Abstract
We study the evolution of inversions that capture locally adapted alleles when two populations are exchanging migrants or hybridizing. By suppressing recombination between the loci, a new inversion can spread. Neither drift nor coadaptation between the alleles (epistasis) is needed, so this local adaptation mechanism may apply to a broader range of genetic and demographic situations than alternative hypotheses that have been widely discussed. The mechanism can explain many features observed in inversion systems. It will drive an inversion to high frequency if there is no countervailing force, which could explain fixed differences observed between populations and species. An inversion can be stabilized at an intermediate frequency if it also happens to capture one or more deleterious recessive mutations, which could explain polymorphisms that are common in some species. This polymorphism can cycle in frequency with the changing selective advantage of the locally favored alleles. The mechanism can establish underdominant inversions that decrease heterokaryotype fitness by several percent if the cause of fitness loss is structural, while if the cause is genic there is no limit to the strength of underdominance that can result. The mechanism is expected to cause loci responsible for adaptive species-specific differences to map to inversions, as seen in recent QTL studies. We discuss data that support the hypothesis, review other mechanisms for inversion evolution, and suggest possible tests.
CHROMOSOMAL inversions are found as fixed differences between species and as polymorphisms within species in many groups of animals and plants. In some groups, speciation is associated with inversions and other changes in the karyotype (White 1978, Chap. 3). The forces responsible for establishing inversions remain obscure, however, as does their evolutionary significance. The main importance of inversions might lie in their ability to produce genetic isolation between populations and species. Inversions can create postzygotic barriers when they reduce the fecundity of heterokaryotypes (chromosomal heterozygotes) (White 1978, Chap. 6; King 1993, Chap. 6).
Alternatively, the main evolutionary importance of inversions might come from the fact that they suppress recombination in heterokaryotypes (Sturtevant 1917; Roberts 1976). This view was championed by Dobzhansky (1947, 1954,1970), who argued that inversions represent sets of coadapted alleles. His verbal arguments were followed by the development of a substantial body of theory on recombination modifiers, beginning with Nei (1967). A major conclusion from that work is that, for a single population in a constant environment, fitness interactions between loci (epistasis) will generally favor the evolution of decreased recombination (Feldman et al. 1997). When populations are connected by migration, selection can favor loosely linked modifiers that decrease recombination between loci involved in local adaptation, even in the absence of epistasis (Charlesworth and Charlesworth 1979; Pylkov et al. 1998; Lenormand and Otto 2000). This result suggests that inversions could be established by a similar mechanism. Simulations by Trickett and Butlin (1994) show that this can indeed happen.
Inversions have two genetic features that may make them particularly favorable agents for decreasing recombination between sets of locally adapted genes. An inversion is very tightly linked to the loci whose recombination rates it changes. This means that evolutionary forces acting on inversions may be much stronger than those for unlinked modifiers of recombination. Second, an inversion's effect on recombination is underdominant: it suppresses only recombination in heterokaryotypes. Once an inversion is established in a population, recombination again occurs at substantial rates. Thus that region of the chromosome is not doomed to the deleterious effects that ensue when recombination is completely and permanently shut down (Barton and Charlesworth 1998; Charlesworth and Charlesworth 2000).
In this article we study the conditions in which an inversion will spread if it carries a set of locally adapted alleles. We see that the chance that a new inversion happens to capture an advantageous haplotype can be very high. If it does, selection will favor its invasion. The scenario applies equally to species that hybridize since, from a population genetic perspective, hybridization and immigration are equivalent.
Two aspects of the local adaptation mechanism imply that it could contribute to the evolution of inversions under quite biologically general conditions. First, no epistasis is required. The loci must to be individually adapted to local conditions, but no coadaptation between them is needed. Second, it does not depend on drift and so can operate in populations of any size. Thus it provides an alternative (or complement) to the standard hypotheses based on epistasis, drift, and/or meiotic drive (e.g., Dobzhansky 1951; White 1978; Lande 1979).
The basic idea is simple. Consider a population that is receiving immigrants carrying alleles at two or more loci that are disadvantageous under the local conditions. The population is at equilibrium. That means that each copy of a locally adapted allele leaves one descendant on average. These alleles find themselves on different genetic backgrounds, sometimes with locally adapted alleles at the other loci, but sometimes with disadvantageous migrant alleles. An inversion now appears in a chromosome that happens to be carrying only the locally favorable alleles. This new chromosome does not recombine with others. Consequently, a copy of the locally favored allele that it carries at one locus never suffers the disadvantage of being found on the same chromosome with deleterious immigrant alleles at the other loci. This gene therefore has higher fitness than competing copies of the allele at the same locus that are carried on recombining chromosomes. Consequently, the allele on the inversion spreads, and the inversion spreads with it. This scenario is plausible in light of the evidence for local adaptation of a number of inversion polymorphisms in Drosophila (Dobzhansky 1970; Krimbas and Powell 1992; Balanyà et al. 2003; Etges and Levitan 2004).
We begin by quantifying this verbal argument with a model of two haploid loci that are initially loosely linked. The results are then extended to consider other factors such as more loci, partial linkage, diploidy, the geographical setting, and underdominance. In the discussion, we put the local adaptation mechanism in perspective by reviewing alternative hypotheses, discussing the relevant data, and suggesting futher empirical tests.
MODELS AND RESULTS
A simple and robust rule for determining when an inversion will spread is shown schematically in Figure 1. Consider a population that is at a migration–selection balance. In each generation, recombination causes the frequencies of some haplotypes to decrease and others to increase. Consider an inversion whose fitness is determined only by the alleles it carries (that is, there is no additional selection caused, for example, by meiotic problems in heterokaryotypes). If the inversion captures a haplotype whose frequency is decreased by recombination, then selection favors its spread. This result is quite general: it applies regardless of whether mating is random, whether mutation or other forces are acting, or whether selection is during the haploid or the diploid phase.
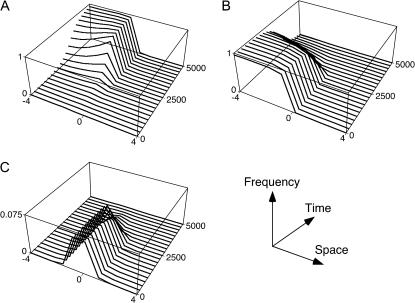
Figure 1.
Schematic for the conditions in which an inversion will invade. The vertical axis is the frequency of a haplotype through a generation, relative to its frequency in zygotes: (1) in zygotes, (2) in juveniles after migration, (3) in adults after selection, and (4) in zygotes in the following generation. (Top) The pattern for an ancestral haplotype captured by an inversion, which then spreads because it avoids the recombination load (bottom).
The mechanism can also be understood in terms of the migration load. Every copy of an immigrant allele that enters the population must be killed off to maintain the equilibrium. If the population is near linkage equilibrium (which happens when recombination is much stronger than migration), an individual will rarely carry more than one of these alleles. Thus in each generation there must be nm selective deaths, where n is the number of loci involved in local adaptation and m is the migration rate. But the situation is very different if an inversion that carries the locally favored alleles at these loci is at high frequency. The immigrant alleles are then unable to recombine with the locally favored alleles, and so each selective death will eliminate n immigrant alleles. This is a group-selection argument and therefore not a rigorous demonstration of how or when an inversion will be established, but it does make the evolutionary forces easy to visualize.
These qualitative analyses raise a series of questions. How rapidly will the inversion spread? How likely is it to capture a fortuitous combination of alleles? How is the outcome affected by genetic details such as the number of loci, the selection coefficients, and ploidy? Can the mechanism establish underdominant inversions? To answer these questions, a bit of math is needed.
Two haploid loci with multiplicative effects:
We can see the detailed workings of this mechanism clearly with the simplest possible case. Two loci have multiplicative fitness effects during the haploid phase of the life cycle (meaning that there is no epistasis). The population is large enough that drift can be neglected, and mating is random.
At each locus, a locally adapted allele has a fitness advantage s over an alternative allele that is introduced by one-way migration into the population at rate m. We assume that migration is weak relative to selection (m ≪ s) to ensure that the locally adapted allele is not lost by swamping, because if it is the immigrant allele becomes fixed everywhere and the locus becomes irrelevant to this process. To simplify the algebra, we also assume that selection is weak (s ≪ 1) and that recombination is strong relative to migration (m ≪ r). These assumptions imply that at migration–selection equilibrium, the frequency of the locally adapted allele at each locus is 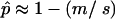 and that the linkage disequilibrium is O(m) (that is, no larger than m times a constant factor).
and that the linkage disequilibrium is O(m) (that is, no larger than m times a constant factor).
Now an inversion appears that captures the locally adapted alleles at both loci. For simplicity, assume initially that the inversion's fitness is determined only by those alleles (that is, there are no other fitness effects) and that the inversion completely suppresses recombination when it appears as a heterokaryotype. The inversion then evolves as a single allele with fitness WI = (1 + s)2. Each generation, migration decreases its frequency by a proportion m, and then selection increases its frequency by a factor equal to its relative fitness. The result is that the expected frequency of a rare inversion in the next generation is equal to its current frequency multiplied by a factor
 |
(1) |
where the approximation neglects terms that are  .
.
Equation 1 shows that the expected rate that the inversion spreads is simply equal to the migration rate. It is intriguing that, although the spread is driven by selection, the rate of spread is independent of the selection coefficient s. The inversion spreads because it is free of the load of immigrant alleles. At equilibrium, selection removes maladapted immigrant alleles at the rate that they enter the population. Thus the migration load is set by m and is independent of the strength of selection. The situation is analogous to the mutation load, which is set by the mutation rate and is independent of the selection coefficient (Haldane 1937; Muller 1950).
More loci:
The result from the last section previews a more general relation that relates the rate of spread to the number of locally adapted alleles that the inversion carries. Consider the situations in which an inversion spans zero, one, and two loci that are at a migration–selection balance (Figure 2). The frequency of an inversion that captures zero loci (that is, only selectively neutral DNA) decreases at a rate m per generation because one-way immigration dilutes it from the population. If the inversion captures one locus and that locus carries the locally adapted allele, then it is evolutionarily neutral because selection exactly offsets migration. With two loci, both of which carry the advantageous allele, Equation 1 shows that the rate of increase is m. These three cases suggest a general rule: the inversion spreads at a rate (n − 1)m, where n is the number of loci that the inversion captured, provided it captures the locally adapted alleles at all the loci.
Figure 2.
The same as in Figure 1, but showing the specific cases in which the inversion captures the locally adapted alleles at zero, one, and two loci.
A simple extension of Equation 1 confirms that rule and leads to a new insight. The inversion can now span any number n of loci that have multiplicative fitness effects, it can carry migrant alleles at some of these loci, and selection coefficients can vary between loci. The set of all loci spanned by the inversion is written I, and the subset of them that carry the locally adapted allele is denoted L. The frequency of the inversion is now multiplied in each generation by
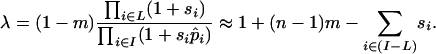 |
(2) |
In the first step, the product in the denominator is over all the loci that the inversion involves, while the product in the numerator is over only those loci where the inversion carries the locally adapted allele. The summation in the last expression is over those loci at which the inversion carries the immigrant allele (a set that we denote as I − L). The approximation in the second step drops terms that are 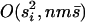 , where
, where  is the mean selection coefficient.
is the mean selection coefficient.
This result confirms the speculation that an inversion with locally adapted alleles at all n loci spreads a rate (n − 1)m. It also provides another intuitive rationale for why the inversion spreads, as illustrated in Figure 2. The inversion's frequency is stable if it captures one locus that carries a locally favored allele, because then selection offsets migration exactly. Each additional locus with a locally adapted allele gives the inversion a fitness boost above that baseline. The boost is equal to how much the frequency of that additional locally adapted allele is increased by selection. At a migration–selection balance, that increment is equal to the migration rate, independent of the selection coefficient. Thus after the first locus, each additional locus carried by the inversion increases its rate of spread by an increment m, and so λ = 1 + (n − 1)m.
Equation 2 also shows that a new inversion that spans just a small number of loci involved in local adaptation will not spread if it carries any immigrant alleles. Specifically, invasion requires that the sum of selection coefficients for the loci carrying immigrant alleles be less than (n − 1)m. That condition will generally not be met when migration is weak relative to selection, as we are assuming. Consequently, to simplify the discussion we assume for the rest of the article that the inversion carries locally adapted alleles at all of its n loci.
Effects of linkage:
Earlier we made the assumption that linkage is not too tight relative to migration before the inversion appears. This assumption is a concern, however, because an inversion typically involves only a relatively small chromosome segment. The rate that the inversion spreads is equal to the rate that recombination decreases the frequency of the ancestral haplotype before the inversion appears (see Figure 1). Since lowering the initial recombination rate must decrease that rate, tighter initial linkage causes the inversion to spread more slowly. In the extreme case where initially there is no recombination between a pair of the loci captured by the inversion, the effective number of loci n that appears in Equation 2 is reduced by 1. In short, Equation 2 gives the upper limit for the rate of spread; this rate applies when migration is weak relative to recombination in the ancestral population (m ≪ r). At the lower limit, the inversion is evolutionarily neutral (λ = 1) if all its loci are completely linked (r = 0) because the effective number of loci is then n = 1.
To get a sense of what happens in intermediate situations, consider a simple case: in the inversion that carries locally adapted alleles at all n loci, these loci have equal effects on fitness s and are equally spaced at an interval r. Again assuming that linkage is initially weak (r ≫ s), a simple approximation for the population's mean fitness can be calculated using the quasi-linkage approximation developed in Kirkpatrick et al. (2002). That, together with the first step of Equation 1, shows that the inversion's frequency is multiplied in each generation by
 |
(3) |
The term in brackets represents the effect of linkage. Its value is 0 with complete linkage (r = 0) and approaches 1 when the recombination rate between adjacent loci is much greater than (n − 1)ms. The inversion therefore is favored so long as the population initially has at least some recombination in the chromosome region that the inversion spans. But when the number of loci involved is small (two or three, say), the force favoring the inversion becomes very weak (much smaller than the migration rate m) if the loci are so tightly linked that the recombination rate between them is much smaller than ms.
Up to this point we have assumed that the migrants are fixed for an allele that is disadvantageous in the local population. If migrants are polymorphic and some carry locally favored alleles, then the inversion's selective advantage is decreased. This is easy to see from the first steps of Equations 1 and 2. With polymorphic migrants, the equilibrium frequencies of locally adapted alleles before the inversion appears (the 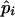 ) are higher, with the result that the rate of spread λ is smaller. When migration is weak relative to selection, however, this effect is very small because the locally adapted alleles are so near fixation that they cannot increase much in frequency.
) are higher, with the result that the rate of spread λ is smaller. When migration is weak relative to selection, however, this effect is very small because the locally adapted alleles are so near fixation that they cannot increase much in frequency.
The fate of a new inversion:
How likely is it that an inversion will be lucky enough to capture just locally adapted alleles at all of its loci? It can be shown that the haplotype carrying just those alleles will be near fixation before the inversion occurs if nm is much less than the harmonic mean of the selection coefficients. Thus an inversion will typically capture high-fitness alleles at all its loci when migration is weak relative to selection.
While a new inversion is still very rare, it is at risk of being lost by chance even if the expected rate of spread λ > 1. When the number of offspring is Poisson distributed, the probability that a new inversion survives random loss is approximately twice its expected rate of spread, or 2m(n − 1) (see Haldane 1927). There is an upper limit, however, to how much migration can promote the invasion. If the migration rate exceeds si/(1 + si), the locally adapted allele at locus i will be lost by swamping and so it will not contribute to the invasion. An inversion can spread only when it occurs in a region of the chromosome that experiences local selection pressures that are strong enough to prevent swamping.
If the inversion is not lost by chance, it will compete evolutionarily with other haplotypes that also carry locally adapted alleles. The inversion has a fitness advantage over all of those haplotypes with fewer locally adapted alleles, and it will spread at their expense. The inversion is equal in fitness to other haplotypes that also have no migrant alleles. Those haplotypes, however, are continually recombining with others that do carry immigrant alleles and so they produce descendant chromosomes with lower fitness. Since the inversion does not suffer this recombination load, it has an evolutionary advantage over its ancestral recombining haplotype.
The new inversion will therefore ultimately displace all haplotypes except the one introduced by migration. A final equilibrium is reached where the inversion is in a migration–selection balance with a frequency of  . This is very close to fixation and at a higher frequency than any of the locally adapted alleles were before the inversion appeared. If immigration from the other population is disrupted, the inversion will spread all the way to fixation. Thus the local adaptation mechanism can fix alternative inversions in different populations or species.
. This is very close to fixation and at a higher frequency than any of the locally adapted alleles were before the inversion appeared. If immigration from the other population is disrupted, the inversion will spread all the way to fixation. Thus the local adaptation mechanism can fix alternative inversions in different populations or species.
Inversions in clines and hybrid zones:
The general argument extends to other geographical settings. Consider, for example, a spatially continuous population in which the population is regulated at a constant density everywhere. Assume that in a transect across the habitat, the environment changes abruptly at spatial point x = 0. Allele 1 at each locus is selected against everywhere to the left of this point, while to the right it is favored. The selection coefficient at locus i is again denoted si. Individuals disperse randomly, and the mean square distance between where an individual and its mother were born is written σ2. Figure 3 shows an example of what happens if an inversion appears anywhere and captures the locally adapted alleles. It will be favored by selection, and (if it avoids chance loss while very rare) it will eventually replace all the ancestral haplotypes because they suffer recombination with alleles from the other side of the ecotone.
Figure 3.
Simulation of the invasion of an inversion in a cline. Two haploid loci with multiplicative fitness effects are initially at a migration–selection equilibrium in a stepping-stone population with 10 demes. At each locus, one allele has fitness 1 + s to the left of the midpoint and fitness 1 − s to the right. An inversion that captures the locally favored haplotype is introduced in the central deme at an initial frequency of 0.01. (A) The invasion of the inversion. (B) The extinction of the ancestral recombining haplotype that carries the same alleles as the inversion. (C) The extinction of recombinant haplotypes as the inversion invades. Parameters are s = 0.025, m = 0.1, and r = 0.5. Note the change in the vertical scale in C.
Ultimately the entire population will consist of just two haplotypes that cannot recombine with each other: the inversion and a haplotype with the ancestral gene arrangement that carries only the alleles favored on the other side of the ecotone. Before the inversion appears, the width of the cline for locus i will be proportional to 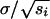 if linkage disequilibrium between the loci is not large (Slatkin 1973). When the inversion has been established, the cline again behaves as if it consists of just two alleles, but now their fitness effects are those of their entire haplotypes. Thus the width of the cline will decrease to
if linkage disequilibrium between the loci is not large (Slatkin 1973). When the inversion has been established, the cline again behaves as if it consists of just two alleles, but now their fitness effects are those of their entire haplotypes. Thus the width of the cline will decrease to  , where
, where 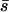 is the mean selection coefficient. This shows that an inversion sharpens the genetic boundaries between parapatric populations that straddle an ecotone, as expected from the basic theory of multilocus clines (Slatkin 1975). Higher migration rates and weaker selection promote polymorphism of the inversion over a larger geographical area.
is the mean selection coefficient. This shows that an inversion sharpens the genetic boundaries between parapatric populations that straddle an ecotone, as expected from the basic theory of multilocus clines (Slatkin 1975). Higher migration rates and weaker selection promote polymorphism of the inversion over a larger geographical area.
A new inversion will spread more rapidly (and have less chance of random loss while rare) if it captures locally adapted alleles in a population near the hybrid zone, where maladapted genotypes against which it competes are most common. (Close to the center of a hybrid zone, on the other hand, there is a greater chance that a new inversion will capture some maladapted alleles.) Far from the hybrid zone, the inversion is favored only very weakly, because the frequency of foreign alleles declines approximately exponentially away from the ecotone boundary. If it suffers even a weak direct fitness cost, that may prevent it from becoming fixed everywhere that its alleles are favored.
In other cases, however, inversions may be favored over much greater geographical areas. Examples include situations where two species hybridize over a large part of their geographical range and when an ecotonal transition is gradual so that clines at the adapting loci are broad.
Diploidy:
Many chromosomes of interest are selected during the diploid phase of the life cycle. Returning to the case of a single population that receives migrants fixed for a foreign allele at a rate m, Equation 1 still applies if WI is replaced by 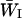 , the mean fitness of the inversion in diploid genotypes. Write the fitness advantage of a homozygote for the locally adapted allele at locus i as si and that of a heterozygote as hisi and assume that alleles are not strongly dominant or recessive (that is,
, the mean fitness of the inversion in diploid genotypes. Write the fitness advantage of a homozygote for the locally adapted allele at locus i as si and that of a heterozygote as hisi and assume that alleles are not strongly dominant or recessive (that is, 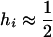 ). Then in each generation the frequency of the inversion is multiplied by
). Then in each generation the frequency of the inversion is multiplied by
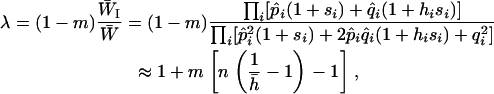 |
(4) |
where  , and
, and  is the average dominance coefficient. [The final approximation drops terms that are
is the average dominance coefficient. [The final approximation drops terms that are 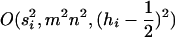 .]
.]
This result shows how dominance affects the outcome. When the alleles are codominant ( ), the results are the same as for the haploid case. When foreign alleles are partly recessive (
), the results are the same as for the haploid case. When foreign alleles are partly recessive ( ), the inversion spreads more quickly and is less likely to be lost when rare, while the reverse holds true when foreign alleles are partly dominant (
), the inversion spreads more quickly and is less likely to be lost when rare, while the reverse holds true when foreign alleles are partly dominant ( ).
).
Underdominant inversions:
Some inversions that are found as fixed differences between species have underdominant fitness effects when they appear as heterokaryotypes in hybrids (White 1973, Chap. 11). Some evolutionists have argued that this observation implies that these inversions spread by some force other than selection, notably drift and/or meiotic drive (e.g., White 1978; Lande 1979; King 1993). Others suggest that the underdominant effects may represent incompatibilities that accumulate after inversions are established in allopatric or parapatric populations (Coyne and Orr 2004). There is uncertainty, however, over the common assumption that the underdominance is structural, that is, caused by meiotic problems due to the different orientations of the chromosome segment in heterokaryotypes. Underdominance might instead be genic, meaning that it is caused by incompatible alleles carried by the inversions (Dobzhansky 1937, pp. 263–264; Searle 1993; Coyne and Orr 2004).
The local adaptation mechanism can establish inversions that have either structural or genic underdominance, without any contribution from drift or meiotic drive. First consider structural underdominance. Say that meiotic problems reduce the fitness of heterokaryotypes by a fraction hs. Then the rate of increase in the new inversion is found by simply subtracting hs from the values for λ that we found earlier (Equations 1–4). The inversion will spread if the result is >1. Take, for example, the case of an inversion that captures only locally adapted codominant alleles at n loci. Selection favors the spread of the new inversion if (n − 1)m > hs. The local adaptation mechanism could therefore plausibly establish an inversion that reduces fitness in heterokaryotypes by several percent. Large inversions are more likely to span loci involved in local adaptation but are also more likely to suffer meiotic costs, and the balance between these forces could favor the establishment of certain size classes of inversions. A new inversion that is unconditionally deleterious, for example because of a loss-of-function mutation caused by the inversion break, can also be established by the local adaptation mechanism. The fact that selection can cause a deleterious inversion to spread because it reduces recombination between a favorable combination of genes was pointed out by Bengtsson and Bodmer (1976).
The local adaptation mechanism has much greater scope to fix underdominant inversions if selection against heterokaryotypes is genic. This is because inversion prevents the recombination that generates unfit genotypes, in which incompatible alleles are brought together in the same genome. All that is needed is that the allele favored at (say) locus A in one population be incompatible with the allele favored at locus B in the other. Figure 4 shows two scenarios that lead to this situation. These are nothing more than the conditions proposed by Dobzhansky (1934) and Muller (1939, 1940, 1942) for the evolution of F1 hybrid incompatibilities. The genotypes favored in each population might be established either in allopatry or parapatry. In either event, with genetic exchange between the populations, the conditions are set for a new inversion that captures the locally favored alleles to spread by the local adaptation mechanism. This could happen in either population. From its first appearance, the inversion will show underdominant fitness interactions with the chromosomal arrangement that is most common in the other population. In this case, there is no limit to how low the fitness of heterokaryotypes can be. In the extreme, the local adaptation mechanism could fix an inversion that is sterile or lethal when heterozygous. This possibility does not apply, of course, to all forms of genic incompatibility. If the inversion classes are fixed for alternative alleles at a single locus and these alleles are underdominant, then the situation is as we described for structural underdominance.
Figure 4.
Two scenarios for establishing the conditions that favor the spread of an underdominant inversion in parapatry. The population occupies two habitats, indicated by the stippled and open regions. Alleles A and B are deleterious in combination in both habitats. Allele A is favored in the habitat to the left and allele B in the habitat to the right. The most common genotype is shown in the top corners. Dashed curves show the frequency of Ab and solid curves the frequency of aB. Asterisks show the appearance of a new mutation that spreads by selection in one of the habitats. (Left side) One advantageous mutation appears in each of the two populations. (Right side) Both advantageous mutations appear in the right-hand population. (Both sides, bottom) An inversion appearing in either habitat that captures the most common haplotype will spread by the local adaptation mechanism.
Associative overdominance and inversion polymorphisms:
Some inversions show evidence of balancing selection. A dramatic demonstration come from Dobzhansky (1954), who started replicate laboratory cage populations of Drosophila pseudoobscura with different initial frequencies of an inversion. In some cases, they converged on the same stable polymorphic equilibrium (see also Wallace 1968).
This observation can be explained by considering the fact that an inversion captures a single chromosome segment that can be carrying any number of variants in addition to the locally adapted alleles we have been focusing on. Often those might include recessive deleterious alleles at one or more loci (Muller 1918; Sturtevant and Mather 1938; Ohta 1966). If these other alleles are fully recessive, then the inversion will invade as outlined above. Once it becomes frequent enough that chromosomal homozygotes form at appreciable frequencies, however, the deleterious mutations will be exposed to selection. Although this will not affect the evolution of the new inversion when it is rare, any recessive deleterious mutations carried by the inversion can prevent it from spreading to fixation.
The conditions under which deleterious mutations in the inversion produce overdominance in our model are easy to find. Again consider the simple case where the alleles involved in local adaptation at n loci have independent fitness effects of average size 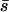 . Deleterious alleles captured by the inversion decrease its relative fitness by hDsD when heterozygous and sD when homozygous. Then the inversion behaves as overdominant when
. Deleterious alleles captured by the inversion decrease its relative fitness by hDsD when heterozygous and sD when homozygous. Then the inversion behaves as overdominant when
 |
(5) |
As we expected intuitively, the conditions are most easily met when the effects of the deleterious mutations are highly recessive (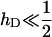 ) and they have a large effect on fitness (sD is not very small).
) and they have a large effect on fitness (sD is not very small).
Epistasis:
We have been discussing cases where there is no epistasis or coadaptation between the alleles captured by an inversion. That assumption emphasizes that the mechanism we are describing does not depend on epistasis and also simplifies the algebra. If epistasis is present, two questions arise: When will the results developed above still give reasonable approximations? And how will the evolutionary outcome be affected when they do not?
Epistasis will either augment or decrease the linkage disequilibria caused by migration. When its contribution is small relative to that of migration, the results above will be good approximations. But what if epistasis is too strong to neglect? The basic framework for visualizing when an inversion will spread remains intact. In particular, Figure 1 and the first step of Equation 1 are valid for any form of epistasis.
To find quantitative results, we need to know the population's mean fitness 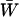 , which can be calculated if specific assumptions are made about the fitnesses. Qualitatively, we anticipate that epistasis will enhance the invasion of an inversion when genotypes that carry two or more locally adapted alleles are even more fit than would be predicted by the effects of the individual alleles. Then epistasis magnifies the disequilibria generated by migration, and so the inversion evades an even bigger recombination load. To put it another way, positive synergism between locally adapted alleles makes an inversion that carries two or more of them even more fit. Conversely, negative epistasis that makes groups of locally adapted alleles less fit works against the invasion of an inversion. Under biologically plausible conditions, however, migration will often make a much stronger contribution to linkage disequilibrium than epistasis (Barton 1986; Pylkov et al. 1998; Lenormand and Otto 2000). In that case, we can neglect epistasis without much loss of accuracy.
, which can be calculated if specific assumptions are made about the fitnesses. Qualitatively, we anticipate that epistasis will enhance the invasion of an inversion when genotypes that carry two or more locally adapted alleles are even more fit than would be predicted by the effects of the individual alleles. Then epistasis magnifies the disequilibria generated by migration, and so the inversion evades an even bigger recombination load. To put it another way, positive synergism between locally adapted alleles makes an inversion that carries two or more of them even more fit. Conversely, negative epistasis that makes groups of locally adapted alleles less fit works against the invasion of an inversion. Under biologically plausible conditions, however, migration will often make a much stronger contribution to linkage disequilibrium than epistasis (Barton 1986; Pylkov et al. 1998; Lenormand and Otto 2000). In that case, we can neglect epistasis without much loss of accuracy.
DISCUSSION
Migration and hybridization set the stage for a mechanism that can cause a new inversion to spread to high frequency. At a minimum, all that is needed is that the inversion capture locally favored alleles at two loci. When many locally adapted loci are involved, the selection favoring inversions can become very strong—of the order of the migration rate times the number of loci. While it is difficult to develop even a rough estimate of the genomic density of loci involved in local adaptation, it is plausible that there are several to many on some chromosomes. A crucial point is that coadaptation (epistasis) between the loci is not needed. This implies that the mechanism may apply to many combinations of loci. It does not depend on drift and so can operate under very general demographic conditions. The local adaptation mechanism therefore appears to be a quite general way that new inversions might be established.
Adding additional biological factors enriches the range of possible outcomes. If the interacting populations are distributed along an environmental gradient, then the inversion can establish a cline. A particularly dramatic example consistent with this prediction comes from D. subobscura. Over the course of just 20 years following the introduction of this species to the New World, several inversions have converged independently in North and South America to latitudinal clines that are strikingly similar to those seen in their native Old World range (Balanyà et al. 2003). If the inversion also happens to capture recessive deleterious alleles at other loci, this can generate associative overdominance that will stabilize the inversion at an intermediate frequency. This situation is consistent with the stable inversion polymorphisms observed, for example, in natural populations of D. persimilis (Coyne et al. 1992) and laboratory populations of D. pseudoobscura (Dobzhansky 1954; Wallace 1968; Ohta 1971). If the fitnesses of alleles involved in local adaptation vary with environmental conditions, as suggested by laboratory experiments on flies (Dobzhansky 1954), then the inversion frequencies could cycle seasonally, as is seen in D. pseudoobscura (Epling et al. 1953; Dobzhansky 1971). If the alleles favored in one population (or species) are incompatible with those favored in the other, inversions will show underdominance, as seen in many interspecific crosses (White 1978; King 1993).
The local adaptation mechanism causes inversions to carry loci responsible for adaptive population- or species-specific differences. Several lines of evidence are consistent with this prediction. Inversion polymorphisms are associated with adaptive intraspecific variation in a number of groups, including Drosophila (Krimbas and Powell 1992), Anopheles mosquitoes (Coluzzi et al. 2002; Cohuet et al. 2004; Tripet et al. 2005), Rhagoletis flies (Feder et al. 2003a,b), and seaweed flies (Butlin et al. 1982; Gilburn and Day 1994, 1999). Recently, it has emerged that much adaptive species-specific variation maps to inversions in sunflowers (Rieseberg et al. 1999; Rieseberg 2001) and Drosophila (Noor et al. 2001a,b). Sterility factors occur in inversions that distinguish two species of Drosophila that are sympatric and that hybridize, but not a pair of species that are allopatric (Brown et al. 2004). Perhaps most intriguing is the observation that Drosophila species that are sympatric, and so can potentially hybridize, differ more often by inversions than species that are allopatric (Noor et al. 2001a).
Thus many of the major evolutionary features of inversions—fixed differences between populations and species, clines, stable polymorphisms within species, cycling frequencies, underdominance in crosses between species, and the association of inversions with genes responsible for adaptive traits within and between species—are consistent with the local adaptation mechanism. While the other mechanisms that we review below are surely also at work, it appears that the local adaptation mechanism could play an important role in many of the patterns observed in nature. This causes decreased recombination between populations, which in turn can contribute to the formation of species (Darlington 1936; Felsenstein 1981; Butlin 2005).
The local adaptation mechanism can work in many biogeographical settings. If the contact between two populations occurs along a narrow hybrid zone, then the strongest forces favoring a new inversion will be found inside this zone. It may therefore seem improbable that the new inversion would be able to escape from the zone (Rieseberg 2001). This verdict may be premature, however, for three reasons. First, as our simulations show (Figure 3), a new inversion can indeed escape the hybrid zone and spread throughout the range of an incipient species. Second, the argument assumes that hybrid zones are either in demographic equilibrium with the rest of the species' ranges or perhaps demographic sinks. All else being equal, hybrid zones will tend to migrate toward areas that are population sinks (Barton and Hewitt 1989). Some hybrid zones may in fact lie in ecotone regions where the total carrying capacity is higher than that elsewhere, causing them to be demographic sources that export new favorable genetic combinations to the rest of a species' range. Finally, not all hybridizing species are in contact only along a narrow zone. For example, the range of D. persimilis lies entirely within the range of D. pseudoobscura. These species hybridize, differ with respect to chromosomal rearrangements, and show evidence of reinforcement (Noor 1995; Ortiz-Barrientos et al. 2004). The point here is that a narrow hybrid zone is not the only geographical context in which species come into genetic contact: inversions can be selected throughout much (or even all) of a species' range.
Our examples have emphasized examples in which fitnesses vary in space. But in fact the local adaptation mechanism can be driven by any kind of selection that favors different allele frequencies in different populations that are in genetic contact. An important category of such genes is those that show incompatibilities when they appear in the genetic background of the other population. That is, pleiotropic interactions can be the basis for the local adaptation needed to drive the invasion of an inversion. Possible examples come from sunflowers and flies, where inversions that distinguish closely related species carry alleles that cause sterility when introgressed into the other species (Rieseberg et al. 1999; Noor et al. 2001a,b).
Other kinds of genes can also contribute to the local adaptation mechanism. Loci involved in habitat choice are one example. Genes contributing to sexual displays can participate when different displays are favored in different populations. In fact, a locus can contribute to the establishment of an inversion even if the locus itself is only under indirect selection. For example, reinforcement can cause mating preferences to diverge between populations when the alleles affecting the preferences have no direct effect on fitness (Liou and Price 1994; Kirkpatrick and Servedio 1999). From the perspective of a new inversion, the selective boost that a mating preference allele gets from reinforcement is indistinguishable from what it might get from any other kind of selective advantage. This scenario is consistent with the association species-specific mating preferences with inversions that distinguish D. pseudoobscura and D. persimilis (Noor et al. 2001a).
If the local adaptation mechanism is so powerful, why are inversions not everywhere? One possibility is that they in fact are, and that their frequency has been greatly underestimated. Karyotypic methods for detecting inversions are reasonably powerful in flies because of the high resolution provided by salivary chromosomes, but are much less effective in almost all other groups of organisms. Sequence-based comparisons of fixed inversion differences between species are just now becoming possible, and early results do indeed suggest that there have been more inversions fixed than had been previously thought (Bourque et al. 2004; Zhao et al. 2004; Mikkelsen et al. 2005). Several biological factors may limit how often the local adaptation mechanism can come into play. One is a limit on the rate of occurrence of inversions that do not have strongly deleterious direct effects on fitness. Rough estimates suggest the genomic rate of spontaneous inversions in mammals may be on the order of 10−3 (Lande 1979; King 1993). One might expect that the vast majority of these are deleterious, but no data on this point are available. Separate from the effects of the breakpoints, inversion heterozygotes can have reduced fertility because of the production of aneuploid gametes (White 1973; King 1993). While some groups like flies and mammals have compensatory mechanisms, the reduced fitness in those that do not may generate another kind of selection against most new inversions. Finally, the mechanism may be limited by the number of genomic regions that have multiple loci involved in local adaptation.
Throughout the development of the models we made the simplifying assumption that recombination is completely suppressed in heterokaryotypes. Recombination can occur at very low levels, however, as the result of double crossovers within the inversion and gene conversion (Andolfatto et al. 2001). The opportunities for recombination between chromosomes carrying different inversions will be rare if the inversion spreads through the local population quickly, in which case our simplifying assumption provides a good approximation. Roughly speaking, this situation holds when the migration (or hybridization) rate is much larger than the recombination rate in heterokaryotes.
The local adaptation mechanism for the evolution of inversions is based on forces that have already been identified by theoreticians. Li and Nei (1974) were the first to point out that migration generates linkage disequilibrium when populations differ in allele frequencies. Charlesworth and Charlesworth (1979) used simulations to study the evolution of recombination between two loci that have clines produced by a migration–selection balance with multiplicative selection (no epistasis). They found that when the clines coincide, corresponding to the situation in our models, an unlinked modifier that decreases recombination between the selected loci spreads. Trickett and Butlin (1994) simulated explicitly the evolution of an inversion that captures locally adapted alleles in a two-population system. Although their main interest was in speciation by assortative mating, they observed the inversion spread whenever it captured two locally adapted alleles. Pylkov et al. (1998) and Lenormand and Otto (2000) studied the evolution of loosely linked modifiers that alter recombination between a pair of loci at a local adaptation balance. Both studies found that, with weak or no epistasis, decreased recombination is always favored, which is consistent with our results. They also found that the evolutionary force favoring the modifier can decrease as the migration rate increases. That is because when migration is sufficiently strong relative to selection, the allele frequency differences between populations decrease to the point where little disequilibrium is produced and so the force favoring recombination declines. In our simple analytic models, by contrast, the rate of spread of the inversion is proportional to the migration rate (Equations 1–4). That is because we assumed that migration is much weaker than selection, and presumably the same effect would be seen in our models at higher migration rates. Since inversions are completely linked to the loci under direct selection, the evolutionary forces acting on them are much stronger than those on the modifiers they studied.
Dobzhansky (1954, 1970) and later workers (e.g., Turner 1967a,b; Wasserman 1968; Alvarez and Zapata 1997) have promoted the view that inversions represent sets of coadapted genes. That is, each inversion carries a set of alleles that have positive epistatic fitness interactions. Under the selection–mutation mechanism described here, by contrast, no epistasis is required. It operates because the inversion binds together alleles that are individually favored in a local population, with the result that each allele is always associated with the superior genetic background provided by the (potentially independent) fitness advantage of alleles at the other loci. That is, the alleles carried by the inversion are individually adapted, rather than coadapted.
Mechanisms that establish inversions:
The local adaptation mechanism studied here is one of several families of mechanisms that can cause a new inversion to spread. Some will cause it to spread to fixation, while others will lead either to a neutral or to a selectively maintained polymorphism. The mechanisms are summarized in Table 1.
TABLE 1.
Mechanisms that cause new inversions to spread
| Mechanism | Fate of inversion |
|---|---|
| Indirect effects: recombination suppression | |
| Local adaptation | Near fixation |
| Epistatic selection | Fixation |
| Deleterious mutation | Neutral polymorphism |
| Drift acting on an underdominant inversion | Fixation or loss |
| Direct positive effects of the chromosomal lesion | |
| Effects of breakpoints | Fixation or stable polymorphism |
| Position effects | Fixation or stable polymorphism |
| Meiotic drive | Fixation or stable polymorphism |
| Hitchhiking with a spreading advantageous mutation | Neutral polymorphism |
The first family of mechanisms that can cause an inversion to spread depends on the effects that inversions have on recombination. The local adaptation mechanism discussed here is one of these. Another is when an inversion captures a set of epistatically favored alleles in a single population. The conditions for that to occur, however, are fairly delicate. They require both that the selection coefficients are able to maintain polymorphism with linkage disequilibrium between the selected loci and that the inversion captures a high-fitness genotype, which may not be at high frequency in the population (Kimura 1956; Charlesworth and Charlesworth 1973; Bengtsson and Bodmer 1976). When both epistasis and gene flow contribute to linkage disequilibrium, as can happen (for example) in a hybrid zone, epistasis will typically be a much weaker force (Barton 1986; Pylkov et al. 1998; Lenormand and Otto 2000).
Recombination suppression also favors a new inversion if it happens to capture a chromosome segment that is relatively free of deleterious mutations (Ohta 1966; Nei et al. 1967; Ohta and Kojima 1968). This advantage is transitory, however, because the advantage dissipates as the inversion itself begins to accumulate deleterious mutations. That occurs on a timescale of 1/(hs), which may typically be on the order of dozens to hundreds of generations. If the inversion initially captures one or more deleterious alleles at these loci when it first appears, it will ultimately equilibrate at a fitness below that of the ancestral rearrangement, and it is certain to be lost from a large population. On the other hand, if it happens to capture only favorable alleles, then its fitness ultimately converges to that of the ancestral chromosomal arrangement and at that point becomes selectively neutral. During the transient phase while it has a fitness advantage, however, the number of copies of the new inversion can be boosted by a factor on the order of Exp{U/hs}, where U is the total deleterious mutation rate in the chromosome segment and hs is the fitness of mutant heterozygotes (Nei et al. 1967). For example, a large inversion in Drosophila might plausibly have a mutation rate of U = 0.1, and hs might typically be ∼0.01. Those values would boost the number of copies of a mutation-free inversion by a factor of >105. The deleterious mutation mechanism is most powerful when the inversion spans many loci and occurs in a small population. With the aid of drift, it might cause a new inversion to spread to fixation.
The second family of mechanisms is the most widely discussed and the most controversial. Here random genetic drift fixes inversions that have underdominant fitness effects. This possibility has attracted attention because some inversions found as fixed differences between species show underdominant fitness effects in hybrid crosses, suggesting that the derived form did not originally spread by selection (White 1978; King 1993). Theoretical analysis shows that this mechanism requires quite special conditions. Inversions that have negative fitness effects can be established by drift only if effective population sizes are small for substantial evolutionary periods, selection against the heterokaryotypes is very weak, or meiotic drive favors the new inversion (Wright 1941; Bengtsson and Bodmer 1976; Lande 1979, 1985; Hedrick 1981; Walsh 1982; Barton and Charlesworth 1984; Barton and Rouhani 1991). This mechanism may be particularly important in groups that self or are otherwise highly inbred (Coyne and Orr 2004; L. Rieseberg, personal communication). We return to the issue of underdominant inversions below.
The third family of mechanisms involves the effects of the chromosomal lesion itself. The break in the chromosome may result in a fortuitous selective advantage, either by disrupting a gene where the breakpoint occurs or by changing gene expression in the chromosomal neighborhood. This kind of “position effect” has been documented in the inversion In(3L)Payne in Drosophila (Wesley and Eanes 1994). On the other hand, the results from searches for gene disruption caused by the inversions that distinguish humans and chimps have to date been negative (Kehrer-Sawatzki et al. 2002, 2005). As most mutations seem to show intermediate dominance, fitness effects caused by the breakpoint itself will typically favor fixation or loss of the new inversion, but of course it is possible that the fitness effects will be overdominant and lead to a stable polymorphism. Another way that chromosomal rearrangements like inversions can become established is through distortions they cause in meiosis. There is evidence that some chromosomal rearrangements cause meiotic drive, which can cause an inversion to spread to fixation (White 1978; King 1993; Searle 1993). Alternatively, if a drive suppressor arises, the outcome can be either a stable or a neutral polymorphism (Hartl 1970, 1975; Prout et al. 1973; Thomson and Feldman 1974; Liberman 1976).
Fourth, if a new inversion is lucky enough to capture an advantageous allele that is spreading in the population, it will hitchhike to higher frequency. Once the new allele reaches an equilibrium, either polymorphic or at fixation, the hitchhiking advantage disappears. On average, the inversion's frequency will be boosted by a factor equal to the change in the frequency of the advantageous allele from the moment that the inversion occurs. If at that point the inversion's fitness is very close to the mean fitness of other chromosomes carrying the advantageous allele, the inversion will then drift in frequency between 0 (that is, loss) and the equilibrium frequency of the allele (which is 1 if the allele spread to fixation, but <1 if it reached a stable polymorphism). On the other hand, alleles at other loci captured by inversion may give the inversion lower or higher fitness than the mean of other copies of the advantageous mutation. These effects could favor the inversion to be lost or to spread further within the allelic class, respectively. Overall, it seems unlikely that the hitchhiking mechanism is a common factor in inversion evolution. That is because to have important effects, the inversion has to capture the advantageous allele while it is still rare, which is an improbable event.
Mechanisms that maintain inversion polymorphisms:
Motivated largely by data from Drosophila, there has been a lot of discussion of how inversion polymorphisms might be maintained. Dobzhansky (1947, 1951, 1970) proposed that inversions carry sets of coadapted alleles that confer high fitness when heterozygous. Haldane (1957) modeled this situation and found the conditions in which inversions will be maintained at a stable equilibrium and fixed for alternative alleles. This outcome requires that at least two loci carried by the inversions experience overdominant selection. Further, there must be positive epistasis such that the fitness of heterokaryotypes is higher than expected from the contributions of the individual loci. van Valen and Levins (1968) used a phenomenological version of this model to generate predictions for length distribution of inversions. Other workers found sufficient conditions for a stable equilibrium in which there is polymorphism within each inversion class (Deakin 1972; Charlesworth 1974; Deakin and Teague 1974).
Wasserman (1968) suggested a variant of Dobzhansky's idea of coadapted gene complexes. Here each class of inversions consists of a set of coadapted haplotypes that have reduced fitness when they recombine with other haplotypes in the same inversion class. When an inversion class increases in frequency, it occurs more often in homokaryotypes. This causes the haplotypes within that inversion class to recombine more frequently, breaking up the coadapted alleles and reducing the fitness of that inversion class. The result is analogous to negative frequency-dependent selection acting on the frequencies of the inversion classes, maintaining the polymorphism. Partial mathematical analyses of this verbal model were provided by Wasserman (1968) and Alvarez and Zapata (1997).
The theoretical difficulty with all the hypotheses based on coadapted genes is that they require a delicate balance of fitness effects. Although a full analysis has not been done, it would seem that the requirements for Wasserman's hypothesis would be even more severe than those for Dobzhansky's. Further, it is not clear what historical sequence of evolutionary events would produce inversions with the required fitness properties.
Two other ideas for the maintenance of inversion polymorphisms are frequency-dependent selection and fluctuating selection pressures (Wright and Dobzhansky 1946; Lewontin and White 1960; Alvarez-Castro and Alvarez 2005). While frequency-dependent and fluctuating selection can certainly maintain polymorphism at a single locus, it is not clear how an inversion polymorphism can be maintained that way for substantial evolutionary periods. If there is a small amount of recombination (or gene conversion) in heterokaryotypes, that will break down the disequilibrium between the locus that is the target of balancing selection and the inversion.
The local adaptation scenario studied in this article appears to be a more plausible mechanism for maintaining inversion polymorphisms. The prerequisite is genetic variation maintained by a balance between migration and selection at two or more loci, which occurs under quite general conditions. That situation suffices to favor the spread of a new inversion. That in turn can produce a stable polymorphism maintained either by a migration–selection balance or by selection against deleterious recessives also carried by the new inversion.
Underdominant inversions:
Some inversions reduce fitness when heterozygous and so contribute to postzygotic isolation between species. This suggests that they may have played a role in the speciation process (White 1978; Sites and Moritz 1987; King 1993). Our results show that the local adaptation mechanism can establish underdominant inversions. Its ability to do that, however, depends on whether underdominance is caused by structural or genic incompatibilities between the inversions. If selection against heterokaryotypes has structural causes, that is, resulting from problems in pairing during meiosis, then the local adaptation mechanism probably cannot establish inversions that reduce fitness by more than several percent. On the other hand, if the incompatibility is genic, that is, caused by interactions between alleles within the inversions, then in theory the mechanism can establish an inversion even when heterokaryotypes are lethal or sterile.
The main cause of structural problems arises when a single crossover occurs inside an inversion during meiosis. The resulting gametes carry major deletions, insertions, or an unbalanced set of chromosomes, which could cause heterokaryotypes to suffer a large fitness loss (White 1973). Delneri et al. (2003) provide an elegant experimental demonstration of this effect in yeast. In other cases, however, a variety of mechanisms greatly reduce this cost (Lande 1979; King 1993; Searle 1993). In both flies and mammals there are examples of pericentric inversions that seem to have little or no effect on the fitness of heterokaryotypes (Sites and Moritz 1987; Coyne et al. 1993), and in Drosophila there are many examples of paracentric inversion polymorphisms that are in fact overdominant (Dobzhansky 1970; Krimbas and Powell 1992). The point here is simply that inversions have a broad spectrum of structural effects on fitness when heterozygous and are not always strongly underdominant (White 1978, Chap. 6; King 1993). Thus the local adaptation mechanism could establish an inversion even when there is some selection against heterokaryotypes from structural causes.
Although underdominance is often assumed to be structural, in fact it may be partly or largely genic (Searle 1993; Coyne and Orr 2004, Chap. 7). In this case, the scope of the local adaptation mechanism to fix an underdominant inversion is much greater. We saw earlier that Dobzhansky–Muller incompatibilities can drive the spread of an inversion by the local adaptation mechanism. These kinds of incompatibilities seem to be common between species (Coyne and Orr 2004, Chap. 7), which suggests that they could also make a substantial contribution to the incompatibilities seen between some inversions.
Regardless of whether incompatibilities are structural or genic, the local adaptation mechanism is appealing because it does not require the very strong drift or meiotic drive envisioned by other hypotheses. Thus it might contribute to the spread of underdominant inversions over a much broader range of biological conditions than those other mechanisms. But in many cases they may work together. Consider what happens when an inversion is underdominant for structural reasons. Local adaptation happens, but is not strong enough to offset selection against the heterokaryotypes. The rate of fixation by drift of new underdominant chromosomal rearrangements is extremely sensitive to the strength of selection against heterokaryotypes (Wright 1941; Bengtsson and Bodmer 1976; Lande 1979). The rate is greatly accelerated when there is fitness variation between chromosomes (Lande 1984). The local adaptation mechanism provides that variation. It can also reduce the strength of selection against heterokaryotypes to the point where drift can cause an underdominant inversion to spread.
Inversions and speciation genes:
One of the most striking empirical patterns to emerge in recent studies of speciation is that loci responsible for species-specific differences often map to inversions (Rieseberg et al. 1999; Rieseberg 2001; Noor et al. 2001a,b; Feder et al. 2003b). That observation led Noor et al. (2001a) and Rieseberg (2001) to propose the following hypothesis. The inversions that distinguish species are established by some (unspecified) mechanism, perhaps in allopatry. Once the species are in contact, colinear parts of the genome tend to be homogenized by hybridization. Inverted regions, on the other hand, are protected from introgression because they suppress recombination. Additional differences will also tend accumulate subsequently in the inverted regions (Navarro and Barton 2003b).
The local adaptation mechanism turns this sequence of events on its head. Now the inversion invades secondarily, as a result of genic differences caused by local adaptation. In this scenario, QTL responsible for species differences are the cause, rather than the consequence, of the establishment of the inversion.
Noor et al. (2001a) noted that sympatric species pairs of Drosophila tend to be fixed for different inversions more often than allopatric pairs. If current sympatry correlates with the frequency of hybridization in the past, then that pattern is also consistent with the local adaptation hypothesis. Even if the local adaptation mechanism establishes an inversion, however, that region of the genome will tend to accumulate further genetic differences between the populations than will colinear parts of the genome (Barton and Bengtsson 1986; Navarro and Barton 2003b).
There is some evidence that segments of the human genome that have structural rearrangements (such as inversions) tend to carry loci that are under positive directional selection (Navarro and Barton 2003a; Lu et al. 2003; but see Mikkelsen et al. 2005). This correlation, if verified, might be explained by the local adaptation mechanism. There is regional variation across the genome in rates of amino acid substitution (Mikkelsen et al. 2005). It is plausible that rapidly evolving parts of the genome also tend to be often involved in local adaptation and thus are susceptible to triggering the spread of a new inversion by the local adaptation mechanism.
The mechanism might also contribute to the correlation observed between rates of speciation and rates of karyotypic change (Wilson et al. 1975; Bush et al. 1977; Bengtsson 1980). Speciation can be driven by adaptation to novel habitats (Schluter 2000). That, in turn, sets the stage for chromosomal evolution via a local adaptation mechanism.
Candidate examples and predictions:
We have seen that many features of the genetic and natural history of inversions are consistent with the local adaptation mechanism set out in this article. These are relatively weak tests of the hypothesis, however, since each of the observations is also compatible with other mechanisms. More direct tests focus on particular inversions and the alleles they carry.
An example of an inversion whose spread might be explained by the local adaptation mechanism is In(2L)t, a polymorphic inversion on the second chromosome of D. melanogaster that has been studied intensively (Lemeunier and Aulard 1992). Molecular evidence shows that the inversion has a unique origin dating to ∼105 years ago (Andolfatto et al. 1999). The inversion is adapted to warmer habitats and shows parallel geographical clines on different continents (Knibb 1982; Oakeshott et al. 1982; Bénassi et al. 1993). Experiments confirm that the inversion has fitness effects that are environmentally sensitive and polygenic (van Delden and Kamping 1991).
The inversion spans Adh, a polymorphic locus with clines that appear to reflect a migration–selection balance (Berry and Kreitman 1993). The Adh polymorphism is much older than the inversion. Almost all In(2L)t chromosomes carry the Adh-S allele (Lemeunier and Aulard 1992; Veuille et al. 1998). Although recombination is suppressed in heterokaryotypes, it is not eliminated. The age of the inversion is at least 100-fold greater than the expected half-life of linkage disequilibrium between the inversion and Adh (Andolfatto et al. 1999), and so their association seems to be actively maintained by some force, for example, selection or migration.
The proximal breakpoint of the inversion is close (within ∼5.1 kb) to an open reading frame (Andolfatto et al. 1999). It is thus plausible that the inversion affects patterns of gene expression, a result known from other inversions (Wesley and Eanes 1994). A survey of polymorphism in the vicinity of the breakpoint shows a pattern reminiscent of Adh: it appears that one or more sites have been under balancing selection for much longer than the age of the inversion and further that haplotype frequencies vary geographically (Andolfatto and Kreitman 2000).
How do these facts fit to theory? They do not support the “protection from introgression” hypothesis proposed by Rieseberg (2001) and Noor et al. (2001a), in which genetic differences between populations that are inside an inversion persist because reduced recombination inhibits introgression. There is enough recombination in heterokaryotypes to have erased the association between the inversion and the Adh polymorphism since the inversion appeared. Nor do the data support the “inversion-first” hypothesis of Navarro and Barton (2003b), in which genic differences build up inside inversions after the inversions are established. The selected sites at Adh and the inversion breakpoint predate the origin of the In(2L)t.
The facts from this case are, however, consistent with the local adaptation mechanism. The scenario here is that before the inversion appeared there were clines at Adh and at one or more nearby loci, perhaps including a site very close to the breakpoint. The inversion then captured the variants at these sites that are adapted to warmer habitats. The breakpoint itself may also have generated a mutation favored in those environments. We emphasize that no epistasis needs to be invoked; the captured alleles might be selected independently and even by unrelated environmental factors. The inversion then spread in low latitudes under the mechanism described above. It is prevented from rising to higher frequencies in the tropics either because of selection against deleterious recessives that it carries or because immigration from temperate populations is too strong.
This scenario could be tested with additional data. We might expect that the ancestral situation that provided the preconditions for the inversion to spread may still persist in the standard (uninverted) class of chromosomes. Looking among localities, there should be positive covariance among Standard chromosomes between allele frequencies at the locally adapted loci spanned by the inversion. Within localities, there should be positive linkage disequilibrium in standard chromosomes between the alleles carried by the inversion. With most inversions, it is difficult to know where to look for these loci. With In(2L)t, however, the Adh Fast/Slow site and the breakpoint region are obvious candidates (Andolfatto and Kreitman 2000). Another is αGpdh, which also lies within the inversion and shows evidence of clines maintained by selection (van Delden and Kamping 1989). Other inversions have patterns reminiscent of In(2L)t. For example, the 2J inversion in D. buzzati shows evidence for adaptive geographical variation and linkage disequilibrium between the inversion and an ancient genic polymorphism that it spans (Gomez and Hasson 2003).
Looking beyond inversions, other kinds of chromosomal rearrangements also put new combinations of genes into tight linkage and so could be favored by similar evolutionary forces. Thus there may be several contexts in which to study the role of local adaptation in chromosomal evolution, giving us a rare opportunity to test theories for the evolution of recombination.
Acknowledgments
We thank Janice Britton-Davidian, Brian Charlesworth, David Hall, Yannis Michalakis, Mohamed Noor, Sally Otto, Trevor Price, Michel Raymond, Loren Rieseberg, and Monty Slatkin for helpful discussions. This research was supported by National Science Foundation grants DEB-9973221 and EF-0328594 and by National Environmental Research Council grant NER/A/S.2002/00857.
References
- Alavarez, G., and C. Zapata, 1997. Conditions for protected inversion polymorphism under supergene selection. Genetics 146: 717–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alavarez-Castro, J. M., and G. Alvarez, 2005. Models of general frequency-dependent selection and mating-interaction effects and the analysis of selection patterns in Drosophila inversion polymorphisms. Genetics 170: 1167–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto, P., F. Depaulis and A. Navarro, 2001. Inversion polymorphisms and nucleotide variability in Drosophila. Genet. Res. 77: 1–8. [DOI] [PubMed] [Google Scholar]
- Andolfatto, P., and M. Kreitman, 2000. Molecular variation at the In(2L)t proximal breakpoint site in natural populations of Drosophila melanogaster and D. simulans. Genetics 154: 1681–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto, P., J. D. Wall and M. Kreitman, 1999. Unusual haplotype structure at the proximal breakpoint of In(2L)t in a natural population of Drosophila melanogaster. Genetics 153: 1297–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balanyà, J., L. Serra, G. W. Gilchrist, R. B. Huey, P. Pascual et al., 2003. Evolutionary pace of chromosomal polymorphism in colonizing populations of Drosophila subobscura: an evolutionary time series. Evolution 57: 1837–1845. [DOI] [PubMed] [Google Scholar]
- Barton, N. H., 1986. The effects of linkage and density-dependent regulation on gene flow. Heredity 57: 415–426. [DOI] [PubMed] [Google Scholar]
- Barton, N. H., and B. O. Bengtsson, 1986. The barrier to genetic exchange between hybridizing populations. Heredity 57: 357–376. [DOI] [PubMed] [Google Scholar]
- Barton, N. H., and B. Charlesworth, 1984. Genetic revolutions, founder effects, and speciation. Annu. Rev. Ecol. Syst. 15: 133–164. [Google Scholar]
- Barton, N. H., and B. Charlesworth, 1998. Why sex and recombination? Science 281: 1986–1990. [PubMed] [Google Scholar]
- Barton, N. H., and G. M. Hewitt, 1989. Adaptation, speciation and hybrid zones. Nature 341: 497–503. [DOI] [PubMed] [Google Scholar]
- Barton, N. H., and S. Rouhani, 1991. The probability of fixation of a new karyotype in a continuous population. Evolution 45: 499–517. [DOI] [PubMed] [Google Scholar]
- Bengtsson, B. O., 1980. Rates of karyotype evolution in placental mammals. Hereditas 92: 37–47. [DOI] [PubMed] [Google Scholar]
- Bengtsson, B. O., and W. F. Bodmer, 1976. On the increase of chromosomal mutations under random mating. Theor. Popul. Biol. 9: 260–281. [DOI] [PubMed] [Google Scholar]
- Bénassi, V., S. Aulard, S. Mazeau and M. Veuille, 1993. Molecular variation of Adh and P6 genes in an African population of Drosophila melanogaster and its relation to chromosomal inversions. Genetics 134: 789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry, A., and M. Kreitman, 1993. Molecular analysis of an allozyme cline: alcohol dehydrogenase in Drosophila melanogaster on the East coast of North America. Genetics 134: 869–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque, G., P. A. Pevzner and G. Tesler, 2004. Reconstructing the genomic architecture of ancestral mammals: lessons from human, mouse, and rat genomes. Genome Res. 14: 507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, K. M., L. M. Burk, L. M. Henagan and M. A. F. Noor, 2004. A test of the chromosomal rearrangement model of speciation in Drosophila pseudoobscura. Evolution 58: 1856–1860. [DOI] [PubMed] [Google Scholar]
- Bush, G. L., S. M. Case, A. C. Wilson and J. L. Patton, 1977. Rapid speciation and chromosomal evolution in mammals. Proc. Natl. Acad. Sci. USA 74: 3942–3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butlin, R. K., 2005. Recombination and speciation. Mol. Ecol. 14: 2621–2635. [DOI] [PubMed] [Google Scholar]
- Butlin, R. K., I. L. Read and T. H. Day, 1982. The effects of a chromosomal inversion on adult size and male mating success in the seaweed fly, Coelopa frigida. Heredity 49: 51–62. [Google Scholar]
- Charlesworth, B., 1974. Inversion polymorphism in a two-locus genetic system. Genet. Res. 23: 259–280. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B, and D. Charlesworth, 1973. Selection of new inversions in multilocus genetic systems. Genet. Res. 21: 167–183. [Google Scholar]
- Charlesworth, D., and B. Charlesworth, 1979. Selection on recombination in clines. Genetics 91: 581–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B, and D. Charlesworth, 2000. The degeneration of Y chromosomes. Philos. Trans. R. Soc. Lond. B 355: 1563–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohuet, A., I. Dia, F. Simard, M. Raymond and D. Fontenille, 2004. Population structure of the malaria vector Anopheles funestus in Senegal based on microsatellite and cytogenetic data. Insect Mol. Biol. 13: 251–258. [DOI] [PubMed] [Google Scholar]
- Coluzzi, M., A. Sabatini, A. della Torre, M. A. Di Deco and V. Petrarca, 2002. A polytene chromosome analysis of the Anopheles gambiae species complex. Science 298: 1415–1418. [DOI] [PubMed] [Google Scholar]
- Coyne, J. A., W. Meyers, A. P. Crittenden and P. Sniegowski, 1993. The fertility effects of pericentric inversions in Drosophila melanogaster. Genetics 134: 487–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne, J. A., B. C. Moore, J. A. Moore, J. R. Powell and C. E. Taylor, 1992. Temporal stability of third-chromosome inversion frequencies in Drosophila persimilis and D. pseudoobscura. Evolution 46: 1558–1563. [DOI] [PubMed] [Google Scholar]
- Coyne, J. A., and H. A. Orr, 2004. Speciation. Sinauer, Sunderland, MA.
- Darlington, C. D., 1936. The limitation of crossing over in Oenothera. J. Genet. 32: 343–352. [Google Scholar]
- Deakin, M. A. B., 1972. A model for inversion polymorphism. J. Theor. Biol. 35: 191–212. [DOI] [PubMed] [Google Scholar]
- Deakin, M. A. B., and R. B. Teague, 1974. A generalized model of inversion polymorphism. J. Theor. Biol. 48: 105–123. [DOI] [PubMed] [Google Scholar]
- Delneri, D., I. Colson, S. Grammenoudi, I. N. Roberts, E. J. Louis et al., 2003. Engineering evolution to study speciation in yeasts. Nature 422: 68–72. [DOI] [PubMed] [Google Scholar]
- Dobzhansky, T., 1934. Studies on hybrid sterility. I. Spermatogenesis in pure and hybrid Drosophila pseudoobscura. Z. Zellforsch. Microsk. Anat. 21: 169–221. [Google Scholar]
- Dobzhansky, T., 1937. Genetics and the Origin of Species, Ed. 1. Columbia University Press, New York.
- Dobzhansky, T., 1947. Genetics of natural populations. XIV. A response of certain gene arrangements in the third chromosome of Drosophila pseudoobscura to natural selection. Genetics 32: 142–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky, T., 1951. Genetics and the Origin of Species, Ed. 3. Columbia University Press, New York.
- Dobzhansky, T., 1954. Evolution as a creative process. Proceedings of the 9th International Congress on Genetics, Vol. 1, Bellagio, Italy, pp. 435–449.
- Dobzhansky, T., 1970. Genetics of the Evolutionary Process. Columbia University Press, New York.
- Dobzhansky, T., 1971. Evolutionary oscillation in Drosophila pseudoobscura, pp. 109–133 in Ecological Genetics and Evolution, edited by R. Creed. Blackwell Scientific, Oxford.
- Epling, C., D. F. Mitchell and R. H. T. Mattoni, 1953. On the role of inversions in wild populations of Drosophila pseudoobscura. Evolution 7: 342–365. [Google Scholar]
- Etges, W. J., and M. Levitan, 2004. Palaeoclimatic variation, adaptation and biogeography of inversion polymorphisms in natural populations of Drosophila robusta. Biol. J. Linn. Soc. 81: 395–411. [Google Scholar]
- Feder, J. L., S. H. Berlocher, J. B. Roethele, H. Darnbroski, J. J. Smith et al., 2003. a Allopatric genetic origins for sympatric host-plant shifts and race formation in Rhagoletis. Proc. Natl. Acad. Sci. USA 100: 10314–10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder, J. L., F. B. Roethele, K. Filchak, J. Niedbalski and J. Romero-Severson, 2003. b Evidence for inversion polymorphism related to sympatric host race formation in the apple maggot fly, Rhagoletis pomonella. Genetics 163: 939–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman, M. W., S. P. Otto and F. B. Christiansen, 1997. Population genetic perspectives on the evolution of recombination. Annu. Rev. Genet. 30: 261–295. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J., 1981. Skepticism towards Santa Rosalia, or why are there so few kinds of animals? Evolution 35: 124–138. [DOI] [PubMed] [Google Scholar]
- Gilburn, A. S., and T. H. Day, 1994. Sexual dimorphism, sexual selection and the alpha-beta chromosomal inversion polymorphism in the seaweed fly, Coelopa frigida. Proc. R. Soc. Lond. Ser. B 257: 303–309. [Google Scholar]
- Gilburn, A. S., and T. H. Day, 1999. Female mating behaviour, sexual selection and chromosome I inversion karyotype in the seaweed fly, Coelopa frigida. Heredity 82: 276–281. [DOI] [PubMed] [Google Scholar]
- Gomez, G. A., and E. Hasson, 2003. Transpecific polymorphisms in an inversion linked esterase locus in Drosophila buzzatii. Mol. Biol. Evol. 20: 410–423. [DOI] [PubMed] [Google Scholar]
- Haldane, J. B. S., 1927. The mathematical theory of natural and artificial selection. V. Selection and mutation. Proc. Camb. Philos. Soc. 23: 838–844. [Google Scholar]
- Haldane, J. B. S., 1937. The effect of variation on fitness. Am. Nat. 71: 337–349. [Google Scholar]
- Haldane, J. B. S., 1957. The conditions for coadaptation in polymorphism for inversions. J. Genet. 55: 218–225. [Google Scholar]
- Hartl, D. L., 1970. Analysis of a general population genetic model of meiotic drive. Evolution 24: 538–545. [DOI] [PubMed] [Google Scholar]
- Hartl, D. L., 1975. Modifier theory and meiotic drive. Theor. Popul. Biol. 7: 168–174. [DOI] [PubMed] [Google Scholar]
- Hedrick, P. W., 1981. The establishment of chromosomal variants. Evolution 35: 322–332. [DOI] [PubMed] [Google Scholar]
- Kehrer-Sawatzki, H., B. Schreiner, S. Tanzer, M. Platzer, S. Muller et al., 2002. Molecular characterization of the pericentric inversion that causes differences between chimpanzee chromosome 19 and human chromosome 17. Am. J. Hum. Genet. 71: 375–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrer-Sawatzki, H., C. A. Sandig, V. Goidts and H. Hameister, 2005. Breakpoint analysis of the pericentric inversion between chimpanzee chromosome 10 and the homologous chromosome 12 in humans. Cytogenet. Genome Res. 108: 91–97. [DOI] [PubMed] [Google Scholar]
- Kimura, M., 1956. A model of a genetic system which leads to closer linkage by natural selection. Evolution 10: 278–287. [Google Scholar]
- King, M., 1993. Species Evolution: The Role of Chromosomal Change. Cambridge University Press, Cambridge, UK.
- Kirkpatrick, M., T. Johnson and N. Barton, 2002. General models of multilocus evolution. Genetics 161: 1727–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick, M., and M. Servedio, 1999. The reinforcement of mating preferences on an island. Genetics 151: 865–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knibb, W. R., 1982. Chromosome inversion polymorphisms in Drosophila melanogaster. 2. Geographic clines and climatic associations in Australasia, North America and Asia. Genetica 58: 213–221. [Google Scholar]
- Krimbas, C. B., and J. R. Powell, 1992. Drosophila Inversion Polymorphism. CRC Press, London.
- Lande, R., 1979. Effective deme sizes during long-term evolution estimated from rates of chromosomal rearrangement. Evolution 33: 234–251. [DOI] [PubMed] [Google Scholar]
- Lande, R., 1984. The expected fixation rate of chromosomal inversions. Evolution 38: 743–752. [DOI] [PubMed] [Google Scholar]
- Lande, R., 1985. The fixation of chromosomal rearrangements in a subdivided population with local extinction and colonization. Heredity 54: 323–332. [DOI] [PubMed] [Google Scholar]
- Lemeunier, F., and S. Aulard, 1992. Inversion polymorphism in Drosophila melanogaster, pp. 339–405 in Drosophila Inversion Polymorphism, edited by C. B. Krimbas and J. R. Powell. CRC Press, Boca Raton, FL.
- Lenormand, T., and S. P. Otto, 2000. The evolution of recombination in a heterogeneous environment. Genetics 156: 423–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin, R. C., and M. J. D. White, 1960. Interaction between inversion polymorphisms of two chromosome pairs in the grasshopper, Moraba scurra. Evolution 14: 116–129. [Google Scholar]
- Li, W.-H., and M. Nei, 1974. Stable linkage disequilibrium without epistasis in subdivided populations. Theor. Popul. Biol. 6: 173–183. [DOI] [PubMed] [Google Scholar]
- Liberman, U., 1976. Modifier theory of meiotic drive: Is Mendelian segregation stable? Theor. Popul. Biol. 10: 127–132. [DOI] [PubMed] [Google Scholar]
- Liou, L. W., and T. D. Price, 1994. Speciation by reinforcement of premating isolation. Evolution 48: 1451–1459. [DOI] [PubMed] [Google Scholar]
- Lu, J., W.-H. Li and C.-I. Wu, 2003. Comment on “Chromosomal speciation and molecular divergence—accelerated evolution in rearranged chromosomes”. Science 302: 988–989. [DOI] [PubMed] [Google Scholar]
- Mikkelsen, T. S., L. W. Hillier, E. E. Eichler, M. C. Zody, D. B. Jaffe et al., 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437: 69–87. [DOI] [PubMed] [Google Scholar]
- Muller, H. J., 1918. Genetic variability, twin hybrids and constant hybrids, in a case of balanced lethal factors. Genetics 3: 422–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, H. J., 1939. Reversibility in evolution considered from the standpoint of genetics. Biol. Rev. Camb. Philos. Soc. 14: 261–280. [Google Scholar]
- Muller, H. J., 1940. Bearing of the Drosophila work on systematics, pp. 185–268 in The New Systematics, edited by J. S. Huxley. Claredon Press, Oxford.
- Muller, H. J., 1942. Isolating mechanisms, evolution, and temperature. Biol. Symp. 6: 71–125. [Google Scholar]
- Muller, H. J., 1950. Our load of mutations. Am. J. Hum. Genet. 2: 111–176. [PMC free article] [PubMed] [Google Scholar]
- Navarro, A., and N. H. Barton, 2003. a Chromosomal speciation and molecular divergence: accelerated evolution in rearranged chromosomes. Science 300: 321–324. [DOI] [PubMed] [Google Scholar]
- Navarro, A., and N. H. Barton, 2003. b Accumulating postzygotic isolation genes in parapatry: a new twist on chromosomal speciation. Evolution 57: 447–459. [DOI] [PubMed] [Google Scholar]
- Nei, M., 1967. Modification of linkage intensity by natural selection. Genetics 57: 625–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei, M., K. Kojima and H. Schaffer, 1967. Frequency changes of new inversions under mutation-selection equilibria. Genetics 57: 741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor, M. A. F., 1995. Speciation driven by natural selection in Drosophila. Nature 375: 674–675. [DOI] [PubMed] [Google Scholar]
- Noor, M. A. F., K. L. Grams, L. A. Bertucci and J. Reiland, 2001. a Chromosomal inversions and the reproductive isolation of species. Proc. Natl. Acad. Sci. USA 98: 12084–12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor, M. A. F., K. L. Grams, L. A. Bertucci, Y. Almendarez, J. Reiland et al., 2001. b The genetics of reproductive isolation and the potential for genetic exchange between Drosophila pseudoobscura and D. persimilis via backcross hybrid males. Evolution 55: 512–521. [DOI] [PubMed] [Google Scholar]
- Oakeshott, J. G., J. B. Gibson, P. R. Anderson, W. R. Knibb, D. G. Anderson et al., 1982. Alcohol dehydrogenase and glycerol-3-phosphate dehydrogenase clines in Drosophila melanogaster on different continents. Evolution 36: 86–96. [DOI] [PubMed] [Google Scholar]
- Ohta, T., 1966. A theoretical study of stochastic survival of inversion chromosomes. Ph.D. Thesis, North Carolina State University, Raleigh, NC.
- Ohta, T., 1971. Associative overdominance caused by linked detrimental mutations. Genet. Res. 18: 277–286. [PubMed] [Google Scholar]
- Ohta, T., and K. Kojima, 1968. Survival probabilities of new inversions in large populations. Biometrics 24: 501–516. [PubMed] [Google Scholar]
- Ortiz-Barrientos, D., B. A. Counterman and M. A. F. Noor, 2004. The genetics of speciation by reinforcement. PLoS Biol. 2: e416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prout, T., J. Bundgaard and S. Bryant, 1973. Population genetics of modifiers of meiotic drive I. The solution of a special case and some general implications. Theor. Popul. Biol. 4: 446–465. [DOI] [PubMed] [Google Scholar]
- Pylkov, K. V., L. A. Zhivotovsky and M. W. Feldman, 1998. Migration versus mutation in the evolution of recombination under multilocus selection. Genet. Res. 71: 247–256. [DOI] [PubMed] [Google Scholar]
- Rieseberg, L. H., J. Whitton and K. Gardner, 1999. Hybrid zones and the genetic architecture of a barrier to gene flow between two sunflower species. Genetics 152: 713–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg, L. H., 2001. Chromosomal rearrangements and speciation. Trends Ecol. Evol. 16: 351–358. [DOI] [PubMed] [Google Scholar]
- Roberts, P. A., 1976. The genetics of chromosomal aberration, pp. 67–184 in The Genetics and Biology of Drosophila, edited by M. Ashburner and E. Novitski. Academic Press, New York.
- Schluter, D., 2000. The Ecology of Adaptive Radiation. Oxford University Press, Oxford.
- Searle, J. B., 1993. Chromosomal hybrid zones in eutherian mammals, pp. 309–353 in Hybrid Zones and the Evolutionary Process, edited by R. G. Harrison. Oxford University Press, New York.
- Sites, J. W., and C. Moritz, 1987. Chromosomal evolution and speciation revisited. Syst. Zool. 36: 153–174. [Google Scholar]
- Slatkin, M., 1973. Gene flow and selection in a cline. Genetics 75: 733–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin, M., 1975. Gene flow and selection in a two-locus system. Genetics 81: 787–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant, A. H., 1917. Genetic factors affecting the strength of linkage in Drosophila. Proc. Nat. Acad. Sci. USA 3: 555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant, A. H., and K. Mather, 1938. The interrelations of inversions, heterosis and recombination. Am. Nat. 72: 447–452. [Google Scholar]
- Thomson, G. J., and M. W. Feldman, 1974. Population genetics of modifiers of meiotic drive. II. Linkage modification in the segregation distortion system. Theor. Popul. Biol. 5: 155–162. [DOI] [PubMed] [Google Scholar]
- Trickett, A. J., and R. K. Butlin, 1994. Recombination suppressors and the evolution of new species. Heredity 73: 339–345. [DOI] [PubMed] [Google Scholar]
- Tripet, F., G. Dolo and G. C. Lanzaro, 2005. Multilevel analyses of genetic differentiation in Anopheles gambiae s.s. reveal patterns of gene flow important for malaria-fighting mosquito projects. Genetics 169: 313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, J. R. G., 1967. a The evolution of supergenes. Am. Nat. 101: 195–221. [Google Scholar]
- Turner, J. R. G., 1967. b Why does the genotype not congeal? Evolution 21: 645–656. [DOI] [PubMed] [Google Scholar]
- van Delden, W., and A. Kamping, 1989. The association between the polymorphisms at the Adh and αGpdh loci and the In(2L)t inversion in Drosophila melanogaster in relation to temperature. Evolution 43: 775–793. [DOI] [PubMed] [Google Scholar]
- van Delden, W., and A. Kamping, 1991. Changes in relative fitness with temperature among 2nd chromosome arrangements in Drosophila melanogaster. Genetics 127: 507–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Valen, L., and R. Levins, 1968. The origins of inversion polymorphisms. Am. Nat. 102: 5–24. [Google Scholar]
- Veuille, M., V. Bénassi, S. Aulard and F. Depaulis, 1998. Allele-specific population structure of Drosophila melanogaster alcohol dehydrogenase at the molecular level. Genetics 149: 971–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace, B., 1968. Topics in Population Genetics. W. W. Norton, New York.
- Walsh, J. B., 1982. Rate of accumulation of reproductive isolation by chromosomal rearrangements. Am. Nat. 120: 510–532. [Google Scholar]
- Wasserman, M., 1968. Recombination-induced chromosomal heterosis. Genetics 58: 125–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley, C. S., and W. F. Eanes, 1994. Isolation and analysis of the breakpoint sequences of chromosome inversion In(3L)Payne in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 91: 3132–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, A. C., G. L. Bush, S. M. Case and M. C. King, 1975. Social structuring of mammalian populations and rate of chromosomal evolution. Proc. Natl. Acad. Sci. USA 72: 5061–5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, M. J. D., 1973. Animal Cytology and Evolution. Cambridge University Press, Cambridge, UK.
- White, M. J. D., 1978. Modes of Speciation. W. H. Freeman, San Francisco.
- Wright, S., 1941. On the probability of fixation of reciprocal translocations. Am. Nat. 75: 513–522. [Google Scholar]
- Wright, S., and T. Dobzhansky, 1946. Genetics of natural populations. XII. Experimental reproduction of some of the changes caused by natural selection in certain populations of Drosophila pseudoobscura. Genetics 31: 125–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, S. Y., J. Shetty, L. H. Hou, A. Delcher, B. L. Zhu et al., 2004. Human, mouse, and rat genome large-scale rearrangements: stability versus speciation. Genome Res. 14: 1851–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]