieter 1989) and contain an 816-bp RNR2 upstream sequence between ClaI and NcoI, followed by a 168-bp NcoI–NdeI fragment from pMH193 that encodes for a (Myc)3 epitope preceding the RNR2 coding sequence (Huang and Elledge 1997). pMH813 contains the wild-type RNR2 ORF; pMH812 and pMH832 contain the rnr2(KRAΔ) (or -3Δ) and rnr2(KRA-to-AAA) (or -3A) mutations, respectively. pMH1470 contains the unfused GFP that is under the control of a mutated, constitutively active RNR3 promoter in which all three binding sites for the repressor Crt1 have been removed (Huang et al. 1998).
Antibodies:
Polyclonal anti-Rnr2 and anti-Rnr4 antibodies were described previously (Yao et al. 2003). Monoclonal anti-Myc (9E10) and anti-HA (12CA5) were purchased from Roche Applied Sciences, and anti-HA (16B12) was from Covance Innovative Antibodies. HRP-, FITC-, and Cy3-conjugated goat anti-mouse and goat anti-rabbit antibodies were purchased from Jackson ImmunoResearch Labs. Polyclonal anti-Zwf1 (glucose-6-phosphate dehydrogenase) was purchased from Sigma.
Indirect immunofluorescence:
Fluorescence and DIC microscopy were performed using an E-800 microscope (Nikon). Images were acquired with a CoolSNAP-HQ 12-bit monochrome digital camera (Roper Scientific) using the Metamorph imaging system (Universal Imaging). Yeast cells were fixed with 4% formaldehyde in 0.1 m potassium phosphate buffer (pH 6.5) at room temperature for 15 min and subsequently treated with 10 μg/ml of zymolyase 100,000T (ICN) in a solution of 0.1 m potassium phosphate buffer (pH 7.0) and 1.2 m sorbitol at 37° for 10–15 min. All other incubations were done at room temperature in a solution of phosphate-buffered saline and 1% BSA. Cells were incubated with primary antibodies at a dilution of 1:100–1:200 (monoclonal) or 1:1000–1:10,000 (polyclonal) for 3 hr, washed extensively with the PBS/1% BSA solution, and incubated with FITC- or Cy3-conjugated secondary antibodies at a 1:200 dilution for 1.5 hr. DNA was visualized by a 3 min incubation of cells with 1 μg/ml of 4′,6-diamidino-2-phenylindole (DAPI).
Protein extraction, immunoprecipitation, and Western blotting:
Yeast cells were harvested from early- to mid-log phase cultures (1–2 × 107 cells/ml). For Western blots, protein extracts were prepared using the trichloroacetic acid (TCA) method. Briefly, cells harvested from a 10-ml culture were washed with 2 ml of 20% TCA, resuspended in 100 μl of 20% TCA, and disrupted with glass beads on a BeadBeater (BioSpec Products). Each sample was centrifuged at 800 × g for 10 min and the supernatant was discarded. The pellets were resuspended in 100 μl of a 2× SDS sample buffer plus 50 μl of 1 m Tris base to adjust the pH and sonicated at 20% output level (Branson Sonifier 250) for 5 sec. Samples were heated at 100° for 5 min, followed by centrifugation at 800 × g for 10 min. The supernatants were resolved by 8–10% SDS–PAGE. For immunoprecipitation, protein extracts were prepared by the glass bead disruption method in buffer B (50 mm HEPES KOH pH 7.5, 140 mm NaCl, 1 mm EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 1 mm phenylmethylsulfonal fluoride, 1 μg/ml each of leupeptin and pepstatin A) and centrifuged at 13,400 × g for 15 min to remove debris. Total yeast protein extracts (500 μg for each sample) were diluted to a total volume of 500 μl with buffer B and incubated with 5–10 μg of anti-HA (12CA5) or anti-Myc (9E10) monoclonal antibodies overnight at 4°. The antibody–protein complexes were precipitated by absorption to protein A-Sepharose beads for 3 hr at 4° and washed three times using a high-salt buffer B (200 mm NaCl) at room temperature. The immunoprecipitates were resolved by 8% SDS–PAGE. For immunoblotting, proteins were transferred to nitrocellulose membranes after electrophoresis. The membranes were incubated with primary antibodies for ≥2 hr followed by secondary antibody incubation for 1 hr, both at room temperature. Blots were developed with peroxidase-labeled secondary antibodies at a dilution of 1:10,000 using an enhanced chemiluminescence substrate (Perkin-Elmer). Primary antibodies were used at the following dilutions: 12CA5 at 1:1000; 9E10 at 1:1000; anti-Rnr2 and anti-Rnr4 at 1:10,000; and anti-Zwf1 at 1:100,000.
RESULTS
Rnr2 and Rnr4 are actively imported into the nucleus:
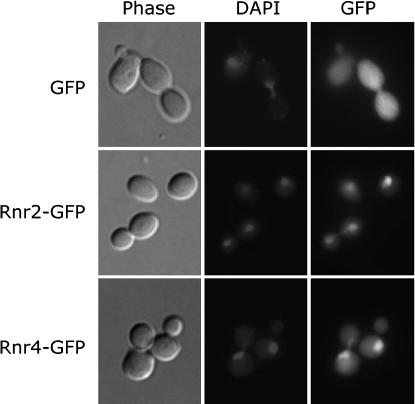
Recent studies suggest that the Rnr2:Rnr4 heterodimer is the active form of the S. cerevisiae RNR small subunit (Huang and Elledge 1997; Chabes et al. 2000; Ge et al. 2001; Perlstein et al. 2005). Both Rnr2 and Rnr4 are predominantly localized to the nucleus during most phases of the cell cycle except for the S phase (Yao et al. 2003). The molecular weights of Rnr2 (46 kDa) and Rnr4 (40 kDa) are within the proposed ∼40–60 kDa cutoff range for unrestrained, passive diffusion through the nuclear pore complex (Mattaj and Englmeier 1998; Gorlich and Kutay 1999; Weis 2003). To determine whether the nuclear localization of Rnr2 and Rnr4 involves active nuclear import, we fused the sequence encoding for the 27-kDa GFP to the end of the RNR2 and RNR4 ORFs and introduced these GFP fusion plasmids into the wild-type cells to track the subcellular localization of the resulting fusion proteins. A plasmid expressing the unfused GFP from a mutated, constitutively active RNR3 promoter was introduced into the wild-type cells as a control. In cells expressing the unfused GFP, the GFP signal was ubiquitously diffused throughout the cell (Figure 1, top). In contrast, in cells expressing the Rnr2-GFP and Rnr4-GFP fusion proteins, the GFP signals were predominantly localized to the nucleus (Figure 1, middle and bottom). These results suggest that Rnr2 and Rnr4 contain a signal for active nuclear import.
Figure 1.
Both Rnr2 and Rnr4 can target the GFP to the nucleus. All proteins were expressed in the wild-type strain and the GFP signal was observed in live cells. (Top) The subcellular localization of the unfused GFP that was expressed under the control of a mutated, constitutive RNR3 promoter in which all three binding sites for the repressor Crt1 were removed. (Middle and bottom) Subcellular localizations of the Rnr2-GFP and Rnr4-GFP fusion proteins under the control of their respective promoters. Nuclei were visualized by DAPI staining.
Rnr2 and Rnr4 are cotransported between the nucleus and the cytoplasm:
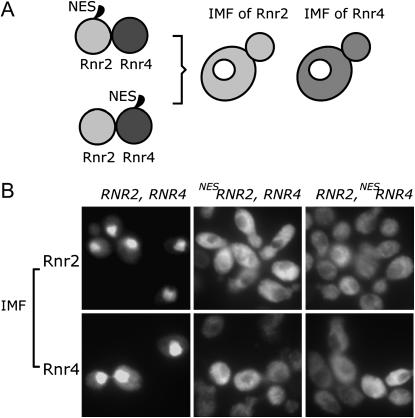
To investigate whether Rnr2 and Rnr4 contain their own nuclear localization signals (NLS) and are thus imported into the nucleus independently of each other, we attempted to map the minimal sequence sufficient for NLS in both proteins by making serial deletions from either the N- or the C-terminus. None of the eight truncated constructs we made leads to stably expressed proteins either on their own or when fused to the GFP (data not shown). We envisioned that one protein of the Rnr2:Rnr4 heterodimer might be directly recognized by the nuclear import machinery, while the other protein could be imported simply by being part of the complex via a piggyback mechanism (Kang et al. 1994; Leslie et al. 2004). To test this hypothesis directly, we tagged the N-terminus of Rnr2 with the NES from the MAP kinase kinase (Fukuda et al. 1996, 1997). A plasmid bearing the NES-RNR2 cassette under the control of the RNR2 promoter was introduced into a rnr2 null strain using a plasmid shuffle strategy (Sikorski and Boeke 1991) so that the only Rnr2 protein produced in these cells was the NES-Rnr2. Cells containing the NES-RNR2 exhibited no difference in either growth rate or sensitivity to the potent RNR inhibitor HU in comparison to the wild-type cells (data not shown). Thus, the NES tag does not appear to perturb the RNR holoenzyme function. Immunofluorescence staining revealed that the NES-Rnr2 protein was exclusively localized in the cytoplasm (Figure 2), indicating that the NES functioned properly. If the nuclear import of Rnr4 is independent of that of Rnr2, then the endogenous Rnr4 protein should still be predominantly localized to the nucleus. Instead, we found that the endogenous Rnr4 protein in these NES-Rnr2-expressing cells was predominantly localized to the cytoplasm (Figure 2). We did the reciprocal experiment to generate an rnr4 null strain expressing a RNR4 promoter-driven NES-RNR4, and found that both the NES-Rnr4 and the endogenous Rnr2 proteins were predominantly localized to the cytoplasm (Figure 2). Taken together, these results support the notion that Rnr2 and Rnr4 are concertedly transported across the nuclear envelope as one protein complex.
Figure 2.
Tagging either Rnr2 or Rnr4 with a nuclear export signal (NES) causes both proteins to be localized to the cytoplasm. (A) A schematic showing that tagging either Rnr2 or Rnr4 with an NES leads to cytoplasmic localization of both proteins. (B) Immunofluorescence (IMF) staining images of Rnr2 (top) and Rnr4 (bottom) in cells that contain no NES tag (left), cells that express an NES-tagged Rnr2 as the only RNR2 product (middle), and cells that express an NES-tagged Rnr4 as the only RNR4 product (right).
Ectopic expression of an NES-Rnr2 affects the subcellular localization pattern of the endogenous Rnr4 but not that of the endogenous Rnr2:
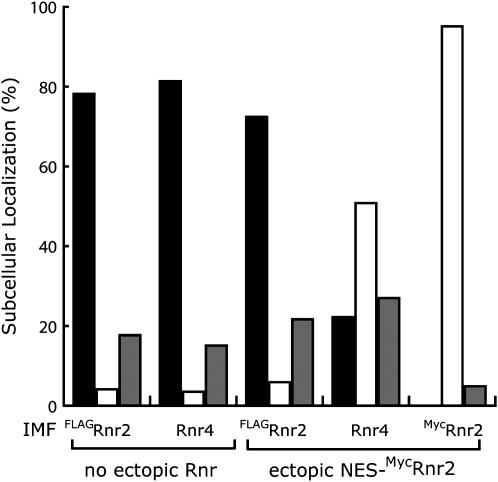
Although Rnr2 and Rnr4 have an ability to form a heterodimer (Huang and Elledge 1997; Chabes et al. 2000; Ge et al. 2001; Voegtli et al. 2001), it is unclear whether the heterodimer is the major form or the Rnr2:Rnr4 heterodimer and the homodimers of Rnr2:Rnr2 and Rnr4:Rnr4 coexist in vivo. We reasoned that if the Rnr2:Rnr4 heterodimer and the Rnr2:Rnr2 homodimer were present at comparable levels inside the cell, then an ectopically expressed NES-Rnr2 should perturb the subcellular localization patterns of both the endogenous Rnr2 and the endogenous Rnr4 so that both of the proteins would be partially localized to the cytoplasm. To test this idea we tagged the endogenous RNR2 gene in its own chromosomal locus with a FLAG epitope-encoding sequence at the 5′ end of the RNR2 coding sequence. The strain bearing the chromosomal FLAG-RNR2 showed no difference in growth rate or HU sensitivity in comparison to the wild-type strain (data not shown). Moreover, both the FLAG-Rnr2 and Rnr4 proteins were predominantly localized to the nucleus in the FLAG-RNR2-expressing cells (Figure 3), suggesting that the FLAG-Rnr2 could substitute for the native Rnr2 function. We then introduced into this strain a plasmid containing a (Myc)3-NES-RNR2 cassette that was under the control of the RNR2 promoter so that there were approximately equal amounts of FLAG-Rnr2 and Myc-NES-Rnr2 in these cells. The endogenous FLAG-Rnr2 and the ectopically expressed (Myc)3-NES-Rnr2 were independently detected by immunofluorescence staining using anti-FLAG and anti-Myc antibodies, respectively. The ectopically expressed (Myc)3-NES-Rnr2 was found to be localized predominantly to the cytoplasm (Figure 3, far right), indicating that the NES functioned properly. Under these conditions the endogenous FLAG-Rnr2 remained largely in the nucleus (Figure 3, left). In contrast, the subcellular localization pattern of the endogenous Rnr4 protein was significantly changed; it was predominantly in the cytoplasm among ∼50% of the cells and was ubiquitously distributed between the nucleus and the cytoplasm in ∼20% of the population (Figure 3, right). Similarly, an ectopically expressed NES-tagged Rnr4 changed the localization of the endogenous Rnr2 but not the endogenous Rnr4 (data not shown). These results strongly suggest that the major form of the S. cerevisiae RNR small subunit is the Rnr2:Rnr4 heterodimer instead of the Rnr2:Rnr2 or the Rnr4:Rnr4 homodimers and are also consistent with the notion that the Rnr2:Rnr4 heterodimer is transported as one protein complex.
Figure 3.
Ectopic expression of an NES-tagged Rnr2 perturbs the subcellular localization of the endogenous Rnr4 but not the endogenous Rnr2. All cells used in this assay contain an N-terminally FLAG-tagged RNR2 in the chromosomal RNR2 locus under the control of the endogenous RNR2 promoter; the chromosomal RNR4 locus is not modified. Subcellular localization patterns of the chromosomally encoded FLAGRnr2 and Rnr4 were examined by immunofluorescence staining (IMF) in the absence (no ectopic Rnr) or presence of a plasmid expressing an N-terminally (Myc)3-tagged Rnr2 (ectopic NES- MycRnr2). At least 200 cells were counted for each experiment. The percentage of cells with a distinct localization pattern was represented as follows: the solid bar shows cells exhibiting a predominantly nuclear signal (Nu); the open bar shows cells exhibiting a predominantly cytoplasmic signal (Cyto); the shaded bar shows cells exhibiting no difference (ND) in signal intensities between the nucleus and the cytoplasm.
Mutations at the Rnr2:Rnr4 heterodimer interface affect the subcellular localization of both proteins without disrupting the heterodimeric complex:
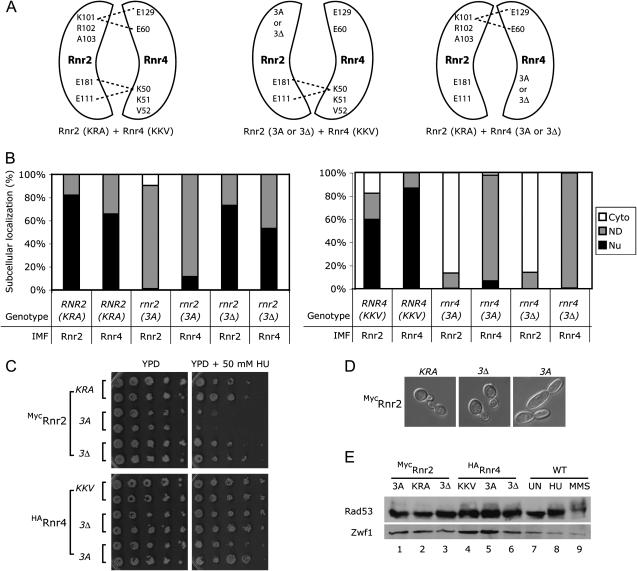
We surmised that if Rnr2 and Rnr4 are transported as one heterodimeric complex, then mutations that affect the heterodimer conformation could disrupt this process. The crystal structure of the Rnr2:Rnr4 heterodimer reveals a cluster of hydrophobic interactions and several hydrogen bonds between charged residues at the Rnr2:Rnr4 interface (Voegtli et al. 2001). The residue lysine101 of Rnr2 forms hydrogen bonds with the residues glutamate 60 and glutamate 129 of Rnr4, while the residue lysine 50 of Rnr4 forms hydrogen bonds with the residues glutamate 111 and glutamate 181 of Rnr2 (Figure 4A). To test our hypothesis, we introduced two types of substitutions at these hydrogen bond-forming, charged residues. One type of mutation was a deletion of three neighboring residues (i.e., 3Δ) including the lysine (KRA in Rnr2 and KKV in Rnr4, Figure 4A); the other was a substitution of all three neighboring residues by alanines (i.e., 3A). All these mutant alleles of RNR2 and RNR4 were introduced into the respective rnr2 and rnr4 null strains so that the only Rnr2 and Rnr4 proteins expressed in these cells were the mutated versions. The strains bearing the rnr2(3Δ), rnr4(3Δ) and rnr4(3A) mutations exhibited normal growth rates (data not shown) and HU resistance relative to a their wild-type counterparts (Figure 4C), suggesting that these mutations did not significantly affect RNR function. The rnr2(3A) mutant cells were more sensitive to HU in comparison to the wild-type [RNR2(KRA)] cells (Figure 4C) and had a more elongated shape than the RNR2(KRA) and rnr2(3Δ) cells (Figure 4D), suggesting that the rnr2(3A) mutation compromised RNR function.
Figure 4.
Mutations at the interface of the Rnr2:Rnr4 heterodimer cause loss of proper nuclear localization of both Rnr2 and Rnr4. (A) A schematic of the charged residues that are involved in hydrogen bond formation at the Rnr2:Rnr4 heterodimer interface, and the mutations introduced at these residues. (B) Quantitative analysis of Rnr2 and Rnr4 subcellular localization patterns in cells expressing different rnr2 and rnr4 interface mutant alleles. Strains shown on the left carry a centromeric plasmid that expresses the N-terminally (Myc)3-tagged RNR2 wild-type (i.e., KRA), the rnr2(KRA-to-AAA) (i.e., 3A), or the rnr2(KRAΔ) (i.e., 3Δ) mutant allele in a rnr2 null background. Strains shown on the right carry a centromeric plasmid that express the N-terminally HA-tagged RNR4 wild-type (i.e., KKV), the rnr4(KKV-to-AAA) (i.e., 3A), or the rnr4(KKVΔ) (i.e., 3Δ) mutant allele in a rnr4 null background. The subcellular localization patterns of Rnr2 and Rnr4 in each strain were quantified and presented as described in the Figure 3 legend. (C) Comparison of growth of the wild-type and the rnr2 and rnr4 interface mutant strains on the rich-medium YPD plates (left) and YPD plates containing 50 mm HU (right). Serial fivefold dilutions of an early-log phase culture of each strain were spotted onto YPD or YPD + HU plates and incubated at 30°C for 2 days. (D) Comparison of cell morphology among strains expressing the N-terminally (Myc)3-tagged RNR2 wild-type (KRA), or rnr2(KRA-to-AAA) (3A) and rnr2(KRAΔ) (3Δ) mutant alleles as the only Rnr2 protein. (E) Comparison of Rad53 mobility among the wild-type and the rnr2 and rnr4 interface mutant cells. Protein extract was prepared from an early-log phase culture of each strain (lanes 1–7) and the wild-type cells after 2 hr treatment with 150 mm HU and 0.025% MMS (lanes 8 and 9). Protein extract from 5 × 106 cells of each strain was separated by SDS–PAGE and probed on a Western blot using an anti-Rad53 polyclonal antibody.
The rnr2(3Δ) mutation did not cause any significant change in the subcellular localization patterns of either the Rnr2(3Δ) or the endogenous Rnr4 proteins compared to the wild-type allele RNR2(KRA) (Figure 4B, left). Conversely, the rnr2(3A) mutation changed subcellular localizations of both the mutant Rnr2(3A) and the endogenous Rnr4 proteins. Instead of being predominantly nuclear, the two proteins exhibited a more ubiquitous localization pattern between the nucleus and the cytoplasm (Figure 4B, left). Moreover, we found that both the rnr4(3Δ) and the rnr4(3A) mutations drastically changed the subcellular distribution of both the respective mutant Rnr4 proteins and the endogenous Rnr2 protein. In these cells bearing the rnr4 mutations, the mutated Rnr4 proteins were present uniformly between the nucleus and the cytoplasm, while the endogenous Rnr2 protein was predominantly in the cytoplasm (Figure 4B, right). Interestingly, the rnr2(3Δ) mutation exhibited a much milder effect on the subcellular localization patterns of Rnr2 and Rnr4 when compared to rnr4(3Δ). These results are consistent with the hypothesis that the Rnr2:Rnr4 heterodimer is recognized and transported into the nucleus by the nucleus–cytoplasm transport machinery as one entity.
It should be emphasized that these rnr2 and rnr4 mutants may compromise the RNR enzyme function and consequently activate the DNA damage and replication checkpoint, which could lead to subcellular redistribution of both Rnr2 and Rnr4 (Yao et al. 2003). To test this possibility, we examined Rad53 phosphorylation status by detecting its electrophoretic mobility on a Western blot, which has been shown to be an indicator of checkpoint activation (Sanchez et al. 1996; Sun et al. 1996). In cases of both RNR2 (Figure 4E, lanes 1–3) and RNR4 (Figure 4E, lanes 4–6) we detected no significant difference in Rad53 mobility between the wild-type strain (KRA in RNR2, KKV in RNR4) and the two mutant strains (3A and 3Δ). Moreover, the majority of the Rnr2 and Rnr4 proteins in the RNR2(KRA) and RNR4(KKV) wild-type strains are localized in the nucleus (Figure 2B). Thus, the difference in the subcellular localization patterns of the RNR small subunit in these mutants is unlikely to be caused by an activated checkpoint response.
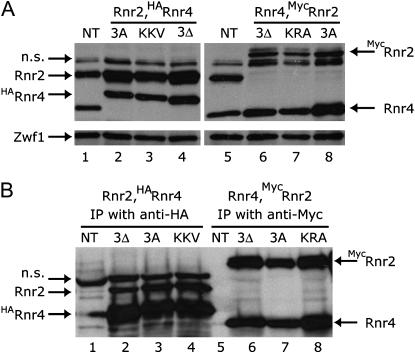
We also considered the possibility that the mutations at the Rnr2:Rnr4 interface might disrupt the formation of the heterodimeric complex, thereby leading to the mislocalization of each protein. To test this notion, we compared the Rnr2:Rnr4 heterodimer formation between the mutant and the wild-type strains by co-immunoprecipitation. We tagged the Rnr4, Rnr4(3Δ), and Rnr4(3A) proteins with an HA epitope, and the Rnr2, Rnr2(3Δ), and Rnr2(3A) proteins with a (Myc)3 epitope, all at the N-terminus. The levels of the (Myc)3-tagged Rnr2 and the HA-tagged Rnr4 proteins were comparable to the respective endogenous Rnr2 and Rnr4 levels in the wild-type strain (Figure 5A). The endogenous Rnr2 levels in the HA-RNR4-expressing strains were higher than that in the untagged wild-type strain (Figure 5A, lanes 2–4 vs. lane 1). Similarly, the endogenous Rnr4 levels in the (Myc)3-RNR2-expressing strains were also higher than those in the untagged wild-type strain (Figure 5A, lanes 6–8 vs. lane 5). However, the endogenous Rnr2 levels among the HA-tagged RNR4 variants were comparable and so were the endogenous Rnr4 levels among the (Myc)3-tagged RNR2 variants. The only exception was the (Myc)3-rnr2(3A) mutant, which appeared to have a slightly higher Rnr4 protein level (Figure 5A, lane 8 vs. lanes 5–7). No significant difference was found in the levels of the Rnr2:Rnr4 heterodimeric complex among any of these strains, as judged by co-immunoprecipitation assays (Figure 5B, lanes 2–4 and lanes 6–8). Thus, we conclude that the difference in RNR small subunit localization patterns caused by these mutations at the Rnr2:Rnr4 interface cannot be attributed to disruption of the Rnr2:Rnr4 heterodimer formation in vivo.
Figure 5.
The Rnr2:Rnr4 interface mutations do not disrupt heterodimer formation in vivo. (A) Comparison of the Rnr2 and Rnr4 protein levels between the wild-type and the rnr2 and rnr4 mutant strains. Genotype description of the strains were the same as in Figure 4B. NT is a wild-type strain that contains no epitope tag. An early log phase culture of each strain was used for protein extract preparation in buffer B, and 10 μg of each protein extract was separated by SDS–PAGE and probed on a Western blot for Rnr2 and Rnr4. Glucose-6-phosphate dehydrogenase (Zwf1) was probed on the same blot as a loading control. The bands shown from the top to the bottom of the blot are (Myc)3-Rnr2, Rnr2, HA-Rnr4, and Rnr4, respectively. The position of an unidentified protein that exhibits nonspecific (n.s.) reaction with the secondary antibody is also indicated. (B) Comparison of Rnr2-Rnr4 coimmunoprecipitation between the wild-type and the rnr2 and rnr4 mutant strains. Protein extracts were prepared as described in A. For each immunoprecipitation, 500 μg of protein extract was incubated with 5–10 μg anti-HA or anti-Myc monoclonal antibodies followed by protein-A beads absorption. After washing, the immune complexes were eluted from the beads, separated by SDS–PAGE, and probed for Rnr2 and Rnr4 on a Western blot. NT refers to a nontagged, wild-type strain. The positions of the (Myc)3-Rnr2, Rnr2, HA-Rnr4 and Rnr4 are indicated. The immune complexes brought down by the anti-HA monoclonal antibody contain an unidentified protein that exhibits nonspecific (n.s.) reaction with the secondary antibody.
DNA damage-induced redistribution of Rnr4 involves both blockage of nuclear import and active nuclear export:
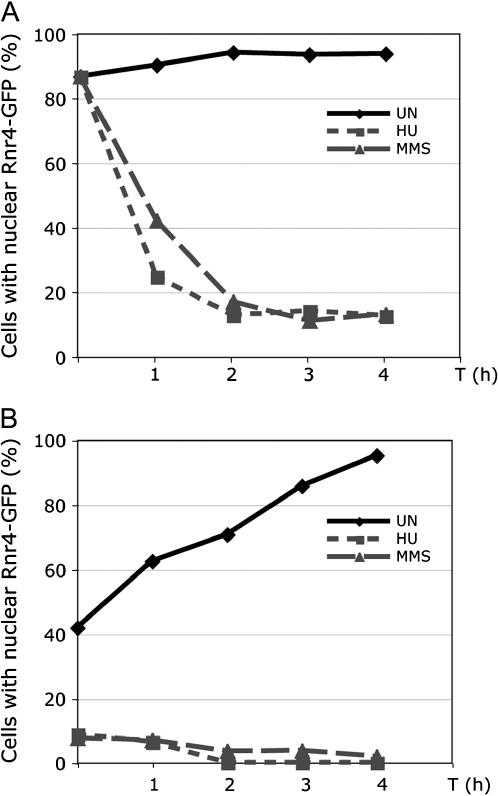
We envisioned two possible mechanisms of DNA damage-induced RNR small subunit redistribution. One possibility is that the newly synthesized Rnr2 and Rnr4 proteins may be blocked from being imported to the nucleus. Alternatively, the previously synthesized proteins that have already been imported into the nucleus may be actively exported back to the cytoplasm. These two mechanisms are not mutually exclusive and both may contribute to the redistribution of Rnr2 and Rnr4. To test these two possibilities, we generated a GAL1–RNR4–GFP construct in which the expression of the Rnr4-GFP fusion protein was under the control of the GAL1 promoter, which could be quickly turned on and off by switching the carbon source (data not shown). In media containing glucose (repressed state) or raffinose (derepressed state) no visible GFP signal was observed, suggesting that no significant amount of Rnr4-GFP was made. Upon galactose addition, GFP signal became visible within ∼30 min and accumulated first in the cytoplasm and later in both the nucleus and the cytoplasm. The Rnr4-GFP protein level peaked ∼90–120 min after galactose induction as determined by Western blot (data not shown), at which point the promoter was shut off by the addition of glucose. After glucose addition, the GFP signal became gradually enriched in the nucleus. By the 120-min time point after promoter shutoff >90% of the population had predominantly nuclear GFP, indicating that most of the Rnr4-GFP was localized to the nucleus (Figure 6A, time zero). When HU and MMS were added at this time point (2 hr after promoter shutoff), the Rnr4-GFP in the majority of the cells became redistributed to the cytoplasm within 2 hr, while the Rnr4-GFP in untreated cells remained in the nucleus (Figure 6A). Because there is no new Rnr4-GFP protein synthesis 2 hr postpromoter shutoff (data not shown), the HU- and MMS-induced redistribution of Rnr4-GFP is likely caused by an active export of the previously synthesized and nuclearly localized Rnr4-GFP protein from the nucleus back to the cytoplasm.
Figure 6.
Redistribution of preexisting and newly synthesized Rnr4-GFP after HU and MMS treatment. (A) Preexisting Rnr4-GFP is exported out of the nucleus in cells treated with HU and MMS. Cells harboring the GAL1-RNR4-GFP plasmid were grown to early-log phase in a raffinose-containing medium. GAL1 promoter-driven expression of the Rnr4-GFP fusion protein was induced for 90 min in a galactose-containing medium before being shut off by glucose addition. At 2 hr after promoter shutoff (time zero), the culture was split into three parts: one was untreated (UN), one treated with HU to a final concentration of 150 mm, and the third treated with MMS to a final concentration of 0.025%. The localization of Rnr4-GFP was visualized in live cells. At each indicated time point, 200 cells were counted and the percentage of cells containing a predominantly nuclear GFP signal was presented. (B) Nuclear import of newly synthesized Rnr4-GFP is blocked in cells treated with HU and MMS. Cells harboring the GAL1-RNR4GFP plasmid were grown to early log phase in a raffinose-containing medium. GAL1 promoter-driven expression of the Rnr4-GFP fusion protein was induced for 45 min by galactose addition and the culture was split into three parts: one was untreated (UN), one treated with HU to a final concentration of 150 mm, and the third treated with MMS to a final concentration of 0.025%. All three cultures were incubated for an additional 45 min in the galactose-containing medium before being switched to a glucose-containing medium to shut off the GAL1 promoter (time zero). The localization of Rnr4-GFP was counted at the indicated time points and presented as described in A.
In a separate assay, we added HU and MMS while the GAL promoter was still on, and followed Rnr4-GFP localization after promoter shutoff in the presence of HU and MMS. We found that Rnr4-GFP remained in the cytoplasm in the HU- or MMS-treated cells, as opposed to being predominantly localized to the nucleus in the untreated cells after promoter shutoff (Figure 6B), which suggests that the newly synthesized Rnr4-GFP protein is prevented from being imported into the nucleus upon HU and MMS treatment. Taken together, our findings support that hypothesis that the DNA damage checkpoint-mediated nucleus-to-cytoplasm redistribution of Rnr4 involves both active nuclear export and blockage of nuclear import.
DISCUSSION
The RNR enzyme plays an essential role in the biosynthesis of dNTPs for DNA replication, recombination, and repair processes in both the nucleus and the mitochondria. Failure in proper regulation of RNR activity can lead to severe outcomes, such as genome instability and cell death (Desany et al. 1998; Zhao et al. 1998; Ouspenski et al. 1999; Chabes et al. 2003). The critical importance of maintaining adequate and balanced dNTP pools in vivo is also manifested in the complex and multilayered mechanisms cells exploit to regulate the RNR activity. Recent studies from several laboratories have demonstrated that the DNA damage-induced subcellular redistribution of the RNR subunits plays an important role in regulating RNR activity (Liu et al. 2003; Xue et al. 2003; Yao et al. 2003; Lincker et al. 2004). The active form of the S. cerevisiae RNR small subunit is likely to be an Rnr2:Rnr4 heterodimer on the basis of both biochemical and genetic studies (Huang and Elledge 1997; Chabes et al. 2000; Ge et al. 2001). In this work we provide in vivo evidence that the Rnr2:Rnr4 heterodimer is the major form of the RNR small subunit. We have shown that excluding either Rnr2 or Rnr4 from the nucleus by NES tagging leads to cytoplasmic localization of both proteins. Moreover, ectopic expression of an NES-Rnr2 disrupts the nuclear localization of the endogenous Rnr4 but not the endogenous Rnr2. We have also demonstrated that mutations at the Rnr2:Rnr4 interface can perturb the subcellular localization patterns of the two proteins, even though the heterodimer can still form. Hence, the Rnr2:Rnr4 heterodimer is not only the major form of RNR small subunit in S. cerevisiae but also cotransported as one protein complex between the nucleus and the cytoplasm.
Proteins that shuttle between the nucleus and cytoplasm usually contain both NLS and NES (Weis 2003). Proteins that lack an NLS also can be imported through association with an NLS-containing protein via the so-called piggyback mechanism (Kang et al. 1994; Leslie et al. 2004). In some cases, only a single subunit of the protein complex contains NLS and is responsible for the nuclear import of the entire complex (Maridor et al. 1993; Pereira et al. 1998; Subramaniam and Johnson 2004; Wendler et al. 2004). In other cases, the nuclear targeting signal is composed of sequences from more than one subunit of the protein complex. For example, the replication initiator protein complex Mcm2-7 has been shown to colocalize as one complex (Nguyen et al. 2000). Recently, Liku and co-workers have reported that the nuclear import of the Mcm2-7 complex is mediated by two partial NLS on two of the six subunits, Mcm2 and Mcm3 (Liku et al. 2005). Each NLS alone is necessary but not sufficient for nuclear localization of the Mcm2-7 complex (Liku et al. 2005). Interestingly, the two NLS can efficiently target a heterologous cytoplasmic protein to the nucleus when they are positioned in cis (Liku et al. 2005). It would thus appear that a functional nuclear-targeting signal can be distributed on more than one subunit of the same protein complex, which makes complex formation a prerequisite for nuclear import (Liku et al. 2005). It is possible that the targeting signal for nuclear import of Rnr2 and Rnr4 is also distributed on both proteins and becomes a sufficient NLS only when they dimerize. The mutations we introduced at the Rnr2:Rnr4 interface, although having no apparent effects on the Rnr2:Rnr4 heterodimer formation, may cause subtle local conformational changes in the heterodimer, thereby perturbing the proper recognition of the NLS by importin(s).
How does the subcellular redistribution of Rnr2:Rnr4 occur? Our finding that both Rnr2 and Rnr4 can target the GFP to the nucleus suggests that their nuclear localizations are a result of active import rather than passive diffusion. The question remains, however, whether there is a nuclear anchoring protein that prevents Rnr2:Rnr4 from being exported back to the cytoplasm, as proposed for the role of Spd1 in S. pombe (Liu et al. 2003). In S. cerevisiae, both active nuclear import and nuclear anchoring appear to be involved in the nuclear localization of the RNR small subunit. Thus, the DNA damage-induced redistribution could result from disruption of both processes. Indeed, HU and MMS treatments not only prevent the newly synthesized Rnr4-GFP from being imported into the nucleus but also cause the preexisting nuclear Rnr4-GFP to re-localize to the cytoplasm. This relocalization process appears to be partially dependent on the exportin Crm1, as the inhibitor of NES-dependent transport leptomycin b (LMB) (Nishi et al. 1994; Fukuda et al. 1997) does not completely block such a process in the crm1(T539C) strain that is LMB-sensitive (Neville and Rosbash 1999; X. An, Z. Zhang, K.Yang and M. Huang, unpublished data). Thus both blockage of import and activation of export appear to play a role in the subcellular redistribution of Rnr2 and Rnr4 upon genotoxic stress. Interestingly, activation of nuclear export is also required for DNA damage-induced RNR small subunit redistribution in S. pombe (Liu et al. 2003). Unraveling the molecular details of the nuclear import and export processes of Rnr2:Rnr4 will lead to a detailed mechanistic understanding of the DNA damage checkpoint-mediated RNR redistribution. These studies may also provide new molecular target(s) for RNR inhibition.
Acknowledgments
We thank JoAnne Stubbe and Debra Perlstein for helpful discussions. This work was supported by a National Institute of Health grant (CA-095207) and an American Society of Cancer grant (RSG0305001) to M.H.
References
- Allen, J. B., Z. Zhou, W. Siede, E. C. Friedberg and S. J. Elledge, 1994. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 8: 2401–2415. [DOI] [PubMed] [Google Scholar]
- Amin, N. S., M. N. Nguyen, S. Oh and R. D. Kolodner, 2001. exo1-Dependent mutator mutations: model system for studying functional interactions in mismatch repair. Mol. Cell. Biol. 21: 5142–5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondar, T., A. Ponomarev and P. Raychaudhuri, 2004. Ddb1 is required for the proteolysis of the Schizosaccharomyces pombe replication inhibitor Spd1 during S phase and after DNA damage. J. Biol. Chem. 279: 9937–9943. [DOI] [PubMed] [Google Scholar]
- Borgne, A., and P. Nurse, 2000. The Spd1p S phase inhibitor can activate the DNA replication checkpoint pathway in fission yeast. J. Cell Sci. 113(Pt 23): 4341–4350. [DOI] [PubMed] [Google Scholar]
- Burke, D., D. Dawson and T. Stearns, 2000. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Chabes, A., V. Domkin and L. Thelander, 1999. Yeast Sml1, a protein inhibitor of ribonucleotide reductase. J. Biol. Chem. 274: 36679–36683. [DOI] [PubMed] [Google Scholar]
- Chabes, A., V. Domkin, G. Larsson, A. Liu, A. Graslund et al., 2000. Yeast ribonucleotide reductase has a heterodimeric iron-radical-containing subunit. Proc. Natl. Acad. Sci. USA 97: 2474–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabes, A., B. Georgieva, V. Domkin, X. Zhao, R. Rothstein et al., 2003. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell 112: 391–401. [DOI] [PubMed] [Google Scholar]
- Desany, B. A., A. A. Alcasabas, J. B. Bachant and S. J. Elledge, 1998. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 12: 2956–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge, S. J., and R. W. Davis, 1987. Identification and isolation of the gene encoding the small subunit of ribonucleotide reductase from Saccharomyces cerevisiae: DNA damage-inducible gene required for mitotic viability. Mol. Cell. Biol. 7: 2783–2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda, M., I. Gotoh, Y. Gotoh and E. Nishida, 1996. Cytoplasmic localization of mitogen-activated protein kinase kinase directed by its NH2-terminal, leucine-rich short amino acid sequence, which acts as a nuclear export signal. J. Biol. Chem. 271: 20024–20028. [DOI] [PubMed] [Google Scholar]
- Fukuda, M., S. Asano, T. Nakamura, M. Adachi, M. Yoshida et al., 1997. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390: 308–311. [DOI] [PubMed] [Google Scholar]
- Ge, J., D. L. Perlstein, H. H. Nguyen, G. Bar, R. G. Griffin et al., 2001. Why multiple small subunits (Y2 and Y4) for yeast ribonucleotide reductase? Toward understanding the role of Y4. Proc. Natl. Acad. Sci. USA 98: 10067–10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich, D., and U. Kutay, 1999. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15: 607–660. [DOI] [PubMed] [Google Scholar]
- Holmberg, C., O. Fleck, H. A. Hansen, C. Liu, R. Slaaby et al., 2005. Ddb1 controls genome stability and meiosis in fission yeast. Genes Dev. 19: 853–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, M., and S. J. Elledge, 1997. Identification of RNR4, encoding a second essential small subunit of ribonucleotide reductase in Saccharomyces cerevisiae. Mol. Cell. Biol. 17: 6105–6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, M., Z. Zhou and S. J. Elledge, 1998. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 94: 595–605. [DOI] [PubMed] [Google Scholar]
- Jordan, A., and P. Reichard, 1998. Ribonucleotide reductases. Annu. Rev. Biochem. 67: 71–98. [DOI] [PubMed] [Google Scholar]
- Kang, K. I., J. Devin, F. Cadepond, N. Jibard, A. Guiochon-Mantel et al., 1994. In vivo functional protein-protein interaction: nuclear targeted hsp90 shifts cytoplasmic steroid receptor mutants into the nucleus. Proc. Natl. Acad. Sci. USA 91: 340–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashlan, O. B., C. P. Scott, J. D. Lear and B. S. Cooperman, 2002. A comprehensive model for the allosteric regulation of mammalian ribonucleotide reductase. Functional consequences of ATP- and dATP-induced oligomerization of the large subunit. Biochemistry 41: 462–474. [DOI] [PubMed] [Google Scholar]
- Leslie, D. M., W. Zhang, B. L. Timney, B. T. Chait, M. P. Rout et al., 2004. Characterization of karyopherin cargoes reveals unique mechanisms of Kap121p-mediated nuclear import. Mol. Cell. Biol. 24: 8487–8503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liku, M. E., V. Q. Nguyen, A. W. Rosales, K. Irie and J. J. Li, 2005. CDK phosphorylation of a novel NLS-NES module distributed between two subunits of the Mcm2–7 complex prevents chromosomal rereplication. Mol. Biol. Cell 16: 5026–5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincker, F., G. Philipps and M. E. Chaboute, 2004. UV-C response of the ribonucleotide reductase large subunit involves both E2F-mediated gene transcriptional regulation and protein subcellular relocalization in tobacco cells. Nucleic Acids Res. 32: 1430–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C., K. A. Powell, K. Mundt, L. Wu, A. M. Carr et al., 2003. Cop9/signalosome subunits and Pcu4 regulate ribonucleotide reductase by both checkpoint-dependent and -independent mechanisms. Genes Dev. 17: 1130–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maridor, G., P. Gallant, R. Golsteyn and E. A. Nigg, 1993. Nuclear localization of vertebrate cyclin A correlates with its ability to form complexes with cdk catalytic subunits. J Cell Sci. 106(Pt 2): 535–544. [DOI] [PubMed] [Google Scholar]
- Mattaj, I. W., and L. Englmeier, 1998. Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 67: 265–306. [DOI] [PubMed] [Google Scholar]
- Mumberg, D., R. Muller and M. Funk, 1994. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 22: 5767–5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neville, M., and M. Rosbash, 1999. The NES-Crm1p export pathway is not a major mRNA export route in Saccharomyces cerevisiae. EMBO J. 18: 3746–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, V. Q., C. Co, K. Irie and J. J. Li, 2000. Clb/Cdc28 kinases promote nuclear export of the replication initiator proteins Mcm2–7. Curr. Biol. 10: 195–205. [DOI] [PubMed] [Google Scholar]
- Nishi, K., M. Yoshida, D. Fujiwara, M. Nishikawa, S. Horinouchi et al., 1994. Leptomycin B targets a regulatory cascade of crm1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J. Biol. Chem. 269: 6320–6324. [PubMed] [Google Scholar]
- Ouspenski, I. I., S. J. Elledge and B. R. Brinkley, 1999. New yeast genes important for chromosome integrity and segregation identified by dosage effects on genome stability. Nucleic Acids Res. 27: 3001–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, G., M. Knop and E. Schiebel, 1998. Spc98p directs the yeast gamma-tubulin complex into the nucleus and is subject to cell cycle-dependent phosphorylation on the nuclear side of the spindle pole body. Mol. Biol. Cell 9: 775–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlstein, D. L., J. Ge, A. D. Ortigosa, J. H. Robblee, Z. Zhang et al., 2005. The active form of the Saccharomyces cerevisiae ribonucleotide reductase small subunit is a heterodimer in vitro and in vivo. Biochemistry 44: 15366–15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichard, P., 1993. From RNA to DNA, why so many ribonucleotide reductases? Science 260: 1773–1777. [DOI] [PubMed] [Google Scholar]
- Sanchez, Y., B. A. Desany, W. J. Jones, Q. Liu, B. Wang et al., 1996. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271: 357–360. [DOI] [PubMed] [Google Scholar]
- Sikorski, R. S., and P. Hieter, 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R. S., and J. D. Boeke, 1991. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 194: 302–318. [DOI] [PubMed] [Google Scholar]
- Smith, M., 1985. In vitro mutagenesis. Annu. Rev. Genet. 19: 423–462. [DOI] [PubMed] [Google Scholar]
- Sommerhalter, M., W. C. Voegtli, D. L. Perlstein, J. Ge, J. Stubbe et al., 2004. Structures of the yeast ribonucleotide reductase Rnr2 and Rnr4 homodimers. Biochemistry 43: 7736–7742. [DOI] [PubMed] [Google Scholar]
- Stubbe, J., J. Ge and C. S. Yee, 2001. The evolution of ribonucleotide reduction revisited. Trends Biochem. Sci. 26: 93–99. [DOI] [PubMed] [Google Scholar]
- Subramaniam, P. S., and H. M. Johnson, 2004. The IFNAR1 subunit of the type I IFN receptor complex contains a functional nuclear localization sequence. FEBS Lett. 578: 207–210. [DOI] [PubMed] [Google Scholar]
- Sun, Z., D. S. Fay, F. Marini, M. Foiani and D. F. Stern, 1996. Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes Dev. 10: 395–406. [DOI] [PubMed] [Google Scholar]
- Thelander, L., and P. Reichard, 1979. Reduction of ribonucleotides. Annu. Rev. Biochem. 48: 133–158. [DOI] [PubMed] [Google Scholar]
- Voegtli, W. C., J. Ge, D. L. Perlstein, J. Stubbe and A. C. Rosenzweig, 2001. Structure of the yeast ribonucleotide reductase Y2Y4 heterodimer. Proc. Natl. Acad. Sci. USA 98: 10073–10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P. J., A. Chabes, R. Casagrande, X. C. Tian, L. Thelander et al., 1997. Rnr4p, a novel ribonucleotide reductase small-subunit protein. Mol. Cell Biol. 17: 6114–6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis, K., 2003. Regulating access to the genome. Nucleocytoplasmic transport throughout the cell cycle. Cell 112: 441–451. [DOI] [PubMed] [Google Scholar]
- Wendler, P., A. Lehmann, K. Janek, S. Baumgart and C. Enenkel, 2004. The bipartite nuclear localization sequence of Rpn2 is required for nuclear import of proteasomal base complexes via karyopherin alphabeta and proteasome functions. J. Biol. Chem. 279: 37751–37762. [DOI] [PubMed] [Google Scholar]
- Woollard, A., G. Basi and P. Nurse, 1996. A novel S phase inhibitor in fission yeast. Embo. J. 15: 4603–4612. [PMC free article] [PubMed] [Google Scholar]
- Xue, L., B. Zhou, X. Liu, W. Qiu, Z. Jin et al., 2003. Wild-type p53 regulates human ribonucleotide reductase by protein-protein interaction with p53R2 as well as hRRM2 subunits. Cancer Res. 63: 980–986. [PubMed] [Google Scholar]
- Yao, R., Z. Zhang, X. An, B. Bucci, D. L. Perlstein et al., 2003. Subcellular localization of yeast ribonucleotide reductase regulated by the DNA replication and damage checkpoint pathways. Proc. Natl. Acad. Sci. USA 100: 6628–6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X., and R. Rothstein, 2002. The Dun1 checkpoint kinase phosphorylates and regulates the ribonucleotide reductase inhibitor Sml1. Proc. Natl. Acad. Sci. USA 99: 3746–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X., E. G. Muller and R. Rothstein, 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell 2: 329–340. [DOI] [PubMed] [Google Scholar]
- Zhou, Z., and S. J. Elledge, 1992. Isolation of crt mutants constitutive for transcription of the DNA damage inducible gene RNR3 in Saccharomyces cerevisiae. Genetics 131: 851–866. [DOI] [PMC free article] [PubMed] [Google Scholar]