Abstract
Schizosaccharomyces pombe senses environmental glucose through a cAMP-signaling pathway, activating cAMP-dependent protein kinase A (PKA). This requires nine git (glucose insensitive transcription) genes that encode adenylate cyclase, the PKA catalytic subunit, and seven “upstream” proteins required for glucose-triggered adenylate cyclase activation, including three heterotrimeric G-protein subunits and its associated receptor. We describe here the cloning and characterization of the git1+ gene. Git1 is distantly related to a small group of uncharacterized fungal proteins, including a second S. pombe protein that is not functionally redundant with Git1, as well as to members of the UNC-13/Munc13 protein family. Mutations in git1+ demonstrate functional roles for the two most highly conserved regions of the protein, the C2 domain and the MHD2 Munc homology domain. Cells lacking Git1 are viable, but display phenotypes associated with cAMP-signaling defects, even in strains expressing a mutationally activated Gα-subunit, which activates adenylate cyclase. These cells possess reduced basal cAMP levels and fail to mount a cAMP response to glucose. In addition, Git1 and adenylate cyclase physically interact and partially colocalize in the cell. Thus, Git1 is a critical component of the S. pombe glucose/cAMP pathway.
NUTRIENT signaling is essential for unicellular organisms, which must regulate their growth and metabolism in response to the available nutrients in their environment. Glucose is the preferred carbon source for bacteria and fungi, such that glucose-signaling pathways exist to control the expression of genes involved in carbon source metabolism and sexual development for various organisms (Lengeler et al. 2000; Thevelein et al. 2000; Titgemeyer and Hillen 2002). Yeasts and other fungi also utilize glucose sensing to regulate cellular morphology and growth, which is required for invasive growth and, for some organisms, virulence. For example, glucose is a major component of the dialyzable fraction of serum that induces germ-tube formation, leading to invasive growth in the human pathogen Candida albicans (Hudson et al. 2004). Similarly, haploid invasive growth in the budding yeast Saccharomyces cerevisiae requires both the sensing of a nitrogen-poor environment and an abundant fermentable carbon source (Lengeler et al. 2000; Harashima and Heitman 2004). Recently, the fission yeast Schizosaccharomyces pombe has also been shown to switch from a unicellular yeast growth mode to a hyphal invasive growth mode in response to nitrogen-poor conditions, but only in strains with an intact cAMP-signaling pathway, which is required for glucose detection (Amoah-Buahin et al. 2005). Thus, there is considerable conservation among yeasts and fungi with regard to their biological response to environmental glucose.
S. cerevisiae cells sense glucose through at least three signaling pathways that control different aspects of the biological response to environmental glucose. Extracellular glucose is detected by Snf3 and Rgt2, 12-transmembrane-domain proteins that resemble hexose transporters, leading to the regulation of the Rgt1 transcriptional repressor, which regulates expression of hexose transporter genes (Johnston and Kim 2005). In addition, glucose uptake and metabolism under glucose-rich conditions inhibit the Snf1/AMP kinase, which leads to repression of target genes by the Mig1, Nrg1, and Nrg2 transcriptional repressors (Treitel et al. 1998; Kuchin et al. 2002). Finally, glucose detection also produces a transient cAMP signal. This involves the Gpr1 G-protein-coupled receptor (GPCR), a 7-transmembrane-domain protein that activates the Gpa2 Gα-subunit protein (Xue et al. 1998; Yun et al. 1998; Lorenz et al. 2000; Lemaire et al. 2004). Glucose detection also activates Ras proteins (Colombo et al. 2004), which together with Gpa2, stimulate cAMP production by adenylate cyclase (Thevelein and de Winde 1999; Thevelein et al. 2005).
Glucose detection in S. pombe is largely carried out by a cAMP-signaling pathway to regulate processes such as sexual development and gluconeogenesis (Hoffman 2005a,b). A stress-activated protein kinase (SAPK) pathway is also regulated by glucose starvation; however, in the absence of a functioning PKA pathway, the basal level of activity of the Spc1/Sty1 SAPK is sufficient to allow high levels of transcription of genes that are subject to glucose repression (Stettler et al. 1996). Conversely, in cells expressing elevated levels of PKA activity, starvation signaling through the SAPK pathway is unable to derepress transcription of glucose-repressed genes (DeVoti et al. 1991; Stettler et al. 1996; Janoo et al. 2001; Stiefel et al. 2004).
In S. pombe, glucose detection leads to a transient cAMP signal due to the activation of adenylate cyclase (Byrne and Hoffman 1993). The genes required for S. pombe adenylate cyclase activation compose at least two functional groups on the basis of genetic interactions involving a mutationally activated form of the gpa2+ gene. On the basis of biochemical studies of similarly altered Gα-subunits in other systems, the gpa2R176H allele encodes a protein presumed to be defective in GTPase activity. This defect reduces GTP hydrolysis of the bound guanine nucleotide, causing the Gα-subunit to remain in the active, GTP-bound state longer. The gpa2R176H allele suppresses the loss of the Git3 GPCR, the Git5 Gβ, or the Git11 Gγ (Welton and Hoffman 2000; Landry and Hoffman 2001), consistent with a model in which the Gβγ dimer is required for efficient coupling of the Gα to the receptor to allow Gα activation. On the other hand, the gpa2R176H allele fails to suppress mutations in git1+, git7+, or git10+, suggesting that these genes encode proteins that may activate adenylate cyclase in a G-protein-independent manner. Alternatively, these proteins may be required for the stabilization or localization of Gpa2 or adenylate cyclase, or are somehow otherwise required for Gpa2-mediated activation of adenylate cyclase.
In this study, we describe the cloning and characterization of the git1+ gene. Surprisingly, while the other proteins of the S. pombe glucose/cAMP pathway have obvious homologs in other eukaryotes, Git1 is only distantly related to a conserved fungal protein family of unknown function as well as the UNC-13/Munc13 protein family involved in synaptic vesicle exocytosis. All of these proteins contain a similar C2 domain near their carboxy-terminus, along with two additional domains known as Munc13-homology domains 1 and 2 (Koch et al. 2000). C2 domains are often utilized by proteins to bind phospholipids in a calcium-dependent manner; however, this domain can also function in a calcium-independent manner and mediate interactions with other proteins (Rizo and Sudhof 1998).
We show that Git1 is not essential for S. pombe viability, but is required both for maintaining basal cAMP levels and for producing a glucose-triggered cAMP response. The Git1 C2 domain is required for function as well as for the regulation of Git1 protein levels, which are reduced in cells growing in a glucose-rich environment. Additional mutant alleles suggest that the C-terminal MHD2 domain is also necessary for function. Finally, we show that Git1 physically interacts with and colocalizes with the Git2/Cyr1 adenylate cyclase enzyme, although Git1 does not appear to be required for adenylate cyclase stability or its steady-state localization. Thus, we have identified in S. pombe a novel regulator of adenylate cyclase activation that plays a critical role in glucose/cAMP sensing.
MATERIALS AND METHODS
S.pombe strains and growth media:
S. pombe strains used in this study are listed in Table 1. The fbp1∷ura4+ and ura4∷fbp1-lacZ reporter constructs have been described previously (Hoffman and Winston 1990). Rich, defined, and 5-FOA-containing growth media have been previously described (Hoffman and Winston 1990). The git1∷his3+ allele is an insertion of plasmid pAF1 into codon 608 of git1+, while git1∷LEU2+ (git1Δ) is a deletion of the entire git1+ ORF.
TABLE 1.
Strain list
| Strain | Genotype |
|---|---|
| 972 | h− |
| CHP7 | h+ fbp1∷ura4+ ura4∷fbp1-lacZ leu1-32 ade6-M210 his7-366 git2-7 |
| CHP9 | h+ fbp1∷ura4+ ura4∷fbp1-lacZ leu1-32 ade6-M210 his7-366 git1-45 |
| CHP45 | h+ fbp1∷ura4+ ura4∷fbp1-lacZ leu1-32 ade6-M210 his7-366 git1-9 |
| CHP46 | h+ fbp1∷ura4+ ura4∷fbp1-lacZ leu1-32 ade6-M210 his7-366 git1-9 |
| CHP61 | h+ fbp1∷ura4+ ura4∷fbp1-lacZ leu1-32 ade6-M210 his7-366 git2-61 |
| CHP79 | h+ fbp1∷ura4+ ura4∷fbp1-lacZ leu1-32 ade6-M210 his7-366 git1-79 |
| CHP99 | h+ fbp1∷ura4+ ura4∷fbp1-lacZ leu1-32 ade6-M210 his7-366 git1-99 |
| CHP216 | h−fbp1∷ura4+ ura4∷fbp1-lacZ leu1-32 ade6-M216 his7-366 git2-216 |
| CHP287 | h−fbp1∷ura4+ ura4∷fbp1-lacZ leu1-32 ade6-M216 his7-366 git1-287 |
| CHP568 | h+ fbp1∷ura4+ ura4∷fbp1-lacZ leu1-32 ade6-M210 his3-D1 git1-1 |
| CHP569 | h−fbp1∷ura4+ ura4∷fbp1-lacZ leu1-32 ade6-M216 his3-D1 lys1-131 git1-1 |
| CHP578 | h+ fbp1∷ura4+ ura4∷fbp1-lacZ leu1-32 ade6-M210 his3-D1 |
| CHP795 | h90fbp1∷ura4+ ura4∷fbp1-lacZ leu1-32 ade6-M216 his7-366 |
| CHP856 | h+fbp1∷ura4+ ura4∷fbp1-lacZ leu1-32 ade6-M210 his3-D1 git1∷his3 |
| CHP857 | h+ fbp1∷ura4+ ura4∷fbp1-lacZ leu1-32 ade6-M210 his3-D1 git1∷his3+ |
| CHP861 | h+fbp1∷ura4+ ura4∷fbp1-lacZ leu1-32 ade6-M210 his3-D1 gpa2∷his3+ |
| CHP889 | h− fbp1∷ura4+ ura4∷fbp1-lacZ leu1-32 ade6-M216 his3-D1 gpa2R176H |
| CHP896 | h90ura4∷fbp1-lacZ leu1-32 ade6-M210 git1Δ∷LEU2+ |
| CHP954 | h+ fbp1∷ura4+ ura4∷fbp1-lacZ leu1-32 git1Δ∷LEU2+ |
| CHP973 | h− leu1-32 git1Δ∷LEU2+ |
| DIP59 | h+ ura4∷fbp1-lacZ leu1-32 git2-13myc∷kan |
| FWP72 | h−fbp1∷ura4+ ura4∷fbp1-lacZ leu1-32 |
| FWP77 | h+ ura4∷fbp1-lacZ leu1-32 |
| FWP79 | h−ura4∷fbp1-lacZ leu1-32 ade6-M216 |
| FWP87 | h+ fbp1∷ura4+ ura4∷fbp1-lacZ leu1-32 |
| FWP89 | h+ fbp1∷ura4+ ura4∷fbp1-lacZ leu1-32 ade6-M216 |
| FWP95 | h−fbp1∷ura4+ ura4∷fbp1-lacZ leu1-32 |
| FWP96 | h−fbp1∷ura4+ ura4∷fbp1-lacZ leu1-32 ade6-M210 |
| FWP111 | h−fbp1∷ura4+ ura4∷fbp1-lacZ leu1-32 ade6-M216 his7-366 git1-1 |
| FWP112 | h−fbp1∷ura4+ ura4∷fbp1-lacZ leu1-32 ade6-M216 his7-366 |
| FWP134 | h+ ura4∷fbp1-lacZ leu1-32 ade6-M210 git1-1 |
| FWP188 | h− fbp1∷ura4+ ura4∷fbp1-lacZ leu1-32 ade6-M216 his7-366 git2-1∷LEU2+ |
| RKP2 | h− fbp1∷ura4+ ura4∷fbp1-lacZ leu1-32 git1Δ∷LEU2+ |
| RKP9 | h+ fbp1∷ura4+ ura4∷fbp1-lacZ leu1-32 his3-D1 git1Δ∷LEU2+gpa2R176H |
| RKP15 | h−fbp1∷ura4+ ura4∷fbp1-lacZ leu1-32 SPAC11E3.02cΔ∷LEU2+ |
| RKP19 | h−leu1-32 git1-V5his6∷LEU2+his3+ |
| RKP29 | h+ ura4∷fbp1-lacZ leu1-32 git1-V5his6∷LEU2+his3+git2-13myc∷kan |
| RKP44 | h+ fbp1∷ura4+ ura4∷fbp1-lacZ leu1-32 git1Δ∷LEU2+git2-13myc∷kan |
| RKP45 | h−fbp1∷ura4+ ura4∷fbp1-lacZ leu1-32 git1Δ∷LEU2+git2-13myc∷kan |
| RKP62 | h+ fbp1∷ura4+ ura4∷fbp1-lacZ leu1-32 git1Δ∷LEU2+SPAC11E3.02cΔ∷LEU2+ |
Recombinant DNA methods:
Escherichia coli transformations were carried out using ElectroTen-Blue or XL1-Blue electroporation-competent cells (Stratagene, La Jolla, CA) or TOP 10 chemically competent cells (Invitrogen, San Diego). Yeast transformations were carried out as previously described (Bähler et al. 1998). The Failsafe PCR system (Epicentre Technologies, Madison, WI), AccuPrime Taq DNA polymerase system (Invitrogen), and PfuTurbo DNA polymerase (Stratagene) were used for PCR according to the manufacturers' protocols. Oligonucleotides were from Integrated DNA Technologies. Isolation of DNA from S. pombe was achieved by the Smash and Grab method (Hoffman and Winston 1987). DNA sequencing was performed using custom oligonucleotides and the CEQ DTCS-Quick Start kit (Beckman Coulter) according to the manufacturer's directions.
Identification of the git1+ gene:
A nonhomologous plasmid integration and rescue method was used to identify git1+ (Hoffman and Welton 2000). S. pombe strain CHP578 was transformed to His+ with KpnI or NgoMIV-linearized plasmid pAF1 (his3+) DNA. Pools of His+ colonies were collected after 5 days growth from each transformation plate. Approximately 106 cells were spread onto SC–ura, and colonies that formed were further tested for elevated β-galactosidase activity. Integrated plasmids were rescued into E. coli by EcoRI digestion of genomic DNA and ligation to circularize the DNA. Rescued plasmids were sequenced using primer BS250 (5′-TTTTTGGGGTCGAGGTGC-3′) to determine the genomic site of plasmid insertion.
Plasmid constructions:
Oligonucleotides git1-for (5′-ATGGGATTCACTAGTGTTGAACC-3′) and git1STOP-rev (5′-CTAACTACTCACTCTTTTGACATCCTTAG-3′) were used in a PCR reaction on wild-type S. pombe genomic DNA to amplify the git1+ ORF, which was cloned into vector pNMT41 (Invitrogen) to create plasmid pRSK3. A second PCR reaction using oligonucleotide git1-for with git1V5-rev (5′-ACTACTCACTCTTTTGACATCCTTAGAC-3′) produced a PCR product lacking the git1+ STOP codon, such that cloning into pNMT41 to create plasmid pRSK4 results in a translational fusion to a C-terminal V5 tag (Southern et al. 1991) followed by a hexahistidine tag (git1-V5his6). Fourteen codons within the C2 domain of git1+ were removed by site-directed mutagenesis using the QuickChange site-directed mutagenesis kit (Stratagene) and oligonucleotides git1ΔC2-for (5′-CTGAGTCGGAATTTTGTGTACATACGTATCAGATGGAGATTAC-3′) and git1ΔC2-rev (5′-GTAATCTCCATCTGATACGTATGTACACAAAATTCCGACTCAG-3′) on pRSK3 and pRSK4 to create pRSK8 (git1ΔC2) and pRSK9 (git1ΔC2-V5his6), respectively.
Epitope tagging of git1+ and git2+ chromosomal loci:
The git1-V5his6 allele was introduced into the git1+ genomic locus so that it is expressed from the git1+ promoter by linearizing plasmid pRSK4 (git1-V5his6) with BspEI at nucleotide 711 of the git1+ ORF and by transforming CHP857 (git1∷his3+) to Leu+. Homologous insertion of the plasmid was confirmed by PCR. The git2-13myc allele was constructed by the PCR-based method of Bähler et al. (1998). Homologous insertion of the PCR product into the git2+ locus of strain FWP77 was confirmed by PCR. The tagged protein was shown to be functional by β-galactosidase assay of fbp1-lacZ expression in glucose-grown cells.
β-Galactosidase assays of fbp1-lacZ expression:
Cells were cultured overnight under repressing conditions (8% glucose) in yeast extract (YEL) or S. pombe minimal (PM; for transformants) liquid medium. Subcultures were grown for 24 hr to a final density of ∼1 × 107 cells/ml. Protein lysates were prepared and assayed as previously described (Nocero et al. 1994).
Deletion of git1+ and SPAC11E3.02c:
The git1+ and SPAC11E3.02c genes were deleted by a two-step PCR approach (Wang et al. 2004) and confirmed by PCR.
Protein isolation and immunoblotting:
Strains were cultured in appropriate liquid media to ∼1 × 107 cells/ml. Total protein extracts were prepared by TCA precipitation as previously described (Volland et al. 1994). Protein extracts were separated by SDS–PAGE and transferred to a polyvinylidene difluoride membrane. Immunodetection of myc-tagged and V5-tagged proteins was carried out using mouse α-myc (Santa Cruz Biotechnology) and mouse α-V5 (Invitrogen) primary antibodies and peroxidase-labeled goat α-mouse IgG secondary antibody (Kirkegaard & Perry Laboratories). Visualization was carried out by using LumiGLO chemiluminescence (Kirkegaard & Perry Laboratories). Cdc2 was detected using rabbit polyclonal IgG against Cdc2 p34 PSTAIRE (Santa Cruz Biotechnology) in combination with horseradish-peroxidase-conjugated secondary antibody.
Co-immunoprecipitations:
S. pombe strains DIP59, RKP19, RKP29, and FWP72 were grown to exponential phase and broken in NP-40 buffer [6 mm Na2HPO4, 4 mm NaH2PO4 H2O, 1% NONIDET P-40, 150 mm NaCl, 2 mm EDTA, 50 mm NaF, 0.1 mm Na3VO4, 16.7 μm PMSF, and 1 tablet of protease inhibitor (Boehringer Mannheim, Indianapolis) in 50 ml of buffer] using acid-washed glass beads in a Bead Beater. A total of 500 μl of cell lysate was incubated with 2.5 μg of α-V5 or α-Myc antibody for >1 hr at 4° on a rotator. Fifty microliters of Protein G Sepharose 4 Fast Flow was added in a 1:1 ratio with NP-40 buffer and incubated on a rotator for 1 hr or more. Precipitated immune complexes were isolated by microcentrifugation for 20 sec. The pellets were washed four times with NP-40 buffer and once with 50 mm Tris pH 8. Pellets were resuspended in 30 μl Laemmli buffer and heated for 3 min at 95°. Beads were pelleted by microcentrifugation for 20 sec, and the supernatants were removed for analysis.
Immunofluorescence microscopy:
Strains were grown in appropriate liquid media to 2–4 × 106 cells/ml before fixing with paraformaldehyde as previously described (Hagan and Hyams 1988) with slight modifications. Cell walls were digested with 0.5 mg/ml 100T Zymolyase. V5-tagged proteins were detected using α-V5 primary antibody diluted 1:100 in PEMBAL and visualized by incubating with Alexa Fluor 488-labeled goat α-mouse secondary antibody diluted 1:50 in PEMBAL overnight. Rabbit polyclonal IgG against c-Myc was used together with Alexa Fluor 594-labeled goat α-rabbit IgG to visualize the 13Myc-tagged protein. Images were visualized and captured using a Zeiss Axioplan 2 microscope with an Orca-ER CCD camera and Openlab software.
RESULTS
Cloning of the S.pombe git1+ gene:
The S. pombe git1+ gene was identified and cloned using a plasmid insertional mutagenesis screen developed to overcome problems associated with the high background of multicopy suppressors encountered during library screens in fission yeast (Hoffman and Welton 2000). This screen involves the random insertion of linearized plasmid pAF1 (his3+) into strain CHP578 (his3Δ), and a subsequent screen of these His+ transformants for Git− strains that are able to form colonies on glucose-rich medium lacking uracil due to the loss of glucose repression of an fbp1-ura4+ reporter gene (see materials and methods). Two independent Git− mutants were identified and the integrated plasmid in each strain was rescued into E. coli such that some genomic DNA from the site of insertion was cloned. Sequence analysis of the rescued plasmids showed that one insertion is in the Gα-encoding gpa2+ gene (Isshiki et al. 1992; Nocero et al. 1994) while the second insertion is in the gene designated SPBC21C3.20c in the S. pombe genomic sequence (Wood et al. 2002). An earlier genetic screen had identified spontaneous Git− mutant strains carrying mutations affecting a collection of genes, git1+–git10+, which confer the ability to form colonies on SC–ura medium as seen in these insertion mutant strains. Git− mutant strains also display 5-FOA-sensitive (5-FOAS) growth (Hoffman and Winston 1990), unlike the 5-FOA-resistant (5-FOAR) phenotype of cells able to glucose repress expression of the fbp1-ura4+ reporter. Eight of these git genes play a significant role in fbp1+ repression, with only git1+ and git10+ remaining to be cloned. We therefore carried out linkage and complementation tests to determine whether SPBC21C3.20c represents either git1+ or git10+. The pAF1 insertion allele of SPBC21C3.20c fails to complement a git1-1 mutant allele in the diploid strains while complementing a git2 deletion (Figure 1) and a git10-201 mutant allele (data not shown). Consistent with this, all 56 tetrads produced from a cross of a SPBC21C3.20c-insertion mutant (CHP857) with a git1-1 mutant (CHP569) were parental ditypes (0:4 5-FOAR:5-FOAS progeny), indicating that the insertion occurred within 0.9 cM of the git1-1 mutation. We conclude that git1+ is SPBC21C3.20c.
Figure 1.
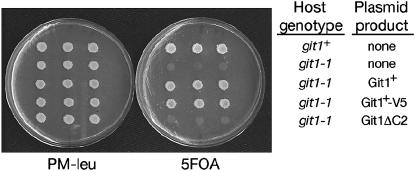
Plasmid pAF1 (his3+) insertion mutation is in the git1+ complementation group. Diploid strains were constructed using intragenically complementing ade6-M210 and ade6-M216 mutant alleles to form Ade+ diploids. Diploid strains were spotted onto 5-FOA medium and grown for 3 days at 30°. 5-FOAR growth reflects the wild-type ability to glucose repress transcription of an fbp1-ura4+ reporter gene.
Using sequence information from the S. pombe genome project (Wood et al. 2002), we designed PCR primers to amplify and clone git1+, creating plasmid pRSK3. As expected, plasmid pRSK3 restores glucose repression of fbp1-ura4 expression to strain CHP568 (git1-1; Figure 2), conferring 5-FOAR growth. These transformants also glucose repress fbp1-lacZ expression as judged by β-galactosidase assays (Table 2). Similar results were observed when the plasmid-expressed Git1 protein contained a C-terminal V5-6his tag, indicating that the Git1+-V5his6 protein expressed by plasmid pRSK4 is functional (Table 2, Figure 2). Plasmid pRSK3 was also introduced into strains carrying mutations in git3+ (GPCR), gpa2+ (Gα), git7+ (Sgt1 family member), git10+, or pka1+ (catalytic subunit of PKA). These transformants remain 5-FOA sensitive, indicating that git1+ cannot act as a multicopy suppressor of mutations in any of these genes (data not shown). In contrast, plasmid pRSK3 partially suppresses certain mutations in the git2+/cyr1+ adenylate cyclase gene. The git2-7 and git2-216 mutations previously have been shown to intragenically complement the git2-61 mutation (Hoffman and Winston 1990, 1991). Surprisingly, multicopy git1+ restores some degree of 5-FOAR growth to host strains carrying any of these three mutations (Figure 3) and reduces fbp1-lacZ expression, but does not fully restore it to the glucose-repressed levels seen in wild-type transformants. [β-Galactosidase values for FWP112 (git+) transformants carrying the pNMT empty vector were 21 ± 3 units, while transformants carrying pRSK3 were 15 ± 5.] The git2-216 transformants carrying pRSK3 display an apparent contradiction in terms of their 5-FOAR growth, yet relatively high β-galactosidase activity (Figure 3). This may be due to greater heterogeneity in these transformants with regard to fbp1-driven expression. A subset of cells may repress fbp1-ura4+ expression to a level permitting growth on 5-FOA, while other cells of the same transformant may express the fbp1-lacZ reporter at a level high enough to produce the observed average β-galactosidase value for the culture that is similar to or greater than that seen in some 5-FOAS transformants.
Figure 2.
Complementation of a git1-1 mutation by plasmid-expressed git1+. Strains FWP72 (git1+) and CHP568 (git1-1) were transformed to Leu+ with pNMT41 (empty vector), pRSK3 (git1+), pRSK4 (git1-V5his6), or pRSK8 (git1ΔC2). Three independent transformants of each host strain and plasmid combination indicated in the figure were spotted to PM–leu and then replica plated after 2 days to either PM–leu or 5-FOA media. Plates were photographed after 3 days at 30°.
TABLE 2.
Glucose repression of fbp1-lacZ expression in transformants
| Strain (relevant genotype) | Plasmid (expressed gene) | β-Gal activity |
|---|---|---|
| FWP72 (git1+) | pNMT41 (none) | 10 ± 1 |
| CHP568 (git1-1) | pNMT41 (none) | 1313 ± 16 |
| CHP568 (git1-1) | pRSK3 (git1+) | 37 ± 6 |
| CHP568 (git1-1) | pRSK4 (git1-V5his6) | 31 ± 4 |
| CHP568 (git1-1) | pRSK8 (git1ΔC2) | 729 ± 32 |
| FWP72 (git1+) | pRSK8 (git1ΔC2) | 47 ± 10 |
β-Galactosidase activity was determined from at least two independent transformants as described in materials and methods. The values represent the average ± standard deviation and are given as specific activity per milligram of soluble protein.
Figure 3.
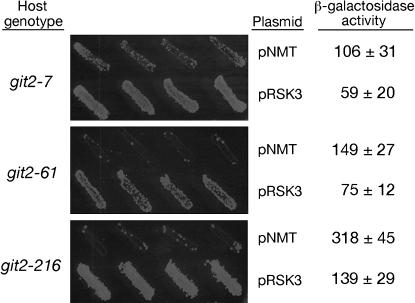
Multicopy suppression of git2− mutations by plasmid-expressed git1+. Host strains CHP7 (git2-7), CHP61 (git2-61), and CHP216 (git2-216) were transformed to Leu+ with pNMT41 (empty vector) or pRSK3 (git1+). Four independent transformants of each host strain and plasmid combination indicated in the figure were plated onto EMM–leu and then replica plated after 1 day to either EMM–leu or 5-FOA media. 5-FOA plates were photographed after 5 days at 30°. All transformants grew equally well on EMM–leu medium (not shown). β-Galactosidase activity was determined as described in materials and methods. The values given represent specific activity ± standard deviation from three independent transformants.
Git1+ is a C2 domain-containing protein:
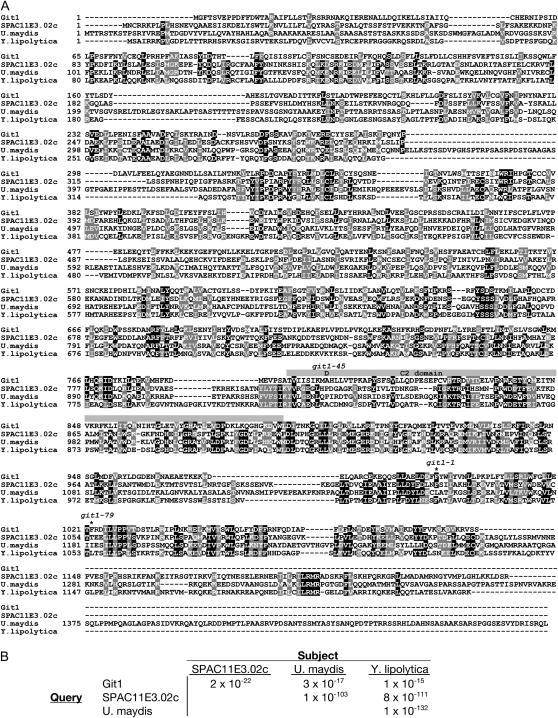
The git1+ gene encodes a 1098-amino-acid protein with only modest sequence similarity to other proteins in the GenBank database (Figure 4A). A BLASTP search (Altschul et al. 1990) using the predicted Git1 protein as the query sequence (with no filter) reveals a group of eight fungal proteins that align with E-values in the range of 10−23–10−12. However, these proteins align to each other with E-values ≤10−52 (Figure 4B), suggesting that Git1 function may be distinct from that of these other proteins. One of these proteins, SPAC11E3.02c, is also from S. pombe, while the remaining seven proteins are from various fungi. A BLASTP search using SPAC11E3.02c as the query sequence reveals a somewhat larger family of proteins that includes S. cerevisiae Yor296w (E-value 5 × 10−37), a protein of unknown function. Yor296w is not returned in a BLASTP search using Git1 as the query protein, consistent with the impression that Git1 is only distantly related to the SPAC11E3.02c/Yor296w protein family. Finally, the SPAC11E3.02c-based search reveals that the UNC-13/Munc13 protein family is very distantly related to these fungal proteins (E-values of 10−4–10−6). In fact, SPAC11E3.02c and Yor296w had been previously identified as possessing both a C2 domain related to that of the UNC-13/Munc13 protein family and two conserved regions referred to as Munc13-homology domains 1 and 2 (MHD1 and MHD2; Koch et al. 2000). MHD1 and MHD2 are also moderately conserved in Git1, representing residues 585–700 and 980–1098, respectively (Figure 4A).
Figure 4.
Alignment of Git1 with SPAC11E3.02c and related proteins from Ustilago maydis and Yarrowia lipolytica. (A) Alignment of S. pombe proteins Git1 (NP_596600) and SPAC11E3.02c (NP_594926) with related proteins from U. maydis (XP_400071) and Y. lipolytica (XP_501312) using the ClustalW sequence alignment program (Thompson et al. 1994) and displayed using BOXSHADE. Identities are shown against a solid background and similarities are shown against a shaded background. A shaded box above the sequences denotes the predicted C2 domain of Git1. The alanine-to-aspartic-acid missense mutation in git1-45 is shown above residue 799 of the Git1 sequence. Asterisks above the Git1 sequence at residues 995 and 1022 denote the last correctly translated residues expressed from alleles git1-1 and git1-79, respectively. (B) E-values associated with alignments from unfiltered BLASTP searches involving the proteins shown in A.
To determine the relative roles of git1+ and SPAC11E3.02c in glucose signaling, we deleted each gene from strain FWP72 and examined the effect on the fbp1-driven reporter constructs. The git1 deletion (git1Δ) confers 5-FOAS growth, similar to that seen in Figure 1, and elevated β-galactosidase expression (Table 3) in glucose-grown cells due to the constitutive expression of the fbp1-ura4 and fbp1-lacZ reporters. Conversely, the SPAC11E3.02c deletion has no effect on expression of either reporter and does not exacerbate the defect in fbp1 regulation in a git1Δ strain (Table 3 and data not shown). Thus, SPAC11E3.02c is not functionally redundant with Git1, and we assume that the proteins closely related to SPAC11E3.02c are not functional homologs of Git1+.
TABLE 3.
Defect in glucose repression of fbp1-lacZ expression in mutant strains
| β-Galactosidase activity
|
|||
|---|---|---|---|
| Strain | Relevant genotype | 8% glucose | 8% glucose + cAMP |
| FWP72 | Wild type | 15 ± 8 | ND |
| RKP2 | git1Δ | 834 ± 128 | 12 ± 2 |
| RKP15 | SPAC11E3.02cΔ | 13 ± 5 | ND |
| RKP62a | git1Δ SPAC11E3.02cΔ | 776 ± 81 | ND |
| CHP889 | gpa2R176H | 18 ± 10 | ND |
| RKP9 | git1Δ gpa2R176H | 705 ± 123 | ND |
Cells were grown in YEL to exponential phase under repressing conditions (8% glucose) and assayed as described in materials and methods. The values given represent specific activity ± standard deviation from three or four independent cultures of each strain. ND, not determined.
RKP62 values are based on an average of assays of strains RKP62, RKP66, and RKP67, which are genetically identical.
Both the Git1 C2 domain and MHD2 are required for function:
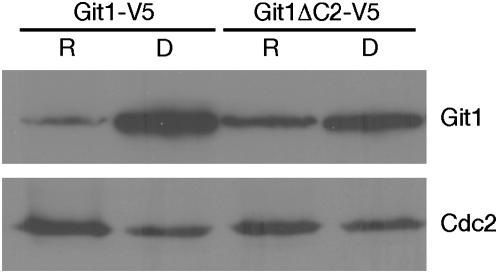
As described above, Git1 contains a putative C2 domain (amino acids 791–889) as well as two potential Munc homology domains (Figure 4A). C2 domains are found in proteins involved in signal transduction such as protein kinase C where they bind phospholipids in a calcium-dependent manner. However, some C2 domains in other proteins neither are calcium dependent nor bind phospholipids (Rizo and Sudhof 1998). To determine if the Git1 C2 domain is required for Git1 function, we used site-directed mutagenesis to delete codons 826–839, predicted to encode a pair of antiparallel β-strands from the git1+ ORF and tested whether this protein can provide Git1 function when expressed from a plasmid. As shown in Figure 2 and Table 2, the Git1ΔC2 protein is largely nonfunctional, failing to restore 5-FOAR growth and glucose repression of fbp1-lacZ expression to a git1-1 host strain, although a modest reduction of fbp1-lacZ expression is observed. In addition, Git1ΔC2 does not confer a dominant-negative phenotype when expressed in FWP72 (git1+) cells (Table 2). To determine whether the loss of function is due to destabilization of the protein, we expressed both Git1+-V5 and Git1ΔC2-V5 proteins from plasmids pRSK4 and pRSK9 in FWP72 (git1+) cells. We observe that the Git1+-V5 protein level is regulated by the carbon source, with 11.5-fold less protein present in glucose-grown cells than in glucose-starved cells (Figure 5). While the Git1ΔC2-V5 protein level is also regulated by glucose, there is only a threefold reduction in glucose-grown cells (Figure 5). As Git1ΔC2-V5 expressed from plasmid pRSK9 is readily detectable, the loss of the C2 domain does not simply destabilize the protein, suggesting a functional role for the Git1 C2 domain. In addition, the presence of the C2 domain appears to be important for the post-translational regulation controlling the level of Git1 in the cell as a function of glucose signaling.
Figure 5.
Git1 protein levels are regulated by glucose conditions. Western blot analysis of protein extracts from FWP72 (Git+) transformants carrying plasmid pRSK4 (Git1+-V5) or plasmid pRSK9 (Git1ΔC2-V5). Cells were grown under glucose-repressing (R) or derepressing (D; glucose-starved) conditions. Immunoblots were carried out using either α-V5 or α-Cdc2 (as a loading control) antibody as described in materials and methods.
The git1+ gene was notable in our original selection for spontaneous mutations that conferred constitutive fbp1+ transcription (Hoffman and Winston 1990) in that almost 40% of the independently isolated mutations were in git1+. While Git1 is a moderately large protein of 1098 residues, this cannot fully account for the number of git1− mutants identified. A DNA sequence analysis of these strains explains the high frequency of mutations and supports the idea that the C2 domain and the MHD2 domain are important for Git1 function. The git1-1 allele contains a +1 frameshift in which a sequence of eight thymidine nucleotides in the coding strand acquires an additional thymidine, leading to the loss of the C-terminal 103 residues of the protein (Figure 4A). This suggests a functional role for the MHD2 domain, although we have not determined whether the truncated protein is stable. In addition, PCR analysis of the original 73 git1 mutant alleles found that 57 alleles possess an additional base pair in a 64-bp region that includes the git1-1 mutation, while direct sequencing of git1-9, git1-46, and git1-99 confirms that they possess the same frameshift as in git1-1 (data not shown). As these mutant strains were isolated independently, this frameshift is a high-frequency event relative to other spontaneous mutations, explaining the large number of git1− strains isolated in the original study. An additional allele, git1-287, contains a −1 frameshift in this same run of thymidine residues. It is worth noting that high-frequency frameshift mutations have also been identified in A-T-rich sequences within the S. cerevisiae IRA1 and IRA2 genes (Halme et al. 2004).
Of the remaining git1− mutant alleles, git1-79 and git1-82 contain a nonsense mutation causing the loss of the C-terminal 76 residues of the protein, further supporting the notion that the MHD2 domain is important for Git1 function. Several other frameshift or nonsense mutations cause larger truncations of the protein and are thus uninformative. Only the git1-45 allele contains a missense mutation, changing an alanine to an aspartic acid at residue 799 within the C2 domain. β-Galactosidase assays of git1-79 and git1-45 mutant strains demonstrate that fbp1-lacZ expression is derepressed to a similar degree in a git1Δ strain (data not shown), indicating that both the C2 and the MHD2 domains are critical for Git1 function.
Analysis of git1Δ strains:
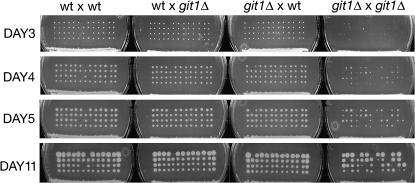
To fully assess the role of Git1 in fbp1+ transcriptional regulation and glucose detection, we constructed a complete deletion of the git1+ gene (git1Δ) and characterized the effect of this deletion on several cAMP-regulated processes. As the loss of several genes in the PKA pathway results in a spore germination delay (Welton and Hoffman 2000), we examined whether git1Δ spores display a similar delay. As shown in Figure 6, in git1+ × git1Δ crosses both git1+ and git1Δ spores germinate to form clearly visible colonies by 3 days. However, in a git1Δ × git1Δ cross, spore germination is more stochastic with relatively few colonies forming within 3 days. Most colonies grew to a visible size within 4–5 days, while others took over a week to form (Figure 6). These differences are due to delays in germination rather than reduced growth rates as judged by daily microscopic examination of the spores and colonies on the dissection plates. In addition, exponential phase cells of strain 972 (git1+) display a doubling time of 2.47 ± 0.03 hr in YEL medium and 3.13 ± 0.13 hr in EMM medium, while CHP973 (git1Δ) cells display a doubling time of 2.62 ± 0.08 hr in YEL medium and 4.32 ± 0.79 hr in EMM medium. The lack of a germination delay in git1Δ spores from a git1+ × git1Δ cross suggests either that sufficient Git1 for germination is packaged in the spore during sporulation or that Git1 carries out its function on a target that is then packaged into the spore.
Figure 6.
Spore germination delay in git1Δ by git1Δ cross. Thirteen tetrads from each cross were dissected on yeast extract agar plates and incubated at 30°. Photographs were taken on days 3, 4, 5, and 11. Crosses (from left to right) are of strains FWP95 (git1+) × FWP87 (git1+), FWP95 × CHP954 (git1Δ), RKP2 (git1Δ) × FWP87, and RKP2 × CHP954.
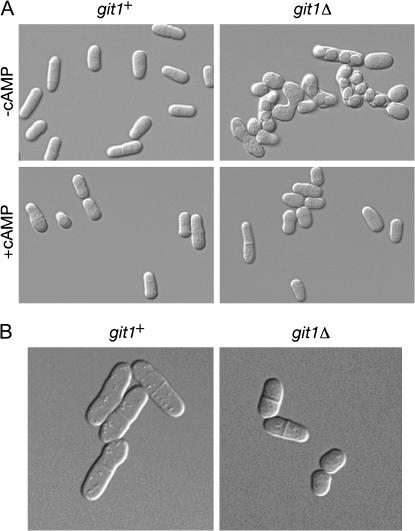
The git1Δ strains were examined for other phenotypes associated with defects in the cAMP pathway. These include a reduced cell length at the time of septation (Jin et al. 1995) and the acquisition of starvation-independent conjugation and sporulation (Dal Santo et al. 1996; Landry et al. 2000; Welton and Hoffman 2000; Landry and Hoffman 2001; Schadick et al. 2002), along with the previously described loss of glucose repression of transcription of an fbp1-lacZ reporter (Hoffman and Winston 1990, 1991). As shown in Figure 7A, homothallic git1Δ cells conjugate and sporulate in nutrient-rich medium. Addition of 5 mm cAMP suppresses the conjugation phenotype (Figure 7A), consistent with ability of exogenous cAMP to suppress the constitutive fbp1-lacZ expression in RKP2 (git1Δ) cells (Table 2). We also measured the cell length of wild-type and git1Δ cells that display septa in exponential-phase cultures. By measuring the first 10 septated cells observed by microscopy (see Figure 7B for examples of such cells), we found that strain 972 (git1+) cells produce septa at an average length of 15.6 ± 1.6 μm, while strain CHP973 (git1Δ) cells produce septa at an average length of 11.7 ± 0.8 μm. Thus, loss of Git1 leads to a significant reduction in the length of septated cells.
Figure 7.
git1Δ cells display defects associated with the loss of cAMP signaling. (A) Homothallic (h90) strains CHP795 (git1+) and CHP896 (git1Δ) were grown to exponential phase in PM liquid medium (at 37° to inhibit conjugation) and then diluted to 106 cells/ml in PM liquid medium in the presence or absence of 5 mm cAMP. These cells were incubated for 24 hr at 30° without shaking and photographed. (B) Strains 972 (git1+) and CHP973 (git1Δ) were grown to exponential phase in YEL. Cells displaying septa were measured and photographed.
To directly test for a defect in cAMP signaling, we assayed the cAMP response to glucose in wild-type and git1Δ cells (Table 4). Relative to wild-type cells, the git1Δ cells display both a modest reduction in basal cAMP levels under glucose-starvation conditions and a total loss of a cAMP response to glucose addition. Therefore, Git1 is essential to glucose/cAMP signaling in S. pombe.
TABLE 4.
cAMP response to glucose in git1+ and git1Δ cells
| cAMP (pmol)/mg protein
|
|||||
|---|---|---|---|---|---|
| Strain | Genotype | T = 0′ | T = 1′ | T = 5′ | T = 10′ |
| FWP79 | git1+ | 3.6 ± 0.6 | 13.9 ± 1.4 | 16.2 ± 5.1 | 16.8 ± 3.9 |
| CHP973 | git1Δ | 2.7 ± 0.6 | 2.6 ± 0.4 | 2.6 ± 0.6 | 2.7 ± 0.7 |
Assays were carried out on glucose-starved cultures (T = 0′), as well as on the same cells, 1, 5, and 10 min after the addition of glucose to a final concentration of 100 mm as previously described (Byrne and Hoffman 1993) using the cAMP [125I]Biotrak Assay System (Amersham Biosciences).
Git1 physically interacts with adenylate cyclase, but is not required for adenylate cyclase levels or localization:
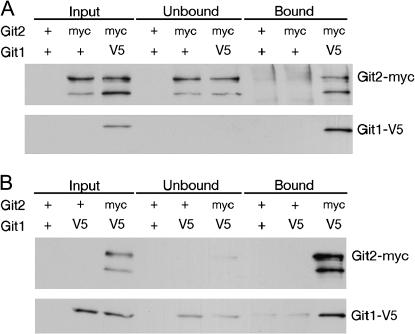
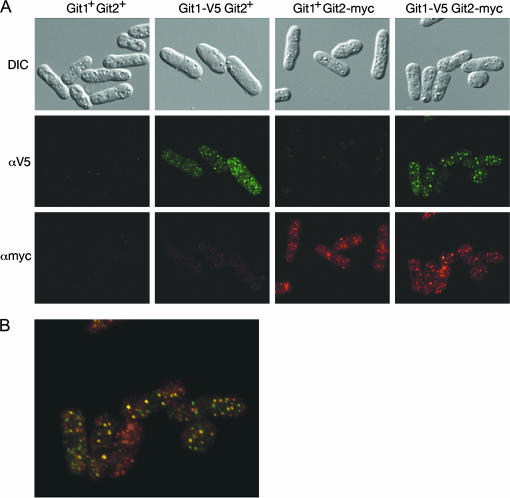
To investigate of the role of Git1 in the glucose/cAMP pathway, we examined whether or not Git1 binds the Git2 adenylate cyclase enzyme. Protein extracts from cells expressing myc-tagged Git2 and V5-tagged Git1 were subjected to co-immunoprecipitations. In reciprocal experiments, we found that immunoprecipitation of Git1-V5 leads to coprecipitation of Git2-myc (Figure 8A) and that immunoprecipitation of Git2-myc leads to coprecipitation of Git1-V5 (Figure 8B). Consistent with a physical interaction between Git1 and adenylate cyclase, indirect immunofluorescence of Git1-V5 and Git2-myc demonstrates that both proteins display a punctate cytoplasmic staining pattern and partially colocalize within cells (Figure 9).
Figure 8.
Git1 and Git2 (adenylate cyclase) physically interact. (A) Immunoblot of protein extracts from α-V5 immunoprecipitation. Protein extracts were prepared from strains expressing tagged (Git2-myc and Git1-V5) or untagged proteins (+) as indicated. Crude extracts, along with fractions that bound or failed to bind α-V5 antibodies, were probed with α-myc to detect Git2-myc and with α-V5 to detect Git1-V5. (B) Immunoblot of protein extracts from α-myc immunoprecipitation. Protein extracts were prepared from strains expressing tagged or untagged proteins as indicated. Crude extracts, along with fractions that bound or failed to bind α-myc antibodies were probed with α-V5 to detect Git1-V5 and with α-myc to detect Git2-myc.
Figure 9.
Partial colocalization of Git1 and Git2. (A) DIC and fluorescent images of cells expressing tagged and untagged forms of Git1 and Git2 (adenylate cyclase) as indicated. Fluorescent images were taken with the same exposure time for each sample. (B) Merged fluorescent image from A of cells expressing Git1-V5 and Git2-myc.
As Git1 is required for glucose/cAMP signaling even in cells expressing the mutationally activated Gpa2R176H Gα-subunit (Table 3), we examined whether Git1 controls either adenylate cyclase stability or localization. Indirect immunofluorescence of Git2-myc produces a punctate staining pattern in a git1Δ strain that is indistinguishable from that seen in git1+ cells with regard to localization and intensity (data not shown). Similarly, immunoblots of extracts made from git1Δ cells expressing Git2-myc show little or no change in the level of Git2-myc in both glucose-grown and glucose-starved cells (data not shown). Therefore, the loss of Git1 does not lead to the destabilization of adenylate cyclase or to a gross change in adenylate cyclase localization within the cell. As discussed below, it remains possible that Git1 is involved in a transient localization event required for adenylate cyclase activation.
DISCUSSION
The fission yeast S. pombe utilizes a cAMP-signaling pathway as the primary mechanism for sensing environmental glucose to regulate transcription of genes involved in gluconeogenesis and sexual development. In this study, we describe the cloning and characterization of the git1+ gene, which is required for glucose-triggered adenylate cyclase activation.
The most surprising aspect of this work is the identity of git1+ itself, as it lacks any obvious functional homologs in other organisms. Prior to this study, work in S. cerevisiae and S. pombe glucose/cAMP signaling had uncovered pathways whose components display considerable conservation, although several key differences exist (see review by Hoffman 2005a and references therein). Both yeasts express seven-transmembrane GPCRs that activate G-proteins, which are involved in adenylate cyclase activation. However, the S. cerevisiae Gpr1 GPCR is coupled to the Gpa2 Gα-subunit that lacks a traditional Gβγ dimer, interacting instead with a Gβγ mimic that negatively regulates the pathway (Harashima and Heitman 2002; Batlle et al. 2003). In addition, glucose signaling through Ras proteins occurs in concert with Gpa2-mediated signaling to play a central role in S. cerevisiae adenylate cyclase activation. In contrast, the S. pombe Git3 GPCR functions together with a heterotrimeric G-protein, whose Gpa2 Gα-subunit directly binds an N-terminal region of adenylate cyclase (Ivey and Hoffman 2005) that is distinct from the Ras-activation domain in the S. cerevisiae enzyme (Suzuki et al. 1993), although another study suggests that S. pombe Gpa2 acts through an adenylate cyclase domain that is homologous to the Ras-activation domain (Ogihara et al. 2004). In addition, the only Ras homolog in S. pombe plays no role in adenylate cyclase activation. Finally, S. cerevisiae Sgt1 and S. pombe Git7 are homologous cochaperone proteins involved in cAMP signaling (Dubacq et al. 2002; Schadick et al. 2002), while carrying out other essential functions as well. The Git1 protein is unusual in that while it is critical for glucose-triggered adenylate cyclase activation in S. pombe, it may be functionally unique to fission yeast.
As seen in Figure 4A, Git1 is distantly related to a small set of proteins that includes a second S. pombe protein, SPAC11E3.02c. Deletion of the SPAC11E3.02c gene has no effect on fbp1-lacZ expression and no additive effect when combined with a git1Δ allele; therefore this gene does not function in the glucose/cAMP pathway. As the remaining proteins of this family are much more closely related to SPAC11E3.02c than to Git1+ (Figure 4B), we assume that they represent functional homologs of SPAC11E3.02c and not Git1. The requirement for a novel protein such as Git1 by S. pombe adenylate cyclase is unexpected due to the high degree of structural conservation among yeast and fungal adenylate cyclase enzymes and may point to a unique regulatory mechanism in fission yeast adenylate cyclase. Alternatively, the Git1 function may be carried out in other fungi by a protein with little sequence similarity to Git1.
While the 1098-amino-acid Git1 protein displays some similarity to a family of large fungal proteins, ranging from 1185 to 1472 residues, the majority of the conserved sequences are found in the C-terminal half of Git1. This includes a 100-amino-acid C2 domain (Rizo and Sudhof 1998), which may promote an interaction with phospholipids, flanked by two additional regions of conservation. Interestingly, two proteins in this family, S. pombe SPAC11E3.02c and S. cerevisiae Yor296w, were previously cited in a study identifying a family of proteins related to the Munc13 protein, which is involved in vesicle priming for neurotransmitter secretion (Koch et al. 2000). Along with the C2 domain, these proteins were defined by the presence of MHD1 and MHD2, which correspond to the sequences before and after the C2 domain in the fungal proteins. Recently, it has been shown that the MHD1–MHD2–C2 domain region (the MHD2 domain precedes the C2 domain in the Munc13 proteins) is sufficient for function of both Caenorhabditis elegans UNC-13 (Madison et al. 2005) and mouse Munc13 (Stevens et al. 2005). Sequence analysis of git1− mutant alleles, along with site-directed mutagenesis of the C2 domain, demonstrates that the C2 and MHD2 domains are required for Git1 function. Therefore, while Git1 may be functionally distinct from these fungal and metazoan proteins, Git1 and these other proteins may act through a similar biochemical mechanism.
The level of Git1 protein is lower in glucose-grown cells than in glucose-starved cells (Figure 5), which resembles the glucose-mediated regulation of other components of the pathway. We previously showed that transcription of cgs1+ (PKA regulatory subunit) and pka1+ (PKA catalytic subunit) is repressed by glucose in a PKA-dependent manner (Stiefel et al. 2004). In addition, there is significantly less Git3 GPCR and adenylate cyclase protein in glucose-grown cells than in glucose-starved cells (D. Chandler-Militello, L. Wang and C. S. Hoffman, unpublished results). Removal of the C2 domain in the Git1ΔC2-V5 protein abolishes much of this regulation, even in a wild-type strain that retains a functional cAMP pathway (Figure 5). Thus, the C2 domain may be required to assemble Git1 into a complex whose abundance is the target of feedback regulation by PKA.
Co-immunoprecipitation experiments using myc-tagged adenylate cyclase and the Git1-V5 protein demonstrate that these proteins either directly or indirectly interact (Figure 8). In contrast, Git7 (Schadick et al. 2002) and Git1 do not co-immunoprecipitate, suggesting that these two proteins carry out distinct functions in the cAMP pathway (M. Alamery and C. S. Hoffman, unpublished results). Consistent with the co-precipitation results, indirect immunofluorescence detection of Git1 and Git2 results in a punctate staining pattern with some colocalization (Figure 9). Thus, Git1 appears to be present in a complex with adenylate cyclase, playing a direct role in adenylate cyclase activation. Such a direct role for Git1 is consistent with the reduction in basal cAMP levels and the total absence of a cAMP response observed in git1Δ cells (Table 4). This is also consistent with the observation that Git1 is required for fbp1 repression, even in a strain expressing a mutationally activated form of the Gpa2 Gα, which also binds adenylate cyclase (Table 3). A direct role for Git1 in adenylate cyclase activation is also supported by the ability of Git1 overexpression to partially suppress mutations in the git2+ adenylate cyclase gene (Figure 3). Prompted by the physical and genetic interactions between Git1 and adenylate cyclase, we examined whether Git1 is required either for adenylate cyclase localization or for stability, but found no significant change in adenylate cyclase levels or localization in a git1Δ strain (data not shown). However, our localization experiments show little or no adenylate cyclase associated with the plasma membrane (Figure 9), which is where the Git3 GPCR is localized (D. Chandler-Militello, D. A. Kelly and C. S. Hoffman, unpublished results). While this appears to differ from the situation in budding yeast, biochemical studies of adenylate cyclase in S. cerevisiae show that ∼50% of the protein is soluble (Mitts et al. 1990). There has not been a similar analysis of budding yeast adenylate cyclase localization by microscopy, presumably due to the low abundance of the protein. In fact, we were able to detect our tagged protein due to the use of a 13myc epitope (GFP fusions expressed from the genomic locus do not produce a visible signal; K. Cummings, L. Wang, and C. S. Hoffman, unpublished results). It remains possible that Git1 is required to shuttle adenylate cyclase to the plasma membrane to allow Gpa2-mediated activation, which could be related to the Munc13 role in priming vesicles for membrane fusion. Future experiments will address the mechanism by which Git1 facilitates adenylate cyclase activation in response to glucose detection in fission yeast.
Acknowledgments
We thank David Burgess for advice on microscopy and members of the Hoffman lab for insightful discussions and critical evaluation of the manuscript. This work was supported by National Institutes of Health grant R01 GM46226 to C.S.H.
References
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers and D. J. Lipman, 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Amoah-Buahin, E., N. Bone and J. Armstrong, 2005. Hyphal growth in the fission yeast Schizosaccharomyces pombe. Eukaryot. Cell 4: 1287–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie, 3rd et al., 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951. [DOI] [PubMed] [Google Scholar]
- Batlle, M., A. Lu, D. A. Green, Y. Xue and J. P. Hirsch, 2003. Krh1p and Krh2p act downstream of the Gpa2p Gα subunit to negatively regulate haploid invasive growth. J. Cell Sci. 116: 701–710. [DOI] [PubMed] [Google Scholar]
- Byrne, S. M., and C. S. Hoffman, 1993. Six git genes encode a glucose-induced adenylate cyclase activation pathway in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 105: 1095–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo, S., D. Ronchetti, J. M. Thevelein, J. Winderickx and E. Martegani, 2004. Activation state of Ras2 protein and glucose-induced signalling in Saccharomyces cerevisiae. J. Biol. Chem. 279: 46715–46722. [DOI] [PubMed] [Google Scholar]
- Dal Santo, P., B. Blanchard and C. S. Hoffman, 1996. The Schizosaccharomyces pombe pyp1 protein tyrosine phosphatase negatively regulates nutrient monitoring pathways. J. Cell Sci. 109: 1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVoti, J., G. Seydoux, D. Beach and M. McLeod, 1991. Interaction between ran1+ protein kinase and cAMP dependent protein kinase as negative regulators of fission yeast meiosis. EMBO J. 10: 3759–3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubacq, C., R. Guerois, R. Courbeyrette, K. Kitagawa and C. Mann, 2002. Sgt1p contributes to cyclic AMP pathway activity and physically interacts with the adenylyl cyclase Cyr1p/Cdc35p in budding yeast. Eukaryot. Cell 1: 568–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan, I. M., and J. S. Hyams, 1988. The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 89: 343–357. [DOI] [PubMed] [Google Scholar]
- Halme, A., S. Bumgarner, C. Styles and G. R. Fink, 2004. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell 116: 405–415. [DOI] [PubMed] [Google Scholar]
- Harashima, T., and J. Heitman, 2002. The Gα protein Gpa2 controls yeast differentiation by interacting with kelch repeat proteins that mimic Gβ subunits. Mol. Cell 10: 163–173. [DOI] [PubMed] [Google Scholar]
- Harashima, T., and J. Heitman, 2004. Nutrient control of dimorphic growth in Saccharomyces cerevisiae, pp. 131–169 in Nutrient Induced Responses in Eukaryotic Cells, edited by J. Winderickx and P. M. Taylor. Springer-Verlag, Berlin/Heidelberg, Germany/New York.
- Hoffman, C. S., 2005. a Except in every detail: comparing and contrasting G-protein signaling in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Eukaryot. Cell 4: 495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman, C. S., 2005. b Glucose sensing via the protein kinase A pathway in Schizosaccharomyces pombe. Biochem. Soc. Trans. 33: 257–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman, C. S., and R. Welton, 2000. Mutagenesis and gene cloning in Schizosaccharomyces pombe via nonhomologous plasmid integration and rescue. Biotechniques 28: 532–536, 538, 540. [DOI] [PubMed] [Google Scholar]
- Hoffman, C. S., and F. Winston, 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57: 267–272. [DOI] [PubMed] [Google Scholar]
- Hoffman, C. S., and F. Winston, 1990. Isolation and characterization of mutants constitutive for expression of the fbp1 gene of Schizosaccharomyces pombe. Genetics 124: 807–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman, C. S., and F. Winston, 1991. Glucose repression of transcription of the Schizosaccharomyces pombe fbp1 gene occurs by a cAMP signaling pathway. Genes Dev. 5: 561–571. [DOI] [PubMed] [Google Scholar]
- Hudson, D. A., Q. L. Sciascia, R. J. Sanders, G. E. Norris, P. J. Edwards et al., 2004. Identification of the dialysable serum inducer of germ-tube formation in Candida albicans. Microbiology 150: 3041–3049. [DOI] [PubMed] [Google Scholar]
- Isshiki, T., N. Mochizuki, T. Maeda and M. Yamamoto, 1992. Characterization of a fission yeast gene, gpa2, that encodes a Gα subunit involved in the monitoring of nutrition. Genes Dev. 6: 2455–2462. [DOI] [PubMed] [Google Scholar]
- Ivey, F. D., and C. S. Hoffman, 2005. Direct activation of fission yeast adenylate cyclase by the Gpa2 Gα of the glucose signaling pathway. Proc. Natl. Acad. Sci. USA 102: 6108–6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janoo, R. T., L. A. Neely, B. R. Braun, S. K. Whitehall and C. S. Hoffman, 2001. Transcriptional regulators of the Schizosaccharomyces pombe fbp1 gene include two redundant Tup1p-like corepressors and the CCAAT binding factor activation complex. Genetics 157: 1205–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, M., M. Fujita, B. M. Culley, E. Apolinario, M. Yamamoto et al., 1995. sck1, a high copy number suppressor of defects in the cAMP-dependent protein kinase pathway in fission yeast, encodes a protein homologous to the Saccharomyces cerevisiae SCH9 kinase. Genetics 140: 457–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, M., and J. H. Kim, 2005. Glucose as a hormone: receptor-mediated glucose sensing in the yeast Saccharomyces cerevisiae. Biochem. Soc. Trans. 33: 247–252. [DOI] [PubMed] [Google Scholar]
- Koch, H., K. Hofmann and N. Brose, 2000. Definition of Munc13-homology-domains and characterization of a novel ubiquitously expressed Munc13 isoform. Biochem. J. 349: 247–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchin, S., V. K. Vyas and M. Carlson, 2002. Snf1 protein kinase and the repressors Nrg1 and Nrg2 regulate FLO11, haploid invasive growth, and diploid pseudohyphal differentiation. Mol. Cell. Biol. 22: 3994–4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry, S., and C. S. Hoffman, 2001. The git5 Gβ and git11 Gγ form an atypical Gβγ dimer acting in the fission yeast glucose/cAMP pathway. Genetics 157: 1159–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry, S., M. T. Pettit, E. Apolinario and C. S. Hoffman, 2000. The fission yeast git5 gene encodes a Gβ subunit required for glucose-triggered adenylate cyclase activation. Genetics 154: 1463–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire, K., S. Van de Velde, P. Van Dijck and J. M. Thevelein, 2004. Glucose and sucrose act as agonist and mannose as antagonist ligands of the G protein-coupled receptor Gpr1 in the yeast Saccharomyces cerevisiae. Mol. Cell 16: 293–299. [DOI] [PubMed] [Google Scholar]
- Lengeler, K. B., R. C. Davidson, C. D'Souza, T. Harashima, W. C. Shen et al., 2000. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64: 746–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz, M. C., X. Pan, T. Harashima, M. E. Cardenas, Y. Xue et al., 2000. The G protein-coupled receptor Gpr1 is a nutrient sensor that regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Genetics 154: 609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison, J. M., S. Nurrish and J. M. Kaplan, 2005. UNC-13 interaction with syntaxin is required for synaptic transmission. Curr. Biol. 15: 2236–2242. [DOI] [PubMed] [Google Scholar]
- Mitts, M. R., D. B. Grant and W. Heideman, 1990. Adenylate cyclase in Saccharomyces cerevisiae is a peripheral membrane protein. Mol. Cell. Biol. 10: 3873–3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nocero, M., T. Isshiki, M. Yamamoto and C. S. Hoffman, 1994. Glucose repression of fbp1 transcription of Schizosaccharomyces pombe is partially regulated by adenylate cyclase activation by a G protein α subunit encoded by gpa2 (git8). Genetics 138: 39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogihara, H., F. Shima, K. Naito, T. Asato, K. Kariya et al., 2004. Direct activation of fission yeast adenylyl cyclase by heterotrimeric G protein gpa2. Kobe J. Med. Sci. 50: 111–121. [PubMed] [Google Scholar]
- Rizo, J., and T. C. Sudhof, 1998. C2-domains, structure and function of a universal Ca2+-binding domain. J. Biol. Chem. 273: 15879–15882. [DOI] [PubMed] [Google Scholar]
- Schadick, K., H. M. Fourcade, P. Boumenot, J. J. Seitz, J. L. Morrell et al., 2002. Schizosaccharomyces pombe Git7p, a member of the Saccharomyces cerevisiae Sgtlp family, is required for glucose and cyclic AMP signaling, cell wall integrity, and septation. Eukaryot. Cell 1: 558–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern, J. A., D. F. Young, F. Heaney, W. K. Baumgartner and R. E. Randall, 1991. Identification of an epitope on the P and V proteins of simian virus 5 that distinguishes between two isolates with different biological characteristics. J. Gen. Virol. 72: 1551–1557. [DOI] [PubMed] [Google Scholar]
- Stettler, S., E. Warbrick, S. Prochnik, S. Mackie and P. Fantes, 1996. The wis1 signal transduction pathway is required for expression of cAMP-repressed genes in fission yeast. J. Cell Sci. 109: 1927–1935. [DOI] [PubMed] [Google Scholar]
- Stevens, D. R., Z. X. Wu, U. Matti, H. J. Junge, C. Schirra et al., 2005. Identification of the minimal protein domain required for priming activity of Munc13–1. Curr. Biol. 15: 2243–2248. [DOI] [PubMed] [Google Scholar]
- Stiefel, J., L. Wang, D. A. Kelly, R. T. K. Janoo, J. Seitz et al., 2004. Suppressors of an adenylate cyclase deletion in the fission yeast Schizosaccharomyces pombe. Eukaryot. Cell 3: 610–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, N., K. Tsujino, T. Minato, Y. Nishida, T. Okada et al., 1993. Antibody mimicking the action of RAS proteins on yeast adenylyl cyclase: implication for RAS-effector interaction. Mol. Cell. Biol. 13: 769–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevelein, J. M., and J. H. de Winde, 1999. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 33: 904–918. [DOI] [PubMed] [Google Scholar]
- Thevelein, J. M., L. Cauwenberg, S. Colombo, J. H. De Winde, M. Donation et al., 2000. Nutrient-induced signal transduction through the protein kinase A pathway and its role in the control of metabolism, stress resistance, and growth in yeast. Enzyme Microb. Technol. 26: 819–825. [DOI] [PubMed] [Google Scholar]
- Thevelein, J. M., R. Gelade, I. Holsbeeks, O. Lagatie, Y. Popova et al., 2005. Nutrient sensing systems for rapid activation of the protein kinase A pathway in yeast. Biochem. Soc. Trans. 33: 253–256. [DOI] [PubMed] [Google Scholar]
- Thompson, J. D., D. G. Higgins and T. J. Gibson, 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titgemeyer, F., and W. Hillen, 2002. Global control of sugar metabolism: a gram-positive solution. Antonie Van Leeuwenhoek 82: 59–71. [PubMed] [Google Scholar]
- Treitel, M. A., S. Kuchin and M. Carlson, 1998. Snf1 protein kinase regulates phosphorylation of the Mig1 repressor in Saccharomyces cerevisiae. Mol. Cell. Biol. 18: 6273–6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volland, C., D. Urban-Grimal, G. Geraud and R. Haguenauer-Tsapis, 1994. Endocytosis and degradation of the yeast uracil permease under adverse conditions. J. Biol. Chem. 269: 9833–9841. [PubMed] [Google Scholar]
- Wang, L., R. Kao, F. D. Ivey and C. S. Hoffman, 2004. Strategies for gene disruptions and plasmid constructions in fission yeast. Methods 33: 199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welton, R. M., and C. S. Hoffman, 2000. Glucose monitoring in fission yeast via the gpa2 Gα, the git5 Gβ, and the git3 putative glucose receptor. Genetics 156: 513–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, V., R. Gwilliam, M. A. Rajandream, M. Lyne, R. Lyne et al., 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415: 871–880. [DOI] [PubMed] [Google Scholar]
- Xue, Y., M. Batlle and J. P. Hirsch, 1998. GPR1 encodes a putative G protein-coupled receptor that associates with the Gpa2p Gα subunit and functions in a Ras-independent pathway. EMBO J. 17: 1996–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun, C. W., H. Tamaki, R. Nakayama, K. Yamamoto and H. Kumagai, 1998. Gpr1p, a putative G-protein coupled receptor, regulates glucose-dependent cellular cAMP level in yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 252: 29–33. [DOI] [PubMed] [Google Scholar]