Abstract
An SSR-based linkage map was constructed in Brassica rapa. It includes 113 SSR, 87 RFLP, and 62 RAPD markers. It consists of 10 linkage groups with a total distance of 1005.5 cM and an average distance of 3.7 cM. SSRs are distributed throughout the linkage groups at an average of 8.7 cM. Synteny between B. rapa and a model plant, Arabidopsis thaliana, was analyzed. A number of small genomic segments of A. thaliana were scattered throughout an entire B. rapa linkage map. This points out the complex genomic rearrangements during the course of evolution in Cruciferae. A 282.5-cM region in the B. rapa map was in synteny with A. thaliana. Of the three QTL (Crr1, Crr2, and Crr4) for clubroot resistance identified, synteny analysis revealed that two major QTL regions, Crr1 and Crr2, overlapped in a small region of Arabidopsis chromosome 4. This region belongs to one of the disease-resistance gene clusters (MRCs) in the A. thaliana genome. These results suggest that the resistance genes for clubroot originated from a member of the MRCs in a common ancestral genome and subsequently were distributed to the different regions they now inhabit in the process of evolution.
GENUS Brassica is cultivated in most parts of the world. It includes various important agronomical crops such as oilseed rape, cabbage, broccoli, Chinese cabbage, black mustard, and other leafy vegetables. Due to the wide choice of edible forms in the genus, extensive morphological variations such as the head, enlarged root, enlarged stem, color variation, and oil content have been selected for, these parts serving as cultivated foods for human beings. Furthermore, the genetic relation to Brassica has been well studied and is referred to as U's triangle (U 1935). The genomes of three diploid species, Brassica rapa, B. nigra, and B. oleracea, have been designated as A, B, and C, respectively, while those of the amphidiploids, B. juncea, B. napus, and B. carinata, have been designated as AB, AC, and BC, respectively (Iwabuchi et al. 1991; Lagercrantz and Lydiate 1996; Snowdon et al. 1997). Because such genetic and morphological variation has occurred as the result of a natural hybridization of species, the remarkable diversity of Brassica is an excellent example for understanding how genetic and morphological variation have developed during the evolution of plant genomes.
Recent studies of the model plant Arabidopsis thaliana have yielded new insights into plant genetics and genomics. A pattern of chromosomal colinearity has been identified between Arabidopsis and other flowering plants (Grant et al. 2000; Rossberg et al. 2001). Genomic synteny has been found to be well conserved among a broad array of species, even though genomic size in plant exhibits a wide diversity, e.g., 125 Mb for A. thaliana to 125 Gb for Fritillaria assyriaca (Bennett and Smith 1976). Physically and functionally conserved gene(s), known as orthologs and paralogs, were also identified (Axelsson et al. 2001). Because Brassica and Arabidopsis are in the same family, the Cruciferae, the level of synteny between them provides a good opportunity to study how genetic and morphological variation has developed during the evolution of the genome, including the endurance of certain genetic structures in Arabidopsis and related Brassica species. Such investigation may lead to a better understanding of plant genetics, including molecular biological and physiological aspects that have emerged via evolution.
In past decades, genetic maps of Brassica have been constructed by means of RFLPs, AFLPs, and RAPDs to better understand their genetic makeup (Slocum et al. 1990; Landry et al. 1991; Kianian and Quiros 1992; Ferreira et al. 1994; Uzunova et al. 1995). These studies have contributed to analyses of complicated quantitative traits and to comparisons of the organization of the chromosomes of Brassica. Since the DNA sequences of homologous genes in the related taxa are quite similar, clones have been extensively used as RFLP markers to elucidate the genomic colinearity between species (Osborn and Lukens 2003). The degree of genomic conservation has been elucidated by comparative linkage mapping, and conserved genomic segments have been identified by micro- and macrosynteny studies in various Brassica species (Cavell et al. 1998; Lagercrantz 1998; O'Neill and Bancroft 2000; Ryder et al. 2001). However, because of the high rate of genomic segment replication, RFLP has been shown at times to detect more than one locus in Brassica genomes (Lagercrantz and Lydiate 1996). Although duplication/triplication of genomic segments permits the evolution of genes, it also leads to considerable complexity in evaluating genomic colinearity within species. For these reasons, a linkage map based on more precise molecular markers that would allow discrimination between homologous and homeologous regions is required for an accurate comparative analysis in Brassica.
Clubroot is one of the most serious diseases that afflict Brassica crops (Crute et al. 1980). Plasmodiophora brassicae, the causal agent of clubroot, is a soil-borne, obligate pathogen. The pathogen can infect most Brassica species such as oilseed rape, cabbage, Chinese cabbage, and turnip. Several mapping studies have already been reported on B. oleracea, B. rapa, and B. napus, and the major QTL for clubroot resistance have been reported (Figdore et al.1993; Grandclément and Thomas 1996; Kuginuki et al. 1997; Voorrips et al. 1997; Suwabe et al. 2003). Despite these efforts, little information has been made available to allow a comprehensive understanding of the clubroot resistance seen in Brassica. An improved understanding of the genetic mechanisms and the evolution of clubroot resistance would be one of the examples for carrying out genetic analysis of disease resistance in Brassica.
In this article, we describe (i) the construction of a genetic linkage map of B. rapa based mainly on SSR markers, (ii) the genomic synteny between B. rapa and A. thaliana by comparative mapping based on the SSRs in the present map, (iii) QTL analysis for clubroot resistance in two different pathogen isolates, and then we discuss the origin and evolution of clubroot resistance in Brassica by a combination of the QTL and synteny analyses of A. thaliana. The result is obtained by comparative genomics, utilizing linkage maps in species possessing complex genomes, and the well established information on model plants.
MATERIALS AND METHODS
Plant materials:
A clubroot-resistant (CR) doubled-haploid (DH) line, G004, and a susceptible (CS) DH Chinese cabbage parental line, nou 7 (A9709), were used as the parents for the mapping population (Suwabe et al. 2003). An F2 population consisting of 94 lines was selected randomly and F3 seeds were obtained from the bud self-pollination of each F2 line for clubroot-resistance analysis. Young leaves of each F2 line were freeze dried and stored at −20° until DNA isolation. Three cultivars of Chinese cabbage, “CR Ryutoku” (Watanabe Seed Co., Japan), “Utage 70” (Nozaki Seed Co., Japan) (CR even though they exhibit a breakdown of resistance to “Wakayama-01”), and “Muso” (CS; Takii Seed Co., Kyoto, Japan) were used as controls in the clubroot-resistance analysis. All plants were grown in a greenhouse.
Molecular markers:
Complete information on the SSR markers is described in Suwabe et al. (2002, 2004). A total of 343 SSR markers were used for the construction of the map. RFLP markers were kindly provided C. Quiros, University of California (Davis, CA) (Harada et al. 1988), and some of the markers were developed originally at the National Institute of Vegetable and Tea Science (NIVTS). RAPD primers were obtained from BEX (Common's primer set, Tokyo), Operon Technologies (Alameda, CA), University of British Columbia (Vancouver, BC, Canada), and Wako Pure Chemical Industries (Osaka, Japan). Some of the primers were originally developed at NIVTS.
To certify the genomic correlations in focused regions within B. rapa and A. thaliana (see the details in the results section), SNP and/or indel markers were developed using the information in the Arabidopsis database (TAIR: http://www.arabidopsis.org). Single-copy genes in the region of focus in the Arabidopsis genome were selected, and the primers were designed from these genes' nucleotide sequences to amplify the products to ∼500 bp. After the confirmation of successful amplification in A. thaliana, the PCR products amplified in A9709 and G004 were sequenced and compared with each other in terms of the nucleotide sequences for SNPs and/or indels.
Genetic analysis:
For map construction, all molecular markers showing polymorphisms between A9709 and G004 were selected and applied to segregation analysis in the F2 population.
The SNP and/or indel markers between A9709 and G004 were also applied to this segregation analysis in the F2 population.
Linkage analysis and map construction:
Segregation of each marker in the F2 population was analyzed by a chi-square test for “goodness-of-fit” to a 1:2:1 (codominant marker) or 3:1 (dominant marker) ratio. Linkage analysis of markers was performed using MAPMAKER/EXP 3.0 (Lander et al. 1987). A framework map of codominant markers was constructed by assignment to linkage groups by the “group” and “order” commands with a LOD score of 8.0. Dominant markers were inserted into the intervals in the framework map by the “try” command. The “ripple” command was used to confirm the order of markers in each linkage group. All genetic distances are expressed in centimorgan values as derived by the Kosambi function (Kosambi 1944).
Comparison of SSR loci between B. rapa and A. thaliana:
The nucleotide sequences of each SSR locus in B. rapa, 123–1129 bp with an average of 484 bp, were aligned with Arabidopsis genome sequences by BLASTN in TAIR. The methodology for claiming homologous SSR loci was the following: (i) upper and lower nucleotide sequences flanking the SSR were aligned independently, because repeat numbers of SSRs differ greatly between B. rapa and A. thaliana (Suwabe et al. 2004), (ii) the results of each alignment for an SSR locus were compared, and (iii) SSR loci with a consistent homology of flanking upper and lower regions were realigned with the entire nucleotide sequences of SSR loci to calculate a homology value. We regarded as homologous SSR loci that had flanking upper and lower genomic regions assigned to the same region in the A. thaliana genome with a threshold value of E < 10−10. When the E-value was between 10−10 and 10−5, corresponding loci were manually confirmed by the presence of SSR sequences and the homology value of the flanking region. We regarded regions having neighboring SSR loci that were relatively conserved between B. rapa and A. thaliana as a homologous synteny region.
QTL analysis for clubroot resistance:
The P. brassicae isolates Wakayama-01 and “Ano-01” were used in this study (Kuginuki et al. 1997; Suwabe et al. 2003). Wakayama-01 has the highest virulence in our pathogen stock (see the details in Suwabe et al. 2003). In contrast, Ano-01 was the lowest, able to infect Muso but not CR Ryutoku or Utage 70. Because of the intermediate reactions of Williams' differential hosts (Williams 1966), the race numbers of the present isolates could not be determined. The inoculum was propagated throughout the infection test and clubs in the infected roots were stored at −20° until required. Resting spores were purified from the clubs according to Williams (1966) and used in the test for clubroot resistance. The tests for clubroot resistance and data evaluation are described in a previous report (Suwabe et al. 2003). The root symptoms of each plant were evaluated as follows: grade 0, no symptoms; grade 1, a few small, separate globular clubs on the lateral roots; grade 2, intermediate symptoms; grade 3, severe clubs on the main roots. A disease index (ID) was constructed from the results as the mean grade for 16 F3 seedlings, and the mean ID for each F2 line was expressed from two independent tests (a total of 32 seedlings). QTL analysis was performed using MAPMAKER/QTL 1.1 with a LOD threshold of 2.0.
RESULTS
Marker analysis:
A total of 200 codominant markers (113 SSRs and 87 RFLPs), exhibiting polymorphisms between the parental lines, were screened for framework markers in the linkage map. Most of the markers segregated with the 1:2:1 Mendelian ratio in the F2 population; however, 15.5% (31/200) of the markers deviated significantly (P < 0.05) from this ratio. In addition, 62 RAPD markers with polymorphisms between the parental lines were screened, and in total 262 markers were ultimately applied to the map.
Construction of the linkage map:
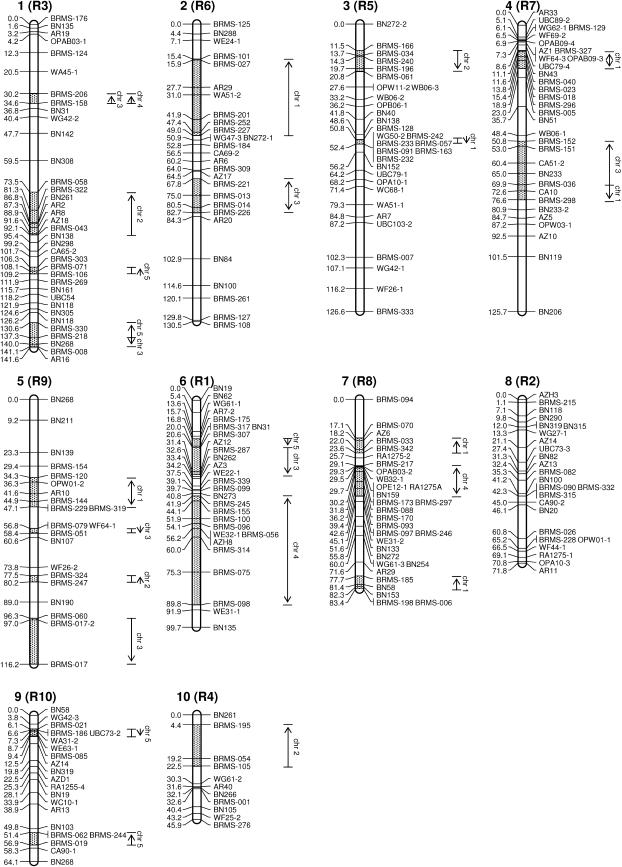
The resulting map is composed of 10 linkage groups covering 1005.5 cM (Figure 1). The largest linkage group consists of 36 loci and is a length of 141.6 cM, and the smallest is 11 loci with a length of 45.9 cM (Table 1). Because the B. rapa karyotype is composed of 10 chromosomes, these 10 linkage groups seem to correspond to the chromosomes of B. rapa. The distances between markers varied from 0 to 24.1 cM, with an average distance of 3.7 cM. SSR markers were distributed throughout the linkage groups with an average distance of 8.7 cM. From the genome size of B. rapa, 550 Mb (Arumuganathan and Earle 1991), the average relationship between the physical and genetic distances is ∼1 cM = 500 kb.
Figure 1.
Linkage map of B. rapa and comparative map with A. thaliana. The 10 linkage groups are represented by vertical bars. Recombination distances are given in Kosambi centimorgans on the left side of the linkage groups, and the locus names are given on the right side. The number in parentheses at the top of each linkage group, R1–R10, indicates the internationally agreed chromosomal nomenclature of B. rapa. The synteny regions with A. thaliana (stippled areas) are shown to the right of the B. rapa linkage groups as vertical lines with chromosome numbers (chr 1 to chr 5). The arrowheads indicate the order of the corresponding region in the A. thaliana chromosomes. Double-headed arrows indicate the recombinant nature in the corresponding regions of B. rapa and A. thaliana. Marker types are indicated by first one to four letters: BRMS, SSR; A and BN, RFLP; and CA, OP, RA, UBC, and W, RAPD marker.
TABLE 1.
Characteristic of the B. rapa linkage map
| Linkage group
|
Length (cM)
|
No. of markers
|
|||
|---|---|---|---|---|---|
| SSR | RFLP | RAPD | Total | ||
| 1 | 141.6 | 14 | 17 | 5 | 36 |
| 2 | 130.5 | 15 | 8 | 4 | 27 |
| 3 | 126.6 | 14 | 5 | 12 | 31 |
| 4 | 125.7 | 11 | 10 | 11 | 32 |
| 5 | 116.2 | 12 | 6 | 3 | 21 |
| 6 | 99.7 | 14 | 10 | 4 | 28 |
| 7 | 83.4 | 15 | 8 | 7 | 30 |
| 8 | 71.8 | 7 | 11 | 7 | 25 |
| 9 | 64.1 | 6 | 8 | 7 | 21 |
| 10 | 45.9 | 5 | 4 | 2 | 11 |
| Total | 1005.5 | 113 | 87 | 62 | 262 |
Synteny analysis between B. rapa and A. thaliana:
To identify the synteny-possessing region(s) of B. rapa and A. thaliana, homologous SSR loci in the A. thaliana genome were sought using the B. rapa nucleotide sequences flanking SSRs. In 113 SSR loci of the linkage map, 74 were identified as having corresponding loci in the A. thaliana genome, while the remaining 39 could not be aligned, although a low level of homology (E > 10−5) was observed in several genomic regions in A. thaliana (Table 2). In homologous SSR loci, 52.7% (39/74) composed a single homologous region in the A. thaliana genome, while the other loci represented >2 loci, most of them corresponding to 2 or 3 loci, in the A. thaliana genome. The majority of the homologous SSR loci in A. thaliana included the same SSR motifs, although most had shorter repeats than those in B. rapa, and the flanking sequences likewise exhibited a high rate of homology.
TABLE 2.
Homologous SSR loci in B. rapa and A. thaliana
| SSR marker
|
A. thaliana
|
||
|---|---|---|---|
| Chromosome | BAC clone | E-value | |
| BRMS-001 | 5 | F3F24. F21E10 | 0.81 |
| BRMS-005 | 5 | MUD21, MSN2 | 4.00E-07 |
| BRMS-006 | 1 | T25K16 | 4.00E-39 |
| 4 | F1104 | 1.00E-08 | |
| 3 | F21F14 | 4.00E-05 | |
| BRMS-007 | 3 | K2019 | 0.008 |
| BRMS-008 | 3 | K1G2 | 0.11 |
| BRMS-013 | 2 | T16F16 | 0.063 |
| BRMS-014 | 3 | F1I16 | 0.043 |
| 2 | T2G17 | 0.043 | |
| BRMS-017 | 3 | K20I9 | 4.00E-08 |
| BRMS-018 | 2 | F24L7 | 1.5 |
| BRMS-019 | 5 | F21E1, F8F6 | 4.00E-65 |
| BRMS-021 | 5 | MBG8 | 6.00E-07 |
| BRMS-023 | 1 | T2P11 | 3.00E-13 |
| BRMS-026 | 1 | F3N23 | 0.014 |
| BRMS-027 | 1 | F10F5 | 6.00E-13 |
| BRMS-033 | 1 | F5D21, F19C24 | 8.00E-34 |
| BRMS-034 | 2 | F18O19 | 7.00E-43 |
| BRMS-036 | 1 | F12A21 | 6.00E-55 |
| 1 | F21J9 | 2.00E-23 | |
| 3 | F20C19 | 2.00E-08 | |
| 1 | F3F19 | 5.00E-06 | |
| BRMS-040 | 1 | F2J7 | 2.00E-19 |
| 1 | F24J5 | 7.00E-10 | |
| BRMS-043 | 2 | F4P9 | 0.035 |
| BRMS-051 | 3 | T5N23 | 2.00E-34 |
| BRMS-054 | 2 | F4P9 | 2.00E-05 |
| 2 | T9J23 | 6.00E-04 | |
| 2 | F17A22 | 6.00E-04 | |
| BRMS-056 | 5 | K18I23, MOP10 | 0.009 |
| BRMS-057 | 5 | MUF9 | 0.31 |
| BRMS-058 | 2 | T6D20 | 5.00E-42 |
| 4 | F22L13, F20D10 | 3.00E-28 | |
| BRMS-060 | 3 | K20I9 | 4.00E-09 |
| BRMS-061 | 2 | F4L23 | 5.00E-12 |
| BRMS-062 | 2 | T27E13 | 2.00E-33 |
| 5 | MHF15 | 2.00E-15 | |
| 5 | T2I1 | 1.00E-06 | |
| BRMS-070 | 1 | F20D21 | 7.00E-41 |
| BRMS-071 | 5 | T6G21 | 0.49 |
| BRMS-075 | 2 | T28M21, T5I7 | 4.00E-04 |
| BRMS-079 | 3 | MOD1 | 6.00E-52 |
| 3 | T17F15 | 1.00E-50 | |
| 3 | T26G12 | 5.00E-34 | |
| 4 | AP21, AP22 | 3.00E-04 | |
| BRMS-082 | 2 | F17K2 | 0.001 |
| BRMS-085 | 5 | MRI1, MTI20 | 6.00E-19 |
| BRMS-088 | 4 | C6L9 | 0.009 |
| 4 | T1J24 | 0.15 | |
| BRMS-090 | 2 | T32G6, F4P9 | 0.88 |
| 5 | K9B18, MUF9 | 0.88 | |
| BRMS-091 | 4 | F1C12 | 0.27 |
| 5 | T1G16 | 0.27 | |
| BRMS-093 | 5 | T1M15 | 0.024 |
| BRMS-094 | 5 | MXC20 | 5.00E-28 |
| BRMS-096 | 4 | T5K18 | 3.00E-07 |
| BRMS-097 | 4 | T30C3 | 0.028 |
| BRMS-098 | 4 | T19K4 | E-104 |
| BRMS-099 | 1 | F13A11, F2H10 | 5.00E-04 |
| BRMS-100 | 4 | F17L22 | 5.00E-13 |
| BRMS-101 | 1 | F10F5 | 9.00E-34 |
| BRMS-105 | 2 | F4P9 | 3.00E-07 |
| BRMS-106 | 5 | MAC 12 | 2.00E-07 |
| BRMS-108 | 3 | T4C21 | 0.89 |
| 2 | T103 | 0.89 | |
| 4 | F4C21 | 0.89 | |
| 3 | F1M23 | 0.89 | |
| BRMS-120 | 1 | F8K4 | 9.00E-15 |
| BRMS-124 | 2 | T32G6 | 0.18 |
| 5 | F7K24 | 0.18 | |
| 5 | T24G5 | 0.18 | |
| 1 | F5M6 | 0.18 | |
| BRMS-125 | 4 | F18A5 | 0.19 |
| 5 | F22D1 | 0.19 | |
| 4 | T1J24 | 0.19 | |
| BRMS-127 | 3 | T4C21 | 0.58 |
| 4 | F4C21 | 0.58 | |
| BRMS-128 | 2 | T32F6 | 2.00E-23 |
| 1 | YUP8H12 | 4.00E-12 | |
| BRMS-129 | 4 | T7B11 | 1.00E-17 |
| 5 | MKD15 | 3.00E-15 | |
| 1 | T14P4 | 1.00E-14 | |
| 3 | T17J13 | 8.00E-13 | |
| 5 | K11I1, F10E10 | 7.00E-04 | |
| BRMS-144 | 1 | F1N18 | 0.006 |
| BRMS-151 | 3 | MAA21 | 8.00E-25 |
| BRMS-152 | 3 | NAA21 | 3.00E-33 |
| BRMS-154 | 2 | T30D6 | 4.00E-16 |
| BRMS-155 | 1 | T23E23 | 0.05 |
| BRMS-158 | 3 | MJG19(T7) | 2.00E-20 |
| 4 | T27D20(T3) | 1.00E-06 | |
| BRMS-163 | 1 | T12O21 | 5.00E-26 |
| BRMS-166 | 2 | F7D19, F23E6 | 2.00E-40 |
| BRMS-170 | 4 | L23H3 | 3.00E-17 |
| BRMS-173 | 4 | T6K21 | 2.00E-22 |
| BRMS-175 | 5 | T32M21, T19N18 | 7.00E-15 |
| BRMS-176 | 5 | T32B03 | 8.00E-36 |
| 3 | T28G19, F21A14 | 5.00E-31 | |
| BRMS-184 | 3 | MPE11 | 2.00E-05 |
| BRMS-185 | 1 | T21E18 | 5.00E-22 |
| 2 | T28P16 | 2.00E-06 | |
| BRMS-186 | 5 | F5O24 | 0.44 |
| BRMS-195 | 3 | F18O21, F27K19 | 4.00E-19 |
| 2 | T3G21, T7M7 | 4.00E-16 | |
| BRMS-196 | 2 | T14P1 | 4.00E-19 |
| BRMS-198 | 5 | MJE4 | 4.00E-05 |
| 1 | T14P4 (′3 only) | 1.00E-16 | |
| BRMS-201 | 1 | F7H2 | 4.00E-75 |
| BRMS-206 | 3 | MUH15 | 5.00E-33 |
| 4 | FCAALL | 3.00E-13 | |
| BRMS-214 | 4 | F17A13 | 8.00E-67 |
| BRMS-215 | 5 | T2L20 | 1.00E-27 |
| 1 | F8D11, F13A11 | 3.00E-07 | |
| 1 | F2G19, F8G22 | 2.00E-04 | |
| BRMS-217 | 4 | F6I18, MAL21, F16B22 | 0.15 |
| BRMS-218 | 5 | NED24, F8F6 | 4.00E-44 |
| 3 | T22K18 | 3.00E-20 | |
| BRMS-221 | 3 | MVA11 | 0.16 |
| BRMS-226 | 3 | K17E12 | 2.00E-67 |
| BRMS-227 | 1 | T26J12 | 0.017 |
| BRMS-228 | 5 | MSL3 | 0.003 |
| BRMS-229 | 1 | T4K22 | 4.00E-35 |
| BRMS-232 | 4 | T13J8 | 0.49 |
| BRMS-233 | 5 | T25O11 | 0.027 |
| BRMS-240 | 2 | F18O19 | 6.00E-43 |
| BRMS-242 | 3 | F4B12 | 0.26 |
| BRMS-244 | 5 | T28J14 | 2.00E-35 |
| 5 | MRI1 | 2.00E-04 | |
| BRMS-245 | 4 | FCAALL | 2.00E-54 |
| 5 | MQL5 | 1.00E-12 | |
| BRMS-246 | 4 | T19F6 | 4.00E-21 |
| BRMS-247 | 2 | F27L4 | 4.00E-25 |
| 4 | AP21, AP22 | 2.00E-05 | |
| BRMS-252 | 1 | F6A14 | 3.00E-18 |
| BRMS-261 | 5 | MLN1 | 7.00E-16 |
| BRMS-269 | 2 | T1B3 | 0.024 |
| 1 | F9I5 | 0.024 | |
| BRMS-276 | 5 | K15O15 | 1.00E-40 |
| BRMS-287 | 3 | MCB17 | 8.00E-50 |
| BRMS-296 | 1 | F5M15 | 2.00E-20 |
| BRMS-297 | 4 | T6K21 | 3.00E-23 |
| BRMS-298 | 1 | T17F3 | 1.00E-64 |
| 1 | F20P5 | 1.00E-12 | |
| BRMS-303 | 5 | MNC17 | 1.00E-33 |
| BRMS-307 | 3 | T21B14, T23B7 | 5.00E-32 |
| 5 | MPH15 | 3.00E-15 | |
| BRMS-309 | 5 | K7J8 | 0.015 |
| BRMS-314 | 4 | F17A13 | 7.00E-67 |
| BRMS-315 | 5 | MRO11 (T3) | 0.065 |
| 5 | K5J14 (T7) | 0.019 | |
| BRMS-317 | 3 | T23B7, T21B14 | 2.00E-31 |
| 5 | MPH15 | 9.00E-15 | |
| BRMS-319 | 1 | F1N18 | 3.00E-35 |
| BRMS-322 | 2 | F6E13 | 1.00E-12 |
| BRMS-324 | 2 | F17H15, F3N11 | 3.00E-10 |
| BRMS-327 | 3 | F24K9 | 0.13 |
| BRMS-330 | 5 | MYH9 | 0.26 |
| BRMS-332 | 2 | T22F11, F23J3, T24P13 | 1.2 |
| BRMS-333 | 3 | T4P13 | 1.00E-31 |
| 1 | F10C21, T16O9 | 5.00E-06 | |
| BRMS-339 | 3 | F9K21, T14D3 | 0.002 |
| BRMS-342 | 1 | T28K15 | 4.00E-26 |
| 1 | F5O11 | 1.00E-19 | |
Five chromosomes of A. thaliana were separated into short segments and dispersed in various regions in the B. rapa map, although no continuous synteny region was observed in linkage group 8 (Figure 1, Table 2). Inversions, tandem duplications, and deletions were identified within some of the synteny regions. The longest synteny region was identified in linkage group 6, between BRMS-245 and BRMS-098 for a length of 47.9 cM, and the shortest regions were found in linkage groups 3 and 5, between BRMS-128 and BRMS-163 and between BRMS-079 and BRMS-051 for a length of 1.6 cM. In total, regions ranging up to 282.5 cM in length in the B. rapa map were identified to have synteny to A. thaliana. In addition to these synteny regions, certain independent SSR loci were found to be homologous to A. thaliana, such as BRMS-215 in linkage group 8 (Table 2), but no correlation was found within the neighboring SSR loci in Arabidopsis (Figure 1). Therefore, the synteny region with A. thaliana in the present B. rapa map is likely underestimated.
QTL analysis for clubroot resistance:
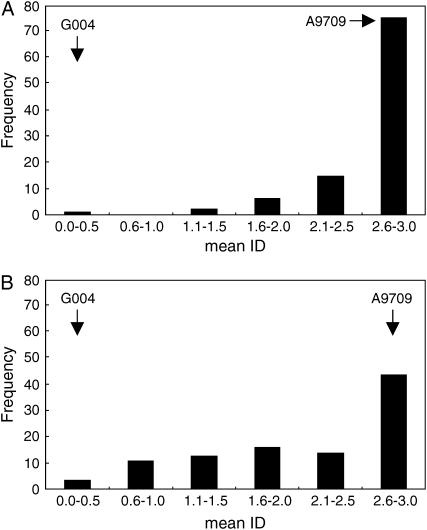
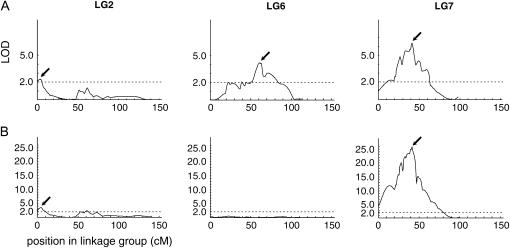
In a previous study, we identified two clubroot resistance loci, Crr1 and Crr2, for a Wakayama-01 isolate, which nearly cosegregated with SSR markers, BRMS-088 and BRMS-096, respectively (Suwabe et al. 2003). In the present linkage map, these two markers were located in linkage groups 7 and 6, respectively. Because detailed information about these two loci, such as a precise position and genetic effect, could not be determined in our previous study, we conducted a QTL analysis for a Wakayama-01 isolate using the present linkage map. The ID of F2 individuals for the Wakayama-01 isolate ranged from “very susceptible” to “resistant,” but very susceptible individuals were more frequent than resistant individuals (Figure 2A). From the analysis, three QTL were identified at a LOD threshold of 2.0 (Figure 3A). Among these, two major QTL in linkage group 7, between BRMS-297 and -088, and in linkage group 6, between BRMS-100 and -096, corresponded to Crr1 and Crr2 (Suwabe et al. 2003), respectively. These accounted for 26.8 and 18.3% of phenotypic variance, respectively (Table 3). In addition, a small QTL found in linkage group 2 accounted for 10.5% of the variance. Because this small QTL was independent of Crr1, Crr2, and Crr3 (Hirai et al. 2003), it is in fact a novel resistance locus and is designated Crr4. The total phenotypic variance of the three QTL was estimated to be 56.4%.
Figure 2.
Frequency distribution of disease index (ID) for clubroot resistance in the F2 population. (A) “Wakayama-01” isolate; (B) “Ano-01” isolate.
Figure 3.
QTL LOD plots for clubroot resistance in B. rapa. The LOD scores for QTL affecting clubroot resistance are presented along with the linkage groups 2, 6, and 7. The LOD threshold of 2.0 is indicated. (A) “Wakayama-01” isolate; (B) “Ano-01” isolate.
TABLE 3.
Position and effect of QTL for clubroot resistance in B. rapa
| Isolate | Linkage group | Marker interval | LOD | % variation |
|---|---|---|---|---|
| Wakayama-01 | 7 | BRMS-297–BRMS-088 | 6.4 | 26.8 |
| 6 | BRMS-100–BRMS-096 | 4.1 | 18.3 | |
| 2 | BN288D–WE24-1 | 2.3 | 10.5 | |
| Ano-01 | 7 | BRMS-297–BRMS-088 | 25.7 | 71.7 |
| 2 | BN288D–WE24-1 | 3.5 | 15.9 |
By analysis of the Ano-01 isolate, the ID of the F2 individuals ranged from very susceptible to resistant (Figure 2B), and two QTL in linkage groups 7 and 2 were identified at a LOD threshold of 2.0 (Figure 3B). The regions of these two QTL were in accordance with Crr1 and Crr4 and accounted for 71.7 and 15.9% of the variance, respectively (Table 3). The total phenotypic variance of these two QTL was estimated to be 74.6%.
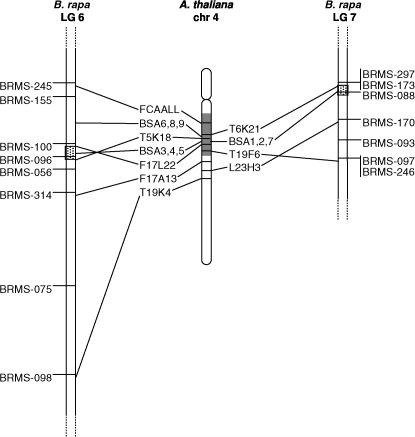
With the synteny map data from B. rapa and A. thaliana, two major QTL regions, Crr1 and Crr2, were found to be aligned to the same genomic region of chromosome 4 in A. thaliana (Figure 4). The region that includes Crr2 corresponds to the Arabidopsis BAC clones T5K18 (BRMS-096) and F17L22 (BRMS-100). In the Crr1 region, BRMS-173 and BRMS-297 correspond to Arabidopsis BAC T6K21, but BRMS-088 displays only a low homology to Arabidopsis BACs. The region corresponding to Crr2 in the Arabidopsis genome ranges to 970 kb and includes 273 open reading frames (ORFs). Of these, 59 ORFs exist as a single-copy gene in the Arabidopsis genome. All primer pairs designed for the 59 ORFs were successful in A. thaliana, and the 30 pairs also successfully amplified the corresponding fragments in the B. rapa mapping parents, A9709 and G004. A total of 12 markers were designed from these fragment sequences and named Brassica SNPs/indels from Arabidopsis (BSA) (Table 4). They included SNPs and/or indels between A9709 and G004. Three markers, BSA 3, BSA4, and BSA5, were mapped to the region of Crr2 in linkage group 6 of B. rapa. Three other markers, BSA1, BSA2, and BSA7, were mapped to the Crr1 region in linkage group 7. Additional BSA markers (BSA6, BSA8, and BSA9) developed from the synteny region of BRMS-100 and BRMS-155 in Arabidopsis were also mapped to the corresponding region in the B. rapa map.
Figure 4.
Synteny map of the Crr1 and Crr2 regions (stippled area) with B. rapa and A. thaliana. The linkage groups (LG) of B. rapa and the chromosome (chr) of A. thaliana are presented as vertical bars. The Arabidopsis BAC clones that correspond to the SSR loci of B. rapa are connected by lines. The shaded region indicates the cluster of disease-resistance genes (MRC-H) in the Arabidopsis chromosome.
TABLE 4.
SNP/indel markers constructed at the homologous Crr2 region in A. thaliana
| Marker | Primer | Product size (bp) | Acc. no. | Predicted homologous protein |
|---|---|---|---|---|
| BSA1 | CAGTCTAAAGGTGATCTTGCAGAGG | 402 | AT4G19550 | Expressed protein |
| ACCTGATGAGCAGCAATAATAACCA | ||||
| BSA2 | TGTTAGACATCAAAGAGGGCTTGAG | 489 | AT4G19900-2 | Glycosyl transferase related |
| TCTCAAAGACGATAATGAACCCAAA | ||||
| BSA3 | CATTTGAGATTAAGATCAGGCGATG | 491 | AT4G20350 | Expressed protein |
| TCGAGTTTGGTCTCTGTGAAAAATC | ||||
| BSA4 | TAACGCTGTTGGTCCAATTCTAGTG | 475 | AT4G20760 | Short-chain dehydrogenase |
| CCACAGGATCTTTCCTCCTACCTAA | Reductase family protein | |||
| BSA5 | ATTCTTTGATATTGAGCCATGTGGT | 589 | AT4G21520 | Transduction family protein |
| TCCAGAGAGATACACAGTTCTCATCA | WD-40 repeat family | |||
| BSA6 | ACTCCTCTGGAAATGCTCTTGAACT | 435 | AT4G21820 | Calmodulin-binding family protein |
| ATTACTGATTGAGGGCCTTCTTGAC | ||||
| BSA7 | AAGCAGAAGGGTTCTTGTCTGAACT | 410 | AT4G21865 | Expressed protein |
| TCACTCTGAGACATGAGGACACTTG | ||||
| BSA8 | GTGATCTTCTTCATGCTGTTGTTCC | 590 | AT4G22930 | Dihydroorotase |
| ACACCATCTTGAGAGTTGGTTGTTG | ||||
| BSA9 | CATTGCCGGTGAAGCTGAAG | 683 | AT4G27430 | COP1-interacting protein 7 |
| CACAAGTCCACAAAGTTAACACACG |
DISCUSSION
The SSR-based linkage map of B. rapa:
The present linkage map consists mainly of SSR markers and 10 linkage groups that correspond to the chromosome number of B. rapa, with an average distance between markers of 3.7 cM (Figure 1). One of the most conspicuous characteristics of the present map is the mapping of the 113 SSR markers. As indicated in the human linkage map (Dib et al. 1996), SSR markers have certain outstanding characteristics: (i) a codominant nature of high information value on genetic analysis; (ii) most can detect a single locus, while RFLP probes have a tendency to detect more than one locus in Brassica; (iii) SSR loci are well conserved among various B. rapa accessions, and most can be applied to related species such as B. oleracea, B. napus, and A. thalinana with high accuracy (Suwabe et al. 2002, 2004); and (iv) their genotype can be detected by a simple PCR technique. That is, in Brassica, SSR markers make it possible to carry out accurate genetic analysis with a simple technique, and moreover, the correspondence of the linkage groups and/or the linkage segments in the different maps can be compared with great clarity. In fact, SSR markers in the present linkage map could be used to assign linkage groups to the internationally agreed chromosomal nomenclature of B. rapa, R1–R10 (Figure 1, G. Teakle and G. King, personal communication). Comparative genomics elucidates not only the chromosomal organization within and between species, but also the evolutional processes of genomes. Several reports have described the conservation and colinearity of small chromosomal segments between various Cruciferous species, such as B. napus and A. thaliana, and have predicted extensive rearrangements of chromosomal segments before/after the differentiation of these species (Conner et al. 1998; Sadowski and Quiros 1998; Quiros et al. 2001; Babula et al. 2003; Lukens et al. 2003). In Brassica, linkage maps have been constructed by RFLP, RAPD, and AFLP markers and contributed to various genetic analyses, although to date alignment of the different maps has been conducted exclusively on the basis of RFLPs.
The comparative map of B. rapa and A. thaliana:
B. rapa and A. thaliana belong to the same family, the Cruciferae. They originated from a common ancestor and differentiated into related genera ∼14.5–20.4 million years ago (Yang et al. 1999). As indicated in previous studies (refer to Osborn and Lukens 2003), a region should exist in common in the chromosomes in these species. Arabidopsis has a relatively small genome (125 Mb), resulting from an exceptionally low rate of repetitive sequences and high gene density (Meyerowitz 1992). In contrast, the genome of B. rapa is far larger (550 Mb) than that of A. thaliana and has a high rate of rearrangements (Kowalski et al. 1994). It is not surprising that the complex rearrangements and overlaps of the Arabidopsis genome exist in the genome of Brassica. The present results reveal that small genomic segments of Arabidopsis are dispersed throughout the entire B. rapa genome (Figure 1). Furthermore, we found no synteny in linkage group 8, although BRMS-215 exhibited a high homology (E < 10−10) to certain parts of chromosomes 1 and 5 in A. thaliana. These results illustrate the complex nature of the chromosomal rearrangements that occurred before/after differentiation of Arabidopsis and Brassica. They also support a previous report that A. thaliana and Brassica originated from a common ancestor and differentiated with certain chromosomal segments conserved (Yang et al. 1999). Together with earlier reports on B. oleracea (Babula et al. 2003), the situation is more complex than has been previously expected in Brassica. These results also support the complexity of the Brassica genome.
Clubroot resistance in B. rapa and its genetic origin:
Crr1 in linkage group 7 and Crr4 in linkage group 2 were effective in the two different pathogen isolates used in this study (Figure 3, Table 3). These results suggest that these two QTL are commonly effective for resistance to clubroot in B. rapa and play key roles in resistance. There are two components to the mechanism of disease resistance. The first is a common pathway that is involved in responses against pathogen attack (Meyers et al. 1999). Many genes would participate in such a cascade, such as genes in the signaling pathways and in the production of resistance factor(s). Although the pathological response mechanisms of Crr1 and Crr4 are not clear at present, key genes for resistance do reside in these two QTL regions. The second component is a determinant for specificity in recognition during plant–pathogen interactions and/or an enhancing factor for the expression of a more potent resistance. In this study, Crr2 was specifically effective against the Wakayama-01 isolate, the most virulent. The expression of resistance to the Ano-01 isolate was effective without Crr2, however (Figure 3, Table 3). One possible explanation for this result is that Crr2 is an enhancer for the resistance expressed by Crr1 and/or Crr4, rather than a determinant for recognition specificity. That is, clubroot resistance in B. rapa is controlled by two components, a common pathway that has a central role in resistance and an additional component that is an enhancer for more powerful resistance.
The two QTL regions containing CR loci, Crr1 and Crr2, were aligned to the same region of chromosome 4 in A. thaliana and overlapped with each other (Figure 4). These results suggest that a progenitor resistance gene(s) existed in a common ancestral genome and resided in the same region of the chromosome, although it is not known whether they are functional against clubroot in either the ancestral species or Arabidopsis. These results suggest two hypotheses about the evolution of the clubroot resistance. One is that clubroot resistance was originally controlled by a single major gene, which differentiated and diverged into two functionally different, duplicate genes, during evolution of the genome in Brassica. If so, it might gain a diversification of the resistance mechanism in response to differences in pathogenicity. This could also lead to an increased complexity of the resistance mechanism. In maize, the rp1 complex for common rust resistance has evolved with a duplication and recombination of genes in the evolution of a family of resistance genes, leading to a diversification of resistance (Collins et al. 1999). Another possibility is that the resistance genes for clubroot were originally clustered in that genomic region of the ancestral genome and distributed into two different genomic regions following chromosomal rearrangements in Brassica. A clustering of disease-resistance genes has been observed in certain genomic regions in A. thaliana (Holub 1997) and also in other plant genomes (Hulbert et al. 2001). A region in chromosome 4 of A. thaliana, a focus of the present study, was previously identified as one of the clusters of disease-resistance genes, termed the major recognition complexes (MRCs) (Holub 1997; Speulman et al. 1998). In this region (MRC-H), disease-resistance genes with characteristic motifs, such as leucine-rich repeats (LRRs) and nucleotide-binding sites (NBSs), are clustered, for example, RPP for a resistance to Peronospora parasitica and RPS for a resistance to Pseudomonas syringae, as well as ACD, acceleration of cell death in response pathogen infection. These genes confer resistance to viral, bacterial, and fungal pathogens. The clustering of resistance genes confers an advantage, because it helps maintain resistance while allowing novel specificities to evolve. The resistance genes for clubroot may be members of just such a resistance gene cluster, and species-specific evolution of the resistance mechanism may have occurred in Brassica. At present, although as yet inconclusive, both models would eventuate in a diversity of resistance mechanisms to clubroot.
In another, diploid Brassica species, B. oleracea, two QTL, CR2a and CR2b have been identified as being involved in clubroot resistance (Landry et al. 1992). These account for 58 and 15% of phenotypic variability. Voorrips et al. (1997) also reported similar results with pb-3 and pb-4. These characteristics in B. oleracea seem to be consistent with the characteristics in B. rapa. Because the genomes of B. rapa and B. oleracea are thought to have evolved from a common ancestor, it is interesting to inquire whether clubroot resistance in B. rapa and B. oleracea might likewise have a common origin and be maintained by similar mechanisms. Further comparative studies of B. rapa and B. oleracea should provide new insights into this hypothesis.
Our results provide evidence for a genetic effect of QTL in clubroot resistance and genomic correlation with Arabidopsis. They could lead to new insights into the complicated mechanisms of clubroot resistance in Brassica. Further genetic analysis using genomic information from Arabidopsis will move us toward a comprehensive understanding of clubroot resistance in Brassica and should yield valuable information for the deliberate breeding of clubroot-resistance crops in Brassica. More generally, they will contribute to the understanding of the emergence of resistance gene clusters in the course of evolution.
Acknowledgments
We thank G. Teakle and G. King for allowing us to cite their work as personal communications. We also thank Y. Kowyama, K. Kakeda, and T. Tsuchiya of Mie University for their valuable advice. We are grateful to K. Tanaka and H. Maeda for their technical assistance and to Pacific Edit for review of the manuscript prior to submission. This work was supported by the Cooperative System for Supporting Priority Research of the Japan Science and Technology Corporation, by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan (Rice Genome Project DM-2105), and also by a grant-in-aid from Ministry of Education, Culture, Sports, Science and Technology, Japan, no. 14360006.
References
- Arumuganathan, K., and E. D. Earle, 1991. Nuclear DNA content of some important plant species. Plant. Mol. Biol. Rep. 9: 211–215. [Google Scholar]
- Axelsson, T., O. Shavorskaya and U. Lagercrantz, 2001. Multiple flowering time QTLs within several Brassica species could be the result of duplicated copies of one ancestral gene. Genome 44: 856–864. [PubMed] [Google Scholar]
- Babula, D., M. Kaczmarek, A. Barakat, M. Delseny, C. F. Quiros et al., 2003. Chromosomal mapping of Brassica oleracea based on ESTs from Arabidopsis thaliana: complexity of the comparative map. Mol. Gen. Genomics. 268: 656–665. [DOI] [PubMed] [Google Scholar]
- Bennett, M. D., and J. B. Smith, 1976. Nuclear DNA amounts in angiosperms. Proc. R. Soc. Lond. Ser. B 274: 227–274. [DOI] [PubMed] [Google Scholar]
- Cavell, A., D. Lydiate, I. A. P. Parkin, C. Dean and M. Trick, 1998. Collinearity between 30-centimorgan segment of Arabidopsis thaliana chromosome 4 and duplicated regions within the Brassica napus genome. Genome 41: 62–69. [PubMed] [Google Scholar]
- Collins, N., J. Drake, M. Ayliffe, Q. Sun, J. Ellis et al., 1999. Molecular characterization of the maize Rp1-D rust resistance haplotype and its mutants. Plant Cell 11: 1365–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner, J. A., P. Conner, M. E. Nashrallah and J. B. Nashrallah, 1998. Comparative mapping of the Brassica S locus region and its homeolog in Arabidopsis. Implications for the evolution of mating systems in the Brassicaceae. Plant Cell 10: 801–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crute, I. R., A. R. Gray, P. Crisp and S. T. Buczacki, 1980. Variation in Plasmodiophora brassicae and resistance to clubroot disease in brassicas and allied crops—a critical review. Plant Breed. Abstr. 50: 91–104. [Google Scholar]
- Dib, C., S. Faure, C. Fizames, D. Samson, N. Drouot et al., 1996. A comprehensive genetic map of human genome based on 5264 microsatellites. Nature 380: 152–154. [DOI] [PubMed] [Google Scholar]
- Ferreira, M. E., P. H. Williams and T. C. Osborn, 1994. RFLP mapping of Brassica napus using doubled haploid lines. Theor. Appl. Genet. 89: 615–621. [DOI] [PubMed] [Google Scholar]
- Figdore, S. S., M. E. Ferreira, M. K. Slocum and P. H. Williams, 1993. Association of RFLP markers with trait loci affecting clubroot resistance and morphological characters in Brassica oleracea L. Euphytica 69: 33–44. [Google Scholar]
- Grandclément, C., and G. Thomas, 1996. Detection and analysis of QTLs based on RAPD markers for polygenic resistance to Plasmodiophora brassicae Woron in Brassica oleracea L. Theor. Appl. Genet. 93: 86–90. [DOI] [PubMed] [Google Scholar]
- Grant, D., P. Cregan and R. C. Shoemaker, 2000. Genome organization in dicots: genome duplication in Arabidopsis and synteny between soybean and Arabidopsis. Proc. Natl. Acad. Sci. USA 97: 4168–4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada, J. J., C. S. Baden and L. Comai, 1988. Spatially regulated genes expressed during seed germination and postgerminative development are activated during embryogeny. Mol. Gen. Genet. 212: 466–473. [Google Scholar]
- Hirai, M., T. Harada, N. Kubo, M. Tsukada, K. Suwabe et al., 2003. A novel locus for clubroot resistance in Brassica rapa and its linkage markers. Theor. Appl. Genet. 108: 639–643. [DOI] [PubMed] [Google Scholar]
- Holub, E. B., 1997. The gene-for-gene relationship in plant-parasite interactions, pp. 5–26 in the Gene-for-Gene Relationship in Plant-Parasite Interactions, edited by I. R. Crute, E. B. Holub and J. J. Burdon. CAB International, Wallingford, UK.
- Hulbert, S. H., C. A. Webb, S. M. Smith and Q. Sun, 2001. Resistance gene complexes: evolution and utilization. Annu. Rev. Phytopathol. 39: 285–312. [DOI] [PubMed] [Google Scholar]
- Iwabuchi, M., K. Itoh and K. Shimamoto, 1991. Molecular and cytological characterization of repetitive DNA sequences in Brassica. Theor. Appl. Genet. 81: 349–355. [DOI] [PubMed] [Google Scholar]
- Kianian, S. F., and C. F. Quiros, 1992. Generation of a Brassica oleracea composite RFLP map—linkage arrangements among various populations and evolutionary implications. Theor. Appl. Genet. 84: 544–554. [DOI] [PubMed] [Google Scholar]
- Kosambi, D. D., 1944. The estimation of map distance from recombination values. Ann. Eugen. 12: 172–175. [Google Scholar]
- Kowalski, S. P., T. H. Lan, K. A. Feldmann and A. H. Paterson, 1994. Comparative mapping of Arabidopsis thaliana and Brassica oleracea chromosomes reveals islands of conserved organization. Genetics 138: 499–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuginuki, Y., H. Ajisaka, M. Yui, H. Yoshikawa, K. Hida et al., 1997. RAPD markers linked to a clubroot-resistance locus in Brassica rapa L. Euphytica 98: 149–154. [Google Scholar]
- Lagercrantz, U., and D. J. Lydiate, 1996. Comparative genome mapping in Brassica. Genetics 144: 1903–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagercrantz, U., 1998. Comparative mapping between Arabidopsis thaliana and Brassica nigra indicates that Brassica genomes have evolved through extensive genome replication accompanied by chromosome fusion and frequent rearrangements. Genetics 150: 1217–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E. C., P. Green, J. Abrahamson, A. Barlow, M. J. Daly et al., 1987. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181. [DOI] [PubMed] [Google Scholar]
- Landry, B. S., N. Hubert, T. Etoh, J. J. Harada and S. E. Lincoln, 1991. A genetic map for Brassica napus based on restriction fragment length polymorphisms detected with expressed DNA sequences. Genome 34: 543–552. [Google Scholar]
- Landry, B. S., N. Hubert, R. Crete, M. Chang, S. E. Lincoln et al., 1992. A genetic map of Brassica oleracea based on RFLP markers detected with expressed DNA sequences and mapping of resistance genes to race 2 of Plasmodiophora brassicae (Woronin). Genome 35: 409–419. [Google Scholar]
- Lukens, L., F. Zou, D. Lydiate, I. Parkin and T. Osborn, 2003. Comparison of a Brassica oleracea genetic map with the genome of Arabidopsis thaliana. Genetics 164: 359–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerowitz, E., 1992. Introduction to the Arabidopsis genome, pp. 102–118 in Methods in Arabidopsis Research, edited by C. Koncz, N. Chua and J. Schell. World Scientific, Singapore.
- Meyers, B. C., A. W. Dickerman, R. W. Michelmore, S. Sivaramakrishnan, B. W. Sobral et al., 1999. Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J. 20: 317–332. [DOI] [PubMed] [Google Scholar]
- O'Neill, C. M., and I. Bancroft, 2000. Comparative physical mapping of segments of the genome of Brassica oleracea var. alboglabra that are homoeologous to sequenced regions of chromosome 4 and 5 of Arabidopsis thaliana. Plant J. 23: 233–243. [DOI] [PubMed] [Google Scholar]
- Osborn, T. C., and L. Lukens, 2003. The molecular genetic basis of flowering time variation in Brassica species. Biotechnol. Agric. For. 52: 69–86. [Google Scholar]
- Quiros, C. F., F. Grellet, J. Sadowski, T. Suzuki, G. Li et al., 2001. Arabidopsis and Brassica comparative genomics: sequence, structure and gene content in the ABI-Rps2-Ck1 chromosomal segment and related regions. Genetics 157: 1321–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossberg, M., K. Theres, A. Acarkan, R. Herrero and T. Schmitt, 2001. Comparative sequence analysis reveals extensive microcolinearity in the lateral suppressor regions of the tomato, Arabidopsis, and Capsella genomes. Plant Cell 13: 979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder, C. D., L. B. Smith, G. R. Teakle and G. J. King, 2001. Contrasting genome organization: two regions of the Brassica oleracea genome compared with collinear regions of the Arabidopsis thaliana genome. Genome 44: 808–817. [PubMed] [Google Scholar]
- Sadowski, J., and C. F. Quiros, 1998. Organization of an Arabidopsis thaliana gene cluster on chromosome 4 including the RPS2 gene, in the Brassica nigra genome. Theor. Appl. Genet. 96: 468–474. [DOI] [PubMed] [Google Scholar]
- Slocum, M. K., S. S. Figdore, W. C. Kennard, J. Y. Suzuki and T. C. Osborn, 1990. Linkage arrangement of restriction fragment length polymorphism loci in Brassica oleracea. Theor. Appl. Genet. 80: 57–64. [DOI] [PubMed] [Google Scholar]
- Snowdon, R. J., W. Kohler, W. Friedt and A. Kohler, 1997. Genomic in situ hybridization in Brassica amphidiploids and interspecific hybrids. Theor. Appl. Genet. 95: 1320–1324. [Google Scholar]
- Speulman, E., D. Bouchez, E. B. Holub and J. L. Beynon, 1998. Disease resistance gene homologs correlate with disease resistance loci of Arabidopsis thaliana. Plant J. 14: 467–474. [DOI] [PubMed] [Google Scholar]
- Suwabe, K., H. Iketani, T. Nunome, T. Kage and M. Hirai, 2002. Isolation and characterization of microsatellites in Brassica rapa L. Theor. Appl. Genet. 104: 1092–1098. [DOI] [PubMed] [Google Scholar]
- Suwabe, K., H. Tsukazaki, H. Iketani, K. Hatakeyama, M. Fujimura et al., 2003. Identification of two loci for resistance to clubroot (Plasmodiophora brassicae Woronin) in Brassica rapa L. Theor. Appl. Genet. 107: 997–1002. [DOI] [PubMed] [Google Scholar]
- Suwabe, K., H. Iketani, T. Nunome, A. Ohyama, M. Hirai et al., 2004. Characterization of microsatellites in Brassica rapa genome and their potential utilization for comparative genomics in Cruciferae. Breed. Sci. 54: 85–90. [Google Scholar]
- U, N., 1935. Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn. J. Bot. 7: 389–452. [Google Scholar]
- Uzunova, M., W. Ecke, K. Weissleder and G. Robbelen, 1995. Mapping the genome of rapeseed (Brassica napus L.). 1. Construction of an RFLP linkage map and localization of QTLs for seed glucosinolate content. Theor. Appl. Genet. 90: 194–204. [DOI] [PubMed] [Google Scholar]
- Voorrips, R. E., M. C. Jongerious and H. J. Kanne, 1997. Mapping of two genes for resistance to clubroot (Plasmodiophora brassicae) in a population of doubled haploid lines of Brassica oleracea by means of RFLP and AFLP markers. Theor. Appl. Genet. 94: 75–82. [DOI] [PubMed] [Google Scholar]
- Williams, P. H., 1966. A system for the determination of races of Plasmodiophora brassicae that infect cabbage and rutabaga. Phytopathology 56: 624–626. [Google Scholar]
- Yang, Y. W., K. N. Kai, P. Y. Tai and W. H. Li, 1999. Rates of nucleotide substitution in angiosperm mitochondrial DNA sequences and dates of divergence between Brassica and other angiosperm lineages. J. Mol. Evol. 48: 597–604. [DOI] [PubMed] [Google Scholar]