Abstract
Functional left/right asymmetry (“laterality”) is a fundamental feature of many nervous systems, but only very few molecular correlates to functional laterality are known. At least two classes of chemosensory neurons in the nematode Caenorhabditis elegans are functionally lateralized. The gustatory neurons ASE left (ASEL) and ASE right (ASER) are two bilaterally symmetric neurons that sense distinct chemosensory cues and express a distinct set of four known chemoreceptors of the guanylyl cyclase (gcy) gene family. To examine the extent of lateralization of gcy gene expression patterns in the ASE neurons, we have undertaken a genomewide analysis of all gcy genes. We report the existence of a total of 27 gcy genes encoding receptor-type guanylyl cyclases and of 7 gcy genes encoding soluble guanylyl cyclases in the complete genome sequence of C. elegans. We describe the expression pattern of all previously uncharacterized receptor-type guanylyl cyclases and find them to be highly biased but not exclusively restricted to the nervous system. We find that >41% (11/27) of all receptor-type guanylyl cyclases are expressed in the ASE gustatory neurons and that one-third of all gcy genes (9/27) are expressed in a lateral, left/right asymmetric manner in the ASE neurons. The expression of all laterally expressed gcy genes is under the control of a gene regulatory network composed of several transcription factors and miRNAs. The complement of gcy genes in the related nematode C. briggsae differs from C. elegans as evidenced by differences in chromosomal localization, number of gcy genes, and expression patterns. Differences in gcy expression patterns in the ASE neurons of C. briggsae arise from a difference in cis-regulatory elements and trans-acting factors that control ASE laterality. In sum, our results indicate the existence of a surprising multitude of putative chemoreceptors in the gustatory ASE neurons and suggest the existence of a substantial degree of laterality in gustatory signaling mechanisms in nematodes.
THE diversification of neuronal fate and function across the left/right axis of nervous systems is poorly understood but represents a fundamental problem in the neurosciences. This problem is well illustrated by a cursory comparison of structure and function of nervous systems. While the organization of nervous systems is largely bilaterally symmetric on a morphological level, brain functions are often highly lateralized (Hugdahl and Davidson 2003). Functional lateralization is presumably brought about by the diversification of neuronal function on a subanatomical level, such as differential gene expression in bilaterally symmetric structures. Indeed, quantitative comparison of transcript levels has recently revealed left/right asymmetries in gene expression profiles in the human brain (Sun et al. 2005).
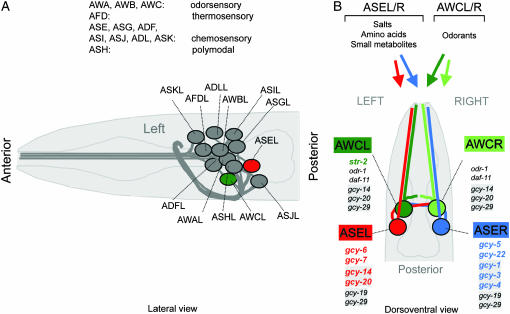
The nematode Caenorhabditis elegans provides a simple model organism to study the lateralization of nervous system function (Hobert et al. 2002). Such lateralization can be observed in the chemosensory system of the nematode. The best-studied chemosensory neurons are a group of 12 classes of neurons called the amphid sensory neurons (Figure 1A). Each class consists of one pair of two bilaterally symmetric and morphologically indistinguishable neurons, most of which are chemosensory neurons (Figure 1B). At least two classes of these chemosensory neurons, the AWC odor sensory neuron class and the ASE gustatory neuron class are functionally lateralized, allowing the animal to sense and discriminate different sensory cues with the left and right neuron (Pierce-Shimomura et al. 2001; Wes and Bargmann 2001) (Figure 1B). Functional lateralization of AWCL/R and ASEL/R correlates with the left/right asymmetric expression of putative chemoreceptors (Yu et al. 1997; Troemel et al. 1999; Chang et al. 2004) but the extent of lateralization of chemoreceptor gene expression in these neurons is still unclear. In the AWCL/R sensory neurons, only one left/right asymmetrically expressed chemoreceptor has been reported, a seven-transmembrane receptor (Troemel et al. 1999) (Figure1B). In the ASEL/R sensory neurons, a total of four asymmetrically expressed, putative chemoreceptors, which all belong to the family of receptor guanylyl cyclases, were known prior to this study (Yu et al. 1997; Johnston et al. 2005) (Figure 1B). gcy-5 and gcy-22 are expressed in the right ASE neuron (ASER), whereas gcy-6 and gcy-7 are expressed exclusively in the left ASE neuron (ASEL).
Figure 1.
An introduction to C. elegans sensory anatomy. (A) A prominent and well-characterized subset of C. elegans sensory neurons, the amphid sensory neurons. As with most other neuron classes, amphid sensory neuron classes consist of one pair of two bilaterally symmetric cells (see also B). Each of the 12 pairs of amphid sensory neurons extends a dendrite to the tip of the nose and an axon into the nerve ring, a nerve bundle where synaptic connections are made (White et al. 1986). Delineated functions of amphid sensory neurons are indicated (Bargmann and Mori 1997). (B) Amphid sensory neuron classes consist of two bilaterally symmetric pairs of neurons, two of which, AWCL/R and ASEL/R, are functionally lateralized. While some other amphid sensory neurons appear to contribute to gustation, ASE is the main gustatory neuron class in C. elegans, mediating responses to salts, amino acids, and small metabolites (Bargmann and Horvitz 1991). ASE mediates not only attractive, but also repulsive, responses to specific chemicals (Sambongi et al. 1999). The salts sodium, chloride, and potassium are sensed in a left/right asymmetric manner, with ASEL sensing sodium, but not chloride and potassium, and ASER sensing chloride and potassium (Pierce-Shimomura et al. 2001). It is not yet known whether other ASE-sensed chemicals may also activate ASEL and ASER differentially. ASE-expressed gcy genes are shown; those newly described in this article are shaded; those that are asymmetric are colored. The AWCL/R neurons can discriminate benzaldehyde and butanone on the basis of the left/right asymmetric expression of str-2 (colored), a G-protein coupled receptor, which is stochastically expressed in either AWCL or AWCR (Troemel et al. 1999; Wes and Bargmann 2001). Newly identified gcy genes in AWC are shaded.
To further analyze the extent of lateralization of the ASE gustatory neurons, we identified the complete set of guanylyl cyclase (gcy) genes in the C. elegans genome and undertook a genomewide analysis of their expression patterns. Previous counts of C. elegans receptor-type guanylyl cyclases were preliminary, given the incomplete nature of the C. elegans genome-sequencing project, but estimated to be in the higher twenties (Yu et al. 1997; Birnby et al. 2000). Expression patterns had been determined for eight receptor-type guanylyl cyclases (Yu et al. 1997; Birnby et al. 2000; L'Etoile and Bargmann 2000). We now report the final count of receptor-type guanylyl cyclases in the complete C. elegans genome to be 27. We present a comparative sequence analysis of all gcy genes and describe the expression patterns of all previously uncharacterized receptor-type guanylyl cyclases using gfp reporter gene fusions. We analyze the mechanisms of the regulation of gcy gene expression in the context of the ASE gustatory neurons, investigate the consequence of removing one ASE-expressed gcy gene on ASE neuron function, and examine the evolutionary divergence of gcy gene structure and expression.
MATERIALS AND METHODS
Strains and transgenes:
The following wild-type and mutant strains were used: N2 wild-type Bristol isolate; Caenorhabditis briggsae AF16 wild-type strain; OH4349 lsy-6(ot71)dpy-11(e224); OH110 lim-6(nr2073) (Hobert et al. 1999); RB1000 gcy-5(ok921), 4× outcrossed; RB1010 gcy-5(ok930), 4× outcrossed; and OH2957 gcy-5(tm897), not outcrossed.
The following transgenes were used: otIs3: Is[gcy-7prom∷gfp; lin-15(+)] (Chang et al. 2003), expressed in ASEL and the excretory canal cell; otIs151: Is[ceh-36prom∷dsRed2; rol-6(d)] (Johnston and Hobert 2003), expressed in ASEL/R and AWCL/R; otIs133: Is[ttx-3promB∷rfp; pNC4.2(unc-4(+)] (Wenick and Hobert 2004), expressed in AIYL/R; oyIs17: Is[gcy-8prom∷gfp; lin-15(+)], expressed in AFDL/R; and oyIs51: Is[srh-142prom∷rfp; lin-15(+)], expressed in ADFL/R, both gifts from Piali Sengupta.
Sequence analysis:
To identify GCY sequences, the sets of predicted proteins for C. elegans and C. briggsae were obtained from the Sanger Institute (http://www.sanger.ac.uk/Projects/C_elegans/WORMBASE/current/wormpep_download.shtml and ftp://ftp.sanger.ac.uk/pub/wormbase/cbriggsae/cb25.agp8/). Representative GCYs were used as position-specific iterated basic local alignment search tool (PSI-BLAST) queries to search the two proteomes. The HMMER 2.3.2 package (Eddy 1998) was used to construct a hidden Markov model from an alignment of GCY catalytic domains and to search for additional GCYs.
The intracellular regions of the transmembrane GCYs and the complete soluble GCY sequences were aligned with T-coffee version 2.03 (Notredame et al. 2000). Maximum parsimony phylogenetic trees were found via heuristic search with PAUP* version 4.0 beta 10 (Swofford 2003). The trees were generated in 100 repeated searches with random addition of taxa to obtain the starting tree. Robustness of the tree partitions was evaluated by constructing a bootstrap consensus tree with 1000 replicates. The trees were visualized with TreeView version 1.6.6 (Page 1996).
Nomenclature of gcy genes and gcy gene predictions:
Most but not all gcy names were previously assigned (http://www.wormbase.org). We named two previously unnamed gcy genes gcy-28 (T01A4.1) and gcy-29 (C04H5.3). Both code for receptor-type proteins. A few gcy genes have been double named in the past. The most current names are (with old names in parentheses): gcy-18 (gcy-26), gcy-20 (gcy-16), gcy-17 (gcy-24), odr-1 (gcy-10), gcy-28 (gcy-38 in WS149), gcy-29 (gcy-39 in WS149).
Since we detected several cases where individual parts of one C. elegans gene are homologous to separate, adjacent predicted C. briggsae genes (gene prediction in Wormbase WS149), we suspected that C. briggsae genes may have been incorrectly predicted. We therefore ran the FGENESH program at http://www.softberry.com (Salamov and Solovyev 2000) on chromosomal regions that contained the following predicted C. briggsae genes (from WS149): CBG07423(CBP15915) + CBG07424(CBP15916) + CBG07425(CBP15917), CBG20867(CBP04902) + CBG20868(CBP04903), and CBG19454(CBP11205) + CBG19453(CBP11204). In each of these three cases, we found that FGENESH predicted only one gene, whose product was homologous over its entire length with putative C. elegans orthologs. However, in two cases, the revised gene prediction overlooked exons predicted in the original prediction, which contained homology to GCY proteins. We therefore assembled alternate gene prediction by hand, on the basis of the similarity to known GCY proteins. We named these revised predictions CBP15915*, CBP04902*, and CBP11205* (see supplemental Figures 1 and 2 at http://www.genetics.org/supplemental/). On the basis of primary sequence homology, we also suspect that one of the two C. briggsae orthologs of C. elegans, gcy-35, has been incorrectly predicted as the two separate genes CBG20390 (CBP04780) and CBG20392 (CBP04781). Similarly, CBG10472 (CBP08561) and CBG10474 (CBP08562) likely constitute one gene. However, neither of these two suspicions could be corroborated by FGENESH.
Generation of gfp reporter gene fusions, transgenic animals, and identification of reporter gene expressing cells:
Most reporter genes were created by PCR fusion (Hobert 2002) and some were generated by subcloning PCR amplicons into pPD95.75 (see Table 1). Primer sequences and resulting transgenic arrays are shown in Table 1. DNA was injected at ∼10–50 ng/μl using either unc-122∷gfp or rol-6 as injection marker. Cell identifications were done on the basis of overall cell position and morphology and was significantly aided by the uses of the following four colabeling procedures.
Most gfp reporters were injected into animals carrying the otIs151 transgene in which ASEL/R and AWCL/R are labeled with DsRed2. Since the otIs151 transgene already contains the rol-6(d) injection marker, we used unc-122∷gfp (Loria et al. 2004) as injection marker for most injections. This marker is expressed in coelomocytes but also yields occasional and mosaic gfp expression within the pharynx, often in the I5 neuron. With the exception of the broadly pharyngeal expression of one gcy gene, we therefore ignored any cell-type-specific pharyngeal expression of gcy reporter genes. Injections into wild-type C. elegans or C. briggsae were done using rol-6(d) as injection marker.
Some gcy∷gfp transgenic animals were crossed with animals carrying the otIs133 transgene in which the AIY interneurons are marked with DsRed2, thereby facilitating cell identification either by red/green overlap (gcy-1 prom∷gfp) or by the determination of relative cell position (gcy-18 prom∷gfp).
In many cases, a subset of the amphid neuron classes of gcy∷gfp transgenic animals was filled with DiI. DiI fills the ASK, ADL, ASI, AWB, ASH, and ASJ in the head in the PHA and PHB neuron classes in the tail (Hedgecock et al. 1985), thereby facilitating cell identification either by an overlap of red and green fluorescence or by the determination of relative cell position. DiI was dissolved in DMF, diluted to 10 μg/ml in M9 or in ddH20 plus 50 mm calcium–acetate to additionally fill the IL2 neurons. Worms were soaked in the DiI solution for at least 1 hr.
Expression of gcy genes in the AFD thermosensory neurons was assessed using the oyIs17 transgene in which the AFD sensory neurons are marked with GFP. Extrachromosomal arrays carrying gcy-18prom∷gfp and gcy-23 prom∷gfp were crossed with oyIs17-containing animals and the number of GFP-expressing amphid sensory neurons was counted. To exclude expression in the closely adjacent ADF neuron class, gfp transgenes were also crossed with an ADF-expressed DsRed2 reporter construct, oyls52, kindly provided by P. Sengupta.
TABLE 1.
PCR primers used to generate reporter constructs
| PCR fusion primersa
|
||||
|---|---|---|---|---|
| Promoter | A primer | A* primer | B primer | Resulting transgenic arraysb |
| gcy-1 | gaagctggagtcaagtgtg | gtgtactacaacaagggactttg | agtcgacctgcaggcatgcaagctgaatatttgcatcgaaaag | otEx2419, otEx2420, otEx2421 |
| gcy-2 | ccattacagggacatcag | ggatacattgaaagtagcg | agtcgacctgcaggcatgcaagctgaccattttgaattaatgattcc | otEx2312, otEx2313, otEx2314 |
| gcy-3 | gagctctcatggatcacg | cacattgaaagtagcgaattag | agtcgacctgcaggcatgcaagctgttttttcaaacaaagatcag | otEx2422, otEx2423 |
| gcy-4 | gaagtttcagtggtacttgc | gattgattcagaattcgagatc | agtcgacctgcaggcatgcaagctgagtatcataattcatggaagtag | otEx2407, otEx2408, otEx2409 |
| gcy-9 | gttatctccgatagccaag | ggttgttgaaagccatatc | agtcgacctgcaggcatgcaagctcggaagtggatagatgg | otEx2307, otEx2308, otEx2541,cotEx2542c |
| gcy-11 | ctgatttgatgccatcac | gatttgtctcttcactggtcac | agtcgacctgcaggcatgcaagctgtccagcatgcttctgc | otEx2304, otEx2305, otEx2306 |
| gcy-13 | gccacaatgacaattatctc | ctcaaaatccattgtgtaagttc | agtcgacctgcaggcatgcaagctccatcctacagaggaggc | otEx2410, otEx2411, otEx2412 |
| gcy-14 | gtgcattaatcagaatgagc | cagaacactgaaagctacc | agtcgacctgcaggcatgcaagctgcacatgttagatgatgaatg | otEx2322, otEx2323, otEx2324 |
| gcy-15 | gttatcctaatagcaactacatcacc | cttcaataatagtttctctcacttg | agtcgacctgcaggcatgcaagctcgcaactgttgcattg | otEx2497 |
| gcy-17 | cagttgaacatctccctgg | caacaacgttcaagctcc | agtcgacctgcaggcatgcaagctcatcatgttcttagagtaggc | otEx2413, otEx2414, otEx2415 |
| gcy-18 | ccgaaagaagtcgagttg | ggtcgagagcagcaaatc | agtcgacctgcaggcatgcaagctcattttctgatgctccgac | otEx2498d |
| gcy-19 | gataccgtgagcaagatgg | gatagattgcattgttgtgg | agtcgacctgcaggcatgcaagctggacattggccatcctatac | otEx2309, otEx2310, otEx2311, otEx2535,fotEx2536f |
| gcy-20 | gatgatttatacccacatacattg | ggagagttttcaatgatttg | agtcgacctgcaggcatgcaagctccgcattgttgaatgaac | otEx2325, otEx2326, otEx2327 |
| gcy-21 | gtaaactgggagtgaaagc | ctcgacgagcattatgtg | agtcgacctgcaggcatgcaagctctgaaagcatagcagaataatatg | otEx2416, otEx2417, otEx2418 |
| gcy-23 | caaacatcttacgcttctcac | ctcgagtgtcgacacg | agtcgacctgcaggcatgcaagctccaacttttacctttaaagttaatg | otEx2499, otEx2500, otEx2501 |
| gcy-25 | caagattcgatttcaaaactg | ccaacctgattttgtgg | agtcgacctgcaggcatgcaagctgagaagcatctccgaatg | otEx2424, otEx2425, otEx2426 |
| gcy-27 | gatactttggaaaagaacaatg | cgaatagacaaccaaacttc | agtcgacctgcaggcatgcaagctgttgtatctcgagaaagactctg | otEx2502,cotEx2503,cotEx2540c |
| gcy-28 | caagacctccaaagttttg | cggtcttgaagcgattc | agtcgacctgcaggcatgcaagctgagcatagcctcataggtac | otEx2504, otEx2505, otEx2537 |
| gcy-29 | cacagacaagcagagcac | gcaaacgagtcccagc | agtcgacctgcaggcatgcaagctgagcattttttactgtttttc | otEx2490, otEx2491, otEx2492 |
| Subcloninge | ||||
| Promoter | 5′ primer | 3′ primer | Resulting transgenic arraysb | |
| C.b.gcy-4 | ttaagcttAAAACGTATCCCTCTTCCT | ttggatcCTAGATGGCTGAGATCTACG | otEx2506,fotEx2507,fotEx2508f | |
| otEx2509, otEx2510, otEx2511 | ||||
| C.b.gcy-19 | ttaagcttGCCGCGGTTGAAAACAAAAATTG | ttggatccAATACTTGGCGAGTAGCCCC | otEx2138,fotEx2139,fotEx2140f | |
| otEx2141, otEx2142 | ||||
Primer nomenclature according to Hobert (2002). All sequences are 5′–3′.
If not indicated otherwise, C.elegans animals containing the otIs151 transgene were injected, using unc-122∷gfp as injection marker.
Injection was done into N2 animals, using rol-6(d) as injection marker.
Injection was done into otIs133 animals, using rol-6(d) as injection marker.
Primers used for subcloning contained restriction sites at their end and were subcloned into pPD95.75.
Injected into C. briggsae, using rol-6(d) as injection marker.
Chemotaxis assays:
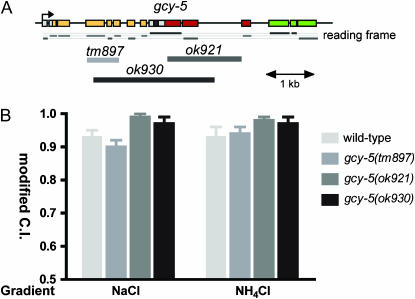
Radial population chemotaxis assays were done as previously described (Chang et al. 2004). Assay plates were 10-cm tissue culture dishes containing 20 g/liter agar, 5 mm potassium phosphate (pH 6.0), 1 mm CaCl2, and 1 mm MgSO4. To set up the chemical gradients on the assay plates, a 10-μl drop of attractant was placed 15 mm from the edge of the plate at the “attractive spot.” A 10-μl drop of ddH20 was placed diametrically opposite and was considered the “negative control spot.” The attractant was allowed to diffuse for 14–16 hr at room temperature. To increase the steepness of the chemical gradient, 4–4.5 hr prior to chemotaxis assay, 4 μl of attractant was added to the “attractive spot” and 4 μl of ddH20 was added to the “negative control spot.” The attractants NaCl and NH4Cl (Sigma, St. Louis) were dissolved in ddH2O to a concentration of 2.5 m and were adjusted to pH 6.0 with either NH4OH or acetic acid. Worms were washed three times in sterile water to remove food and salts. Worms were then placed at the center of the plate and allowed to chemotax for 1 hr. Worms reaching either the attractant peak or the negative control spot (sterile water) were immobilized with sodium azide. Results were quantified by counting worms that were located at (A) the attractant, (B) the center of the plate, or (C) the negative control. Since animals carrying two of the three gcy-5 knockout alleles (ok921 and ok930) did not disperse well from the center of the plate, we calculated a modified chemotaxis index, defined as C.I. = A/(A + C). This index therefore disregards worms that do not reach either the attractant or the negative control spot. It is doubtful that the dispersion defects of ok921 and ok930 alleles are significant since the putative null allele tm897 does not show these defects.
RESULTS
Identification of the complete set of GCY proteins in C. elegans:
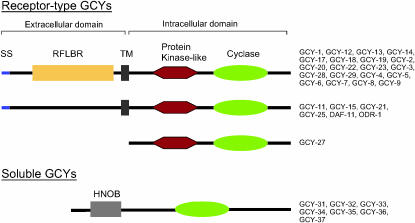
To identify the complete set of guanylyl cyclases in C. elegans, we employed PSI-BLAST to search the latest release of the complete C. elegans genome databases, using a set of known GCY proteins as queries. We identified a total of 34 predicted GCY proteins, several more than reported in previous searches of incomplete genome sequence databases (Yu et al. 1997; Bargmann 1998; Birnby et al. 2000). The identified GCY proteins fall into two distinct families, receptor-type guanylyl cyclases (encoded by 27 genes; henceforth called receptor-type gcy genes) and soluble cyclases (encoded by 7 genes; henceforth called soluble gcy genes). Both families contain a guanylyl cyclase catalytic domain (Figure 2). In several GCY proteins, catalytically important residues are not conserved in the cyclase domain (Yan et al. 1997) (supplemental Figure 1 at http://www.genetics.org/supplemental/) and it has been speculated that, in these cases, heterodimerization with a catalytically active GCY protein ensures activity of the dimer (Morton 2004). Apart from the presence of the cyclase domain, receptor-type and soluble GCY proteins differ significantly (Figure 2). Soluble GCY proteins contain one other characteristic domain, a heme nitric oxide-binding (HNOB) domain. In contrast, receptor-type guanylyl cyclases lack this HNOB domain but always contain an additional protein kinase domain, which is likely to be inactive since it lacks a critical catalytic aspartate residue that is present in the catalytic core of all active protein kinases (Taylor et al. 1992) (HRD motif; supplemental Figure 2 at http://www.genetics.org/supplemental/). In addition to this protein-kinase-like domain, all but one of the C. elegans receptor-type GCY proteins contain a single transmembrane domain and a signal sequence. Moreover, the majority of receptor-type GCY proteins contain a conserved extracellular domain of unknown function that is also present in many mammalian GCY proteins and conserved in amino acid receptors from bacteria to mammals [receptor family ligand-binding region (RFLBR); Figure 2]. The presence of this domain strongly suggests that the C. elegans GCY proteins are indeed ligand-binding receptor proteins.
Figure 2.
Domain structure of GCY proteins. SS, signal sequence; TM, transmembrane domain; RFLBR, receptor family ligand-binding region (PF01094); HNOB, heme nitric oxide-binding domain (PF07700); protein kinase-like, protein kinase domain (PF00069); cyclase, adenylate and guanylate cyclase catalytic domain (PF00211). See supplemental Figure 1 and supplemental Figure 2 at http://www.genetics.org/supplemental/ for the primary sequence alignment of individual domains. We note that several of the receptor-type GCY proteins, such as GCY-11, lack a clear SS at the N terminus but the presence of a clear TM and/or RFLBR domain make us suspect that the absence is due to an incorrectly predicted N terminus of the respective genes and we therefore grouped these genes together with other clear-cut SS/TM-containing proteins. GCY-22 has an unusual and phylogenetically conserved insertion between the transmembrane and protein kinase domain (supplemental Figure 2 at http://www.genetics.org/supplemental/).
The overall domain topology of C. elegans GCY proteins is similar to that of mammalian GCY proteins. Multiple transmembrane-containing GCY proteins, which can be found in unicellular eukaryotes (Wedel and Garbers 2001), are not present in C. elegans.
GCY-27 is an unusual receptor-type GCY protein (Figure 2). While containing all the intracellular signature motifs of receptor-type GCYs, the predicted GCY-27 protein is several hundred amino acids shorter than all other predicted receptor-type GCY proteins and does not contain a predicted signal sequence (SS), transmembrane domain (TM), or other extracellular motifs (Figure 2). In the absence of complete cDNA/EST sequences, we cannot rule out a gene prediction error, but we consider this to be unlikely for two reasons: (a) The upstream gene adjacent to gcy-27 is relatively close (Figure 4), leaving little room for such a prediction oversight, particularly since in other gcy genes the extracellular domains are large and composed of many exons; and (b) a C. briggsae ortholog (described in more detail in a later section) also lacks the extracellular domain. While receptor-type GCY proteins that lack a TM and SS domain have been described before (Morton 2004), they do not appear to contain the protein kinase domain that is present in GCY-27 and all other receptor-type GCY proteins. It is interesting to note that GCY-27 is most closely related to the intracellular domain of ODR-1 (Figure 3A), a transmembrane GCY protein whose extracellular domain (which contains no canonical RFLBP domain) has previously been shown to be dispensable for its function in chemosensory signal transduction (L'Etoile and Bargmann 2000).
Figure 4.
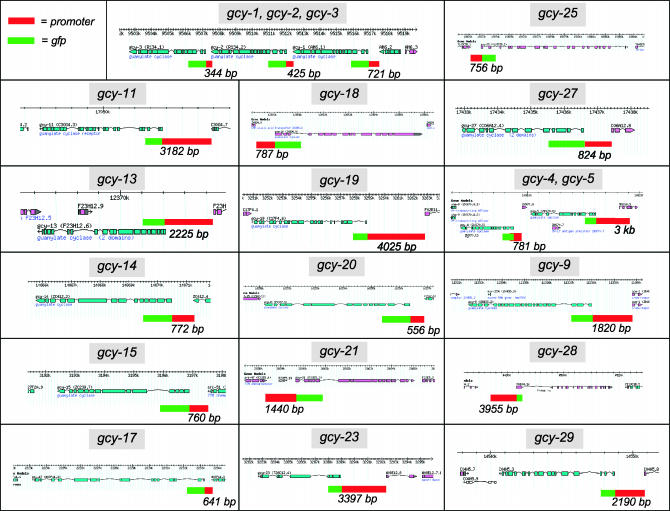
Reporter gene constructs. Representation of genomic loci are adapted from http://www.wormbase.org. Reporter gene constructs are indicated by the 5′ upstream region used (red box), usually the intergenic region up to the next gene, and the gfp coding region (green box; not drawn to scale). See Table 1 for primer sequences and a list of transgenic arrays containing the individual reporter gene constructs.
Figure 3.
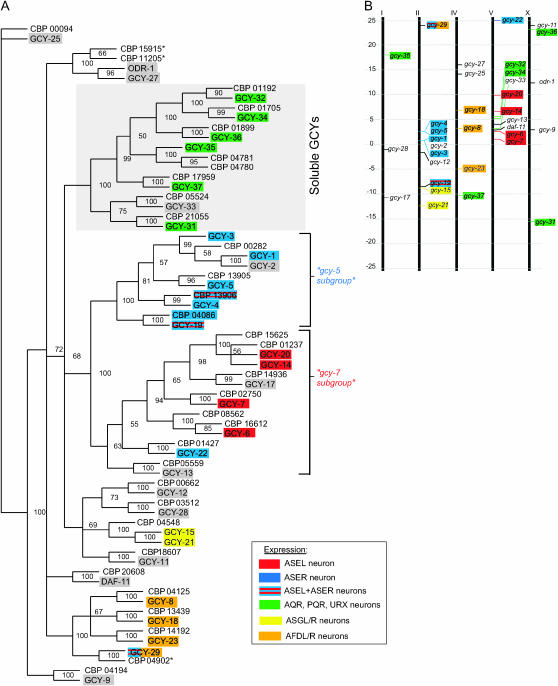
Sequence similarity and chromosomal localization of gcy genes. (A) Phylogenetic tree based on the intracellular domain of the receptor-type guanylyl cyclases and the complete sequences of the soluble guanylyl cyclases. Numbers at the tree nodes are bootstrap values, which indicate the frequency (in percentages) of occurrence of a given partition in the 1000 replicate trees. C. elegans proteins are shaded; C. briggsae proteins all carry the prefix “CBP.” A select number of cells that coexpress multiple gcy genes are indicated by color-coded shading, as indicated in the inset. See materials and methods for comments on the gcy gene names. Note that the CBP15915*, CBP11205*, and CBP04902* proteins used here differ from those in Wormbase WS149 on the basis of an alternative gene prediction that we performed (see materials and methods). (B) Chromosomal localization of gcy genes. See Table 2 for detailed map position. Chromosome III does not contain any gcy genes.
We analyzed the relatedness of soluble and receptor-type GCY proteins in more detail by generating a maximum-parsimony bootstrap tree using the intracellular domain of the receptor-type GCYs and the complete sequences of the soluble GCYs. The soluble GCYs branch separately from the transmembrane-type proteins and are clustered into two subgroups. Many receptor-type GCY proteins also fall into small and well-defined subgroups (Figure 3A).
Notably, in a substantial number of cases, the degree of sequence relation correlates with proximity in the genome sequence (Figure 3B). For example, all six members of one subgroup (“gcy-5 subgroup”) are located within an ∼7-Mb interval on chromosome II, five of these (gcy-1–gcy-5) map to an ∼800-kb interval, and three of them are directly adjacent genes, separated by only a few hundred base pairs (Figure 3B; Figure 4). This suggests that these genes arose by relatively recent gene duplications. Likewise, all seven members of what we term the “gcy-7 subgroup” of gcy genes (Figure 3A) reside on a single chromosome and five of them in a <8-Mb interval (Figure 3B; Figure 4). As we will demonstrate below, the degree of sequence relation and chromosomal location also correlates with similarities in gene expression patterns.
Expression patterns of receptor-type gcy genes:
While the expression of all seven soluble gcy genes has already been described (Yu et al. 1997; Cheung et al. 2004; Gray et al. 2004), the expression patterns of only 8 of the 27 receptor-type gcy genes was previously reported, 6 in the context of a preliminary and incomplete genome analysis (gcy-5, gcy-6, gcy-7, gcy-8, gcy-10/odr-1, gcy-12, and gcy-22) and 2 in the course of a functional analysis (odr-1 and daf-11) (Yu et al. 1997; Birnby et al. 2000; L'Etoile and Bargmann 2000; Johnston et al. 2005) (summarized in Table 2). We generated gfp reporter fusions to the putative cis-regulatory regions of the remaining 19 gcy genes (schematically shown in Figure 4). In all except one case, sequences to the next upstream genes were included in the reporter gene fusions (Figure 4). While such upstream sequences are most likely to harbor gene regulatory elements, it needs to be kept in mind that additional regulatory elements may be located elsewhere and that therefore the expression patterns of the gfp reporter genes can provide only a first approximation of endogenous gene expression profiles. Since gcy genes are likely to primarily function as chemoreceptors in the mature nervous system, we restricted our gene expression analysis of transgenic animals that harbor the respective reporter genes to larval and adult stages. To allow us to reliably detect expression in the ASEL/R gustatory neurons, the neurons in which our laboratory is most interested in, we used a transgenic reporter array in the background in which the ASEL/R neurons are labeled with DsRed2 (otIs151; see materials and methods), thereby enabling us to score for an overlap between green and red fluorescent signals. In most cases, multiple lines were analyzed per construct (see Table 1) and we did not observe any notable differences between individual lines. Reporter gene expression was observed for each of the gcy genes examined. Expression patterns are shown in Figure 5 and summarized in Table 2, which also lists previously described expression patterns. The expression patterns of all receptor-type gcy genes can be summarized as follows.
TABLE 2.
Receptor guanylyl cyclases in C. elegans
| Other namesa
|
Map positionb
|
Expression pattern in adult animals
|
||
|---|---|---|---|---|
| Gene | Previous reports | This report | ||
| daf-11 | B0240.3 | V +3.27 | ASIL/R, ASJL/R, ASKL/R, AWBL/R, AWCL/Rc | — |
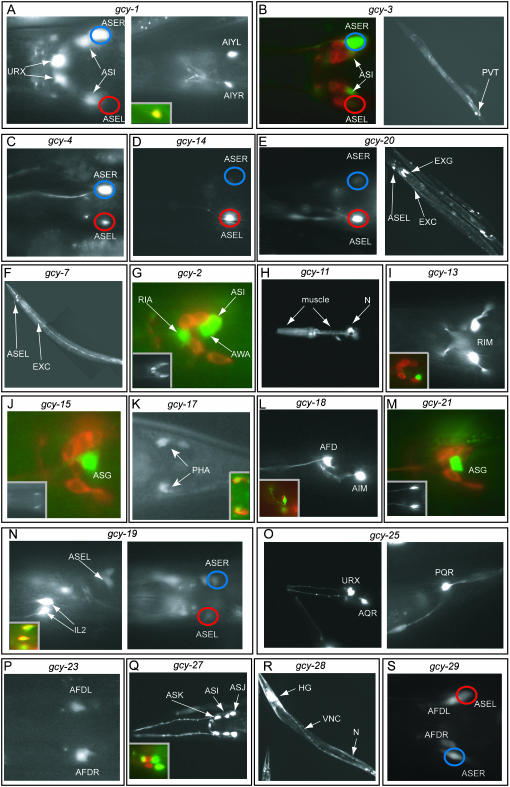
| gcy-1 | AH6.1 | II +1.49 | No signald | ASER, ASIL/R, PVT, URXL/R, AIYL/R, intestine |
| gcy-2 | R134.2 | II +1.48 | — | AWAL/R, ASIL/R, RIAL/R, PVT |
| gcy-3 | R134.1 | II +1.48 | — | ASER, ASIL/R, PVT |
| gcy-4 | ZK970.5 | II +2.31 | No signald | ASER biasedh |
| gcy-5 | ZK970.6 | II +2.32 | ASERd | — |
| gcy-6 | B0024.6 | V +2.46 | ASELd,g | — |
| gcy-7 | F52E1.4 | V +1.37 | ASELd,g | Also expressed in excretory canal cell (only in adults) |
| gcy-8 | C49H3.1 | IV +3.50 | AFDL/Rd | — |
| gcy-9 | ZK455.2 | X +3.0 | No signald | Weak, occasional, and variable expression in non-neuronal tissues |
| gcy-11 | C30G4.3 | X +24.07 | — | Pharyngeal muscle |
| gcy-12 | F08B1.2 | II −1.80 | PHAL/Rd | — |
| gcy-13 | F23H12.6 | V +4.02 | No signald | RIML/R |
| gcy-14 | ZC412.2 | V +6.93 | No signald | ASEL biased,h AWCL/R (faint), PVT |
| gcy-15 | ZC239.7 | II −8.08 | — | ASGL/R (faint) |
| gcy-17 | W03F11.2, gcy-24 | I −10.91 | — | PHAL/R |
| gcy-18 | ZK896.8, gcy-26 | IV +6.48 | — | AFDL/R, AIML/Ri |
| gcy-19 | C17F4.6 | II −7.80 | — | IL2 (strong), ASEL/R (faint),j additional faint sensory neurons (three pairs) |
| gcy-20 | F21H7.9, gcy-16 | V +9.87 | — | ASEL, AWCL/R (faint) excretory gland and canal cells |
| gcy-21 | F22E5.3 | II -12.28 | — | ASGL/R, ADLL/R (faint) |
| gcy-22 | T03D8.5 | V +25.25 | ASERf | — |
| gcy-23 | T26C12.4 | IV -5.01 | — | AFDL/Ri |
| gcy-25 | Y105C5B.2 | IV +14.17 | — | AQR, PQR, URXL/R |
| gcy-27 | C06A12.4 | IV +16.74 | — | ASKL/R, ASIL/R, ASJL/R |
| gcy-28 | T01A4.1 | I −1.17 | — | Many head neurons, ventral cord and tail neurons, body-wall muscle, hypodermis, somatic gonad, intestinek |
| gcy-29 | C04H5.3 | II +23.04 | — | ASEL/R, AWCL/R, AVKL/R, AFDL/R, few variable other neurons (weak) |
| odr-1 | R01E6.1, gcy-10 | X +12.7 | ASIL/R, ASJL/R, ASKL/R, AWBL/R, AWCL/Rd,e | — |
Previously characterized and newly determined receptor gcy expression patterns are summarized. —, indicates that expression was not analyzed.
See materials and methods for comments on gene nomenclature.
From http://www.wormbase.org.
Yu et al. (1997). We suppose that we detected clear expression in several cases where no signal was observed by Yu et al. (1997) since we (1) used a gfp variant that is much brighter than the old gfp version used by Yu et al. (1997) and (2) since our reporter constructs may encompass more cis-regulatory sequences than those of Yu et al. (1997).
Johnston et al, (2005). Additional weak expression that fades in adults is observed in two additional, unidentified head neurons.
As described in Johnston et al. (2005) gcy-6 and gcy-7 are embryonically expressed in both ASEL and ASER and only become restricted to ASEL postembryonically. A similar scenario may apply for other ASEL-expressed gcy genes, but has not been explicitly examined.
“ASER biased” incorporates two categories of expression patterns in a given transgenic line: expression only in ASER in some animals and stronger expression in ASER than in ASEL in other animals. The opposite holds for ASEL-biased expression. Such weak and occasional expression in the other cell could be caused by array-overexpression artifacts and we therefore do not want to emphasize that biased expression is fundamentally different from exclusive expression.
After the initial submission of this article for publication, Inada et al. (2006) also described the expression pattern of gcy-18 and gcy-23 in the AFD neurons.
Expression in ASEL/R is very dim and not completely penetrant and potential biases to ASEL or ASER are therefore difficult to determine.
Expression in all tissue types is very mosaic. Expression in the ASE and AWC neuron classes could not be observed, but as with any other reporter construct described in this table it is possible that additional regulatory elements not contained within the respective constructs may yield expression in these neurons.
Figure 5.
Expression patterns of gcy reporter gene fusions. Transgenic animals expressing gfp reporter gene fusions are shown. Images are of representative animals from several independent lines (see Table 1 for a list of transgenic reporter arrays used). Most transgenic animals are scored in the late larval and adult stage and contain otIs151 in the background to facilitate the identification of the ASE neurons (see materials and methods); blue circles indicate ASER; red circles indicate ASEL. The quantification of the left/right asymmetric expression in ASE is shown in Figure 6. (A) gcy-1prom∷gfp. Dorsal view (left) and ventral view (right) of two different focal plains of the head region. The inset in the right panel shows the overlap of the gfp signal with otIs133, an AIY-expressed rfp marker (lateral view). (B) gcy-3prom∷gfp. Dorsal view of the head region (left) of an animal whose amphid sensory neurons have been filled with DiI. (Right) A full-length worm with expression in the PVT interneuron. (C) gcy-4prom∷gfp. Dorsal view of the head region. (D) gcy-14prom∷gfp. Dorsal view of the head region. (E) gcy-20prom∷gfp. Dorsal view of the head region (left). A full-length worm with expression in the excretory system (right). EXG, excretory gland cell; EXC, excretory canal cell. (F) gcy-7prom∷gfp. Lateral view. Expression in ASEL, but not expression in the excretory canal cell (EXC), has been previously reported (Yu et al. 1997). (G) gcy-2prom∷gfp. Lateral view of the head region. A defined subset of the amphid neurons are filled with DiI to allow for easier assessment of cell position. (Inset) A dorsal view illustrating bilateral symmetry of gfp-expressing cells. (H) gcy-11prom∷gfp. Lateral view of the head region. The strong neuronal expression (N) in the pharynx is likely due to the injection marker, but pharyngeal muscle expression is due to the reporter gene. (I) gcy-13prom∷gfp. Dorsal view of the head region. (Inset) A lateral view of a DiI-filled animal. (J) gcy-15prom∷gfp. Lateral view of the head region. A defined subset of the amphid neurons are filled with DiI to allow for easier assessment of cell position. (Inset) A ventral view to illustrate bilateral symmetry. (K) gcy-17prom∷gfp. Dorsal view of the tail region. (Inset) A DiI-filled animal. (L) gcy-18prom∷gfp. Lateral view of the head region. (Inset) A lateral view of gfp expression in relation to the rfp expression from otIs133, an AIY-specific cell marker. (M) gcy-21prom∷gfp. Lateral view of the head region of a DiI-filled animal. (Inset) A ventral view illustrating bilateral symmetry. (N) gcy-19prom∷gfp. Lateral view (left) and dorsal view (right) of the head region. (Inset, left) The overlap of the gfp signal with DiI-filled IL2 neurons. (O) gcy-25prom∷gfp. (Left) An oblique view of the head region. (Right) A lateral view of the tail region. (P) gcy-23prom∷gfp. Ventral view of the head region. (Q) gcy-27prom∷gfp. Dorsal view of the head region. (Inset) A lateral view of a DiI-filled animal. (R) gcy-28prom∷gfp. VNC, ventral nerve cord; HG, head ganglia; N, non-neuronal cells. The animal is mosaic and does not show muscle expression. (S) gcy-29prom∷gfp. Ventral view of the head region. The reporter is also expressed in AWCL/R and in AVKL/R (not shown in this animal).
Broad vs. cell type specific:
With the exception of the broadly but not ubiquitously expressed gcy-28 gene, all receptor-type gcy genes are expressed in a tissue- and cell-type-restricted manner (Table 2; Figure 5).
Cell types:
The expression of receptor-type gcy genes is strongly biased toward the nervous system. With the exception of two non-neuronally expressed gcy genes (gcy-9 and gcy-11), all gcy genes are expressed in a restricted subset of neurons. Of 27 genes, 21 are exclusively expressed in the nervous system and 4 are expressed in restricted sets of both neuronal and non-neuronal cells (Table 2; Figure 5).
Expression within the nervous system:
Within the nervous system, most but not all, C. elegans gcy genes are expressed in sensory neurons. All except two pairs (ASH and ADF) of the 12 amphid sensory neuron classes shown in Figure 1A express at least one gcy gene. Unlike what the previous analyses of eight gcy genes seemed to indicate (Yu et al. 1997; Birnby et al. 2000; L'Etoile and Bargmann 2000; Johnston et al. 2005), neuronal gcy gene expression is, however, not restricted to sensory neurons. Nonsensory neurons that express gcy genes include the AIY, AIM, AVK, RIA, and PVT interneuron classes and the RIM neuron, which is both a motor neuron and an interneuron. In addition, as mentioned above, many neurons in the nervous system, including, for example, ventral cord motor neurons, express the widely expressed gcy-28 gene. As summarized in Table 3, 25 of 27 of the C. elegans receptor-type gcy genes are expressed in the nervous system, 25 of 27 are expressed in various types of sensory neurons plus other neurons, 15 of 27 are expressed exclusively in sensory neurons, and 9 of 27 are restricted to single neuron classes. The expression of mammalian gcy genes, of which there are only 7, show roughly comparable patterns; some are expressed in non-neuronal cells and those that are expressed in neurons are strongly biased toward expression in sensory structures (Wedel and Garbers 2001).
TABLE 3.
Summary of cell-type specificity of gcy reporter gene expression
| gcy gene expression pattern | No. |
|---|---|
| Non-neuronal only | 2/27 |
| Nervous system | 25/27 |
| Nervous system only | 21/27 |
| Sensory neurons and other neurons | 25/27 |
| Sensory neurons only | 15/27 |
| Single neuron class specific | 9/27 |
Coexpression:
A notable general feature of C. elegans gcy gene expression profiles, both receptor and non-receptor type, is that a small number of neuron classes coexpress a substantial number of gcy genes (Figure 5; summarized in Table 4). The most striking examples are the ASE gustatory neuron class, which expresses a total of 11 gcy genes, more than one-third of all receptor-type gcy genes. In addition, 6 gcy genes are coexpressed in the ASI chemosensory neurons, 5 gcy genes are coexpressed in AWC olfactory neurons, 4 are coexpressed in the AFD thermosensory neurons, and 2 are coexpressed in the ASG and PHA phasmid sensory neurons, respectively (Table 4). We note that after the initial submission of this article for publication, Inada et al. (2006) also described the coexpression of three gcy genes in the AFD neurons. In addition to the overlap in individual neurons, there are also a few examples of gcy genes that show similar combinations of cellular expression profiles. gcy-7 and gcy-20 are coexpressed in ASEL and in the excretory canal cell. Moreover, as previously reported, daf-19 and odr-1 are expressed in precisely the same subset of amphid sensory neurons and 6 soluble gcy genes are coexpressed in the AQR/PQR and URX neurons (Yu et al. 1997; Birnby et al. 2000; L'Etoile and Bargmann 2000; Cheung et al. 2004; Gray et al. 2004). A single receptor-type gcy gene, gcy-25, complements the expression of the 6 soluble gcy genes in the AQR/PQR and URX neurons (Figure 5O; Table 4).
TABLE 4.
Summary of coexpressed gcy reporter genes
| Sensory neuron class
|
Coexpressed gcy genes
|
|
|---|---|---|
| No. | Name | |
| ASE | 11 | gcy-1, gcy-3, gcy-4, gcy-5, gcy-6, gcy-7, gcy-14, gcy-19, gcy-20, gcy-22, gcy-29 |
| AQR, PQR, URX | 7 | gcy-25, six soluble gcy genes |
| ASI | 6 | daf-11, gcy-1, gcy-2, gcy-3, gcy-27, odr-1 |
| AWC | 5 | daf-11, odr-1, gcy-14, gcy-20, gcy-29 |
| AFD | 4 | gcy-8, gcy-18, gcy-23, gcy-29 |
| ASG | 2 | gcy-15, gcy-21 |
| PHA | 2 | gcy-12, gcy-17 |
Left/right asymmetric expression:
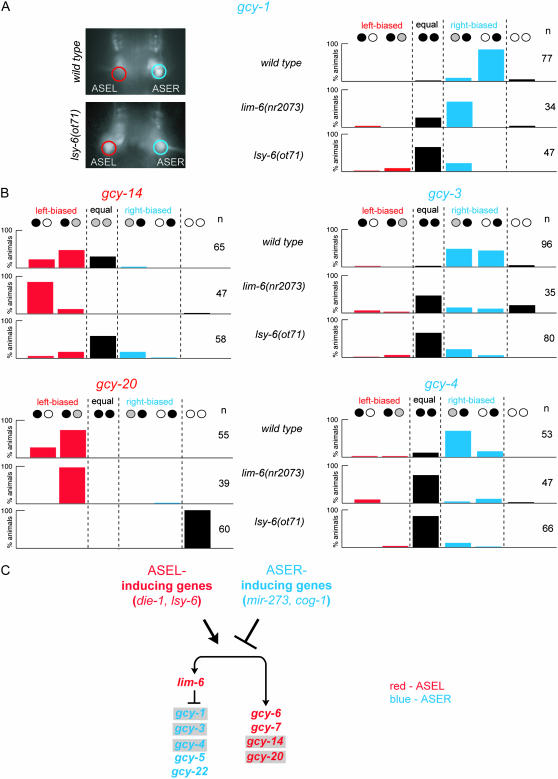
Two previous studies identified a total of four left/right asymmetrically expressed gcy genes in the ASE gustatory neuron class (gcy-5, gcy-6, gcy-7, and gcy-22; Table 2) (Yu et al. 1997; Johnston et al. 2005). We have identified five more gcy genes that are expressed in a left/right asymmetric manner in the ASE neuron class. gcy-1, gcy-3, and gcy-4 are expressed predominantly or exclusively in ASER, while gcy-14 and gcy-20 are expressed predominantly or exclusively in ASEL (Figure 5; data are quantified in Figure 6). Together with the previously reported expression patterns, a total of four gcy genes are lateralized to ASEL and five gcy genes are lateralized to ASER (summarized in Figure 1B). One-third (9/27) of all gcy genes are therefore laterally expressed in the ASE neurons. There are subtle differences in the degree of laterality of ASE-expressed gcy genes. Five of the nine L/R asymmetric gcy genes are exclusively expressed in ASER (gcy-1, gcy-5, gcy-22) or ASEL (gcy-6, gcy-7) (Johnston et al. 2005) (Figure 6A). The other four gcy genes are biased only to ASER (gcy-3 and gcy-4) or ASEL (gcy-14 and gcy-20) (Figure 6B); for example, while expression of gcy-4 is almost always stronger in ASER than in ASEL, there is often faint, but visible, expression in ASEL (Figure 6B). Since C. elegans transgenes harbor multiple copies of reporter gene constructs (Mello et al. 2001), the relevance of such relatively subtle quantitative details is difficult to assess. However, we note that each individual array of every reporter construct behaves in a similar way (see Table 1 for number of arrays scored), arguing that these observations are not due to transgene variance.
Figure 6.
Regulation of the expression of asymmetric gcy genes. (A and B) Quantification of the asymmetry of ASE-expressed gcy genes in wild-type and mutant backgrounds. (A) Quantification and a representative example of gcy-1prom∷gfp reporter gene-carrying animals. (B) Quantification of the results obtained with the other asymmetrically expressed and previously uncharacterized gcy reporter gene constructs. Black, white, and gray circles indicate relative expression levels of gfp in the ASEL and ASER neurons. Data shown are for one representative array each (otEx2419 for gcy-1prom∷gfp, otEx2423 for gcy-3prom∷gfp, otEx2409 for gcy-4prom∷gfp, otEx2322 for gcy-14prom∷gfp, and otEx2327 for gcy-20prom∷gfp). These arrays were each crossed into the indicated mutant backgrounds. Comparable results were obtained with several independent arrays (not shown). As in other figures, red indicates ASEL expression and blue indicates ASER expression. (C) Summary of the gene regulatory interactions. Gray shading indicates genes identified in this article. For more details on the ASEL and ASER inducers, see Johnston et al. (2005).
gcy genes that are asymmetrically expressed in either ASER or ASEL are not asymmetrically expressed in other sensory neurons. For example, the gcy-1 and gcy-3 genes, two ASER-expressed genes, are expressed bilaterally in other pairs of neurons.
In addition to the asymmetrically expressed gcy genes, we found two bilaterally, albeit weakly, expressed gcy genes in ASEL/R, gcy-19 and gcy-29, increasing the percentage of ASE-expressed gcy genes to 41% (11/27) of all gcy genes. The possible lack of regulatory elements in gfp reporter genes may lead to the oversight of perhaps even more ASE-expressed gcy genes.
We have not observed any other obvious left/right asymmetric gcy gene patterns in bilaterally symmetric neurons, including the AWCL/R neurons (which we could easily identify with the DsRed2-expressing otIs151 transgene). The AWCL/R neurons are the only other known neuron pair displaying functional laterality (Wes and Bargmann 2001) and although they express five gcy genes (Table 4), none of them is obviously lateralized.
Similarity of chromosomal position, primary sequence, and gene expression patterns:
Strikingly, the coexpression of gcy genes in ASEL or ASER correlates extensively with the primary sequence similarity and chromosomal location of the gcy genes. The ASER-expressed gcy-1, gcy-3, gcy-4, and gcy-5 genes fall into one sequence subgroup (Figure 3A) and localize within ∼800 kb and two of them are directly adjacent to one another (Figure 3B; Figure 4). Notably, their coexpression is not due simply to a joint cis-regulatory element; as our reporter constructs clearly demonstrate (Figure 4), each gene contains separable cis-regulatory elements.
Similarly, all four ASEL-expressed gcy genes (gcy-6, gcy-7, gcy-14, gcy-20) fall into one sequence subgroup (Figure 3A) and localize at the center of chromosome V (Figure 3B). These observations argue that gene duplication events that lead to the generation of these paralogs also duplicated their regulatory regions. The only exception to this pattern is the gcy-22 gene, which is predominantly expressed in ASER, but, in terms of primary sequence identity, clusters more closely with the ASEL-expressed genes than with the ASER-expressed genes (Figure 3).
A similar correlation of sequence relation and gene expression can also be observed in non-ASE-expressed gcy genes. The gcy-8, gcy-18, and gcy-23 genes are related by sequence (Figure 3A), localize within a 15.6-Mb interval (Figure 3B), and are all coexpressed in the AFD sensory neurons (Figure 5). Additionally, the AFD-expressed gcy-29 gene is closely related to gcy-8, gcy-18, and gcy-23 by primary sequence, but it localizes to a different chromosome. Finally, the sequence-related gcy-15 and gcy-21 genes (Figure 3A) are in close chromosomal proximity (∼530 kb; Figure 3B) and are coexpressed in the ASG chemosensory neurons (Figure 5).
Taken together, all these similarities indicate that related gcy genes arose by local gene duplication events that duplicated not only the protein-coding region but also their cis-regulatory control regions.
Regulation of left/right asymmetric expression of gcy genes:
How is the laterality of the newly characterized ASEL/R-expressed gcy genes controlled? We have previously identified a complex network of transcription factors and microRNAs (miRNAs) that control ASE laterality after animals have passed through an initial hybrid precursor state (Chang et al. 2003, 2004; Johnston and Hobert 2003, 2005; Johnston et al. 2005). These regulatory factors fall into two broad categories: (1) factors that control the activity of a bistable feedback loop that determines whether an ASE neuron adopts the ASEL or ASER state and (2) factors that act outside the regulatory loop to determine specific subsets of terminal differentiation features. To test whether the newly identified, asymmetrically expressed gcy genes are subject to regulation by this network or controlled by a different set of regulatory factors, we analyzed gfp reporter expression profiles in two null mutant backgrounds that are representative for each category, lsy-6 and lim-6. Animals that lack the lsy-6 miRNA display a complete switch from the ASEL fate to the ASER fate (Johnston and Hobert 2003) and animals that lack the lim-6 LIM homeobox gene fail to activate a defined subset of ASEL features and fail to repress a subset of ASER features (Hobert et al. 1999; Johnston et al. 2005). We find that all newly identified gcy genes are components of the ASEL and ASER state that is controlled by the lsy-6-dependent regulatory feedback loop. The ASER-specific gcy genes are ectopically activated in ASEL upon loss of the ASEL inducer lsy-6, and the ASEL-specific gcy genes are lost in the ASEL neuron of lsy-6 null mutant animals (Figure 6). We note that in the case of the ASEL-biased gcy-14 gene, which is normally strongly expressed in ASEL and weakly expressed in ASER, the conversion of the ASEL to the ASER fate does not entail a loss of gcy-14 expression, but rather the bilateralization of weak expression (Figure 6B).
How does the lim-6 LIM homeobox gene contribute to the expression of the newly identified gcy genes? Similarly to the previously described ASEL-specific gcy-6 and gcy-7 genes, we found that the ASEL bias of gcy-14 and gcy-20 are unaffected by loss of lim-6 (Figure 6B), demonstrating that ASEL inducers such as lsy-6 independently regulate the expression of lim-6 and ASEL-specific gcy genes (Figure 6C). Similarly to the previously described ASER-specific gcy-5 and gcy-22 genes, we found that lim-6 represses the expression of the ASER-specific gcy-1, gcy-3, and gcy-4 genes in ASEL (Figure 6, A and B). In summary, all known asymmetrically expressed gcy genes are controlled by the same categories of gene regulatory factors.
Functional analysis of gcy-5:
Three putative loss-of-function alleles have been generated by the C. elegans knockout consortia in one of the nine asymmetrically expressed gcy genes, gcy-5 (Figure 7A). At least one of them is a putative molecular null allele (see legend to Figure 7A). We analyzed all three mutant gcy-5 alleles in a chemotaxis assay that measures the functionality of the ASE neurons (Bargmann and Horvitz 1991). Our previous work has demonstrated that the ASEL neuron preferentially senses sodium, but not chloride, whereas the ASER neuron senses chloride and weakly contributes to sodium sensation (Pierce-Shimomura et al. 2001; Chang et al. 2004). Neither gcy-5 mutant allele shows any significant defects in attraction to sodium or chloride (Figure 7B). We conclude that gcy-5 is not required for generic aspects of ASER development or function.
Figure 7.
Functional analysis of gcy-5. (A) Mutant alleles of gcy-5. Color coding for the domains encoded by the individual exons is shown in Figure 2. Reading frames are indicated to illustrate the effects of the respective deletion alleles. tm897 contains a 691-bp deletion from position 462–1045 of the coding sequence and replacement with 5′-GGGGTAGAAGAGGC. Within the genomic locus, the deletion starts in exon 4 and ends in exon 7. With the deletion and insertion, a frameshift is created, leading to an early stop codon. This allele is therefore a putative null allele. The effect of the other alleles is more difficult to predict since the respective deletions start in exons and end in introns. If one assumes splicing around the half-deleted exons, then the two ok alleles produce large, but in-frame, deletions. (B) Chemotaxis to soluble ions of wild-type and gcy-5 mutant animals. NaCl measures the functionality of both ASEL and ASER. NH4Cl mainly measures ASER function since NH4+ is sensed in a ASE-independent manner [NH4+ sensation is unaffected in che-1 mutants in which ASEL/R fails to develop (Chang et al. 2004)]. All strains were grown and assayed at room temperature (21°–23°). Population chemotaxis assays were performed in a radial gradient of the indicated salt (see materials and methods). Each experiment was done with at least two plates in parallel and for each assay plate at least 20 worms reached either attractant or negative control spot. Three to nine independent experiments were done for each condition. Error bars indicate standard error of the mean. For statistical analysis, a one-way ANOVA was performed for each attractant, with Dunnet's post-test comparing all three alleles to wild-type control data. None of the means were significantly different.
Guanylyl cyclase genes in the nematode C. briggsae:
The availability of the genome sequence of the nematode C. briggsae, which diverged ∼100 million years ago (Stein et al. 2003), provided us with the opportunity to examine the evolution of the gcy gene family. To identify the complete set of guanylyl cyclase genes in C. briggsae, we again employed PSI-BLAST to search the latest release of the complete genome databases of C. briggsae, using a set of known GCY proteins as queries. We identified a total of 33 gcy genes in C. briggsae, one less than in C. elegans (Figure 3A). Compared to C. elegans, C. briggsae contains one additional soluble gcy gene (total = 8) and two fewer receptor-type gcy genes (total = 25). A total of 23 nematode receptor and soluble gcy genes show clear one-to-one ortholog matches between C. elegans and C. briggsae (Figure 3A). All the other gcy genes show species-specific gene duplication events. For example, the C. elegans gcy-6 gene has duplicated to produce two paralogs, while the directly adjacent C. elegans gcy-1, gcy-2, and gcy-3 genes are paralogs with only a single ortholog in C. briggsae (Figure 3A).
We have mentioned above the existence of a receptor-type GCY protein, GCY-27, that lacks a predicted extracellular domain. C. elegans GCY-27 is closely related to the intracellular domain of the receptor-type ODR-1; both proteins are more closely related to each other than to the two closest C. briggsae homologs (Figure 3A). One of these homologs, CBP15915* (see materials and methods for refined gene predictions), is clearly orthologous to ODR-1 due to synteny in the chromosomal region; that is, all predicted proteins in the direct neighborhood of ODR-1 are homologous to the neighboring proteins of the putative C. briggsae ODR-1 ortholog. The protein designated CBP11205* (see materials and methods for refined gene prediction) is the likely ortholog of GCY-27. Both proteins share the unusual feature of not containing an extracellular domain. As in the case of GCY-27, the absence of an extracellular domain in the C. briggsae ortholog is not likely caused by a failure to predict more exons because neighboring genes are in close proximity. We conclude that the extracellular domain of an ancestral protein was lost before the C. briggsae/C. elegans split, giving rise to GCY-27 and its ortholog CBP11205*.
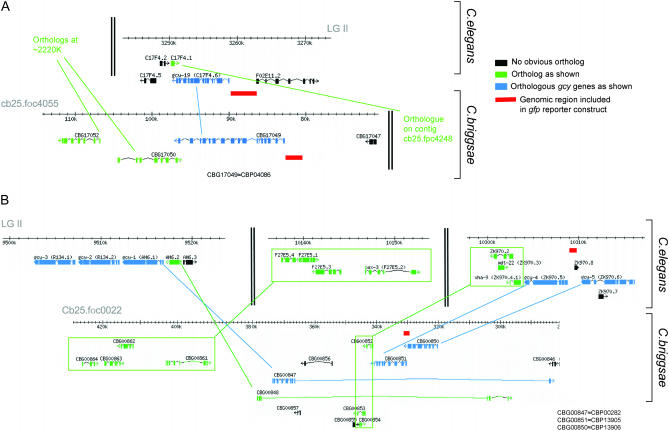
We sought to investigate the issue of orthology and paralogy in more detail by (a) analyzing the degree of synteny among gcy genes and (b) by determining the expression patterns of some orthologous C. briggsae genes. For the syteny analysis, we focused on one subgroup of left/right asymmetrically expressed gcy subfamilies, the “gcy-5 subfamily” (Figure 3A).
We found that the chromosomal arrangement of members of the gcy-5 subfamily differ significantly between C. elegans and C. briggsae. The gcy-19 orthologs are located in distinct environments in C. elegans and C. briggsae (Figure 8A), suggesting a translocation event after the lineage split. The case of the gcy-1, gcy-2, gcy-3, gcy-4, and gcy-5 genes is more complex and provides a fascinating glimpse into evolutionary divergence. While gcy-4 and gcy-5 are direct neighbors in both C. elegans and C. briggsae, they are translocated together with their neighbors to a distinct region in C. briggsae (Figure 8B). Curiously, this distinct region contains the single C. briggsae ortholog of three C. elegans paralogs from the gcy-5 subfamily, gcy-1, gcy-2, and gcy-3 (Figure 8B). Since all paralogous members of the gcy-5 subgroup likely arose by gene duplication, the C. briggsae gene organization is probably more reflective of the ancestral situation than C. elegans. One conceivable scenario is that a common ancestor of C. elegans and C. briggsae contained, like C. briggsae, three adjacent gcy genes (which themselves arose by gene duplication). In the C. elegans lineage, this cluster split up, with gcy-4 and gcy-5 translocating to a distinct chromosomal region and the gcy-1/2/3 ortholog also translocating to a distinct location and then subsequently duplicating to generate gcy-1, gcy-2, and gcy-3 in the C. elegans lineage.
Figure 8.
Analysis of synteny of gcy genes in C. elegans and C. briggsae. Gene predictions were taken from WormBase release WS149 (http://www.wormbase.org). The red line indicates the genomic regions included in gfp reporter gene constructs. The C. elegans reporter constructs are also shown in Figure 4. The size of the C. briggsae gcy-19 construct is 2 kb; the size of the C. briggsae gcy-4 construct is 433 bp (up to the preceding gene).
Evolutionary divergence of left/right asymmetric gcy gene expression patterns:
Using gfp reporter genes generated from genomic C. briggsae DNA, we analyzed the expression of five gcy genes in C. briggsae. We generated gfp fusions to the single C. briggsae ortholog of the C. elegans gcy-1, gcy-2, and gcy-3 genes and the C. briggsae orthologs of the C. elegans gcy-4, gcy-5, gcy-7, and gcy-19 genes. We observed no significant adult expression for C. briggsae gcy-1/2/3 prom∷gfp, C. briggsae gcy-5prom∷gfp, and C. briggsae gcy-7prom∷gfp in several transgenic C. briggsae lines, which is possibly due to the limited size of the reporter gene constructs (due to the size of the intergenic regions, none of the reporter constructs extended to the previous gene; data not shown). We did, however, observe interesting expression patterns for C. briggsae gcy-4prom∷gfp and C. briggsae gcy-19prom∷gfp and focused our analysis on these genes.
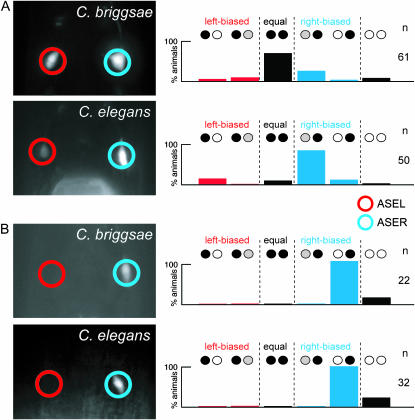
We found that C. briggsae gcy-4prom∷gfp is expressed bilaterally in both ASEL and ASER (Figure 9A). This is in striking contrast to C. elegans gcy-4prom∷gfp, whose expression is biased to ASER in C. elegans (Figure 5C; Figure 6B). Such a difference could be caused by two different mechanisms. The gcy-4 loci may contain distinct cis-regulatory information in C. elegans and C. briggsae, or, alternatively, the cis-regulatory information may be the same, but different trans-acting factors in C. elegans and C. briggsae interpret this information differentially. To distinguish between these possibilities, we injected the C. briggsae gcy-4prom∷gfp construct into C. elegans. If the C. elegans and C. briggsae gcy-4 reporter constructs contained the same cis-regulatory information, C. briggsae gcy-4prom∷gfp should be expressed in the same pattern in C. briggsae and C. elegans (bilateral expression in ASEL and ASER). In contrast, if there are differences in trans-acting factors, then the C. briggsae gcy-4prom∷gfp reporter should be expressed in C. elegans in a manner similar to that of C.elegans gcy-4prom∷gfp (biased to ASER). We found that C. briggsae gcy-4prom∷gfp expression becomes ASER biased when injected into C. elegans, which mimics the expression of C. elegans gcy-4prom∷gfp (Figure 9A). The difference between gcy-4 expression in C. elegans and gcy-4 expression in C. briggsae therefore does not appear to reflect a difference in their cis-regulatory architecture but rather indicates differences in the trans-acting factors that control gcy-4 expression.
Figure 9.
Phylogenetic conservation of gcy gene expression profiles. C. briggsae reporter gene constructs are shown in Figure 8; C. elegans reporter gene constructs are shown in Figure 4. Species names in pictures indicate into which species the respective reporter gene was injected. (A) gcy-4 expression. C. briggsae gcy-4prom∷gfp (CBP13906) is expressed in both ASEL and ASER in C. briggsae (three lines; data for one representative array, otEx2508, are shown), but is expressed predominantly in ASER in C. elegans (three lines; n > 40 each; data for one representative array, otEx2510, are shown). Expression of C. elegans gcy-4prom∷gfp (construct in Figure 4) in C. elegans is also biased to ASER (Figure 5C; Figure 6B). (B) gcy-19 expression. C. briggsae gcy-19prom∷gfp (CBP04086) is expressed exclusively in ASER in C. briggsae (three lines; data for one representative array, otEx2139, are shown) and in C. elegans (three lines; data for one representative array, otEx2141, are shown).
The gcy-19 locus represents another example of evolutionary divergence of left/right asymmetric gene expression. C. elegans gcy-19prom∷gfp is strongly expressed in the IL2 sensory neurons and weakly expressed in both ASEL and ASER and in several other head sensory neurons in C. elegans (Figure 5N). In striking contrast, C. briggsae gcy-19prom∷gfp shows strong and exclusive expression in the C. briggsae ASER neuron, but not in any other head neurons (Figure 9B). In contrast to the case of gcy-4, however, the difference in expression does not appear to be caused by differences in trans-acting factors. When injected into C. elegans, the C. briggsae gcy-19prom∷gfp reporter is still expressed exclusively in ASER (Figure 9B). The cis-regulatory architecture of the C. briggsae gcy-19 locus can therefore be “read out” in the same way by the trans-acting factors in both C. elegans and C. briggsae. Differences in the expression of C. elegans and C. briggsae gcy-19 are therefore more likely caused by a difference in the cis-regulatory architecture of these loci. Since C. elegans and C. briggsae gcy-19 reside in nonsyntenic chromosomal regions (Figure 8A), it appears that the alterations in chromosomal context of these two genes affected not only the neighboring genes of the gcy-19 loci but also their cis-regulatory architecture. As a word of caution, we note the intrinsic limitations of reporter gene constructs, which may not harbor the complete set of cis-regulatory elements, thereby potentially yielding a misleading impression of gene expression patterns.
DISCUSSION
Biochemical properties and functions of receptor-type guanylyl cyclases have been summarized and discussed in several reviews over the past few years (Lucas et al. 2000; Wedel and Garbers 2001; Morton 2004). Recent findings on the function of soluble gcy genes in oxygen sensation (Cheung et al. 2004, 2005; Gray et al. 2004) have also been reviewed (Rankin 2005). We focus here on several specific outcomes of our studies.
The function of nematode receptor-type gcy genes:
C. elegans and C. briggsae contain an unusual number of receptor-type gcy genes. Insects such as Drosophila melanogaster or Anopheles gambiae contain six receptor-type guanylyl cyclases (Morton 2004), mammals contain seven (four orphan and three peptide-binding receptors) (Lucas et al. 2000; Wedel and Garbers 2001), but C. elegans contains 27 and C. briggsae 25 (this study). The physiological function of insect gcy genes is entirely unknown, although the expression of the only two analyzed receptors in sensory neurons (among other neurons) has been noted (Morton 2004). Vertebrate gcy genes are expressed in several different tissue types, including chemosensory neurons (Wedel and Garbers 2001).
We propose that the significant expansion of receptor-type gcy genes in the nematode lineage is a reflection of their employment as chemoreceptors used to assess and navigate through their natural habitat. This hypothesis, which was also put forward by Yu et al. (1997), is mainly based on the observation that almost 90% (24/27) of gcy genes are expressed in sensory neurons (lack of sensory neuron expression of three gcy genes may be caused merely by a lack of the complete set of cis-regulatory elements in the reporter genes used). Moreover, 41% (11/27) of gcy genes are expressed in the main gustatory neuron class of C. elegans, ASE. This neuron class has previously been shown to be functionally lateralized in that it can sense different chemosensory cues (Pierce-Shimomura et al. 2001). Consistent with a role of gcy genes as chemoreceptors, we find that 9/11 ASE-expressed gcy genes are expressed in a left/right asymmetric manner, thereby providing molecular correlates to functional lateralization.
Amino acids are among the several classes of chemicals that can be sensed by the ASE neurons (Bargmann and Horvitz 1991). The presence of a domain in the extracellular parts of many GCY proteins that is homologous to bacterial amino-acid-binding proteins, the RFLBR domain, makes receptor-type GCY proteins good candidates for amino acid receptors. More sensitive assays (Wicks et al. 2000; Faumont et al. 2005; Miller et al. 2005; Faumont and Lockery 2006) than those used in the initial large scale survey of chemosensory cues (Ward 1973) will be required to establish the full spectrum of amino acids and other possible sensory cues that signal through ASE. Such a systematic cataloging of sensory cues needs to be followed by a systematic analysis of strains harboring deletions in gcy genes or misexpressing gcy genes to establish their roles as amino acid receptors.
A role for GCY proteins as salt receptors is also conceivable but highly speculative at present. There is as yet no consensus about the molecular identity of salt receptors in the vertebrate gustatory system. Notably, the crystal structure of the ANP receptor, a mammalian GCY protein that, like most C. elegans GCY proteins, contains an extracellular RFLBR domain, revealed a high-affinity chloride-binding site (van den Akker et al. 2000).
Our attempt to establish a mutant phenotype for a gcy gene, the ASER-expressed gcy-5 gene, has failed so far, but we note that we have tested only one of the two known cues sensed in an ASER-specific manner, namely chloride ions. The failure to detect a mutant phenotype, however, does allow the conclusion that gcy genes such as gcy-5 are unlikely to control a fundamental, nonredundant aspect of the development or overall function of the neuron. It appears more likely that gcy-5 and, by inference, other ASE-expressed gcy genes fulfill a sensory-modality-specific function in ASE, such as being a receptor for a specific class of gustatory cues.
Whereas more than half of C. elegans gcy genes (15/27) are expressed exclusively in sensory neurons, one-third are also expressed in nonsensory neurons, including interneurons and non-neuronal cells, suggesting that GCY proteins also respond to endogenously produced ligands. Since all known ligands for mammalian GCY proteins are peptidergic-signaling molecules (Lucas et al. 2000) and since C. elegans contains scores of neuropeptide-encoding genes (Li et al. 1999), we propose that nonsensory C. elegans GCY proteins may be receptors for peptidergic ligands.
The function of receptor-type GCY proteins may not be restricted to a receptor function. The C. elegans ODR-1 protein does not require its extracellular domain to fulfill its function in transducing odorsensory signals (L'Etoile and Bargmann 2000). Moreover, one GCY protein that we describe here, the ODR-1-related GCY-27 protein, entirely lacks an extracellular domain. These proteins may heterodimerize with ligand-binding GCY receptors to constitute a receptor complex and/or they may serve as second-messenger-producing signaling proteins that are embedded in signal transduction cascades triggered by other receptor systems.
Coexpression of gcy genes:
Another notable feature of gcy gene expression patterns is their degree of coexpression. Six gcy genes are coexpressed in the ASI sensory neuron class, five are coexpressed in ASER, four in ASEL, five in the AWC olfactory neuron class, four in the AFD thermosensory neuron class, and two gcy genes are each coexpressed in the ASG and PHA phasmid sensory neuron classes. In addition, several gcy genes expressed in multiple cell types show similar and nonintuitive combinations of expression patterns. gcy-7 and gcy-20 are coexpressed in ASEL and the excretory cell (two cells of no obvious relation) and daf-11 and odr-1 show a precisely overlapping expression pattern in five amphid sensory neurons (Birnby et al. 2000; L'Etoile and Bargmann 2000). Coexpression of receptor-type GCY proteins raises at least two different possibilities:
The proteins are independent receptors for distinct sensory inputs. While an attractive possibility for the ASE-expressed gcy genes, this is unlikely to be the case for daf-11 and odr-1, which are both independently required for AWC-mediated olfaction to several distinct odorants (Birnby et al. 2000; L'Etoile and Bargmann 2000).
Since GCY proteins dimerize, it is possible that the defined set of GCY proteins of one cell type can homo- and heterodimerize to form an even larger repertoire of dimerized receptor complexes. As previously suggested, such heterodimerization may be obligatory in those cases where a given GCY protein lacks residues that are necessary for cyclase activity (Morton 2004). The cyclase domains of several GCY proteins, including at least GCY-11, DAF-11, and GCY-29, are predicted to be inactive on the basis of the substitution of conserved residues required for activity (supplemental Figure 1 at http://www.genetics.org/supplemental/), but all three genes are coexpressed with other GCY proteins that are predicted to be active (Table 4).
Laterality in the nematode nervous system:
With the identification of a total of nine asymmetrically expressed gcy genes, the ASE neurons present so far the most striking example of a lateralized chemosensory neuron class. As mentioned above, previous work has provided a conceptual framework for the functional relevance of laterality in the ASE neurons (Pierce-Shimomura et al. 2001). We expect that other neuron classes may similarly employ the principle of lateralizing chemosensory function. However, our expression analysis has not revealed further examples of laterality in gcy gene expression profiles in other bilaterally symmetric amphid neurons, thereby leaving only the AWC odorsensory and ASE gustatory neurons as neuron pairs with lateralized functions (Figure 1B). Considering the potential usefulness of lateralizing chemoreceptor function, it is our expectation that the analysis of expression of the hundreds of chemoreceptors of the seven-transmembrane receptor family (Bargmann 1998) will reveal more examples of laterality in the nervous system.
Our analysis of the laterality of gcy gene expression in C. briggsae revealed several striking examples of evolutionary plasticity in the laterality of the gustatory system of nematodes. The variation that we observe appears to be caused by a variation in both cis-acting elements and trans-acting factors, a conclusion that we can draw from our comparison of C. briggsae gcy promoter activity in C. briggsae and C. elegans. Variations in cis-regulatory control have been recognized as a major feature of evolutionary processes (Carroll et al. 2001) but our cross-species analysis also provides strong support for the more conventional view of the evolution of trans-acting factors. The clearly distinct nature of cis-regulatory architecture of at least some gcy genes in C. elegans and C. briggsae is a strong reminder that the use of bioinformatic tools that use phylogenetic conservation to identify cis-regulatory elements in genomic sequences (e.g., Bigelow et al. 2004; for review see Bulyk 2003) may easily lead to false-negative predictions. It is difficult at this point to speculate about differences in the trans-acting factors that control laterality in C. elegans and C. briggsae. We have demonstrated that so far all known left/right asymmetrically expressed terminal differentiation markers in the ASEL/R neurons, including all gcy genes, are under control of a previously described bistable feedback loop that is composed of several transcription factors and miRNAs (Johnston et al. 2005). The activity of transcription factors that are controlled by the output of the loop, for example, the lim-6 LIM homeobox gene (Figure 6B), may have functionally diverged in C. briggsae. This can perhaps be best illustrated with the gcy-4 gene, which in C. elegans is repressed by lim-6 in ASEL. In C. briggsae, gcy-4 is not repressed in ASEL, yet the C. briggsae gcy-4 promoter can be repressed in C. elegans. These observations are consistent with the presence of a lim-6 responsive repressor element in both C. elegans and C. briggsae gcy-4 promoters but with an inability of C. briggsae lim-6 to control this element in C. briggsae.
Mutant screening approaches that identify the complete set of trans-acting factors controlling left/right asymmetric gcy gene expression in C. elegans and experimental promoter dissection approaches that identify cis-regulatory elements through which these trans-acting factors act are currently ongoing in our laboratory and are likely to reveal the molecular control of laterality in the gustatory system of C. elegans and its divergence in C. briggsae.
Acknowledgments
We thank Qi Chen for expert technical assistance; the C. elegans gene knockout consortia at Tokyo Women's Medical University School of Medicine (led by Shohei Mitani) and at the Oklahoma Medical Research Foundation (led by Bob Barstead) for providing mutant strains; the Caenorhabditis Genetics Center (CGC) and members of the worm community for providing strains; Piali Sengupta for help with cell identifications; and members of the Hobert lab and Cori Bargmann for comments on the manuscript. This work was supported by the National Institutes of Health (NIH) Medical Scientist Training Program (to C.O.O.), NIH R01 NS050266-01 (O.H.), and NIH R01 NS39996-05 (O.H.). B.H. and O.H. are Investigators of the Howard Hughes Medical Institute.
References
- Bargmann, C. I., 1998. Neurobiology of the Caenorhabditis elegans genome. Science 282: 2028–2033. [DOI] [PubMed] [Google Scholar]
- Bargmann, C. I., and H. R. Horvitz, 1991. Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron 7: 729–742. [DOI] [PubMed] [Google Scholar]
- Bargmann, C. I., and I. Mori, 1997. Chemotaxis and thermotaxis, pp. 717–738 in C.elegans II, edited by D. L. Riddle, T. Blumenthal, B. J. Meyer and J. R. Priess. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- Bigelow, H. R., A. S. Wenick, A. Wong and O. Hobert, 2004. CisOrtho: a program pipeline for genome-wide identification of transcription factor target genes using phylogenetic footprinting. BMC Bioinformatics 5: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnby, D. A., E. M. Link, J. J. Vowels, H. Tian, P. L. Colacurcio et al., 2000. A transmembrane guanylyl cyclase (DAF-11) and Hsp90 (DAF-21) regulate a common set of chemosensory behaviors in Caenorhabditis elegans. Genetics 155: 85–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulyk, M. L., 2003. Computational prediction of transcription-factor binding site locations. Genome Biol. 5: 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll, S. B., J. K. Grenier and S. D. Weatherbee, 2001. From DNA to Diversity: Molecular Genetics and the Evolution of Animal Design. Blackwell Science, Malden, MA.
- Chang, S., R. J. Johnston, Jr. and O. Hobert, 2003. A transcriptional regulatory cascade that controls left/right asymmetry in chemosensory neurons of C. elegans. Genes Dev. 17: 2123–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, S., R. J. Johnston, C. Frokjaer-Jensen, S. Lockery and O. Hobert, 2004. MicroRNAs act sequentially and asymmetrically to control chemosensory laterality in the nematode. Nature 430: 785–789. [DOI] [PubMed] [Google Scholar]
- Cheung, B. H., F. Arellano-Carbajal, I. Rybicki and M. de Bono, 2004. Soluble guanylate cyclases act in neurons exposed to the body fluid to promote C. elegans aggregation behavior. Curr. Biol. 14: 1105–1111. [DOI] [PubMed] [Google Scholar]
- Cheung, B. H., M. Cohen, C. Rogers, O. Albayram and M. de Bono, 2005. Experience-dependent modulation of C. elegans behavior by ambient oxygen. Curr. Biol. 15: 905–917. [DOI] [PubMed] [Google Scholar]
- Eddy, S. R., 1998. Profile hidden Markov models. Bioinformatics 14: 755–763. [DOI] [PubMed] [Google Scholar]
- Faumont, S., and S. R. Lockery, 2006. The awake behaving worm: simultaneous imaging of neuronal activity and behavior in intact animals at millimeter scale. J. Neurophysiol. 95: 1976–1981. [DOI] [PubMed] [Google Scholar]
- Faumont, S., A. C. Miller and S. R. Lockery, 2005. Chemosensory behavior of semi-restrained Caenorhabditis elegans. J. Neurobiol. 65: 171–178. [DOI] [PubMed] [Google Scholar]
- Gray, J. M., D. S. Karow, H. Lu, A. J. Chang, J. S. Chang et al., 2004. Oxygen sensation and social feeding mediated by a C. elegans guanylate cyclase homologue. Nature 430: 317–322. [DOI] [PubMed] [Google Scholar]
- Hedgecock, E. M., J. G. Culotti, J. N. Thomson and L. A. Perkins, 1985. Axonal guidance mutants of Caenorhabditis elegans identified by filling sensory neurons with fluorescein dyes. Dev. Biol. 111: 158–170. [DOI] [PubMed] [Google Scholar]
- Hobert, O., 2002. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques 32: 728–730. [DOI] [PubMed] [Google Scholar]
- Hobert, O., K. Tessmar and G. Ruvkun, 1999. The Caenorhabditis elegans lim-6 LIM homeobox gene regulates neurite outgrowth and function of particular GABAergic neurons. Development 126: 1547–1562. [DOI] [PubMed] [Google Scholar]
- Hobert, O., R. J. Johnston, Jr. and S. Chang, 2002. Left-right asymmetry in the nervous system: the Caenorhabditis elegans model. Nat. Rev. Neurosci. 3: 629–640. [DOI] [PubMed] [Google Scholar]
- Hugdahl, K., and R. J. Davidson (Editors), 2003. The Asymmetrical Brain. MIT Press, Cambridge, MA.
- Inada, H., H. Ito, J. Satterlee, P. Sengupta, K. Matsumoto et al., 2006. Identification of guanylyl cyclases that function in thermosensory neurons of Caenorhabditis elegans. Genetics 172: 2239–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, R. J., and O. Hobert, 2003. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 426: 845–849. [DOI] [PubMed] [Google Scholar]
- Johnston, R. J., Jr., and O. Hobert, 2005. A novel C. elegans zinc finger transcription factor, lsy-2, required for the cell type-specific expression of the lsy-6 microRNA. Development 132: 5451–5460. [DOI] [PubMed] [Google Scholar]
- Johnston, R. J., Jr., S. Chang, J. F. Etchberger, C. O. Ortiz and O. Hobert, 2005. MicroRNAs acting in a double-negative feedback loop to control a neuronal cell fate decision. Proc. Natl. Acad. Sci. USA 102: 12449–12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Etoile, N. D., and C. I. Bargmann, 2000. Olfaction and odor discrimination are mediated by the C. elegans guanylyl cyclase ODR-1. Neuron 25: 575–586. [DOI] [PubMed] [Google Scholar]
- Li, C., L. S. Nelson, K. Kim, A. Nathoo and A. C. Hart, 1999. Neuropeptide gene families in the nematode Caenorhabditis elegans. Ann. NY Acad. Sci. 897: 239–252. [DOI] [PubMed] [Google Scholar]
- Loria, P. M., J. Hodgkin and O. Hobert, 2004. A conserved postsynaptic transmembrane protein affecting neuromuscular signaling in Caenorhabditis elegans. J. Neurosci. 24: 2191–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas, K. A., G. M. Pitari, S. Kazerounian, I. Ruiz-Stewart, J. Park et al., 2000. Guanylyl cyclases and signaling by cyclic GMP. Pharmacol. Rev. 52: 375–414. [PubMed] [Google Scholar]
- Mello, C. C., J. M. Kramer, D. Stinchcomb and V. Ambros, 1991. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10: 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, A. C., T. R. Thiele, S. Faumont, M. L. Moravec and S. R. Lockery, 2005. Step-response analysis of chemotaxis in Caenorhabditis elegans. J. Neurosci. 25: 3369–3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton, D. B., 2004. Invertebrates yield a plethora of atypical guanylyl cyclases. Mol. Neurobiol. 29: 97–116. [DOI] [PubMed] [Google Scholar]
- Notredame, C., D. G. Higgins and J. Heringa, 2000. T-coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302: 205–217. [DOI] [PubMed] [Google Scholar]
- Page, R. D., 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12: 357–358. [DOI] [PubMed] [Google Scholar]
- Pierce-Shimomura, J. T., S. Faumont, M. R. Gaston, B. J. Pearson and S. R. Lockery, 2001. The homeobox gene lim-6 is required for distinct chemosensory representations in C. elegans. Nature 410: 694–698. [DOI] [PubMed] [Google Scholar]
- Rankin, C. H., 2005. Nematode memory: Now, where was I? Curr. Biol. 15: R374–R375. [DOI] [PubMed] [Google Scholar]
- Salamov, A. A., and V. V. Solovyev, 2000. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 10: 516–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambongi, Y., T. Nagae, Y. Liu, T. Yoshimizu, K. Takeda et al., 1999. Sensing of cadmium and copper ions by externally exposed ADL, ASE, and ASH neurons elicits avoidance response in Caenorhabditis elegans. Neuroreport 10: 753–757. [DOI] [PubMed] [Google Scholar]
- Stein, L. D., Z. Bao, D. Blasiar, T. Blumenthal, M. R. Brent et al., 2003. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 1: E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, T., C. Patoine, A. Abu-Khalil, J. Visvader, E. Sum et al., 2005. Early asymmetry of gene transcription in embryonic human left and right cerebral cortex. Science 308: 1794–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford, D. L., 2003. PAUP: Phylogenetic Analysis Using Parsimony and Other Methods. Sinauer Associates, Sunderland, MA.
- Taylor, S. S., D. R. Knighton, J. Zheng, L. F. Ten Eyck and J. M. Sowadski, 1992. Structural framework for the protein kinase family. Annu. Rev. Cell Biol. 8: 429–462. [DOI] [PubMed] [Google Scholar]
- Troemel, E. R., A. Sagasti and C. I. Bargmann, 1999. Lateral signaling mediated by axon contact and calcium entry regulates asymmetric odorant receptor expression in C. elegans. Cell 99: 387–398. [DOI] [PubMed] [Google Scholar]
- van den Akker, F., X. Zhang, M. Miyagi, X. Huo, K. S. Misono et al., 2000. Structure of the dimerized hormone-binding domain of a guanylyl-cyclase-coupled receptor. Nature 406: 101–104. [DOI] [PubMed] [Google Scholar]
- Ward, S., 1973. Chemotaxis by the nematode Caenorhabditis elegans: identification of attractants and analysis of the response by use of mutants. Proc. Natl. Acad. Sci. USA 70: 817–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedel, B., and D. Garbers, 2001. The guanylyl cyclase family at Y2K. Annu. Rev. Physiol. 63: 215–233. [DOI] [PubMed] [Google Scholar]
- Wenick, A. S., and O. Hobert, 2004. Genomic cis-regulatory architecture and trans-acting regulators of a single interneuron-specific gene battery in C. elegans. Dev. Cell 6: 757–770. [DOI] [PubMed] [Google Scholar]
- Wes, P. D., and C. I. Bargmann, 2001. C. elegans odour discrimination requires asymmetric diversity in olfactory neurons. Nature 410: 698–701. [DOI] [PubMed] [Google Scholar]
- White, J. G., E. Southgate, J. N. Thomson and S. Brenner, 1986. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 314: 1–340. [DOI] [PubMed] [Google Scholar]
- Wicks, S. R., C. J. de Vries, H. G. van Luenen and R. H. Plasterk, 2000. CHE-3, a cytosolic dynein heavy chain, is required for sensory cilia structure and function in Caenorhabditis elegans. Dev. Biol. 221: 295–307. [DOI] [PubMed] [Google Scholar]
- Yan, S. Z., Z. H. Huang, R. S. Shaw and W. J. Tang, 1997. The conserved asparagine and arginine are essential for catalysis of mammalian adenylyl cyclase. J. Biol. Chem. 272: 12342–12349. [DOI] [PubMed] [Google Scholar]
- Yu, S., L. Avery, E. Baude and D. L. Garbers, 1997. Guanylyl cyclase expression in specific sensory neurons: a new family of chemosensory receptors. Proc. Natl. Acad. Sci. USA 94: 3384–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]