Abstract
Histones are essential for the compaction of DNA into chromatin and therefore participate in all chromosomal functions. Specific mutations in HTA1, one of the two Saccharomyces cerevisiae genes encoding histone H2A, have been previously shown to cause chromosome segregation defects, including an increase in ploidy associated with altered pericentromeric chromatin structure, suggesting a role for histone H2A in kinetochore function. To identify proteins that may interact with histone H2A in the control of ploidy and chromosome segregation, we performed a genetic screen for suppressors of the increase-in-ploidy phenotype associated with one of the H2A mutations. We identified five genes, HHT1, MKS1, HDA1, HDA2, and HDA3, four of which encode proteins directly connected to chromatin function: histone H3 and each of the three subunits of the Hda1 histone deacetylase complex. Our results show that Hda3 has functions distinct from Hda2 and Hda1 and that it is required for normal chromosome segregation and cell cycle progression. In addition, HDA3 shows genetic interactions with kinetochore components, emphasizing a role in centromere function, and all three Hda proteins show association with centromeric DNA. These findings suggest that the Hda1 deacetylase complex affects histone function at the centromere and that Hda3 has a distinctive participation in chromosome segregation. Moreover, these suppressors provide the basis for future studies regarding histone function in chromosome segregation.
DURING mitosis the cell undergoes a complex and timely set of events necessary to segregate the replicated chromosomes into daughter cells. An important component of the process is the centromere, the specialized DNA region of the chromosome that serves as anchor to the kinetochore, a large protein structure necessary for attaching sister chromatids to microtubules. Proper biorientation of sister kinetochores is essential for capturing the spindle microtubules that extend from opposite poles (reviewed in Biggins and Walczak 2003; Hauf and Watanabe 2004). Kinetochore attachment to microtubules defines the fate of the mitotic chromosome, since errors or deviations from the normal bipolar attachment can result in chromosome loss or nondisjunction events, leading to aneuploidy, polyploidy, and often cell death.
DNA packaging into chromatin creates many challenges for the cellular processes that require interaction with DNA, such as transcription, DNA replication, repair, recombination, and chromosome segregation. Understanding how the cell manages chromatin is a necessary condition for unraveling the mechanisms underlying these processes. The basic unit of chromatin, the nucleosome, is evolutionarily conserved and composed of histone proteins and DNA. Histones H3 and H4 form a tetramer that is bound by two H2A–H2B dimers to form the histone octamer, to which ∼146 bp of DNA wrap around to form the nucleosome (van Holde 1988). Chromosome segregation relies on a specialized chromatin surrounding the centromere–kinetochore, the pericentric heterochromatin, consisting of specialized nucleosomes that maintain an epigenetic transcriptionally silent state formed by covalently modified histones (Richards and Elgin 2002; Sharp and Kaufman 2003). In contrast with the long heterochromatic regions characteristic of higher eukaryotes, Saccharomyces cerevisiae appears to be devoid of centromeric heterochromatin. Instead, its centromere consists of a minimally functional DNA element (CEN) of 125 bp that is composed of three conserved elements, CDEI, CDEII, and CDEIII, which are bound by kinetochore proteins (Hegemann and Fleig 1993; Cheeseman et al. 2002; McAinsh et al. 2003; Sharp and Kaufman 2003). The CDEI (8 bp) and CDEIII (25 bp) segments are separated by CDEII, 78–86 bp of highly AT-rich DNA. Mutations in CDEI or lack of Cbf1, the CDEI-binding protein, result in a slight defect in chromosome segregation (Cai and Davis 1990; Niedenthal et al. 1993); however, CDEII and CDEIII are essential for centromere activity. Although the CDEII region is somewhat flexible to small alterations, deletion of the complete sequence abolishes centromere function (Gaudet and Fitzgerald-Hayes 1987). CDEIII is a highly conserved but imperfect palindrome that binds to the multiprotein complex CBF3 (Ndc10, Cep3, Ctf13, and Skp1) (Lechner and Carbon 1991; Connelly and Hieter 1996). Mutations in CDEIII that impair CBF3 binding or in any of the essential subunits of CBF3 have severe effects on mitotic chromosome segregation (McGrew et al. 1986, 1989; Saunders et al. 1988; Densmore et al. 1991; Goh and Kilmartin 1993; Sorger et al. 1994). Current models propose the formation of a specialized nucleosome, where two copies of the highly conserved histone H3 variant, Cse4, replace both copies of histone H3 to form a tetramer with H4 (Meluh et al. 1998). Cse4 is the S. cerevisiae counterpart of human CENP-A, a histone H3 isoform exclusively present in centromeric chromatin (Stoler et al. 1995; Keith and Fitzgerald-Hayes 2000).
In spite of the lack of a defined centromeric heterochromatin, many studies suggest that histones perform critical functions in S. cerevisiae mitosis. Centromeric chromatin structure, revealed by nuclease mapping, consists of a nuclease-resistant core flanked by arrays of phased nucleosomes (Bloom and Carbon 1982). Depletion of histone H2B or H4 caused increased nuclease sensitivity in centromeric DNA, as well as in the pericentric nucleosomes (Saunders et al. 1990), and impaired mitotic chromosome segregation (Han et al. 1987; Kim et al. 1988), a defect also seen with increasing histone dosage (Meeks-Wagner and Hartwell 1986). Single amino acid replacements in histone H2A revealed specific defects in chromosome segregation and centromere structure (Pinto and Winston 2000), suggesting a direct role for histones in centromere function. In addition, deletion of the conserved N-terminal domains of H3 and H4 causes a mitotic cell cycle delay often seen in mutants with defects in chromosome segregation (Morgan et al. 1991), and a conditional allele of H4 leads to a G2/M cell cycle arrest and an increased rate of chromosome loss (Smith et al. 1996). Although these studies clearly implicate histones in chromosome segregation, their particular roles are not yet understood.
Previous analysis of two mutations in HTA1, one of the two genes encoding histone H2A, revealed an increase-in-ploidy phenotype, in addition to an increased rate of chromosome loss associated with an altered structure of pericentromeric chromatin (Pinto and Winston 2000). Ploidy increase during mitosis can result from the syntelic attachment of sister kinetochores to microtubules, where sister chromatids become mono-oriented and segregate to one spindle pole (Biggins and Walczak 2003; Hauf and Watanabe 2004). A key role in promoting bioriented kinetochore–microtubule attachment relies upon the Ipl1/Aurora kinase, which was originally identified as a conditional allele that diploidized after shifting to the restrictive temperature (Chan and Botstein 1993; Biggins et al. 1999; Tanaka et al. 2002). If the H2A mutants are defective in some aspect of kinetochore–microtubule attachment, we hypothesize that centromeric histones must encounter specific protein interactions that affect chromosome segregation. To learn more about the proteins that may interact with histone H2A in the control of ploidy and chromosome segregation, we devised a genetic screen designed to isolate suppressors of the increase-in-ploidy phenotype associated with one of the H2A mutations. We identified five genes, four of which encode proteins directly connected to chromatin function: histone H3 and each of the three subunits of the Hda1 histone deacetylase complex. Our results indicate that Hda3 has functions distinct from Hda2 and Hda1 and that it is required for normal chromosome segregation and cell cycle progression. In addition, HDA3 shows genetic interactions with kinetochore components, emphasizing a role in centromere function. Together, the data presented suggest that the Hda1 deacetylase complex affects histone function at the centromere and that Hda3 has a distinctive participation in chromosome segregation.
MATERIALS AND METHODS
Yeast strains, genetic methods, growth, and media:
The S. cerevisiae strains used in this study are listed in Table 1. Unless indicated, strains are isogenic to FY2, originally derived from S288C (Winston et al. 1995). Strain construction and other genetic manipulations were carried out by standard methods (Rose et al. 1990; Guthrie and Fink 1991). Strains carrying deletions of HDA1, HDA2, and HDA3 were constructed by PCR-mediated disruption, using either HIS3 or TRP1 flanked by sequences homologous to the target gene as previously described (Brachmann et al. 1998). PCR primers for hda1Δ were 5′-AGGGAAAGTTGAGCACTGTAA TACGCCGAACAGATTAAGCAGATTGTACTGAGAGTGCAC-3′ and 5′-CTTTATTATTAT TCAACTTTCATAAGGCATGAAGGTTGCCCTGTGCGGTATTTCACACCG'-3′; for hda2Δ, 5′-GAAGGCGTTTAAGTAGTAGTAAAAGTATTTGGCTTCATTAGATTGTACTGAGAG TGCAC-3′ and 5′-CTATATTATACAGGCTACTTCTTTTAGGAAACGTCACATTCT GTCCGGTATTTCACACC-3′; and for hda3Δ, 5′-ATAAAAAGGAAAGTTAGAAGC ACAAACTAATTGCCCTTTAGATTGTACTGAGAGTGCAC-3′ and 5′-TGTATTTAGTATTTTTGAAACTAACGTTTTATTAACACTCTGTGCGGTATTTCACACCG-3′. Confirmation of the correct deletions was made by PCR using primers that hybridize outside the sequences targeted for recombination. All yeast media, including YPD, synthetic minimal, omission media (SC), and media containing 5-fluoroorotic acid (5-FOA) were made as described previously (Rose et al. 1990). Benomyl plates were made by adding benomyl (Sigma, St. Louis) to hot YPD to a final concentration of 15 μg/ml. Canavanine plates contain 60 μg/ml of canavanine sulfate (Sigma). Synchronization of cells in G1 was carried out by adding 1 μg/ml of α-factor to exponentially growing cells. Cultures were incubated at 30° for two generation times, washed twice in α-factor-free medium, and resuspended in fresh YPD. Aliquots were taken at various time points and processed for flow cytometry.
TABLE 1.
S. cerevisiae strains
| Strain | Genotype |
|---|---|
| FY604 | MATα his3Δ200 leu2Δ1 ura3-52 trp1Δ63 (hta2-htb2)Δ∷TRP1 |
| FY1333 | MATα leu2Δ0 ura3Δ0 |
| IPY152 | MATahta1-300 trp1Δ63 (hta2-htb2)Δ∷TRP1 his3Δ200 ade2Δ3∷HIS3∷ade2Δ5 leu2Δ1 ura3-52 [pSAB6] |
| IPY153 | MATα hta1-300 trp1Δ63 (hta2-htb2)Δ∷TRP1 his3Δ200 ade2Δ3∷HIS3∷ade2Δ5 leu2Δ1 ura3-52 [pSAB6] |
| IPY158 | MATaura3Δ0 trp1Δ63 HDA3-3xHA |
| IPY171 | MATahis3Δ200 leu2Δ1 ura3-52 |
| IPY176 | MATahis3Δ200 leu2Δ1 ura3-52 hda3Δ∷HIS3 |
| IPY210 | MATα his3Δ200 leu2Δ1 ura3Δ0 hda2Δ∷URA3 |
| IPY217 | MATahis3Δ200 leu2Δ1 ura3Δ0 hda2Δ∷URA3 |
| IPY224 | MATahis3Δ200 leu2Δ1 ura3-52 hda2Δ∷URA3 hda3Δ∷HIS3 |
| IPY273a | MATahis3Δ200 leu2Δ1 ura3-52 hda3Δ∷HIS3 mif2-3 |
| IPY276a | MATahis3Δ200 leu2Δ1 ura3-52 mif2-3 |
| IPY287 | MATahta1-300 trp1Δ63 (hta2-htb2)Δ∷TRP1 his3Δ200 ade2Δ3∷HIS3∷ade2Δ5 leu2Δ1 ura3-52 hda3-1∷mTn3 |
| IPY295 | MATα leu2Δ0 ura3Δ0 HDA2-3xHA |
| IPY298 | MATahis3Δ200 leu2Δ1 ura3-52 lys2Δ202 hda1Δ∷HIS3 |
| IPY337a | MATα his3Δ200 leu2Δ1 ndc10(ctf)-42 hda3Δ∷HIS3 |
| IPY339a | MATahis3Δ200 leu2Δ1 ndc10(ctf)-42 |
| IPY346a | MATα his3Δ200 or his3-11,15 leu2Δ1 ura3-52 cse4-1 hda3Δ∷HIS3 |
| IPY350a | MATahis3Δ200 or his3-11,15 leu2Δ1 ura3-52 |
| IPY352a | MATahis3Δ200 or his3-11,15 leu2Δ1 ura3-52 hda3Δ∷HIS3 |
| IPY358a | MATα his3Δ200 or his3-11,15 leu2Δ1 ura3-52 cse4-1 |
| IPY360 | MATa/ MATα ura3Δ0/ ura3Δ0 his3Δ200/ his3Δ200 HIS4/his4Δ10∷URA3 trp1Δ63/ trp1Δ63 leu2Δ0/LEU2 LYS2/lys2Δ0 |
| IPY361 | MATa/ MATα ura3Δ0/ ura3Δ0 his3Δ200/ his3Δ200 HIS4/his4Δ10∷URA3 trp1Δ63/ trp1Δ63 leu2Δ0/LEU2 LYS2/lys2Δ0 hda3Δ∷HIS3/ hda3Δ∷TRP1 |
| IPY386 | MATα leu2Δ0 ura3Δ0 HDA1-13xMyc∷kanMX |
| YMB2142b | MATatrp1Δ1 lys2-801 ade2-101 leu2Δ1 ura3-52 + [pCSE4-HA/URA3] |
Strain derived from a cross with a different background, backcrossed three times with FY2 background.
From Crotti and Basrai (2004).
Bacterial strains and plasmids:
Plasmids were amplified and isolated from Escherichia coli strain DH5α, according to standard procedures (Ausubel et al. 1988). pIP66 was constructed by ligating the 550-bp SalI–NotI fragment containing the truncated ADE2 gene from pIP65 (Pinto and Winston 2000) into pRS403 (Sikorski and Hieter 1989).
Suppressor screen:
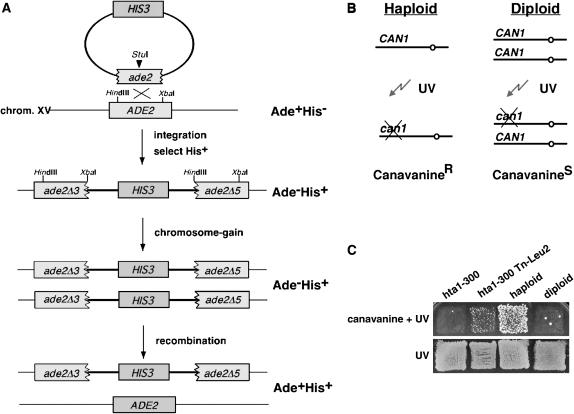
Strains IPY152 and IPY153 carry the hta1-300 allele as the only source of histone H2A (HTA2; the second gene copy is deleted) and a HIS3 marked chromosome XV at the ADE2 locus, described below. Plasmid pSAB6 carries HTA1 in a URA3-ARS-CEN vector and maintains the hta1-300 strains as haploid (Pinto and Winston 2000). The ade2Δ3∷HIS3∷ade2Δ5 was created by integrating the HIS3 gene into the ADE2 locus using the plasmid pIP66 linearized with StuI. This allele, ade2Δ3∷HIS3∷ade2Δ5, serves as a genetic marker for monitoring the copy number of chromosome XV (Chan and Botstein 1993; Pinto and Winston 2000). The integration results in a partial duplication of ade2, rendering the strain phenotypically Ade− His+. Ade+ colonies can be generated by homologous recombination between the duplicated sequences of the disrupted ADE2 gene, in which case the HIS3 marker would be lost. If a marked chromosome duplicates, then His+ Ade+ derivatives can be obtained via a recombination event on one of the two homologs for the marked chromosome. Such events are detected as papillae after replica plating to selective medium (see Figure 1). This assay was used to demonstrate the increase-in-ploidy phenotype caused by the hta1-300 mutation (Pinto and Winston 2000). To screen for suppressors, strains IPY152 and IPY153 were mutagenized by transformation with a miniTn3lacZ-LEU2 library that had been linearized at a unique NotI site to allow for homologous recombination between the genomic library and the yeast genome (Burns et al. 1994). Leu+His+ transformants were replica plated onto SC–Leu–His medium containing 5-FOA to select for cells that had lost the URA3-containing plasmid carrying the wild-type HTA1 gene. The 5-FOARLeu+His+colonies were then assayed for ploidy by replica plating them onto SC–His–Ade and SC–His media. The presence of papillae on medium lacking adenine and histidine (SC–His–Ade) indicates that cells have increased in ploidy. Therefore, cells that remain Ade− His+ would represent cells that might contain a suppressor of the increase-in-ploidy phenotype. Because this screen is based on recombination, a second screen was included to discriminate against Ade− His+ cells that would result from mutations in genes that affect recombination per se. In this assay, ploidy was assayed by monitoring the CAN1 gene as described (Figure 1B; Schild et al. 1981). Since canavanine (Can) resistance is conferred by recessive mutations in the CAN1 gene, the frequency of CanR mutants is much greater among haploids than among diploids or among strains with two copies of chromosome V. Colonies were replicated onto SC–Arg plates either with or without canavanine, the cells were mutagenized by UV irradiation (300 ergs/mm2), and plates were incubated at 30° for 3 days. Cells that remained haploid would form papillae on plates containing canavanine, after exposure to UV irradiation. Thus, colonies that did not grow on SC–His–Ade but formed papillae on SC–Arg + Can plates were retested and crossed back to a hta1-300 ade2Δ3∷HIS3∷ade2Δ5 strain to test for cosegregation of the LEU2 gene in the transposon (Leu+) and the suppressor phenotypes (His+Ade−, CanR). The suppressor genes were identified by retrieving genomic DNA immediately adjacent to the lacZ gene present in the transposon, using plasmid pRSQ-URA3, as described (Burns et al. 1994).
Figure 1.
Assays for monitoring ploidy increase in the hta1 mutants. (A) Genetic events that result from chromosome gain in the strains marked at ade2 on chromosome XV, leading to Ade+His+ papillae. (B) Genetic test for monitoring the copy number of chromosome V. Canavanine-resistant papillae readily appear upon mutagenesis of the CAN1 gene in haploid cells. (C) Example of the canavanine assay. The hta1-300 mutant shows no growth, like a diploid. One of the suppressors isolated (hta1-300 Tn-LEU2) shows an intermediate phenotype, with some papillae like the haploid strain.
Flow cytometry:
DNA content of yeast cells was determined as described, using a Becton Dickinson (San Jose, CA) FACSCalibur instrument (Pinto and Winston 2000).
Chromosome loss and recombination assays:
Diploids homozygous for HDA3 or hda3Δ were marked at the HIS4 locus on the right arm of chromosome III by the integration of URA3 into HIS4, and the resulting strains, IPY360 and IPY361, are HIS4/his4Δ10∷URA3. Construction of the alleles was confirmed by Southern blot analysis. Each strain was grown overnight on SC–Ura, streaked for single colonies on a YPD plate, and incubated at 30° for 2 (HDA3) or 5 (hda3Δ) days. A total of 5–10 colonies were excised from each plate using a sterile scalpel and resuspended in 1 ml of YPD. For the analysis at the restrictive temperature, the cultures were transferred to a 14° shaker for 24 hr. After brief sonication, the cells were counted and dilutions were plated on YPD (for counting) and 5-FOA medium at 30°. 5-FOAR colonies can result from either chromosome III loss or a mitotic recombination event between CEN3 and his4Δ10∷URA3, loosing the URA3 gene. To distinguish between the two events, we assayed the MAT locus, located on the left arm of chromosome III, by testing the 5-FOAR colonies for their mating phenotype. 5-FOAR colonies resulting from chromosome loss will be either MATa or MATα and therefore should mate, whereas mitotic recombinants will remain nonmating MATa/MATα diploid strains. The rates of chromosome III loss and recombination were calculated by the method of the median (Lea and Coulson 1949) from at least two independent experiments.
Northern blot analysis:
Total mRNA was extracted from exponentially growing YPD cultures of IPY171, IPY176, IPY217, and IPY224 as described (Ausubel et al. 1988). Specific probes internal to the coding regions of HDA2, HDA3, and SPT15 were generated by random priming using [α-P32]dATP, according to the manufacturer's instructions (United States Biochemical, Cleveland).
Preparation of yeast nuclei and indirect end-labeling analysis:
Yeast nuclei preparation, MNase digestions, and indirect end-labeling analysis of CEN3 chromatin were performed as described previously (Pinto and Winston 2000).
Chromatin immunoprecipitation:
Strains containing Hda1-13xMyc (IPY386), Hda2-3xHA (IPY295), and Hda3-3xHA (IPY158) epitope tags were constructed using standard procedures (Longtine et al. 1998). A wild-type untagged strain (FY1333) and the tagged strains were used for in vivo crosslinking and chromatin immunoprecipitation (ChIP) as described previously (Crotti and Basrai 2004), using anti-HA (12CA5, BAbCO) or anti-Myc (Santa Cruz) antibodies and Protein A Dynabeads (Dynal, Great Neck, NY). Primers for CEN3, CEN16, and PGK1 are described in Meluh and Koshland (1997). Primers for HMRa are 5′-CTCCACTTCAAGTAAGAGTTTGGG-3′ and 5′-CGCAGTAGAAAGACATATTTCTCTC-3′ Each experiment was repeated at least three times, starting with independent cultures.
RESULTS
Identification of suppressors of the increase-in-ploidy defect caused by the histone H2A allele hta1-300:
To understand how histone H2A participates in centromere function and chromosome segregation, we sought to identify the factors that interact genetically with H2A by initiating a suppressor analysis of the increase-in-ploidy phenotype. Our approach was to use essentially the same genetic assay originally used to measure the ploidy increase in the hta1 mutants, only this time to identify genes that, when mutated, diminished the increase-in-ploidy phenotype (Chan and Botstein 1993; Pinto and Winston 2000). We constructed a strain that carries the hta1-300 allele and a marked chromosome XV at the ADE2 locus. This allele, ade2Δ3∷HIS3∷ade2Δ5, serves as a genetic marker for monitoring the copy number of chromosome XV (Figure 1A). The assay is based on the ability of the cells that have gained an extra copy of the chromosome to undergo homologous recombination between the ade2 sequences on one homolog, without losing the HIS3 marker on the other homolog. Upon recombination, the cell becomes prototrophic for both markers, i.e., Ade+ His+. The presence of papillae on medium lacking adenine and histidine (SC–Ade–His) indicates that cells have increased in ploidy. Therefore, cells that remain Ade− His+ would represent cells that might contain a suppressor of the increase-in-ploidy phenotype. Because this screen is based on recombination, a second screen was included to discriminate against Ade− His+ cells that would result from mutations in genes that affect recombination processes. This assay monitors the copy number of chromosome V (Figure 1B). Since canavanine resistance is conferred by recessive mutations in the CAN1 gene, the frequency of CanR mutants is much greater among haploids than among diploids or among strains with two copies of chromosome V. The formation of papillae on plates containing canavanine, after exposure to UV irradiation, would indicate that the cells are haploid (Schild et al. 1981).
The hta1-300 ade2Δ3∷HIS3∷ade2Δ5 strain was maintained as haploid during the mutagenesis procedure by carrying pSAB6, an episomal copy of HTA1 on a URA3-ARS-CEN plasmid (Hirschhorn et al. 1995). For mutagenesis, we transformed the hta1-300 ade2Δ3∷HIS3∷ade2Δ5 strain with a yeast genomic library that had been randomly mutagenized with a mini-transposon tagged with the yeast LEU2 gene as a marker (mTn3-lacZ-LEU2) (Burns et al. 1994). About 15,000 Leu+His+ transformants were replica plated onto SC–Leu–His medium containing 5-FOA, to select for cells that had lost the URA3-containing plasmid carrying the wild-type HTA1 gene. The 5-FOARLeu+His+ colonies were then assayed for ploidy increase by replica plating them to SC–His–Ade medium (first screen) and SC–Arg + canavanine (second screen). In addition, we screened the transformants for suppression of the cold sensitivity (Cs) associated with the hta1-300 mutation (Pinto and Winston, 2000). Twenty-two colonies that did not grow on the first medium but formed papillae on the second were considered candidates for suppressors of the increase-in-ploidy phenotype of the hta1-300 mutant (Figure 1C). These candidates were crossed back to the hta1-300 ade2Δ3∷HIS3∷ade2Δ5 parental strain to test for cosegregation of the LEU2 gene (Leu+), indicative of the transposon insertion, and the suppressor phenotypes (His+Ade−, CanR). On the basis of their suppressor and dominant Cs+ phenotype, it appeared that eight strains had reverted to the wild-type HTA1 allele and were not followed further. Of the remaining 14 strains, 1 showed 4:0 segregation of the LEU2 marker on all of the spores dissected, indicating that the transposon insertion likely occurred at the endogenous 2μ yeast plasmid, as previously reported (Burns et al. 1994). Seven showed poor sporulation, poor spore viability, or a very weak suppression phenotype. Six strains showed cosegregation of the Leu+, His+Ade−, and CanR phenotypes, and the suppressor alleles were rescued and sequenced; the results are summarized in Table 2.
TABLE 2.
Suppressors of the hta1-300 increase-in-ploidy phenotype
| Gene name | ORF size (bp) | mTn3-lacZ-LEU2 insertion site | Protein | Function |
|---|---|---|---|---|
| HHT1 | 411 | 73 | Histone H3 | Chromatin |
| MKS1 | 1755 | 664 | Mks1p | Regulation of the retrograde pathway |
| PLO1/HDA3 | 1968 | 1644 | Subunit of Hda1 complex | Unknown/necessary for HDA activity |
| PLO1/HDA3 | 1968 | 1542 | Subunit of Hda1 complex | Unknown/necessary for HDA activity |
| PLO2/HDA2 | 2025 | 1638 | Subunit of Hda1 complex | Unknown/necessary for HDA activity |
| HDA1 | 2121 | 1938 | Subunit of Hda1 complex | Histone deacetylase activity (H2B, H3) |
Two suppressor alleles, plo1 and plo2 (for ploidy related), were identified as open reading frames YPR179C and YDR295C. We now know that these genes are identical to HDA3 and HDA2, respectively, encoding two subunits of the histone deacetylase Hda1 complex (Wu et al. 2001). Surprisingly, the next allele sequenced identified a mutation in HDA1, the presumptive catalytic subunit of the complex, and another suppressor revealed a second hda3 allele. Remarkably, the screen independently identified each of the three genes encoding subunits of the Hda1 complex as suppressors of the ploidy increase caused by the hta1-300 mutation. Comparison of the amino acid sequences of HDA3 and HDA2 with the databank revealed partial sequence similarity to the mammalian centromere proteins CENP-E and CENP-F, respectively (data not shown). Human CENP-E is a microtubule-binding protein involved in chromosome alignment during metaphase (Schaar et al. 1997), which interacts directly with CENP-F (Chan et al. 1998). These findings suggest the exciting possibility of a functional connection between the Hda1 complex and the centromere. It was particularly interesting that each of the alleles corresponding to genes of the Hda1 complex had the transposon insertion near the C terminus of the protein (Table 2), suggesting that the alleles could encode truncated proteins.
The finding that a likely deletion of one of the two S. cerevisiae genes that encode histone H3 suppresses the H2A defect is quite striking and provides a way to investigate the effects of histone gene dosage and histone–histone interactions in centromeric chromatin and chromosome segregation. The second gene found, MKS1 (multicopy kinase suppressor), was originally described as a negative regulator of the Ras–cAMP pathway (Matsuura and Anraku 1993). It has been implicated in lysine biosynthesis (Feller et al. 1997), the TOR kinase pathway (Shamji et al. 2000), and as a regulator of Ure2 (Edskes and Wickner 2000). It was later shown to be a phosphoprotein that interacts with Rtg2 in the retrograde pathway, where it functions as a negative regulator of the RTG target genes (Sekito et al. 2002). The latter finding reconciled most of the previous observations about Mks1 function; however, it does not hint of a role as an hta1-300 suppressor. The lack of any obvious connection with histone function or chromosome segregation suggests that the suppression that we observed is likely an indirect effect caused by the mks1∷mTn3 allele.
Suppressors maintain a haploid state in the hta1-300 strains:
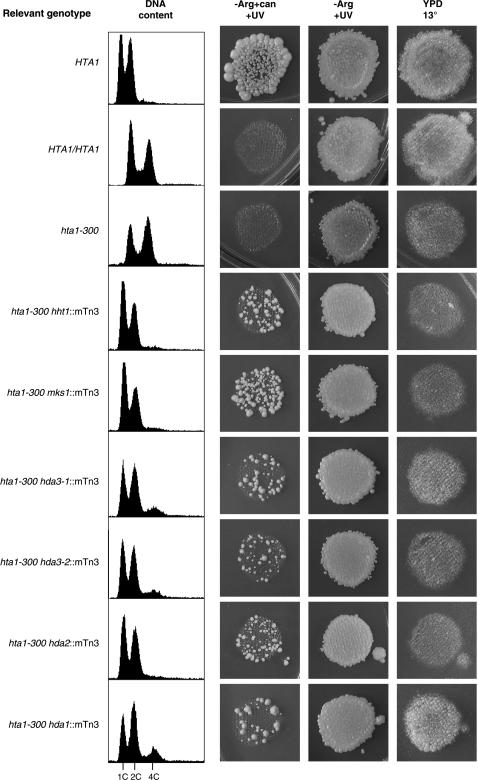
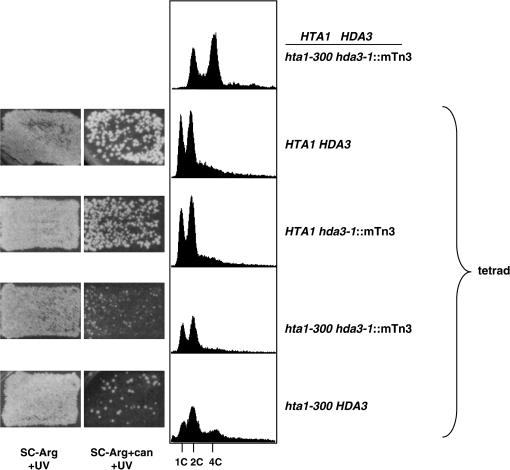
Each of the suppressors identified showed a similar CanR phenotype and DNA content profile. Only hda1∷mTn3 weakly suppressed the Cs− phenotype of hta1-300 (Figure 2). The haploid state maintained by the suppressors is mitotically and meiotically stable. In contrast with the hta1-300 mutants, which become completely diploid over several generations after germination from a heterozygous cross and form triploids upon mating (Pinto and Winston 2000), the suppressor strains showed normal mating, sporulation, and Mendelian segregation of the suppressor alleles and auxotrophic markers. Moreover, in crosses with a wild-type strain, the suppressors were able to keep the newly germinated hta1-300 strains as haploid while the hta1-300 strains begin to show the transition to diploid (Figure 3 and data not shown). In addition, hda1∷mTn3, hda2∷mTn3, and hda3∷mTn3 also suppress the increase in ploidy of the hta1-200 allele (data not shown).
Figure 2.
Suppressors of the hta1-300 increase-in-ploidy phenotype. Phenotype of the six suppressor strains, including ploidy, shown as DNA content determined by flow cytometry (left) and canavanine assay (right), as well as cold sensitivity at 13° (plates were incubated at 13° for 7 days).
Figure 3.
Suppression by hda3-1 is mitotically and meiotically stable. A diploid made between a wild-type HTA1 HDA3 (FY604) and an hta1-300 hda3-1 suppressor strain (IPY287) was sporulated and tetrads were dissected. The ploidy of the germinated spores was analyzed. Canavanine assay (left) and DNA content profiles determined by flow cytometry (right) are shown for a representative tetrad.
Deletion of the Hda1 complex genes reveals a distinct role for Hda3 in cell cycle progression:
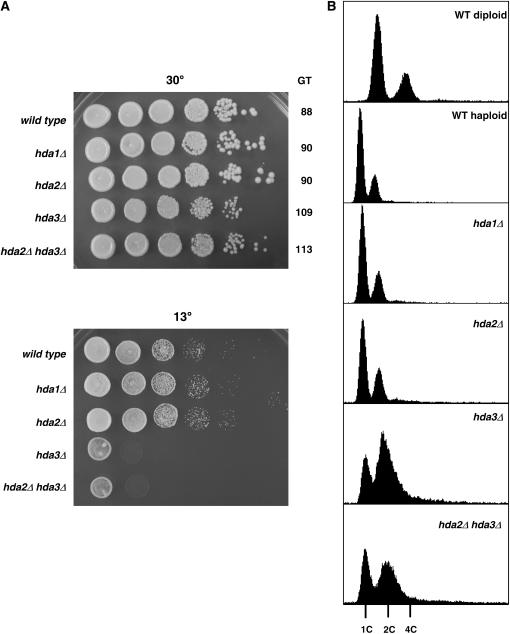
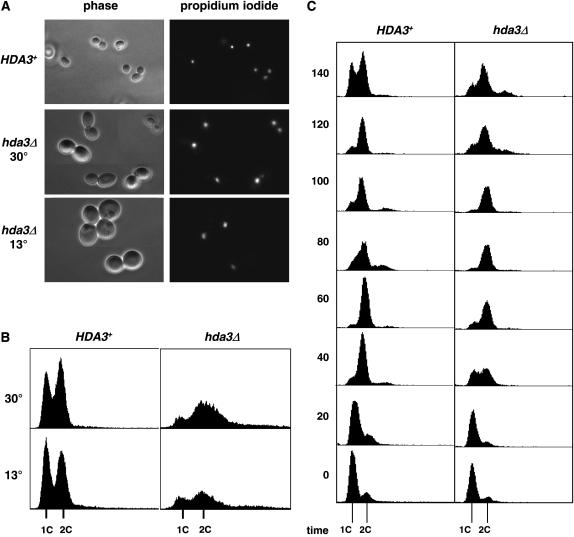
Since all three subunits of the Hda1 complex were identified in the suppressor screen, we concentrated on the analysis of their genes to understand their involvement in the status of cell ploidy and centromere function. Deletion of HDA1 and HDA2 did not show any obvious phenotype; in contrast, hda3Δ strains showed cold sensitivity at 13° and mild slow growth at 30° (Figure 4A). This result was quite unexpected; on the basis of the significant amino acid sequence similarity shared by Hda2 and Hda3 (Wu et al. 2001), we would have expected somewhat similar phenotypes in the deletion strains. The hda2Δ hda3Δ double mutant showed the same slow growth and cold sensitivity of the hda3Δ mutant, consistent with a lack of phenotype observed in hda2Δ strains. These results provide a clear distinction between the hda3Δ and hda3∷mTn3 alleles, since the latter do not show any phenotype on their own (Figure 3 and data not shown). We then analyzed the DNA content of the deletion strains by fluorescent cytometry. Again, the hda3Δ and hda2Δ hda3Δ strains had similar profiles, showing expanded G2/M peaks (Figure 4B). Interestingly, an hta1-300 hda3Δ strain shows suppression of the increase-in-ploidy defect (data not shown), indicating that the phenotypic differences between the hda3Δ and hda3∷mTn3 alleles do not relate to their suppression function. These results suggest that deletion of HDA3 leads to slow cell cycle progression, without affecting cell ploidy, and prompted us to investigate the phenotypes of hda3Δ strains in more detail. At permissive temperature, a significant number of cells appeared large budded with one undivided nucleus at the neck of the bud (Figure 5A) suggesting a mitotic delay that is consistent with the longer generation time of the mutant (109 min; see Figure 4). After a 16-hr shift to 13°, the cells become enlarged, and an increasing number of cells become large budded with a single nucleus. The DNA content profile becomes shallow and the G1 and G2/M peaks more diffused (Figure 5B). To determine whether hda3Δ cells delay at a particular stage of the cell cycle, we synchronized the wild-type and hda3Δ strains in G1 with α-factor, released them in fresh medium at 30°, and took samples for DNA content analysis (Figure 5C). The hda3Δ mutant shows an apparent S-phase delay that continues into a long G2/M phase. Cell count of the nuclear morphology at 60 min after release from G1 arrest showed ∼40% of the wild-type cells in anaphase, with the nucleus dividing between mother and daughter cells. In contrast, only 6% of the hda3Δ cells were at that stage; the majority still contained single nuclei with a small bud, and ∼10% had a single nucleus and a large bud, indicative of a mitotic delay. At 140 min, when the wild-type cells have lost their synchrony and the G1 and G2/M peaks become evident, the hda3Δ mutant remains with a G2/M peak, with a G1 peak beginning to emerge. At this time point, ∼45% of the cells show a divided nucleus, but the cells have not completed cytokinesis. Together, these phenotypes suggest that Hda3 is required for normal cell division and that its function may be required throughout mitosis.
Figure 4.
Deletion of HDA1, HDA2, and HDA3 reveals a unique phenotype for HDA3. Wild-type (IPY171) and otherwise isogenic hda1Δ (IPY298), hda2Δ (IPY217), hda3Δ (IPY176), and hda2Δ hda3Δ (IPY224) strains were grown on liquid YPD. (A) Growth was assessed by spotting 5 μl of serial dilutions (108–103cells/ml) on YPD plates grown at 30° (3 days) and 13° (6 days). The generation time (GT) of the strains grown on YPD at 30° is indicated. (B) DNA content was determined by flow cytometry.
Figure 5.
The hda3Δ mutant shows a defect in mitosis. (A) Wild-type (IPY171) and hda3Δ mutant (IPY176) strains grown at 30° and after 24 hr at 13° were used for cell morphology (phase) and nuclear staining (propidium iodide) analysis by fluorescence microscopy. (B) The same strains were used for DNA content analysis by flow cytometry. (C) Exponentially growing wild-type (IPY171) and hda3Δ (IPY176) cells were synchronized in G1 with α-factor. Cells were washed and released into pheromone-free YPD at 30°. Aliquots were removed at 20-min intervals for DNA content analysis by flow cytometry.
Deletion of HDA3 causes increased chromosome loss:
Mutants that affect progression through mitosis are often defective in maintaining normal chromosome segregation. To monitor chromosome segregation defects, we constructed homozygous diploid strains for HDA3 and hda3Δ that carry a marked chromosome III, which allowed for assaying the frequency of chromosome loss as well as mitotic recombination. Diploids that are heterozygous at the HIS4 locus by integration of URA3 into HIS4 on one homolog were constructed, creating HIS4/his4Δ∷URA3 diploids. Either loss of the chromosome that contains the his4Δ∷URA3 allele or mitotic recombination between CEN3 and HIS4 would result in 5-FOA-resistant cells. Chromosome loss and mitotic recombination can be distinguished by scoring the mating type (see materials and methods). The hda3Δ homozygous diploid showed a significant increase in the frequency of chromosome loss at 30°, ∼100-fold over that of the wild type (Table 3). Interestingly, the frequency of mitotic recombination also increased ∼20-fold. Both chromosome loss and recombination frequencies showed a similar increase at 14°, indicating that exposure to the restrictive temperature does not affect the behavior of the cells, at least in the conditions tested. Alternatively, the results at 14° may be interpreted as coming from cells that survived the temperature shift; therefore the frequencies of chromosome loss and recombination are equivalent to those at 30°. Thus, we conclude that hda3Δ mutants are defective in chromosome segregation, suggesting a function for Hda3 in mitosis.
TABLE 3.
Chromosome III loss and recombination in hda3 mutants
| Chromosome loss frequencya (×10−6)
|
Recombination frequencyb (×10−6)
|
|||
|---|---|---|---|---|
| Genotype | 30° | 14° | 30° | 14° |
| HDA3/HDA3 | 1.2 | 2.6 | 1.5 | 3.7 |
| hda3Δ/hda3Δ | 105 | 65 | 27 | 25 |
Frequencies of mitotic recombination and loss of chromosome III in diploids were determined as described in materials and methods. Frequencies were derived by fluctuation analysis from 5 to 10 individual cultures of each strain. Values shown are derived from one set of data. The experiment was repeated at least once for each condition with similar results. The strains used were IPY360 and IPY361.
The frequency of chromosome loss was determined by scoring for the loss of the URA3 marker (growth on 5-FOA medium) at the his4 locus, located on the right arm of chromosome III in conjunction with the loss of the respective MAT locus (mater) on the left arm of chromosome III.
The frequency of mitotic recombination was determined by scoring for the loss of the URA3 marker (growth on 5-FOA medium) at the his4 locus, located on the right arm of chromosome III, and the maintenance of both MATa/MATα loci (nonmater) on the left arm on chromosome III.
Genetic interactions with kinetochore components:
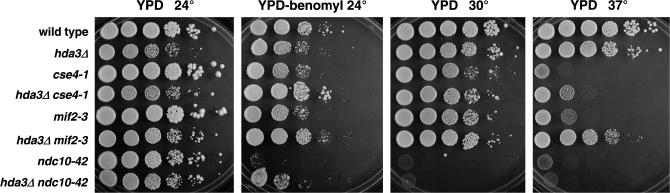
The cold sensitivity, mitotic delay, and increased chromosome loss and recombination phenotypes of hda3Δ strains show great resemblance to those of the hta1-200 and hta1-300 mutants (Pinto and Winston 2000). If Hda3 has a role at the kinetochore, it would likely interact genetically with kinetochore components. To test this hypothesis, we created strains carrying double mutations between hda3Δ and temperature-sensitive alleles of CSE4, MIF2, and NDC10. CSE4 encodes the centromere-specific histone H3 variant (Stoler et al. 1995), and MIF2 encodes an essential centromere protein that has been genetically and biochemically associated with kinetochore function (Meeks-Wagner et al. 1986; Brown et al. 1993; Meluh and Koshland 1995, 1997). NDC10 encodes the p110 subunit of the CBF3 complex, which binds to the CDEIII region of the centromere and is essential for kinetochore assembly (Lechner and Carbon 1991; Goh and Kilmartin 1993). We monitored the growth of the single and double mutants at 24°, a temperature permissive for all strains in YPD and YPD-containing benomyl, a microtubule-depolymerizing agent (Figure 6). At 24°, the hda3Δ cse4-1 and hda3Δ mif2-3 strains did not show any synthetic phenotype; in both cases the double mutant behaved as the hda3Δ single mutant, which grows more slowly than the wild-type strain at permissive temperature. Like many mutants defective in kinetochore function, the ndc10(ctf)-42 allele is sensitive to 15 μg/ml of benomyl. Surprisingly, the hda3Δ ndc10-42 strain showed growth on benomyl-containing plates, indicating that a deletion of HDA3 suppresses the ndc10-42 drug sensitivity. In addition, hda3Δ was able to suppress the temperature sensitivity of cse4-1 and mif2-3, as seen in the hda3Δ cse4-1 and hda3Δ mif2-3 strains grown on YPD at 37°. The suppression of mif2-3 was particularly strong (Figure 6). These findings provide a new connection between Hda3 and the kinetochore and suggest a distinct function for Hda3 in chromosome segregation.
Figure 6.
Suppression of cse4-1, mif2-3, and ndc10-42 by hda3Δ. Double mutants were generated by crosses between hda3Δ strains and kinetochore mutants. All strains were grown at 24°, a temperature permissive for all alleles tested. Serial dilutions (108–103cells/ml) were spotted (5 μl) onto YPD or benomyl plates and incubated at 24° (4 days), 30° (2 days), or 37° (2 days). The genotypes correspond to the following strains: wild type (IPY350), hda3Δ (IPY352), cse4-1 (IPY358), hda3Δ cse4-1 (IPY346), mif2-3 (IPY276), hda3Δ mif2-3 (IPY273), ndc10-42 (IPY339), and hda3Δ ndc10-42 (IPY337).
These genetic interactions are clearly different from those observed between hta1-300 and the kinetochore alleles. The hta1-300 cse4-1, hta1-300 mif2-3, and hta1-300 ndc10-42 strains display synthetic sickness at permissive temperature (Pinto and Winston 2000) and enhanced temperature sensitivity at 37° or 39° (data not shown). Although both HTA1 and HDA3 are implicated in kinetochore function, the differences in their genetic interactions suggest distinct genetic pathways.
HDA2 and HDA3 gene expressions are independent from each other:
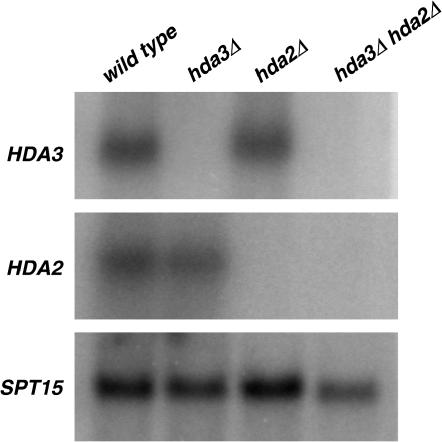
Considering the extensive amino acid sequence and predicted coiled-coil motif similarities between Hda2 and Hda3 (Wu et al. 2001), their common function as subunits of the Hda1 complex, and our finding of both genes being suppressors of the H2A defect, we asked whether the clear difference in phenotypes between hda2Δ and hda3Δ mutants could be caused by compensated gene expression when one of the two genes is deleted, with potential redundancy in function. We performed Northern blot analysis in hda2Δ, hda3Δ, and hda2Δ hda3Δ mutant strains to compare HDA2 and HDA3 transcript levels (Figure 7). The results show similar levels of HDA2 and HDA3 mRNA in hda3Δ, hda2Δ, and wild-type strains, indicating that there is no cross-regulation of their gene expression. In addition, a high-copy-number plasmid expressing HDA2 does not suppress the Cs phenotype of an hda3Δ mutant (data not shown). Thus, our results establish that HDA2 and HDA3 are expressed independently and suggest that their sequence similarity does not translate to exchangeable function.
Figure 7.
Analysis of mRNA levels in hda2Δ, hda3Δ, and hda2Δ hda3Δ mutants. Total RNA was isolated from exponentially growing wild-type (IPY171), hda2Δ (IPY210), hda3Δ (IPY176), and hda2Δ hda3Δ (IPY224) strains. Northen blot analysis was carried out with radiolabeled probes specific for HDA2, HDA3, and SPT15 (used as a loading control).
Centromere chromatin is not detectably affected in hda3 mutants:
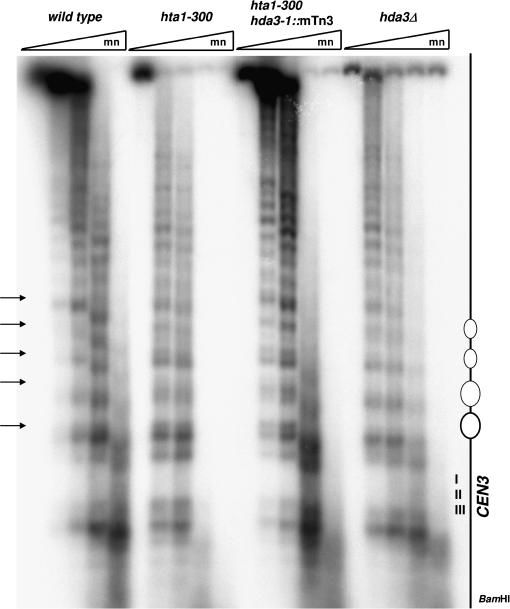
One of the characteristics of the hta1-300 mutant associated with the chromosome segregation defects is an altered pattern of micrococcal nuclease digestion at the nucleosomes flanking the centromere (Pinto and Winston 2000). We therefore wanted to examine the effect of the hda3-1∷mTn3 suppressor on the chromatin structure of CEN3 in an hta1-300 strain. We also included in our analysis an hda3Δ strain to assess the function of Hda3, if any, in the architecture of centromeric chromatin. Nuclei from the wild type, hta1-300, hta1-300 hda3-1∷mTn3, and hda3Δ strains were isolated and treated with micrococcal nuclease, and the purified DNA was subjected to indirect-end-labeling analysis. All strains show the characteristic nuclease-resistant centromeric core flanked by ∼2 kb of an organized array of nucleosomes to the left of CDEI (Figure 8; Bloom and Carbon 1982). However, the hta1-300 hda3-1∷mTn3 strain shows the same altered cleavage pattern seen in the hta1-300 strain, indicating that the hda3-1∷mTn3 suppressor does not act by restoring the normal nucleosome architecture. The hda3Δ strain displayed a pattern of MNase digestion indistinguishable from the wild type, suggesting that Hda3 is not required for the formation of the nuclease-resistant centromere–kinetochore complex and the flanking array of nucleosomes.
Figure 8.
Effect of hda3 alleles on CEN3 chromatin structure. Nuclei were isolated from wild-type (IPY171), hta1-300 (FY988), hta1-300 hda3-1∷mTn3 (IPY287), and hda3Δ (IPY176) stains after growth at 30° on YPD, digested with increasing concentrations of MNase and subjected to indirect end-labeling analysis as described in materials and methods. BamHI-digested DNA was hybridized with a radiolabeled 616-bp DNA fragment adjacent to the restriction site. Positions of the CEN3 nuclease-resistant core and the flanking nucleosomes are indicated in the diagram at the right. Arrows indicate the altered cleavage sites in the hta1-300 mutant.
Hda1, Hda2, and Hda3 associate with centromeric DNA:
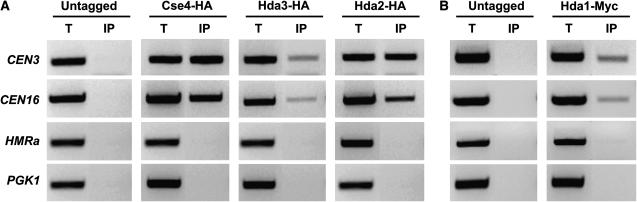
The independent isolation of each Hda subunit as a suppressor of the histone H2A defect along with the data presented above suggest a direct role of the Hda1 histone deacetylase complex at the centromere. To study the association of the Hda proteins with centromeric loci, we performed ChIP analysis. C-terminal tags were created for Hda1 (13xMyc), Hda2 (3xHA), and Hda3 (3xHA) in a wild-type background. Immunoprecipitation of formaldehyde crosslinked cell extracts was carried out with anti-HA or anti-Myc antibodies. The coprecipitated DNA was analyzed by PCR with primer pairs specific for the core centromeric regions of chromosomes III and XVI (CEN3 and CEN16), for a noncentromeric actively transcribed locus (PGK1), and for the heterochromatic locus HMRa (Figure 9). For comparison, centromeric-specific Cse4–HA was included in the analysis. All three Hda proteins interacted with CEN DNA but not with PGK1, which was not expected to be associated with deacetylases. In addition, no DNA was recovered from the heterochromatic locus HMRa. We conclude that the Hda1 deacetylase complex interacts directly with centromeric loci.
Figure 9.
Hda1, Hda2, and Hda3 associate with centromeric chromatin. Formaldehyde crosslinked chromatin was prepared from exponentially growing wild-type strains containing none (untagged; FY1333), HA (Hda2, IPY295; Hda3, IPY158; and Cse4, YMB2142), or Myc (Hda1, IPY386) epitope tags. Strains were immunoprecipitated with anti-HA (A) or anti-Myc (B) antibodies and PCR was performed on total input DNA (T) and immunoprecipitated DNA (IP) to visualize the core centromeric region of CEN3, CEN16, HMRa, and PGK1.
DISCUSSION
Previous characterization of two hta1 alleles, encoding single amino acid replacements in histone H2A, showed that these mutations cause chromosome segregation defects and a ploidy increase from a haploid to a diploid state. On the basis of the analysis of the hta1 mutants, it was proposed that histone H2A is required for proper centromere–kinetochore function (Pinto and Winston 2000). In this work, we performed a genetic screen for suppressors of the increase-in-ploidy phenotype with the goal of identifying proteins that interact with histones in controlling ploidy. We isolated mutants that maintain an hta1-300 strain as haploid. Our results show that the screen was successful in identifying genes that relate to chromatin function. We identified mutations in the three subunits of the Hda1 histone deacetylase complex and demonstrate that Hda3, a component of the Hda1 complex, is required for proper chromosome segregation and mitosis. The involvement of the Hda1 complex in chromosome segregation is strengthened by the direct association of Hda1, Hda2, and Hda3 with centromeric DNA.
We identified five genes, four of them (hht1, hda1, hda2, and hda3) directly associated with chromatin function. HHT1 encodes histone H3, and the hht1∷mTn3 mutation creates a deletion near the 5′-end of the HHT1 coding sequence, likely equivalent to a null allele. Since the second gene copy encoding H3, HHT2, is intact, histone H3 levels would be expected to provide normal cell function, as has been shown in a wild-type background (Smith and Stirling 1988). Therefore, the suppression seen may reflect unknown regulation of histone function on the basis of subtle changes in histone levels, and this allele provides an excellent tool for future studies. The mks1∷mTn3 allele does not offer any clear connection with chromatin; therefore we interpret the suppression as an indirect effect. The remaining suppressors consist of hda1∷mTn3, hda2∷mTn3, and two alleles of hda3∷mTn3. Although we do not consider the screen saturated, the finding of a second hda3 allele stressed the involvement of the Hda1 complex. Hda1, Hda2, and Hda3 were originally isolated as proteins that copurified chromatographically with histone deacetylase activity, and sequencing of the peptides led to the identification of their encoding genes (Carmen et al. 1996; Rundlett et al. 1996; Wu et al. 2001). Hda1 appears to be the catalytic subunit of the complex, and all three subunits interact directly and are necessary for its activity (Wu et al. 2001). We find it intriguing that the four hda alleles that we isolated as suppressors carry the mTn3 insertions near the 3′-end of the genes, likely creating C-terminal truncations. Although the significance of this finding is not clear, it is evident that the hda3∷mTn3 alleles are different from an hda3Δ, since they are neither cold sensitive nor grow more slowly. Thus, the hda3∷mTn3 allele must encode a protein that maintains normal chromosome segregation and mitosis, but has the ability to suppress the ploidy increase caused by the hta1-300 mutation. Biochemical data indicate that Hda1 does not interact with Hda2 in the absence of Hda3, suggesting that Hda3 is required for the formation of the complex (Wu et al. 2001). Whether the intact complex and/or deacetylase activity is required for suppression remains to be established, although our results clearly indicate that the hda suppressors do not affect the normal function of Hda3 in mitosis (see below).
The cold sensitivity, slow growth, and G2/M delay displayed by the hda3Δ mutant resemble the phenotypes of the hta1-200 and hta1-300 mutants and prompted us to investigate whether Hda3 has a role in mitosis. Analysis of the DNA content and nuclear morphology in synchronized hda3Δ cells revealed a mitotic delay, with cells traversing slowly through anaphase and cytokinesis. We did not observe a 4C peak, indicating that the hda3Δ mutants do not diploidize. The wide and shallow 1C and 2C DNA content peaks were consistent with chromosome segregation defects, which were confirmed by the analysis of chromosome loss rates. We found the rate of chromosome loss ∼100-fold higher in the hda3Δ/hda3Δ mutant compared with that of the wild type at 30°, which is higher than the rate observed in the hta1 mutants (Pinto and Winston 2000) but consistent with rates reported for mutations in kinetochore components (Doheny et al. 1993; Meluh and Koshland 1995). Interestingly, mitotic recombination frequency was also higher in the hda3Δ/hda3Δ mutant, a result that could be attributed to the lack of Hda3, although a more general pleiotropic defect due to altered Hda1 deacetylation cannot be ruled out.
The mitotic function described for Hda3 was reinforced by the genetic interactions found between hda3Δ and temperature-sensitive alleles of kinetochore components. The fact that hda3Δ suppresses the sensitivity of ndc10-34 to the microtubule-depolymerizing drug benomyl and the temperature sensitivity of cse4-1 and mif2-3 suggests that the Hda3 role in mitosis may be directly related to the kinetochore. Because Ndc10 has been shown to migrate to the spindle midzone during anaphase and proposed to maintain spindle stability (Bouck and Bloom 2005), one possible explanation for the suppression is that hda3Δ could restore this aspect of Ndc10 function, visualized as benomyl sensitivity in the ndc10-34 allele. Although the mechanism by which hda3Δ suppresses these kinetochore alleles may be different, it is conceivable that the absence of Hda3 stabilizes faulty microtubule attachment in these mutants.
The independent finding of the three subunits of the Hda1 complex as suppressors, and their direct association with centromeric loci, suggests a connection between histone deacetylation by this complex and centromere function. One of the covalent modifications associated with centromeric heterochromatin and silent loci is the hypoacetylation of histones (Richards and Elgin 2002). Fission yeast and higher eukaryotes have defined heterochromatic regions flanking their centromeres. The relatively large centromeres of fission yeast, which resemble those of vertebrates, contain heterochromatic regions marked by specific post-translational modifications of core histones, in particular deacetylation of histones H3 and H4 (Pidoux and Allshire 2005). Inhibition of histone deacetylases causes hyperacetylation of centromeres and defective chromosome segregation (Ekwall et al. 1997). The SIN3–histone deacetylase complex, in addition to its role in the modulation of transcription, has been shown to associate with Schizosaccharomyces pombe centromeres where it regulates silent chromatin (Silverstein et al. 2003). In contrast, the compact genome of S. cerevisiae has a minimal centromeric sequence flanked by relatively close coding sequences, and it has long been presumed that heterochromatin in budding yeast was practically nonexistent. However, budding yeast pericentric chromatin structure consists of well-phased nucleosomal arrays, characteristic of heterochromatic domains (Bloom and Carbon 1982). Recent evidence has pointed to the presence of the known silencing factor Sir1, required for the formation of specialized chromatin structure at the silent mating-type loci HML and HMR (Loo and Rine 1995) at the S. cerevisiae centromere. Sir1, in association with the nucleosome assembly factors CAF-1 and the Hir proteins, contributes to mitotic chromosome stability (Sharp et al. 2002, 2003). Another chromatin-associated factor, Spt4, has been shown to be a functional component of the kinetochore and also participates in the silencing at the HML and HMR loci (Crotti and Basrai 2004). These findings suggest that the budding yeast centromere may indeed have a type of silent heterochromatin, although structured in a way different from the characteristic pericentromeric regions seen in other eukaryotes. Global microarray analyses in S. cerevisiae have provided information about the distinct deacetylation functions of Rpd3 and Hda1 at promoter regions and a function for Hda1 in the deacetylation of subtelomeric domains (Robyr et al. 2002; Ekwall 2005); however, no function has been described for deacetylases at centromeric regions. On the basis of our results, we hypothesize that the Hda1 complex may be responsible for deacetylating histones at or around the centromere. Since the Hda1 complex functions at many different chromosomal sites, it is conceivable that a specific centromeric factor brings the complex to the centromere. Potential candidates include the known factors, such as Sir1, Spt4, or the CAF-1 and Hir histone deposition proteins, or some yet-unknown players. Interestingly, we do not find that Hda proteins are associated with the silent mating-type locus HMRa, where Sir1 and Spt4 bind, suggesting a complex pattern of protein interactions in the establishment of heterochromatic loci. Our findings raise many questions about the function of suppressors and the involvement of deacetylation in the formation of a functional kinetochore. Further analysis of the hda suppressors will provide some answers to these questions and expand our knowledge of the chromatin requirements at the S. cerevisiae centromere.
Acknowledgments
We thank Molly Fitzgerald-Hayes, Leland Hartwell, and Phil Hieter for strains and Munira Basrai for strains and advice on ChIP experiments. We are grateful to Fred Winston and David McNabb for critically reading the manuscript. Special thanks go to Fred Winston for his generosity, support, and encouragement. This work was supported by funds from the Arkansas Biosciences Institute and National Science Foundation grant MCB-0131480 to I.P. I.P. dedicates this work to the memory of Paz Cequiel.
References
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman et al., 1988. Current Protocols in Molecular Biology. Greene Publishing Associates and Wiley-Interscience, New York.
- Biggins, S., and C. E. Walczak, 2003. Captivating capture: how microtubules attach to kinetochores. Curr. Biol. 13: R449–R460. [DOI] [PubMed] [Google Scholar]
- Biggins, S., F. F. Severin, N. Bhalla, I. Sassoon, A. A. Hyman et al., 1999. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 13: 532–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom, K. S., and J. Carbon, 1982. Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell 29: 305–317. [DOI] [PubMed] [Google Scholar]
- Bouck, D. C., and K. S. Bloom, 2005. The kinetochore protein Ndc10p is required for spindle stability and cytokinesis in yeast. Proc. Natl. Acad. Sci. USA 102: 5408–5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li et al., 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132. [DOI] [PubMed] [Google Scholar]
- Brown, M. T., L. Goetsch and L. H. Hartwell, 1993. MIF2 is required for mitotic spindle integrity during anaphase spindle elongation in Saccharomyces cerevisiae. J. Cell Biol. 123: 387–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns, N., B. Grimwade, P. B. Ross-Macdonald, E. Y. Choi, K. Finberg et al., 1994. Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes Dev. 8: 1087–1105. [DOI] [PubMed] [Google Scholar]
- Cai, M., and R. W. Davis, 1990. Yeast centromere binding protein CBF1, of the helix-loop-helix protein family, is required for chromosome stability and methionine prototrophy. Cell 61: 437–446. [DOI] [PubMed] [Google Scholar]
- Carmen, A. A., S. E. Rundlett and M. Grunstein, 1996. HDA1 and HDA3 are components of a yeast histone deacetylase (HDA) complex. J. Biol. Chem. 271: 15837–15844. [DOI] [PubMed] [Google Scholar]
- Chan, C. S., and D. Botstein, 1993. Isolation and characterization of chromosome-gain and increase-in-ploidy mutants in yeast. Genetics 135: 677–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, G. K., B. T. Schaar and T. J. Yen, 1998. Characterization of the kinetochore binding domain of CENP-E reveals interactions with the kinetochore proteins CENP-F and hBUBR1. J. Cell Biol. 143: 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman, I. M., D. G. Drubin and G. Barnes, 2002. Simple centromere, complex kinetochore: linking spindle microtubules and centromeric DNA in budding yeast. J. Cell Biol. 157: 199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly, C., and P. Hieter, 1996. Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell 86: 275–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotti, L. B., and M. A. Basrai, 2004. Functional roles for evolutionarily conserved Spt4p at centromeres and heterochromatin in Saccharomyces cerevisiae. EMBO J. 23: 1804–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Densmore, L., W. E. Payne and M. Fitzgerald-Hayes, 1991. In vivo genomic footprint of a yeast centromere. Mol. Cell. Biol. 11: 154–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doheny, K. F., P. K. Sorger, A. A. Hyman, S. Tugendreich, F. Spencer et al., 1993. Identification of essential components of the S. cerevisiae kinetochore. Cell 73: 761–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edskes, H. K., and R. B. Wickner, 2000. A protein required for prion generation: [URE3] induction requires the Ras-regulated Mks1 protein. Proc. Natl. Acad. Sci. USA 97: 6625–6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwall, K., 2005. Genome-wide analysis of HDAC function. Trends Genet. 21: 608–615. [DOI] [PubMed] [Google Scholar]
- Ekwall, K., T. Olsson, B. M. Turner, G. Cranston and R. C. Allshire, 1997. Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell 91: 1021–1032. [DOI] [PubMed] [Google Scholar]
- Feller, A., F. Ramos, A. Pierard and E. Dubois, 1997. Lys80p of Saccharomyces cerevisiae, previously proposed as a specific repressor of LYS genes, is a pleiotropic regulatory factor identical to Mks1p. Yeast 13: 1337–1346. [DOI] [PubMed] [Google Scholar]
- Gaudet, A., and M. Fitzgerald-Hayes, 1987. Alterations in the adenine-plus-thymine-rich region of CEN3 affect centromere function in Saccharomyces cerevisiae. Mol. Cell. Biol. 7: 68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh, P. Y., and J. V. Kilmartin, 1993. NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol. 121: 503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie, C., and G. R. Fink, 1991. Guide to Yeast Genetics and Molecular Biology. Academic Press, San Diego.
- Han, M., M. Chang, U. J. Kim and M. Grunstein, 1987. Histone H2B repression causes cell-cycle-specific arrest in yeast: effects on chromosomal segregation, replication, and transcription. Cell 48: 589–597. [DOI] [PubMed] [Google Scholar]
- Hauf, S., and Y. Watanabe, 2004. Kinetochore orientation in mitosis and meiosis. Cell 119: 317–327. [DOI] [PubMed] [Google Scholar]
- Hegemann, J. H., and U. N. Fleig, 1993. The centromere of budding yeast. BioEssays 15: 451–460. [DOI] [PubMed] [Google Scholar]
- Hirschhorn, J. N., A. L. Bortvin, S. L. Ricupero-Hovasse and F. Winston, 1995. A new class of histone H2A mutations in Saccharomyces cerevisiae causes specific transcriptional defects in vivo. Mol. Cell. Biol. 15: 1999–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith, K. C., and M. Fitzgerald-Hayes, 2000. CSE4 genetically interacts with the Saccharomyces cerevisiae centromere DNA elements CDE I and CDE II but not CDE III. Implications for the path of the centromere DNA around a cse4p variant nucleosome. Genetics 156: 973–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, U.-J., M. Han, P. Kayne and M. Grunstein, 1988. Effects of histone H4 depletion on the cell cycle and transcription of Saccharomyces cerevisiae. EMBO J. 7: 2211–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea, D. E., and C. A. Coulson, 1949. The distribution of number of mutants in bacterial populations. J. Genet. 49: 264–284. [DOI] [PubMed] [Google Scholar]
- Lechner, J., and J. Carbon, 1991. A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell 64: 717–725. [DOI] [PubMed] [Google Scholar]
- Longtine, M. S., A. McKenzie, III, D. J. Demarini, N. G. Shah, A. Wash et al., 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Loo, S., and J. Rine, 1995. Silencing and heritable domains of gene expression. Annu. Rev. Cell Dev. Biol. 11: 519–548. [DOI] [PubMed] [Google Scholar]
- Matsuura, A., and Y. Anraku, 1993. Characterization of the MKS1 gene, a new negative regulator of the Ras-cyclic AMP pathway in Saccharomyces cerevisiae. Mol. Gen. Genet. 238: 6–16. [DOI] [PubMed] [Google Scholar]
- McAinsh, A. D., J. D. Tytell and P. K. Sorger, 2003. Structure, function, and regulation of budding yeast kinetochores. Annu. Rev. Cell Dev. Biol. 19: 519–539. [DOI] [PubMed] [Google Scholar]
- McGrew, J., B. Diehl and M. Fitzgerald-Hayes, 1986. Single base-pair mutations in centromere element III cause aberrant chromosome segregation in Saccharomyces cerevisiae. Mol. Cell. Biol. 6: 530–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrew, J. T., Z. X. Xiao and M. Fitzgerald-Hayes, 1989. Saccharomyces cerevisiae mutants defective in chromosome segregation. Yeast 5: 271–284. [DOI] [PubMed] [Google Scholar]
- Meeks-Wagner, D., and L. H. Hartwell, 1986. Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission. Cell 44: 43–52. [DOI] [PubMed] [Google Scholar]
- Meeks-Wagner, D., J. S. Wood, B. Garvik and L. H. Hartwell, 1986. Isolation of two genes that affect mitotic chromosome transmission in S. cerevisiae. Cell 44: 53–63. [DOI] [PubMed] [Google Scholar]
- Meluh, P. B., and D. Koshland, 1995. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol. Biol. Cell 6: 793–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh, P. B., and D. Koshland, 1997. Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes Dev. 11: 3401–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh, P. B., P. Yang, L. Glowczewski, D. Koshland and M. M. Smith, 1998. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell 94: 607–613. [DOI] [PubMed] [Google Scholar]
- Morgan, B. A., B. A. Mittman and M. M. Smith, 1991. The highly conserved N-terminal domains of histones H3 and H4 are required for normal cell cycle progression. Mol. Cell. Biol. 11: 4111–4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedenthal, R. K., M. Sen-Gupta, A. Wilmen and J. H. Hegemann, 1993. Cpf1 protein induced bending of yeast centromere DNA element I. Nucleic Acids Res. 21: 4726–4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux, A. L., and R. Allshire, 2005. The role of heterochromatin in centromere function. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360: 569–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto, I., and F. Winston, 2000. Histone H2A is required for normal centromere function in Saccharomyces cerevisiae. EMBO J. 19: 1598–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, E. J., and S. C. Elgin, 2002. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell 108: 489–500. [DOI] [PubMed] [Google Scholar]
- Robyr, D., Y. Suka, I. Xenarios, S. K. Kurdistani, A. Wang et al., 2002. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell 109: 437–446. [DOI] [PubMed] [Google Scholar]
- Rose, M. D., F. Winston and P. Hieter, 1990. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Rundlett, S. E., A. A. Carmen, R. Kobayashi, S. Bavykin, B. M. Turner et al., 1996. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl. Acad. Sci. USA 93: 14503–14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders, M., M. Fitzgerald-Hayes and K. Bloom, 1988. Chromatin structure of altered yeast centromeres. Proc. Natl. Acad. Sci. USA 85: 175–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders, M. J., E. Yeh, M. Grunstein and K. Bloom, 1990. Nucleosome depletion alters the chromatin structure of Saccharomyces cerevisiae centromeres. Mol. Cell. Biol. 10: 5721–5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaar, B. T., G. K. Chan, P. Maddox, E. D. Salmon and T. J. Yen, 1997. CENP-E function at kinetochores is essential for chromosome alignment. J. Cell Biol. 139: 1373–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schild, D., H. N. Ananthaswamy and R. K. Mortimer, 1981. An endomitotic effect of a cell cycle mutation of Saccharomyces cerevisiae. Genetics 97: 551–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekito, T., Z. Liu, J. Thornton and R. A. Butow, 2002. RTG-dependent mitochondria-to-nucleus signaling is regulated by MKS1 and is linked to formation of yeast prion [URE3]. Mol. Biol. Cell 13: 795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamji, A. F., F. G. Kuruvilla and S. L. Schreiber, 2000. Partitioning the transcriptional program induced by rapamycin among the effectors of the Tor proteins. Curr. Biol. 10: 1574–1581. [DOI] [PubMed] [Google Scholar]
- Sharp, J. A., and P. D. Kaufman, 2003. Chromatin proteins are determinants of centromere function. Curr. Top. Microbiol. Immunol. 274: 23–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, J. A., A. A. Franco, M. A. Osley and P. D. Kaufman, 2002. Chromatin assembly factor I and Hir proteins contribute to building functional kinetochores in S. cerevisiae. Genes Dev. 16: 85–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, J. A., D. C. Krawitz, K. A. Gardner, C. A. Fox and P. D. Kaufman, 2003. The budding yeast silencing protein Sir1 is a functional component of centromeric chromatin. Genes Dev. 17: 2356–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R. S., and P. Hieter, 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstein, R. A., W. Richardson, H. Levin, R. Allshire and K. Ekwall, 2003. A new role for the transcriptional corepressor SIN3: regulation of centromeres. Curr. Biol. 13: 68–72. [DOI] [PubMed] [Google Scholar]
- Smith, M. M., and V. B. Stirling, 1988. Histone H3 and H4 gene deletions in Saccharomyces cerevisiae. J. Cell Biol. 106: 557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, M. M., P. Yang, M. S. Santisteban, P. W. Boone, A. T. Goldstein et al., 1996. A novel histone H4 mutant defective in nuclear division and mitotic chromosome transmission. Mol. Cell. Biol. 16: 1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger, P. K., F. F. Severin and A. A. Hyman, 1994. Factors required for the binding of reassembled yeast kinetochores to microtubules in vitro. J. Cell Biol. 127: 995–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler, S., K. C. Keith, K. E. Curnick and M. Fitzgerald-Hayes, 1995. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 9: 573–586. [DOI] [PubMed] [Google Scholar]
- Tanaka, T. U., N. Rachidi, C. Janke, G. Pereira, M. Galova et al., 2002. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell 108: 317–329. [DOI] [PubMed] [Google Scholar]
- Van Holde, K., 1988. Chromatin. Springer-Verlag, New York.
- Winston, F., C. Dollard and S. L. Ricupero-Hovasse, 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11: 53–55. [DOI] [PubMed] [Google Scholar]
- Wu, J., A. A. Carmen, R. Kobayashi, N. Suka and M. Grunstein, 2001. HDA2 and HDA3 are related proteins that interact with and are essential for the activity of the yeast histone deacetylase HDA1. Proc. Natl. Acad. Sci. USA 98: 4391–4396. [DOI] [PMC free article] [PubMed] [Google Scholar]