Abstract
Ataxia telangiectasia (A-T) is an inherited disorder characterized by progressive loss of motor function and susceptibility to cancer. The most prominent clinical feature observed in A-T patients is the degeneration of Purkinje motor neurons. Numerous studies have emphasized the role of the affected gene product, ATM, in the regulation of the DNA damage response. However, in Purkinje cells, the bulk of ATM localizes to the cytoplasm and may play a role in vesicle trafficking. The nature of this function, and its involvement in the pathology underlying A-T, remain unknown. Here we characterize the homolog of ATM (AtmA) in the filamentous fungus Aspergillus nidulans. In addition to its expected role in the DNA damage response, we find that AtmA is also required for polarized hyphal growth. We demonstrate that an atmA mutant fails to generate a stable axis of hyphal polarity. Notably, cytoplasmic microtubules display aberrant cortical interactions at the hyphal tip. Our results suggest that AtmA regulates the function and/or localization of landmark proteins required for the formation of a polarity axis. We propose that a similar function may contribute to the establishment of neuronal polarity.
ATM is a phosphatidyl-3-kinase-related protein kinase (PIKK) that functions as a central regulator of the DNA damage response in eukaryotic cells (Shiloh 2001; McKinnon 2004). Like its homolog ATR, ATM responds to damage such as double-strand breaks (DSBs) by phosphorylating multiple targets that collectively act to maintain genome integrity. Key targets include proteins that direct chromatin modification (histone H2AX), promote DNA repair (BRCA1, BLM), activate cell cycle checkpoints (Chk1, Chk2, Nbs1, FancD2, Mdm2), and trigger apoptosis (p53) (summarized by McKinnon 2004). Many of these proteins contain consensus ATM phosphorylation sites whose importance has been demonstrated in functional studies (O'Neill et al. 2000). Notably, studies in yeast have shown that the ATM homolog Tel1 is one of the first proteins recruited to DSBs, where it appears to mediate formation of protein complexes involved in checkpoint activation and repair (Lisby et al. 2004). Accordingly, it is generally thought that the primary function of ATM is to choreograph the response to DNA damage.
In humans, mutations in ATM cause the devastating neurodegenerative disease ataxia telangiectasia (A-T; summarized by Gatti 1998; Chun and Gatti 2004). The most prominent neurological feature of A-T is progressive cerebellar ataxia, which is triggered by the loss of Purkinje cells within the cerebellum. Although the number of Purkinje cells initially appears normal at birth, the cells begin to degenerate shortly thereafter. ATM is predominantly a nuclear protein (Lakin et al. 1996), but it surprisingly displays prominent cytoplasmic localization in mouse Purkinje cells and cerebellar tissue (Kuljis et al. 1999; Barlow et al. 2000). Additional biochemical studies show that ATM associates with cytoplasmic vesicles (Watters et al. 1997). Both in vitro and in vivo assays demonstrate that ATM interacts with β-adaptins (Lim et al. 1998), which are components of the adaptor complexes involved in clathrin-mediated vesicle transport (Boehm and Bonifacino 2001). Consistent with these observations, A-T cells and mouse Atm−/− mutants display aberrant accumulation of lysosomes (Barlow et al. 2000). Additional cytoplasmic abnormalities observed in A-T cells include the presence of unusually thick actin stress fibers (McKinnon and Burgoyne 1985) and possible defects in exocytosis (O'Connor and Linthicum 1980). Given the large size of ATM, these studies collectively support the notion that ATM has a cytoplasmic function independent of its role in the DNA damage response.
The filamentous fungus Aspergillus nidulans possesses a sophisticated DNA damage response that ensures the maintenance of genome integrity (Goldman et al. 2002; Goldman and Kafer 2004). The ATR homolog UvsBATR is a central component of this response, in which it controls the activation of multiple checkpoints, regulates damage-induced gene expression, and also promotes DNA repair (De Souza et al. 1999; Hofmann and Harris 2000). UvsBATR displays an extensive web of genetic interactions with other proteins involved in the DNA damage response, including the Mre11 complex (ScaANBS1, MreAMRE11, and SldIRAD50), the cdc2-related kinase NpkA, and SepBCTF4 (Fagundes et al. 2004, 2005; Gygax et al. 2005; Malavazi et al. 2005). However, the function of UvsBATR relative to its paralog ATM remains unclear for the filamentous fungi. In particular, ATM and ATR homologs possess both shared and unique functions in the DNA damage responses of the model yeast Saccharomyces cerevisiae (Rouse and Jackson 2002). To determine if a similar relationship exists in A. nidulans, we cloned and initiated the functional characterization of the A. nidulans ATM homolog, AtmAATM. As expected, AtmAATM is intimately involved in the DNA damage response. However, to our surprise, it is also required for the formation of a stable axis of hyphal polarity. On the basis of our observations, we propose that AtmAATM performs a conserved cytoplasmic function that is important for the growth of highly polarized cells.
MATERIALS AND METHODS
Strains, media, and growth conditions:
The strains used in this study are A28 (pabaA6; biA1), A781 (wA2; nimA5), GR5 (pyrG89; wA3; pyroA4), R21 (pabaA1; yA2), IM69 (pyrG89 ;wA3; pyroA1; ΔatmA∷pyrG), and IM69-221 (pyrG89; wA3; nimA5; ΔatmA∷pyrG). Rich media (YG: 2% glucose, 0.5% yeast extract, trace elements) and minimal glucose media (MM: 1% glucose, original high nitrate salts, trace elements, pH 6.5) were prepared using standard approaches (Hill and Kafer 2001; Fagundes et al. 2005). For M-glycerol media, 1.2% glycerol was substituted for glucose in MM. In all media, uridine (5 mm) and uracil (10 mm) were added when necessary for complementation of the pyrG89 marker. Routine genetic approaches were used for strain construction (Kafer 1977, Appendix; available upon request). For the UV-light viability assays, conidiospores were suspended in 0.2% Tween-20 and plated out on YAG plates (YG plus 2% agar; ∼100 conidia/plate). The plates were then irradiated immediately with UV using a UV Stratalinker 1800 (Stratagene, La Jolla, CA) and incubated at 30° for 48 hr to determine UV sensitivity of nondividing cells. To determine UV survival of dividing cells, conidiospores on YAG plates were first allowed to germinate for 4.5 hr at 30°. By this time the germinated spores had entered the cell cycle and were about to undergo the first mitosis. These germlings were UV irradiated on the plates and then similarly incubated at 30° for 48 hr. Viability was determined as the percentage of colonies on treated plates compared to untreated controls.
DNA manipulations and construction of the ΔatmA strain:
DNA manipulations were according to Sambrook and Russell (2001). DNA fragment probes for Southern blots were labeled with [α-32P]dCTP using the RTS Rad prime DNA labeling system kit (GIBCO-BRL, Gaithersburg, MD). PCR primers were designed for amplifying each DNA fragment necessary for PCR-mediated technique by using Primer Express Version 1.0 (Applied Biosystems, Foster City, CA) design software. In the deletion construction, the A. fumigatus pyrG gene was amplified from the pCDA21 plasmid (Chaveroche et al. 2000) and is referred to as the zeopyrG cassette because the amplified fragment also contains the zeocin-resistance gene. The PCR-mediated construction for the atmA gene consisted of three initial amplifications that generated a 5′- and 3′-flanking region of atmA gene and a final fusion PCR. For the DNA fragments containing the flanking regions, genomic DNA was used as a template. The 5′-flanking fragment, which encompasses 2000 bp upstream from the nucleotide +5214 of the atmA genomic sequence, was amplified with the primers ATM1 5′-AATCTTAATGACATGATATGC-3′ and ATM2 5′-CTGAGAATTCCGGCTGAGGAAGCTACCG-3′ (underlined bases indicate the region of homology to the zeo-pyrG cassette). The 3′-flanking region, which encompasses 750 bp downstream of nucleotide +7632 of the atmA genomic sequence, was amplified with the primers ATM3 5′-TGAGGCGAATTCATGAAGCCCAGCGCCTG-3′ and ATM4 5′-TCATGCATAAGCGGCCC-3′ (underlined bases indicate the region of homology to the zeo-pyrG cassette). The zeopyrG cassette was amplified using plasmid pCDA21 as a template and the following primers: ATM5′ZEO 5′-GGTAGCTTCCTCAGCCGGAATTCTCAGTCCTGCT-3′ and ATM3′PYR 5′-AGGCGCTGGGCTTCATGAATTCGCCTCAAACAAT-3′ (where underlined bases indicate the region of homology to the 5′- and 3′-atmA flanking regions, respectively). The final 5167-bp fusion PCR fragment was generated using the three previous DNA fragments as templates and ATM1 and ATM4 as primers. The 50-μl amplification mixture included 1× Platinum Taq DNA Polymerase high-fidelity buffer (Invitrogen, San Diego), 20 pmol of each primer, 0.4 mm deoxynucleotide triphosphate mix, 1.0 unit of Platinum Taq high-fidelity DNA polymerase (Invitrogen), and 500 ng of genomic DNA or 100 ng of plasmid DNA. PCR amplification was carried out in a PTC100 96-well thermal cycler (MJ Research, Watertown, MA), at 94° for 2 min, and 30 times at 94° for 1 min, 52°–60° (depending on the fragment) for 1 min, and 68° for 2–3 min, followed by an extension step at 68° for 10 min. After the reaction, the PCR products were gel purified with a QIAGEN (Chatsworth, CA) PCR clean-up kit following the manufacturer's instructions. Transformation of A. nidulans strain GR5 was according to the procedure of Osmani et al. (1987) using 5 μg of linear DNA fragments. Transformants were scored for their ability to grow on YG medium. Southern analysis demonstrated that the deletion cassette had integrated at the atmA locus.
Protein expression, purification, and phosphorylation assays:
Protein assays were performed by initially growing conidia from the wild type (GR5) and IM69 (ΔatmA) strains in a reciprocal shaker at 37° for 16 hr in liquid YG medium (plus uridine and uracil if necessary). Mycelia were aseptically transferred to fresh YG medium plus 25 μm camptothecin (CPT) for different periods of time at 37°. Each sample was harvested by filtration through a Whatman filter number 1, washed thoroughly with sterile water, quickly frozen in liquid nitrogen, and disrupted by grinding. Total protein was extracted at 4° with extraction buffer (15 mm p-nitrophenylphosphate, 25 mm Tris pH 7.5, 15 mm EGTA, pH 7.5, 15 mm MgCl2) plus protease inhibitors (1 μg/ml leupeptin, 10 μg/ml aprotinin, 5 mm benzamidine, 15 mm PMSF). Protein concentration was determined using a modified Bradford assay (Bio-Rad, Hercules, CA). Twenty-five μg of ScaANBS1∷His6x fusion protein (purified as described in Fagundes et al. 2005) was incubated with 25 μg of GR5 or IM69 total extract in a final volume of 100 μl and incubated for 5 min at 30°. Phosphorylation reaction buffer (0.2 mm ATP, 2.5 mm MgCl2, 2.5 mm MnCl2, 0.5 μl [γ-32P]dATP) was added to this protein mixture and incubated for 1 hr at 30°. The Ni-NTA super flow (QIAGEN) resin was prepared by washing in PBS buffer and equilibrated with ligation buffer (20 mm sodium phosphate, pH 7.6, 500 mm NaCl, 20 mm imidazole, 1 mm PMSF, 1 mm benzamidine). The resin (90 μl) was subsequently combined with 100 μl of the protein mixture and incubated for 1 hr at room temperature. The resin was recovered by centrifugation at 3000 rpm for 1 min, washed three times with the ligation buffer, and finally resuspended in 30 μl of SDS–PAGE sample buffer (62.5 mm Tris–HCL, pH 6.8, 2% SDS, 10% glycerol, 5% β-mercaptoethanol, and 5% bromofenol blue). The proteins were eluted from the resin by heating at 100° for 3 min and separated by SDS–PAGE gels. Gels were stained with Coomassie solution (Coomassie 0.25%, methanol 50%, and acetic acid 30%), dried, and exposed to Kodak (MXG/Plus 35 × 43 cm) films.
Microscopy:
Conidiospores from wild-type and ΔatmA mutant strains were germinated at 37° or 42° on glass coverslips arrayed on the bottom of a plastic 100-mm petri dish containing the appropriate growth medium. Coverslips with adherent hyphae were fixed and stained as previously described (Harris et al. 1994). Nuclei were visualized by staining with Hoechst 33258 or DAPI, and septa were stained using Calcofluor. After staining, coverslips were washed in sterile water and mounted in n-propyl gallate. For lectin staining, coverslips were incubated in prewarmed media containing 5 μg/ml FITC-conjugated wheat germ agglutinin (WGA) for 5 min and then fixed, washed, and mounted. To depolymerize microtubules, hyphae grown for 12 hr in minimal glucose media were exposed to 5 μg/ml benomyl for 2 hr before staining with WGA. Slides were viewed using an Olympus BX51 fluorescent microscope. Images were captured with a Photometrics CoolSnap HQ CCD camera (Roper Scientific) and processed using IPLab software (Scanalytics) and Adobe PhotoShop 6.0. Confocal images were obtained with an Olympus FW500/BX61 confocal laser scanning microscope using the following laser lines: 405 nm for Hoechst 33258 and 488 nm for FITC. Images were captured by direct acquisition with a Z step of 1 μm and were subsequently processed using Adobe PhotoShop 6.0.
Polarity maintenance assay:
Conidiospores from wild-type strain A28 and the ΔatmA mutant IM69 were germinated at 37° for 12 hr on glass coverslips in a petri dish containing MM. Coverslips were transferred to prewarmed MM containing 2 μg/ml cytochalasin A (CA; Sigma; maintained as a 1 mg/ml stock in DMSO). Untreated controls received the corresponding volume of DMSO. Hyphae were incubated at 37° in the presence of CA for 60 min, washed twice in prewarmed media, and then returned to MM for an additional 2 hr. Coverslips were harvested prior to and immediately after the CA incubation, as well as at multiple intervals following release into drug-free media. Hyphae were fixed, stained with Calcofluor and Hoechst 33258, and mounted for imaging by brightfield and fluorescence microscopy.
RESULTS
Cloning and disruption of AtmAATM:
BLAST searches of the recently completed A. nidulans genome sequence (Galagan et al. 2005; http://www.genome.wi.mit.edu/annotation/fungi/aspergillus/) were used to identify a homolog of ATM. These searches resulted in the identification of AN0038.2 (renamed here as AtmAATM; note that the previously characterized UvsBATR is annotated as AN6975.2). The atmA gene possesses nine introns (nucleotide positions 83–133, 177–231, 1594–1648, 1960–2244, 2530–2643, 6503–6548, 6940–6982, 8051–8097, and 9059–9123, respectively) and encodes a predicted 2793-amino-acid protein with 30% identity and 48% similarity to ATM (4e-137).
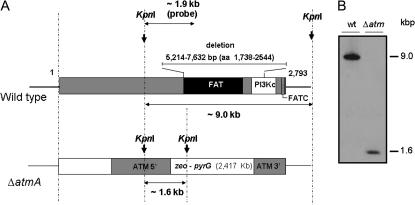
To characterize the function of AtmAATM, an internal deletion was generated using a fusion PCR-based approach (see materials and methods). This 2418-bp deletion removes 806 amino acids that encompass the entire FAT domain and a considerable portion of the predicted catalytic kinase domain (Figure 1A). Moreover, RT–PCR experiments verified that no atmA transcripts were generated from the region upstream of the deletion (data not shown). Protoplasts of A. nidulans strain GR5 were transformed using the fusion PCR product, and several transformants were obtained by their ability to grow in YAG. Allelic replacement of atmA was verified in at least one transformant as confirmed by the analysis of Southern blots (Figure 1B), thereby generating the ΔatmA strain IM69.
Figure 1.
Schematic of AtmA and the deletion strategy. (A) Structure of the A. nidulans ATM homolog. The number of residues is indicated. This protein has three motifs: the FAT and FAT-C domains and the PI3K domain. The two first domains are of unknown functional significance, and the last one contains the phosphatidylinositol 3-kinase motifs that harbor the catalytic site in the active kinases of the family (for a review, see Shiloh 2003). Genomic DNA from both wild-type and ΔatmA strains was isolated and cleaved with the enzyme KpnI; an internal 1963-bp DNA fragment was used as a hybridization probe. This fragment recognizes a single 9.0-kb DNA band in the wild-type strain and a unique 1.6-kb DNA band in the ΔatmA mutant because the zeo-pyrG cassette has a single KpnI site as shown in the Southern blot analysis (B).
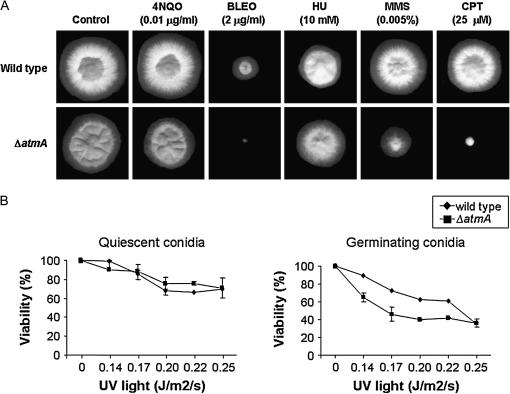
When tested on rich media (i.e., YG), the extent of radial colony growth and the production of asexual spores were not dramatically affected by the loss of AtmA (Figure 2A). However, no viable meiotic ascospores were recovered when IM69 was self-fertilized (data not shown).
Figure 2.
Growth phenotypes of GR5 (wild type) and IM69 (ΔatmA). (A) The strains were grown for 72 hr at 37° in YG medium in the presence or absence of 4-NQO, BLEO, hydroxyurea (HU), MMS, and CPT. (B) Quiescent (left) and germinating (right) conidiospores from different strains were exposed to UV light, and viability was scored after exposure as the percentage of colonies on treated plates compared to untreated controls. To evaluate the differences in UV-light assays, we used one-way ANOVA and Student Newman Keuls posthoc test (Daniel 2005). The results were expressed by the average of four independent experiments and the means plus the standard deviation are shown. Analyses were performed using the software package Sigma Stat (Jandel Scientific, San Rafael, CA) and the statistical significance was set at a = 0.05.
Role of AtmAATM in the DNA damage response:
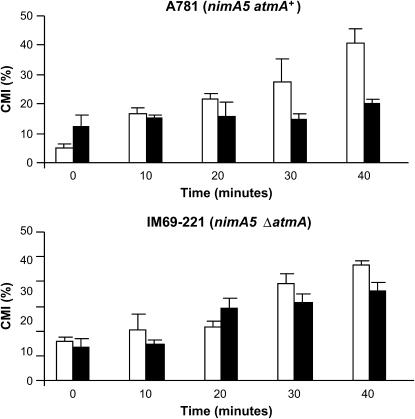
As an initial test of AtmA function in the DNA damage response, the effects of several different DNA-damaging agents on the growth of IM69 were assessed. The ΔatmA strain was more sensitive to several agents, including CPT, bleomycin (BLEO), and methyl methanesulfonate (MMS), whereas it was no more sensitive than wild type to other agents such as hydroxyurea (HU) and 4-nitroquinoline-1-oxide (4-NQO) (Figure 2A). In addition, germinating ΔatmA conidiospores (i.e., mitotically active cells) exhibited a slight increase in sensitivity to UV irradiation, while dormant spores (i.e., mitotically quiescent cells) displayed wild-type levels of resistance (Figure 2B), suggesting that AtmA is important for UV-light-induced DNA damage only during cell cycle progression. To understand the basis for the sensitivity of the ΔatmA mutant to DNA damage, we examined its ability to undergo checkpoint-mediated delay of mitotic entry. The G2/M DNA damage checkpoint prevents entry into mitosis in the presence of damaged DNA (for reviews, see Hartwell and Weinert 1989; Carr 1995). To address the role of AtmA in this checkpoint, we first constructed a nimA5 ΔatmA double-mutant strain (IM69-221). The nimA5 mutation causes a late G2 arrest at the restrictive temperature of 44°, thereby making it possible to synchronize cells in G2 before causing DNA damage. Preliminary tests verified that the nimA5 mutation did not affect the growth of the ΔatmA mutant. MMS (0.0025%) was added to either single-mutant nimA5 atmA+ or double-mutant nimA5 ΔatmA G2-arrested cells for 75 min to cause transient DNA damage and then removed from the culture by replacement with fresh medium as cells were downshifted to release the G2 block. The fresh medium contained 5 μg/ml of nocodazole to cause cell block in mitosis and to allow chromosomal mitotic index (CMI) determination. Addition of MMS to nimA5 atmA+ cells markedly delayed entry into mitosis after return to 30° (Figure 3, top). In contrast, no delay was observed when MMS was added to nimA5 ΔatmA (Figure 3, bottom). These observations highlight the role of AtmA in the G2/M checkpoint that operates to prevent mitosis in the presence of DNA damage.
Figure 3.
AtmA is required for the G2-M DNA damage checkpoint. The CMI was determined for strains A781 (nimA5; top) and IM69-221 (nimA5 ΔatmA; bottom) following release from mitotic arrest into media containing 0.0025% MMS. □, Control; ▪, 0.0025% MMS.
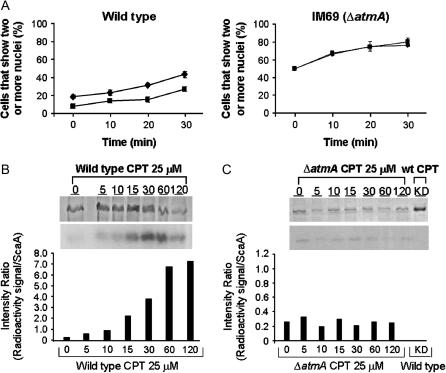
As a second approach to testing the role of AtmA in the DNA damage checkpoint response, we determined the ability of the ΔatmA mutant to undergo radioresistant DNA synthesis (RDS). The failure of A-T cells to undergo transient RDS after irradiation is a characteristic feature of the disease (for review, see D'Amours and Jackson 2002). Wild-type hyphae exposed to the radiomimetic drug BLEO display a transient delay in the kinetics of nuclear division (Figure 4A, left), whereas no such delay is observed in the ΔatmA mutant (Figure 4A, right). This observation implies that exposure to BLEO does not delay DNA synthesis when AtmA function is compromised.
Figure 4.
AtmA is required for the S-phase checkpoint response to DNA damage and phosphorylates ScaANBS1. (A) S-phase checkpoint response of GR5 (wild type; left) and IM69 (ΔatmA; right). Conidiospores were inoculated onto coverslips in YG medium and incubated at 37° for 5 hr. After this period of growth, 1 μg/ml of BLEO (▪) was added to the culture medium and the incubation at 37° continued for an additional 30 min. Control treatments without BLEO (♦) were performed for each strain. Samples were collected every 10 min and the number of nuclei assessed by DAPI staining. (B) Phosphorylation of the ScaANBS1 by GR5 (wild type) and (C) IM69 (ΔatmA) protein extracts. Extracts were prepared and phosphorylation assays performed as described in materials and methods. After the phosphorylation reaction, ScaANBS1 was precipitated with the Ni-NTA super flow (QIAGEN) resin. SDS gels were stained with Coomassie (B and C, top bands), dried, and exposed to Kodak films (B and C, bottom bands). The graphs in B and C represent the intensity ratio, i.e., the densitometry of the ScaANBS1 Coomassie staining divided by the densitometry of the radioactive signal of phosphorylated ScaANBS1. A heat-inactivated (KD) wild-type extract (boiled at 100° for 10 min) was used as a negative control for the ScaANBS1 phosphorylation reaction (Figure C, bottom).
Finally, we tested the capacity of AtmA to phosphorylate a downstream target in response to DNA damage. A general function of ATM homologs is the ability to phosphorylate p95/Nbs1 on serine 343 in vitro and in vivo after ionizing radiation (IR; Gatei et al. 2000; Lim et al. 2000; Wu et al. 2000; Zhao et al. 2000). Considering that DSBs are the main effect of IR, and that CPT induces DSBs, we used the antitopoisomerase I drug in experiments to verify whether the ΔatmA strain would be unable to phosphorylate ScaANBS1. Accordingly, protein extracts prepared from the ΔatmA mutant and a wild-type strain were tested for the ability to phosphorylate the apparent A. nidulans Nbs1 homolog ScaANBS1 upon treatment with CPT (Figure 4B). For the wild-type strains, the intensity ratio (radioactivity signal/amount ScaA) increased with incubation time in the presence of CPT (from 0.8 at 5 min to 7.6 at 120 min). In contrast, extracts from the ΔatmA mutant were unable to phosphorylate ScaANBS1 to the same levels observed in the wild-type strain (Figure 4C). A heat-inactivated extract (KD), i.e., a protein extract from the wild-type grown for 30 min and heated at 100° for 10 min, was used as a negative control for the ScaANBS1 phosphorylation reaction (Figure 4C, bottom).
Taken together, these data strongly suggest that AtmA shares with other ATM homologs a central and conserved role in the DNA damage response.
Role of AtmAATM in the septation checkpoint:
In A. nidulans, the DNA damage checkpoint inhibits septum formation by modulating the activity of NimXCDK1 (Harris and Kraus 1998; De Souza et al. 1999). In particular, chronic exposure of predivisional hyphae to low levels of DNA damage causes increased AnkAWEE1-mediated Tyr-15 phosphorylation of NimXCDK1 (Kraus and Harris 2001), which prevents septation even though nuclear division can occur (Harris and Kraus 1998). Notably, the ability of DNA damage to block septum formation requires UvsBATR function (Harris and Kraus 1998). We evaluated the role of AtmA in the septation checkpoint by determining whether the ΔatmA mutant could undergo septum formation in the presence of a DNA-damaging agent. As expected, prolonged exposure to 0.0025% MMS reduced the level of septation in wild-type hyphae (42% of MMS-treated hyphae possessed septa vs. 82% of untreated controls). However, unlike uvsBATR mutants, septum formation was not restored in the ΔatmA mutant. Indeed, the septation block was much more reduced than in the wild type (2% of MMS-treated ΔatmA hyphae possessed septa vs. 92% of untreated controls). This observation suggests that AtmA does not play a direct role in blocking septum formation upon activation of the DNA damage response.
AtmAATM is required for the formation of a stable polarity axis:
During the analysis of ΔatmA phenotypes, it was noted that the mutant displayed pronounced morphological defects even when grown under normal conditions (i.e., in the absence of DNA damage). The morphogenetic program associated with the germination of A. nidulans conidiospores includes (i) a brief period of isotropic spore swelling, (ii) the establishment of a stable polarity axis, and (iii) the emergence of a polarized germ tube (Momany 2002). This program leads to the formation of a multinucleate hyphal cell that grows solely by apical extension. Landmark proteins that act as positional cues to specify sites of polarized growth have not yet been identified in A. nidulans (Harris and Momany 2004).
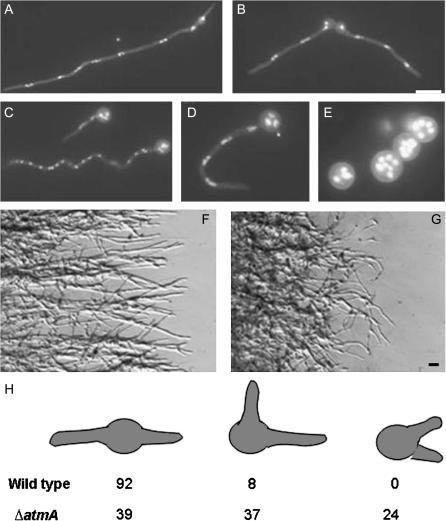
On a standard rich glucose medium (YG), ΔatmA conidiospores produce polarized germ tubes with kinetics similar to wild type. However, the resulting hyphae exhibit zig-zag or curled growth patterns (Figure 5) suggestive of a defect in the microtubule-dependent regulation of hyphal growth. Consistent with this view, the normal bipolar pattern of germ-tube emergence is abolished in the ΔatmA mutant. Whereas >90% of wild-type conidiospores produce a second germ tube opposite from the first, <40% of ΔatmA conidiospores display the same pattern (Figure 5). Previous observations suggest that the bipolar pattern requires an intact cytoskeleton (Harris et al. 1999), which presumably transports a landmark protein to the incipient polarization site. The wild-type growth pattern leads to the formation of mycelial colonies whose edge consists of well-spaced hyphae that grow outward in a linear manner (Figure 5). Because of the defects in growth pattern, ΔatmA colony edges are extremely disorganized and contain hyphae whose growth appears randomly oriented with respect to each other and the colony center (Figure 5). On a poorer minimal glycerol medium (M-glycerol), ΔatmA mutants display a pronounced defect in polarity establishment. Instead of forming a germ tube, they continue to grow in an isotropic manner and produce large round multinucleate spheres (Figure 5E). The aberrant growth patterns on rich media and the failure to establish polarity on poorer media occur despite apparently normal nuclear division and cell growth. Indeed, swollen ΔatmA conidiospores actually contain a greater number of nuclei than wild-type hyphae grown for the same length of time. These results suggest that AtmA might play a role in recruiting the morphogenetic machinery to polarization sites.
Figure 5.
Morphological defects caused by the ΔatmA mutation. Wild-type hyphae grown on YG (A) or M-glycerol (B) media. ΔatmA mutant grown on YG (C and D) or M-glycerol (E) media. Note the zig-zag or curled hyphae in C and D and the failure to establish polarity in E. Wild-type (F) and ΔatmA (G) hyphae growing on the surface of a YG plate. Note that wild-type hyphae radiate outward in a linear fashion, whereas mutant hyphae do not and frequently curl backward. (H) Germination pattern of wild-type and ΔatmA conidiospores. Spores were allowed to germinate on YG media for 11 hr. Spores possessing two germ tubes were classified as displaying (left to right) bipolar, quarterpolar, or random germination patterns. N = 200. Bar, 10 μm.
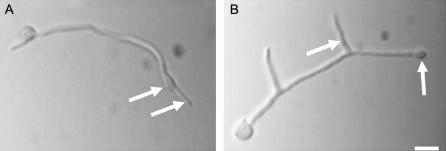
We established an assay that would allow us to quantify the ability of a mutant to retain a stable axis of polarized hyphal growth. This assay is based on the ability of the microfilament-depolymerizing agent cytochalasin A (CA) to cause depolarization of growing hyphal tips. When incubated in the presence of 2 μg/ml CA for 1 hr, wild-type hyphal tips swell in an isotropic manner (Figure 6A). Upon removal of CA and return to untreated media, polarized growth resumes. Most importantly, in 94% of wild-type hyphae tested (n = 300; two independent experiments), growth resumes along the original polarity axis (Figure 6A). This observation suggests that, despite the loss of detectable actin filaments, the morphogenetic machinery is retained at polarization sites for at least 1 hr. When subjected to the same CA treatment regimen, ΔatmA mutants grown in MM also form swollen hyphal tips (Figure 6B). However, upon washout, growth did not resume using the original polarity axis. Instead, in 79% of tested hyphae (n = 300; two independent experiments), a new axis was established in the subapical region of the hyphal tip cell, thereby producing a new lateral branch (Figure 6B). Moreover, 16% of the tested ΔatmA hyphae failed to resume growth, which was never observed in CA-treated wild-type hyphae. These results imply that the morphogenetic machinery is not stably localized at the hyphal tip of ΔatmA mutants. We propose that this reflects the failure to properly utilize positional cues at the hyphal tip or the loss of these cues altogether.
Figure 6.
ΔatmA mutants fail to reestablish a lost polarity axis. Hyphae were incubated with cytochalasin A to disrupt polarized growth and cause tip swelling (arrows). Upon return to drug-free medium, wild-type hyphae resumed polarized growth along the original axis (A, arrow). By contrast, ΔatmA mutants established new axes that became lateral branches (B, arrow). Bar, 10 μm.
AtmAATM regulates microtubule organization at hyphal tips:
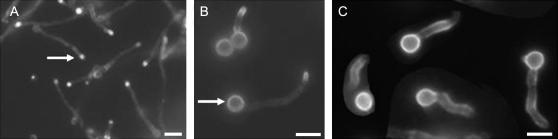
In A. nidulans, the FITC-conjugated lectin WGA can be used to detect sites of cell-wall deposition (Harris et al. 1999). At a concentration of 5 μg/ml, FITC–WGA staining is solely confined to the tips of wild-type hyphae (Figure 7A). In ΔatmA mutants, FITC–WGA stained hyphal tips as well, but prominent spore body staining was also observed (Figure 7B). This observation suggests that a pool of vesicles carrying precursors for cell-wall biosynthesis was inappropriately retained within the spore body. Wild-type hyphae grown in the presence of the microtubule-depolymerizing agent benomyl exhibit a similar intense localization of FITC–WGA to the spore body (Figure 7C), although in this case staining of the hyphal tip is not observed. Accordingly, we propose that a defect in microtubule-dependent vesicle transport underlies the aberrant cell-wall deposition pattern in ΔatmA mutants.
Figure 7.
Growth is not confined to the hyphal tip in ΔatmA mutants. Hyphae were stained with FITC-conjugated wheat germ agglutinin to localize sites of active cell-wall deposition. Growth occurs at the tips of wild-type hyphae (A, arrow). In ΔatmA mutants (B), growth is also observed in the spore body (arrow). In wild-type hyphae that were germinated in the presence of 5 μg/ml benomyl, growth is largely confined to the spore body (C). Bars, 10 μm.
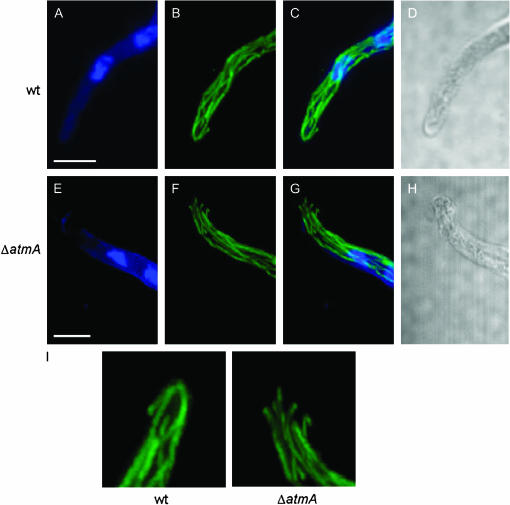
Immunofluorescence microscopy with anti-β-tubulin antibodies was used to examine the state of cytoplasmic microtubules in the ΔatmA mutant. When viewed using laser-scanning confocal microscopy, wild-type hyphae featured longitudinal arrays of cytoplasmic microtubules that terminated at a discrete site within the hyphal tip (Figure 8, A–D). By contrast, in the ΔatmA mutant, microtubule arrays typically did not terminate at a discrete site and instead appeared to be splayed out in a random manner (Figure 8, E–H). Quantification of these phenotypes revealed that 51% of ΔatmA hyphae possessed cytoplasmic microtubules that failed to converge at a discrete site, compared to only 9.5% of wild-type hyphae (n = 100; two independent experiments; Figure 8I). These observations suggest that the morphogenetic defects observed in ΔatmA mutants reflect the inability to specify a site of cortical microtubule capture at hyphal tips.
Figure 8.
Microtubules fail to converge at the hyphal tip in ΔatmA mutants. Wild-type (A–D) and ΔatmA (E–H) hyphae were examined by immunofluorescence microscopy. (A and E) Nuclei visualized using Hoechst 33258. (B and F) Microtubules visualized with an anti-β-tubulin antibody. (C and G) merged image. (D and H) Brightfield images (single section, whereas all other images are maximum projection of Z stacks). (I) The microtubule organization at the hyphal tip is enlarged to highlight the defect. Bars, 5 μm.
DISCUSSION
ATM and ATR are paralogous PIKKs that orchestrate the DNA damage response in eukaryotic cells (Shiloh 2001; McKinnon 2004). These kinases possess both overlapping and distinct roles in the regulation of this response. In A. nidulans, we have previously shown that UvsBATR regulates the DNA damage checkpoint response, inhibits septation in predivisional hyphae that have experienced DNA damage, and controls both the expression and the localization of proteins involved in the processing and repair of DNA damage (Harris and Kraus 1998; Hofmann and Harris 2000; Fagundes et al. 2005). In this study, we present the first analysis of ATM in a filamentous fungus. Like other ATM kinases, we show that AtmAATM modulates DNA damage checkpoint responses and phosphorylates the downstream target ScaANBS1. However, we also demonstrate that AtmAATM has an additional role in the formation of stable axes of polarized hyphal growth. In this respect, AtmA may share a pivotal function with its homologs in other highly polarized cell types such as neurons.
Role of AtmAATM in the DNA damage response:
Our results demonstrate that AtmAATM prevents mitotic entry when DNA is damaged during S or G2 phases of the cell cycle. To some extent, this may account for the DNA damage sensitivity of the ΔatmA mutant, because dormant conidiospores, in which the checkpoint is dispensable, display wild-type levels of resistance to some forms of DNA damage. On the other hand, AtmAATM likely has additional roles in promoting DNA repair. Like other eukaryotes, one such function may be to regulate the activity of the MRN complex (D'Amours and Jackson 2002). Homologs of each component of this complex have been characterized in A. nidulans (i.e., MreA = Mre11, SldI = Rad50, and ScaA = Nbs1) and shown to be involved in multiple facets of the DNA damage response (Bruschi et al. 2000; Semighini et al. 2003; Malavazi et al. 2005). Notably, UvsBATR regulates both the expression and the nuclear localization of ScaANBS1 (Fagundes et al. 2005), whereas AtmAATM controls the level of ScaANBS1 phosphorylation. These distinct regulatory inputs could potentially confer ATR- and ATM-specific functions on the A. nidulans MRN complex. Alternatively, they could act in concert to ensure that the complex is fully activated in response to the presence of damaged DNA. A more detailed analysis of the interactions between the MRN complex and both UvsBATR and AtmAATM is needed to address these possibilities.
UvsBATR inhibits septum formation in predivisional hyphae that have been exposed to low levels of DNA damage (Harris and Kraus 1998). Although this effect is mediated by AnkWEE1-dependent Tyr-15 inhibitory phosphorylation of NimXCDK1 (Kraus and Harris 2001), the mechanism by which UvsBATR may influence the Tyr-15 phosphorylation state of NimXCDK1 has not been determined. By contrast, AtmAATM does not appear to be involved in the inhibition of septum formation. Instead, the ΔatmA mutation exacerbates the septation block. This could simply reflect an increase in the strength of the DNA damage signal caused by loss of AtmAATM. However, an intriguing possibility is that AtmAATM may directly promote septum formation during recovery from the DNA damage response. We have previously shown that this function requires the A. nidulans RecQ/BLM helicase homolog MusNBLM (Hofmann and Harris 2001). Because extensive interactions between ATM and BLM have been characterized in animal cells (Beamish et al. 2002), it is tempting to speculate that similar interactions could trigger recovery from the DNA response in predivisional hyphae.
Role of AtmAATM in polarized hyphal growth:
The observation that AtmAATM is required for the formation of a stable axis of hyphal polarity was unexpected. In yeast cells, ATM function has never been linked to the establishment or maintenance of cell polarity. Moreover, only in human and mouse Purkinje cells has ATM been implicated in functions related to cellular morphogenesis (Barlow et al. 2000). Accordingly, our results raise the interesting possibility that ATM possesses a unique and previously uncharacterized role in the formation of stable polarity axes that is specific to highly polarized cells such as hyphae and neurons. Furthermore, we propose that failure to complete this function causes morphological defects that disrupt neuronal differentiation and thus contribute to the disease pathology of A-T.
Our observations suggest that AtmA is required to specify the growth zone at hyphal tips. In the filamentous fungi, an apical cluster of vesicles and cytoskeletal elements termed the Spitzenkorper plays a critical role in defining the growth zone (summarized by Harris et al. 2005). Notably, interactions between cytoplasmic microtubules and the cell cortex appear to play an important role in maintaining the position of the Spitzenkorper within the middle of the growth zone (Konzack et al. 2005). We propose that AtmA-dependent phosphorylation may influence these interactions, thereby helping to define a discrete zone for the cortical recruitment of cytoplasmic microtubules. The absence of this zone in the ΔatmA mutant would presumably lead to random interactions between microtubules and the cortex at hyphal tips, which would in turn affect Spitzenkorper position and the maintenance of a stable polarity axis. Consistent with this hypothesis, Horio and Oakley (2005) demonstrate that although microtubules are not strictly required for polarized growth, they are rate limiting for the growth of hyphal-tip cells. What proteins at the hyphal tip could be the targets of ATM-dependent regulation? Potential candidates include proteins that associate with the plus ends of cytoplasmic microtubules, such as dynactin, which has been suggested as a possible ATM target (O'Neill et al. 2000), or plus-end-directed kinesins involved in the transport of these proteins. One such kinesin, KipA, has already been shown to regulate Spitzenkorper position (Konzack et al. 2005). Alternatively, homologs of known cortical microtubule-anchoring proteins from budding or fission yeast (i.e., Kar9, Tea1, Tea4) may be ATM targets. Finally, fungal β-adaptins, whose animal homologs are known substrates of ATM, may be AtmA targets that control the trafficking of a landmark protein that specifies a cortical zone for microtubule interactions. Proteomic approaches designed to identify AtmA-interacting partners should ultimately reveal the relevant target.
Acknowledgments
We thank Joe Zhao from the University of Nebraska at Lincoln Center for Biotechnology Microscopy Core Facility for invaluable assistance with the confocal microscope. This research was supported by the Nebraska Research Foundation (S.H.) and Fundação de Amparo a Pesquisa do Estado de São Paulo, and Conselho Nacional de Desenvolvimento Científico e Tecnológico (G.H.G.).
References
- Barlow, C., C. Ribaut-Barassin, T. A. Zwingman, A. J. Pope, K. D. Brown et al., 2000. ATM is a cytoplasmic protein in mouse brain required to prevent lysosomal accumulation. Proc. Natl. Acad. Sci. USA 97: 871–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beamish, H., P. Kedar, H. Kaneko, P. Chen, T. Fukao et al., 2002. Functional link between BLM defective in Bloom's syndrome and the ataxia-telangiectasia mutated protein, ATM. J. Biol. Chem. 277: 30515–30523. [DOI] [PubMed] [Google Scholar]
- Boehm, M., and J. S. Bonifacino, 2001. Adaptins: the final recount. Mol. Biol. Cell 12: 2907–2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruschi, G. C. M., C. C. de Souza, M. R. Z. K. Fagundes, M. A. C. Dani, M. H. S. Goldman et al., 2000. Sensitivity to camptothecin in Aspergillus nidulans identified a novel gene, scaA, related to the cellular DNA damage response. Mol. Genet. Genomics 265: 264–275. [DOI] [PubMed] [Google Scholar]
- Carr, A. M., 1995. DNA structure checkpoints in fission yeast. Semin. Cell Biol. 6: 65–72. [DOI] [PubMed] [Google Scholar]
- Chaveroche, M. K., J. M. Ghigo and C. d'Enfert, 2000. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 28: E97–E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun, H. H., and R. A. Gatti, 2004. Ataxia-telangiectasia, an evolving phenotype. DNA Rep. 3: 1187–1196. [DOI] [PubMed] [Google Scholar]
- Daniel, W. W., 2005. Biostatistics: A Foundation for Analysis in the Health Sciences (Wiley Series in Probability and Statistics). John Wiley & Sons, New York.
- D'Amours, D., and S. P. Jackson, 2002. The Mre11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat. Rev. Mol. Cell Biol. 3: 317–327. [DOI] [PubMed] [Google Scholar]
- DeSouza, C. P., X. S. Ye and S. A. Osmani, 1999. Checkpoint defects leading to premature mitosis also cause endoreplication of DNA in Aspergillus nidulans. Mol. Biol. Cell 10: 3661–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagundes, M. R. Z. K., J. F. Lima, M. Savoldi, I. Malavazi, R. E. Larson et al., 2004. The Aspergillus nidulans npkA gene encodes a Cdc2- related kinase that genetically interacts with the UvsBATR kinase. Genetics 167: 1629–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagundes, M. R. Z. K., C. P. Semighini, I. Malavazi, M. Savoldi, J. F. Lima et al., 2005. Aspergillus nidulans uvsBATR and scaANBS1 genes show genetic interactions during recovery from replication stress and DNA damage. Eukaryot. Cell. 4: 1239–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan, J. E., S. E. Calvo, C. Cuomo, L-J. Ma, J. Wortman et al., 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438: 1105–1115. [DOI] [PubMed] [Google Scholar]
- Gatei, M., D. Young, K. M. Cerosaletti, A. Desai-Mehta, K. Spring et al., 2000. ATM-dependent phosphorylation of nibrin in response to radiation exposure. Nat. Genet. 25: 115–119. [DOI] [PubMed] [Google Scholar]
- Gatti, R. A., 1998. Ataxia-telangiectasia, pp. 275–300 in The Genetic Basis of Human Cancer, edited by B. Vogelstein and K. W. Kinzler. McGraw-Hill, New York.
- Goldman, G. H., and E. Kafer, 2004. Aspergillus nidulans as a model system to characterize the DNA damage response in eukaryotes. Fungal Genet. Biol. 41: 428–442. [DOI] [PubMed] [Google Scholar]
- Goldman, G. H., S. L. McGuire and S. D. Harris, 2002. The DNA damage response in filamentous fungi. Fungal Genet. Biol. 35: 183–195. [DOI] [PubMed] [Google Scholar]
- Gygax, S. E., C. P. Semighini, G. H. Goldman and S. D. Harris, 2005. SepBCTF4 is required for the formation of DNA damage-induced UvsCRAD51 foci in Aspergillus nidulans. Genetics 169: 1391–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, S. D., and P. R. Kraus, 1998. Regulation of septum formation in Aspergillus nidulans by a DNA damage checkpoint pathway. Genetics 148: 1055–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, S. D., and M. Momany, 2004. Polarity in filamentous fungi: moving beyond the yeast paradigm. Fungal Genet. Biol. 41: 391–400. [DOI] [PubMed] [Google Scholar]
- Harris, S. D., J. L. Morrell and J. E. Hamer, 1994. Identification and characterization of Aspergillus nidulans mutants defective in cytokinesis. Genetics 136: 517–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, S. D., A. F. Hofmann, H. W. Tedford and M. P. Lee, 1999. Identification and characterization of genes required for hyphal morphogenesis in the filamentous fungus Aspergillus nidulans. Genetics 151: 1015–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, S. D., N. D. Read, R. W. Roberson, B. Shaw, S. Seiler et al., 2005. Polarisome meets Spitzenkorper: microscopy, genetics, and genomics converge. Eukaryot. Cell. 4: 225–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell, L. H., and T. A. Weinert, 1989. Checkpoints: controls that ensure the order of cell cycle events. Science 246: 629–634. [DOI] [PubMed] [Google Scholar]
- Hill, T. W., and E. Kafer, 2001. Improved protocols for Aspergillus minimal medium: trace element and minimal medium salt stock solutions. Fungal Genet. Newsl. 48: 20–21. [Google Scholar]
- Hofmann, A. F., and S. D. Harris, 2000. The Aspergillus nidulans uvsB gene encodes an ATM-related kinase required for multiple facets of the DNA damage response. Genetics 154: 1577–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann, A. F., and S. D. Harris, 2001. The Aspergillus nidulans musN gene encodes a RecQ helicase that interacts with the PI-3K-related kinase UvsB. Genetics 159: 1595–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horio, T., and B. R. Oakley, 2005. The role of microtubules in rapid hyphal tip growth of Aspergillus nidulans. Mol. Biol. Cell 16: 918–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafer, E., 1977. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv. Genet. 19: 33–131. [DOI] [PubMed] [Google Scholar]
- Konzack, S., P. E. Rischitor, C. Enke and R. Fischer, 2005. The role of the kinesin motor KipA in microtubule organization and polarized growth of Aspergillus nidulans. Mol. Biol. Cell 16: 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus, P. R., and S. D. Harris, 2001. The Aspergillus nidulans snt genes are required for the regulation of septum formation and cell cycle checkpoints. Genetics 159: 557–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuljis, R. O., G. Chen, E. Y. Lee, M. C. Aguila and Y. Xu, 1999. ATM immunolocalization in mouse neuronal endosomes: implications for ataxia telangiectasia. Brain Res. 842: 351–358. [DOI] [PubMed] [Google Scholar]
- Lakin, N. D., P. Weber, T. Stankovic, S. T. Rottinghaus, A. M. Taylor et al., 1996. Analysis of the ATM protein in wild type and ataxia telangiectasia cells. Oncogene 13: 2707–2716. [PubMed] [Google Scholar]
- Lim, D. S., D. G. Kirsch, C. E. Canman, J. H. Ahn, Y. Ziv et al., 1998. ATM binds to β-adaptin in cytoplasmic vesicles. Proc. Natl. Acad. Sci. USA 95: 10146–10151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, D. S., S. T. Kim, B. Xu, R. S. Maser, J. Lin et al., 2000. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature 404: 613–617. [DOI] [PubMed] [Google Scholar]
- Lisby, M., J. H. Barlow, R. C. Burgess and R. Rothstein, 2004. Choreography of the DNA damage response: spatiotemporal relationship among checkpoint and repair proteins. Cell 118: 699–713. [DOI] [PubMed] [Google Scholar]
- Malavazi, I., J. F. Lima, M. R. Z. K. Fagundes, V. P. Efimov, M. H. S. Goldman et al., 2005. The Aspergillus nidulans sldIRAD50 gene interacts with bimEAPC1, a homologue of an anaphase-promoting complex subunit. Mol. Microbiol. 57: 222–237. [DOI] [PubMed] [Google Scholar]
- McKinnon, P. J., 2004. ATM and ataxia telangiectasia. EMBO Rep. 5: 772–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnon, P. J., and L. A. Burgoyne, 1985. Altered cellular morphology and microfilament array in ataxia-telangiectasia fibroblasts. Eur. J. Cell Biol. 39: 161–166. [PubMed] [Google Scholar]
- Momany, M., 2002. Polarity in filamentous fungi: establishment, maintenance and new axes. Curr. Opin. Microbiol. 5: 580–585. [DOI] [PubMed] [Google Scholar]
- O'Connor, R. D., and D. S. Linthicum, 1980. Mitogen receptor redistribution defects and concomitant absence of blastogenesis in ataxia-telangiectasia T lymphocytes. Clin. Immunol. Immunopathol. 15: 66–75. [DOI] [PubMed] [Google Scholar]
- O'Neill, T., A. J. Dwyer, Y. Ziv, D. W. Chan, S. P. Lees-Miller et al., 2000. Utilization of oriented peptide libraries to identify substrate motifs selected by ATM. J. Biol. Chem. 275: 22719–22727. [DOI] [PubMed] [Google Scholar]
- Osmani, S. A., G. S. May and N. R. Morris, 1987. Regulation of the mRNA levels of nimA, a gene required for the G2-M transition in Aspergillus nidulans. J. Cell Biol. 104: 1495–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse, J., and S. P. Jackson, 2002. Interfaces between the detection, signaling, and repair of DNA damage. Science 558: 547–551. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., and D. W. Russell, 2001. Molecular Cloning: A Laboratory Manual, Ed. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Semighini, C. P., M. R. Z. K. Fagundes, J. C. Ferreira, R. C. Pascon, M. H. S. Goldman et al., 2003. Different roles of the Mre11 complex in the DNA damage response in Aspergillus nidulans. Mol. Microbiol. 48: 1693–1709. [DOI] [PubMed] [Google Scholar]
- Shiloh, Y., 2001. ATM and ATR: networking cellular responses to DNA damage. Curr. Opin. Genet. Dev. 11: 71–77. [DOI] [PubMed] [Google Scholar]
- Shiloh, Y., 2003. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer 3: 155–168. [DOI] [PubMed] [Google Scholar]
- Watters, D., K. K. Khanna, H. Beamish, G. Birrell, K. Spring et al., 1997. Cellular localization of the ataxia telangiectasia (ATM) gene product and discrimination between mutated and normal forms. Oncogene 14: 1911–1921. [DOI] [PubMed] [Google Scholar]
- Wu, X., V. Ranganathan, D. S. Weisman, W. F. Heine, D. N. Ciccone et al., 2000. ATM phosphorylation of Nijmegen breakage syndrome protein is required in a DNA damage response. Nature 405: 477–482. [DOI] [PubMed] [Google Scholar]
- Zhao, S., Y. C. Weng, S. S. Yuan, Y. T. Lin, H. C. Hsu et al., 2000. Functional link between ataxia-telangiectasia and Nijmegen breakage syndrome gene products. Nature 405: 473–477. [DOI] [PubMed] [Google Scholar]