Abstract
A major unresolved challenge of evolutionary biology is to determine the nature of the allelic variants of “speciation genes”: those alleles whose interaction produces inviable or infertile interspecific hybrids but does not reduce fitness in pure species. Here we map quantitative trait loci (QTL) affecting fertility of male hybrids between D. yakuba and its recently discovered sibling species, D. santomea. We mapped three to four X chromosome QTL and two autosomal QTL with large effects on the reduced fertility of D. yakuba and D. santomea backcross males. We observed epistasis between the X-linked QTL and also between the X and autosomal QTL. The X chromosome had a disproportionately large effect on hybrid sterility in both reciprocal backcross hybrids. However, the genetics of hybrid sterility differ between D. yakuba and D. santomea backcross males, both in terms of the magnitude of main effects and in the epistatic interactions. The QTL affecting hybrid fertility did not colocalize with QTL affecting sexual isolation in this species pair, but did colocalize with QTL affecting the marked difference in pigmentation between D. yakuba and D. santomea. These results provide the basis for future high-resolution mapping and ultimately, molecular cloning, of the interacting genes that contribute to hybrid sterility.
UNDERSTANDING the genetic basis of speciation—the splitting of a group of interbreeding populations into two reproductively isolated groups—is a major challenge of evolutionary biology. Yet the framework in which we must seek to determine the genetic basis of postzygotic reproductive isolation—the inviability or sterility of interspecific offspring—has been clear from the beginning of the last century. First, hybrid dysfunction must be caused by deleterious epistatic interactions (“incompatibilities”) between genes that function perfectly well in pure-species backgrounds. Dobzhansky (1937) and Muller (1940) proposed a simple two-locus model that explains how such incompatibilities could arise if different alleles at the two loci become fixed in the two species. In this model, the ancestral species has genotype A1A1B1B1, and its two descendant species have genotypes A1A1B2B2 and A2A2B1B1, respectively. There would be no selection against deleterious interactions between A1 and A2 or B1 and B2 in the pure species, but such interactions may exist between alleles A1 and B2 in the separate lineages, causing inviability or sterility of the A1A2B1B2 hybrids. Second, Haldane (1922, p. 101) noted that: “When in the F1 offspring of two different animal races one sex is absent, rare, or sterile, that sex is the heterozygous [heterogametic] sex.” One of the causes of Haldane's rule thus is likely to be incompatibilities involving the sex chromosomes with each other or with autosomes (other factors probably also contribute to Haldane's rule; see Coyne and Orr 2004).
Thus, to understand the genetic architecture of postzygotic reproductive isolation, we must identify the interacting pairs (or more) of genes affecting inviability and fertility. Most progress to date has been made by studying hybridizations of the four Drosophila species, Drosophila melanogaster, D. simulans, D. sechellia, and D. mauritiana (Coyne and Orr 2004). D. simulans, D. mauritiana, and D. sechellia are particularly suitable for the genetic analysis of factors affecting male hybrid fertility, since crosses between D. simulans and either of the other two species yield sterile males but fertile females that can be backcrossed to either species. (Since these species are allopatric and fertile hybrids can be produced, one could argue that full speciation between them has not yet been completed.) Early work used mapping with visible morphological markers to show that genes affecting male fertility in interspecific backcross males mapped to all chromosomes except the fourth (Coyne 1984; Coyne et al. 1991; Coyne and Berry 1994), including the Y chromosome (Johnson et al. 1993; Joly et al. 1997; Dermitzakis et al. 2000), and that the X chromosome had a disproportionately large effect (for review see Coyne and Orr 2004). Further genetic dissection revealed at least three X-linked factors affecting sperm motility in hybrid D. simulans/D. mauritiana and D. simulans/D. sechellia backcross males (Coyne and Charlesworth 1986, 1989). One region with a large effect on hybrid male fertility, closely linked to forked (Coyne and Charlesworth 1986, 1989), later mapped to the Odysseus (Ods) locus (Perez et al. 1993), which was eventually localized to a 8.4-kb region containing a rapidly evolving homeobox gene (Ting et al. 1998). However, the genetic architecture of hybrid male sterility is typically highly polygenic. High-resolution analysis of the other two X-linked factors of large effect revealed at least seven genes with smaller effects on sperm mobility in D. simulans/D. mauritiana hybrid males (Cabot et al. 1994; Davis and Wu 1996). A second factor with small effect, closely linked to Ods, is required for full sterility when this region of D. mauritiana is introgressed into D. simulans (Perez and Wu 1995). Further complexity arises in identifying individual loci since the same genes do not affect hybrid male sterility in reciprocal introgressions of the same region of D. simulans into the D. mauritiana background (Palopoli and Wu 1994).
High-resolution mapping has also shed light on the genetic mechanisms underlying the large effect of the X chromosome on sterility of male hybrids between these species. Autosomal introgressions have largely recessive effects on hybrid male sterility (Hollocher and Wu 1996; True et al. 1996; Tao et al. 2003a), but there are also proportionately more genes affecting hybrid male sterility on the X chromosome than on the autosomes (Tao et al. 2003a). Thus, Haldane's rule for sterility of male hybrids between these species is attributable to both the recessivity of genes causing hybrid incompatibilities and a higher rate of evolution of genes affecting hybrid male sterility, perhaps through sexual selection (reviewed by Laurie 1997; Coyne and Orr 2004).
The extent to which these features of the genetic architecture of loss of fitness in interspecific hybrids are general can be addressed only by equally comprehensive studies of additional pairs of species. The recent discovery of D. santomea, a species endemic to the island of São Tomé (Lachaise et al. 2000), has greatly expanded the opportunities for genetic analysis. Molecular phylogenetic analysis shows that D. yakuba and D. santomea are sister species within the D. melanogaster subgroup (Lachaise et al. 2000; Cariou et al. 2001). This pair represents a speciation event independent of that separating the ancestor of D. melanogaster from that of the D. simulans triad, as well as the two speciation events in which a D. simulans-like ancestor produced its island descendants D. sechellia and D. mauritiana (Lachaise et al. 1988).
D. santomea and D. yakuba show substantial sexual isolation when tested in the laboratory and differ in male genital morphology and in pigmentation (D. yakuba has the black abdominal pigmentation typical of all other species in the D. melanogaster group, while D. santomea lacks any pigmentation (Lachaise et al. 2000; Coyne et al. 2005). In interspecific crosses, F1 male hybrids are sterile but female hybrids are fertile; the latter can be backcrossed to either parental species (Lachaise et al. 2000; Cariou et al. 2001). In addition to behavioral isolation and intrinsic hybrid sterility, the species also show conspecific sperm precedence: when D. yakuba females are multiply inseminated by both conspecific and D. santomea males, they produce very few hybrid progeny, regardless of the order of mating (Chang 2004). Here, we use quantitative trait locus (QTL) mapping to identify chromosomal regions affecting fertility in D. yakuba/D. santomea male backcross hybrids. These data offer a comparison to previous work on other traits within this species pair, comparisons to loci for fertility in other species pairs, and a basis for future high-resolution genetic analysis.
MATERIALS AND METHODS
Drosophila strains:
All flies were maintained in 8-dram vials containing standard cornmeal-agar-Karo media on a 12:12 hr light:dark cycle at 24°. We used the D. yakuba ST and D. santomea STO.4 strains described in the accompanying article (Moehring et al. 2006, this issue) as parental strains for QTL mapping. D. yakuba ST was derived from the isofemale strain D. yakuba Taï18 by eliminating the polymorphic inversion distinguishing it from D. santomea, thus creating a strain homosequential with D. santomea for improved mapping resolution (Moehring et al. 2006).
Crosses:
Previous studies (Lachaise et al. 2000; Coyne et al. 2004) have shown that crosses between these species produce fertile F1 females but sterile F1 males. To produce F1 females, 4-day-old virgin D. yakuba ST females were crossed to D. santomea STO.4 males. Virgin F1 females were then backcrossed (BC) in two ways: (A) to D. yakuba ST males, producing 550 BC males, and (B) to D. santomea STO.4 males, producing 550 BC males. Male backcross hybrids have autosomal loci with one chromosome that is pure (A) D. yakuba or (B) D. santomea while the other chromosome is a mixture of the two parental genomes; X-linked loci and the Y chromosome are either pure (A) D. yakuba or (B) D. santomea.
Fertility assay:
Four-day-old virgin males were scored for fertility by removing their testes and lightly crushing them in Ringer's solution under a coverslip (Coyne et al. 2004). To provide a quantitative measure of fertility, each male was scored for degree of fertility on an eight-point scale of increasing degree of motile sperm formation: 0, testes completely atrophied; 1, testes of normal size, no spermatids or sperm; 2, testes of normal size containing spermatids but no free sperm; 3, testes of normal size containing a few free sperm, but none of them motile; 4, testes of normal size, many free sperm but none of them motile; 5, testes of normal size, a few free sperm of which a few were motile; 6, testes of normal size, a few free sperm, most of which were motile; 7, testes of normal size, many free sperm of which a few were motile; and 8, testes of normal size, many free sperm of which many were motile. A few individuals had one atrophied testis and one larger testis; these contained no sperm or spermatids and thus were scored as “1.” All scoring was done by a single observer.
Molecular markers and genotypes:
Flies were immediately frozen at −80° after they were scored for fertility. The sample then underwent DNA extraction and marker-specific PCR amplification and restriction endonuclease digestion. Each of the 1100 samples was scored for 32 molecular markers that are roughly evenly spaced throughout the genome, as described in the accompanying article by Moehring et al. (2006, this issue). The markers and their cytological (relative to D. melanogaster; Drysdale et al. 2005) and recombination map positions are: y, 1A5, 0.00; per, 3B1–2, 2.62; sog, 13E1, 17.89; v, 9F11, 25.23; rux, 5D2, 53.01; f, 15F4–7, 59.89; bnb, 17D6, 68.61; Hex-A, 8E10, 72.41; AnnX, 19C1, 77.16; su(f), 20E, 79.77; l(2)gl, 21A5, 0.00; Rad1, 23A1, 6.65; RpL27A, 24F3, 13.47; salr, 32E4–F1, 29.49; Rep4, 34B4, 37.29; His3, 39D3–E1, 57.75; barr, 38B1–2, 62.62; Sara, 57E6, 73.70; Ngp, 54C8, 90.25; Kr, 60F5, 150.65; Lsp1γ, 61A6, 0.00; dib, 64A5, 19.74; sfl, 65B3–4, 33.04; Est-6, 69A1, 63.36; Ssl1, 80B2, 90.35; ry, 87D9, 101.96; Rpn5, 83C4, 121.42; AP-50, 94A15–16, 130.17; Mlc1, 98A14–15, 150.90; ymp, 96E, 160.32; krz, 100E3, 176.99; and ci, 102A1–3, 0.00.
QTL mapping:
QTL for fertility were mapped three different ways in each backcross population. First, fertility was treated as a continuous trait using the full range of scores (see above). Second, whether or not spermatids were present was considered as a binary trait since this is the stage to which spermatogenesis proceeds for one direction of the cross (D. yakuba mothers) but not the other (D. santomea mothers). Third, whether or not sperm were present was considered as a binary trait, for comparison to previous work. The assumption of normality in composite interval mapping (CIM) (Zeng 1994) is violated when fertility is considered as a binary trait. However, we showed previously (Moehring et al. 2004) that using an extension of CIM on the basis of logistic regression (Xu and Atchley 1996), which assumes that the binary trait is connected to its continuous underlying liability by a threshold model (Falconer and Mackay 1996), gives the same results as CIM performed on binary data for large sample sizes.
QTL were mapped using CIM, implemented using QTL Cartographer software (Basten et al. 1999), exactly as described in the accompanying article (Moehring et al. 2006, this issue). Empirical experimentwise 5% significance thresholds, which take account of the multiple tests performed and correlations among markers, were determined by permutation (Churchill and Doerge 1994; Doerge and Churchill 1996). The effects of each QTL were estimated as the difference between heterozygous D. santomea/D. yakuba genotypes and homozygous pure species genotypes at the peak likelihood ratio (LR), scaled by the phenotypic standard deviation. The approximate boundaries of regions containing QTL were determined by taking 2-LOD intervals (9.22 LR) surrounding the point of greatest significance and estimating the cytological location of the interval by dividing the cytology within the region according to the observed amount of recombination between flanking markers (for the D. yakuba cytology relative to D. melanogaster, see Moehring et al. 2006, this issue). We evaluated pairwise epistatic interactions using either the marker positioned at the highest LR of each QTL peak or the haplotype of the two markers flanking the QTL peak. Tests for epistasis were calculated for the binary data with a log linear model using PROC CATMOD and for overall fertility with an ANOVA using PROC GLM, using SAS 8.2 software (SAS Institute, Cary, NC).
RESULTS AND DISCUSSION
Fertility of pure species and F1 hybrids:
Table 1 describes the fertility assay of the two pure species and the two reciprocal F1 hybrids. As expected, the pure species were fertile and the hybrid males sterile. In the two pure species, >90% of males had some motile sperm (stages 5–8), with most falling into stage 8 (testes of normal size, many free sperm of which many are motile). In contrast, none of the F1 hybrids had any motile sperm. However, there was a difference in the stage of spermatogenesis attained in the two reciprocal hybrids: nearly all males with a D. santomea mother had testes of normal size but no spermatids or free sperm (stage 1), while male offspring of the reciprocal cross had testes of normal size and spermatids whose differentiation had not progressed to sperm (stage 3). This difference in the degree of spermatogenesis between the two reciprocal hybrids, which is highly significant (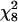 = 182.6, P < 0.001), was also described by Coyne et al. (2004) (Table 3).
= 182.6, P < 0.001), was also described by Coyne et al. (2004) (Table 3).
TABLE 1.
Fertility of males of pure species and F1 hybrids
| Stage of spermatogenesis
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Genotype | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
| S | 0 | 1 | 2 | 3 | 0 | 5 | 10 | 1 | 82 |
| Y | 0 | 0 | 2 | 0 | 0 | 6 | 12 | 5 | 75 |
| F1, S × Y | 6 | 93 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| F1, Y × S | 0 | 4 | 94 | 2 | 0 | 0 | 0 | 0 | 0 |
At least 100 individuals were scored for each genotype. S = D. santomea STO.4 strain; Y = D. yakuba ST strain. For F1 hybrids, the genotype of the female parent is given first. See materials and methods for definitions of spermatogenesis stages.
TABLE 3.
Epistatic interactions between marker regions
| Fertility QTL | Trait | Marker 1 | Marker 2 | P-value |
|---|---|---|---|---|
| D. yakuba BC males | Fertility | per–sog | v–rux | 0.0070* |
| ry | 0.026 | |||
| Rpn5 | 0.05 | |||
| Spermatids | per–sog | I(2)gl | 0.045 | |
| RpL27A | 0.04 | |||
| Ngp | 0.038 | |||
| Est-6 | 0.02 | |||
| v–rux | His3 | 0.016 | ||
| barr | 0.009 | |||
| ry | 0.005 | |||
| Rpn5 | 0.006 | |||
| AP-50 | 0.017 | |||
| ymp | 0.049 | |||
| Sperm | per–sog | HexA–su(f) | 0.0057* | |
| ry | 0.032 | |||
| AP-50 | 0.027 | |||
| Rad1 | 0.024 | |||
| RpL27A | 0.014 | |||
| D. santomea BC males | Fertility | per–sog | Ssl1 | 0.016 |
| v | barr | 0.0013* | ||
| Ssl1 | 0.007 | |||
| bnb | barr | 0.033 | ||
| Ssl1 | 0.026 | |||
| Spermatids | per–sog | rux–f | 0.02 | |
| bnb | 0.0029* | |||
| His3 | 0.0002* | |||
| barr | 0.0002* | |||
| v | rux–f | 0.012* | ||
| bnb | 0.0005 | |||
| salr | 0.0003* | |||
| Rep4 | 0.0002* | |||
| Ngp | 0.0001* | |||
| rux–f | barr | 0.0004* | ||
| bnb | barr | 0.0002* |
Pairwise epistatic interactions were calculated using the marker at the highest LR peak or the haplotype of the two markers flanking a peak. Trait is fertility scored as a continous trait (fertility), the presence/absence of spermatids (spermatids), or the presence/absence of sperm (sperm). * denotes formal significance after Bonferroni correction.
QTL for fertility in D. yakuba backcross males:
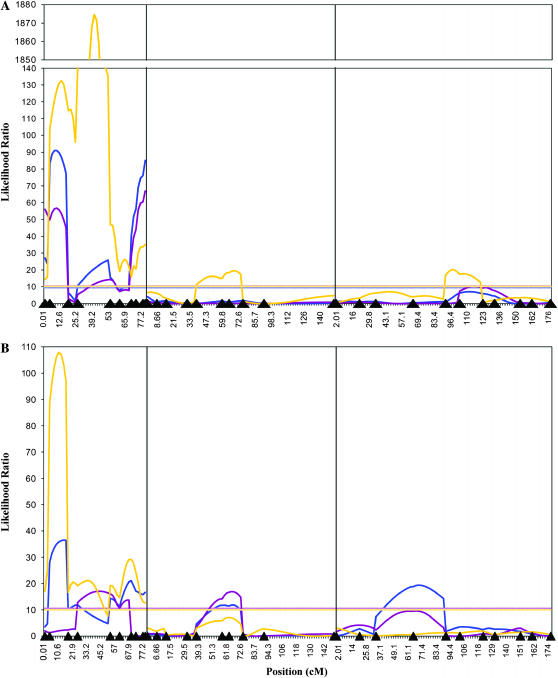
We mapped three X chromosome QTL with large effects on the reduced fertility of D. yakuba backcross males, in the per–sog (3B–13A), v–rux (8D–5D), and AnnX–su(f) (19C–20E) intervals when fertility was considered as a continuous trait. We use the cytological locations when reporting the regions (rather than recombination distance) as the end goal is to locate the genes responsible for hybrid sterility, which will most likely be accomplished through comparisons to the genome of the model system of D. melanogaster, making the cytology, rather than the recombination distance, of relevance. The same three QTL were detected when fertility was considered as a binary trait of presence or absence of sperm, although these QTL tended to map to larger intervals, consistent with the lower power of mapping threshold traits (Xu and Atchley 1996) (Table 2, Figures 1A and 2). We identified six QTL when the presence or absence of spermatids was considered as a dichotomous trait: per–sog (11E–13A), v–rux (7D–7B), f–bnb (15F–18B), su(f) (9A–20E), Rep4–Sara (34B–57E), and Ssl1–ry (82A–83C). Particularly striking is the effect of the v–rux interval, which explains >47% of the variance for this trait.
TABLE 2.
QTL affecting fertility in offspring of crosses between D. yakuba and D. santomea
| BC cross | Trait | QTL | Peak | LR | Effect | Effect/σp | R2 |
|---|---|---|---|---|---|---|---|
| F1 females × D. yakuba males | Fertility (scored 0–8) | 3B–13A | 11E | 91.08 | 1.48 | 0.72 | 0.1178 |
| 8D–5D | 5C | 25.88 | 0.91 | 0.44 | 0.0294 | ||
| 19C–20E | 20E | 85.05 | 1.69 | 0.82 | 0.0978 | ||
| Presence/absence of spermatid | 11E–13A | 12D | 132.49 | 0.43 | 20.11 | 0.1561 | |
| 7D–7B | 7C | 1874.56 | 1.01 | 47.60 | 0.7298 | ||
| 15F–18B | 16F | 26.28 | 0.29 | 13.47 | 0.0332 | ||
| 9A–20E | 20E | 35.14 | 0.24 | 11.46 | 0.0339 | ||
| 34B–57E | 49A | 19.52 | −0.14 | −6.83 | 0.0196 | ||
| 82A–83C | 84B | 20.19 | −0.15 | −6.99 | 0.0215 | ||
| Presence/absence of sperm | 1A–13A | 11E | 56.80 | 0.23 | 14.50 | 0.0865 | |
| 8A–15D | 5D | 14.35 | 0.13 | 8.11 | 0.0173 | ||
| 9D–20E | 20E | 66.95 | 0.29 | 18.41 | 0.0897 | ||
| F1 females × D. santomea males | Fertility (scored 0–8) | 3B–13E | 12F | 36.53 | 0.60 | 0.40 | 0.0592 |
| 14A–18C | 19B | 11.95 | 0.44 | 0.29 | 0.0178 | ||
| 5D–20E | 17F | 21.11 | 0.47 | 0.31 | 0.0317 | ||
| 41F–51E | 49A | 11.84 | 0.33 | 0.21 | 0.0197 | ||
| 65F–80B | 70D | 19.38 | 0.42 | 0.28 | 0.0324 | ||
| Presence/absence of spermatid | 11B–13A | 12B | 107.91 | 0.40 | 21.24 | 0.1849 | |
| 13E–7A | 8C | 21.10 | 0.27 | 14.14 | 0.0504 | ||
| 5C–16A | 5D | 19.26 | 0.18 | 9.46 | 0.0259 | ||
| 16F–9B | 17F | 29.26 | 0.19 | 10.38 | 0.0387 | ||
| Presence/absence of sperm | 19B–17D | 7C | 17.07 | 0.11 | 8.84 | 0.0342 | |
| 45F–57E | 50A | 16.92 | 0.10 | 8.09 | 0.0295 |
QTL regions are estimated from 2 LOD support intervals (P ≤ 0.05) and the cytological locations were extrapolated from recombination rate between markers in comparison to the D. yakuba cytological map. The peak is the cytological location with the highest likelihood ratio (LR). Effects were estimated from the least-squares means of the two genotype classes as [homozygous − heterozygous] and are also listed when scaled by dividing by the phenotypic standard deviation (σp). R2 is the proportion of the variance accounted for the QTL and is estimated by R2 = (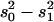 )/s2, where s2 is the variance of the trait,
)/s2, where s2 is the variance of the trait, 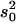 is the sample variance of the residuals, and
is the sample variance of the residuals, and 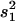 is the variance of the residuals (Basten et al. 1999).
is the variance of the residuals (Basten et al. 1999).
Figure 1.
QTL for the X, second, and third chromosomes affecting fertility in backcross hybrids between D. yakuba and D. santomea. There were no QTL for the small fourth chromosome. (A) F1 females backcrossed to D. yakuba males. The y-axis from likelihood ratio (LR) 140–1850 is truncated. (B) F1 females backcrossed to D. santomea males. Plots are the LR test statistics for overall fertility score (blue), presence of spermatids (yellow), and presence of sperm (purple) as determined by composite interval mapping. The significance thresholds were determined by permutation testing, are represented by correspondingly colored dashed horizontal lines, and are all approximately LR = 10. Marker locations are represented by black triangles on the x-axis and are in the same order from left to right as the order listed in materials and methods: y, per, sog, v, rux, f, bnb, Hex-A, AnnX, su(f), l(2)gl, Rad1, RpL27A, salr, Rep4, His3, barr, Sara, Ngp, Kr, Lsp1γ, dib, sfl, Est-6, Ssl1, ry, Rpn5, AP-50, Mlc1, ymp, and krz. Note that markers are spaced according to recombination distance.
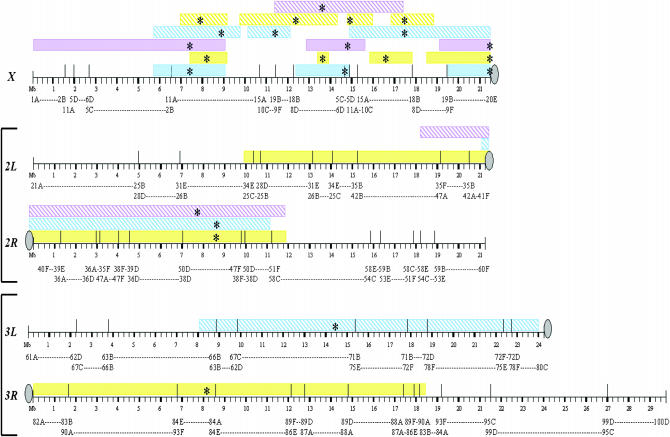
Figure 2.
The significant regions from QTL mapping when compared to D. melanogaster cytology. Short vertical lines below the horizontal are every 250 kb; the short thick lines are every 1 Mb. Tall vertical lines above the horizontal represent the inversion/translocation breakpoints. Colored boxes represent QTL peaks: overall fertility (blue), presence of spermatids (yellow), and presence of sperm (purple) scored for BC males from F1 females × D. yakuba males (solid boxes) and BC males from F1 females × D. santomea males (hatched boxes). * indicates the peak of the QTL. Centromeres are represented by a gray circle. Open red triangles represent markers, with the markers in the same order as listed in materials and methods: y, per, sog, v, rux, f, bnb, Hex-A, AnnX, su(f), l(2)gl, Rad1, RpL27A, salr, Rep4, His3, barr, Sara, Ngp, Kr, Lsp1γ, dib, sfl, Est-6, Ssl1, ry, Rpn5, AP-50, Mlc1, ymp, and krz. Note that markers are spaced according to base pair distance.
Thus at least four genes are responsible for hybrid sterility when considered as a continuous trait or for presence or absence of sperm, since the D. santomea X-linked loci must interact with at least one D. yakuba autosomal or Y-linked locus. In this backcross we mapped autosomal QTL only for the presence of spermatids. If D. santomea X-linked loci interact with D. yakuba Y-linked loci, this interaction could be at least partially responsible for reduced fertility of backcross males. We also note that the third chromosome region from ry-AP-50 (87D9–94A16) approached formal significance (LR = 10.17) for presence of sperm and further that epistatic interactions may occur between X-linked QTL and autosomal QTL that do not themselves have a main effect on fertility.
We tested for epistatic interactions among the QTL (Table 3) in two ways. First, we considered only interactions between QTL having significant main effects on hybrid fertility and used a Bonferroni correction to assess significance. We found a significant interaction for fertility scored on the continuous scale between the QTL in the per–sog interval and the QTL in the v–rux interval. In addition, we observed an epistatic interaction between the QTL in the per–sog interval and the QTL at the base of the X chromosome [HexA–su(f), 9D–20E] for fertility considered as the dichotomous trait of presence/absence of sperm. The backcross design affords us the opportunity to test for X–autosome interactions that may define pairs of loci contributing to hybrid incompatibilities. Therefore, we assessed interactions between the three X chromosome QTL and all autosomal markers, even though autosomal markers were formally significant only for presence of spermatids.
We found interactions between the QTL in the per–sog interval and the ry and Rpn5 markers on chromosome 3 and the RpL27A marker on chromosome 2, for the continuous fertility data. For the dichotomous trait presence of spermatids, interactions were observed between the QTL in the per–sog interval and the l(2)gl, RpL27A, and Ngp markers on chromosome 2 and Est-6 on chromosome 3. Epistatic interactions for this trait were also identified between the v–rux QTL interval and His3, barr, ry, Rpn5, AP-50, and ymp. It is interesting to note that the v–rux interval, which had the highest effect on this trait, also had the greatest number of epistatic interactions. Considering the presence of sperm, we found epistatic interactions between the per–sog interval QTL and the third chromosome markers ry and AP-50 and the second chromosome markers Rad1 and Rpl27A. All of the interactions were in the expected direction, with double heterozygotes (and hemi-heterozygotes for X markers) being the least fertile and double homozygotes (and hemi-homozygotes) the most fertile. Although these interactions would not be significant if corrected for multiple tests, it is possible that the hybrid incompatibilities in this backcross are at least partially attributable to interactions between X chromosome QTL and recessive or partially recessive autosomal QTL.
These results are consistent with those of Coyne et al. (2004), who used visible morphological markers to estimate the relative contributions of the X chromosome and autosomes to the reduced fertility (scored as presence or absence of motile sperm) of D. yakuba backcross males. Coyne et al. (2004) observed a large effect of the X chromosome and showed that this effect was attributable to at least three genes, in the regions between w (at cytological location 3B) and g (12B), g and sn (7D), and sn to the base of the chromosome. These regions exactly match the QTL mapped here by linkage to molecular markers. Coyne et al. (2004) also detected a smaller but significant effect of the second chromosome marker no (notch). However, this effect was in the direction of increased sterility of the homozygous no D. yakuba genomic segment relative to the heterozygous no/+ D. yakuba/D. santomea genotype, possibly implying that homozygosity for no itself causes a reduction of fertility in a hybrid background. If this explanation is true, we would not expect to detect an effect of the no region when using molecular markers to map QTL affecting hybrid sterility, as indeed observed in this study. Finally, Coyne et al. (2004) noted a significant effect of the D. yakuba Y chromosome, consistent with a role of X–Y interactions in hybrid sterility.
QTL for fertility in D. santomea backcross males:
We mapped three X chromosome QTL and two autosomal QTL with large effects on the reduced fertility of D. santomea backcross males, when fertility is considered as a continuous trait (Table 2, Figures 1B and 2). The X chromosome QTL mapped to the per–sog (3B–13E), sog–rux (14A–18C), and rux–su(f) (5D–20E) intervals, and the autosomal QTL mapped to the region from His3–barr (41F–51E) on chromosome 2 and the region from sfl–Ssl1 (65F–80B) on chromosome 3. Again, the X chromosome has a disproportionately large effect on hybrid sterility. Four QTL were detected on the X chromosome when fertility was measured according to whether or not spermatids were present, in the per–sog (11B–13A), v (13E–7A), rux–f (5C–16A), and bnb (16F–9B) regions. We also detected two QTL for hybrid fertility when considering fertility as the threshold trait of presence of sperm, one on the X chromosome in the v–rux interval (19B–17D) and one on chromosome 2 in the Rep4–Sara interval (45F–57E). The results from the different measurements of fertility are in good agreement, since two of the X chromosome QTL overlap, the second chromosome QTL is detected in two of the analyses, and there is a strong suggestion of linkage on chromosome 3 in the threshold model in the sfl–Ssl1 region (LR = 9.56) (Table 2, Figures 1B and 2).
When we evaluated epistatic interactions between QTL with significant main effects on fertility considered as a continuous trait in D. santomea backcross males, we observed an interaction between the X-linked QTL at 14A–18C (near v) and the second chromosome QTL. In addition, we observed multiple interactions between X chromosome QTL and the autosomal QTL when we tested for all possible X–autosome interactions. The QTL in the per–sog interval interacts with the third chromosome QTL, with a peak at Ssl1. The QTL near v interacts with both the second and third chromosome QTL, with peak associations at barr and Ssl1, as does the QTL near bnb, also with peak associations at barr and Ssl1. When fertility is measured as presence of spermatids, we observed that the per–sog QTL had epistatic interactions with both the rux–f and bnb QTL and that v also interacted with both the rux–f and bnb QTL. When testing for all possible X–autosome interactions, the per–sog interval interacted with most of the autosomal regions, with formally significant peaks at His3 and barr; v significantly interacted with salr, Rep4, and Ngp; and both rux-f and bnb had significant interactions with barr. As with the epistatic interactions in D. yakuba backcross males, all of the interactions were in the expected direction, with double heterozygotes (and hemi-heterozygotes) being the least fertile and double homozygotes (and hemi-homozygotes) the most fertile.
Thus, the genetics of hybrid sterility are partially overlapping but not identical between D. yakuba and D. santomea backcross males, in terms of both main effects of and epistatic interactions between QTL (Table 2, Figures 1 and 2). Fine mapping these QTL to smaller intervals will resolve whether the basis of sterility in reciprocal hybridizations between D. yakuba and D. santomea is due to the same genetic loci.
QTL affecting sexual isolation and pigmentation differences between D. yakuba and D. santomea:
In the accompanying article, we mapped QTL affecting the discrimination of D. yakuba males against pure-species D. santomea females by pairing D. yakuba backcross hybrid males with D. santomea females (Moehring et al. 2006, this issue). There is one scenario under which we might expect the same QTL to affect hybrid mating behavior and fertility, and that is if sexual selection is driving the rapid evolution of male hybrid sterility. However, this appears not to be the case. The QTL affecting sexual isolation in D. yakuba male backcross hybrids all mapped to the autosomes (Moehring et al. 2006), while the QTL affecting reduced fertility in D. yakuba male backcross hybrids mapped mainly to the X chromosome (this report).
We also mapped QTL affecting the marked difference in pigmentation between D. yakuba and D. santomea (Carbone et al. 2005). The same four QTL affected variation in pigmentation in D. yakuba and in D. santomea male backcross hybrids: 1A5–13E1 (y–sog), 15F4–20E [f–su(f)], 34B4–57E6 (Rep4–Sara), and 69A1–83C4 (Est6–Rpn5). Strikingly, these QTL overlap with the QTL affecting reduced male hybrid fertility at the tip and the base of the X chromosome and the chromosome 2 locus in D. yakuba and D. santomea backcross males and with the chromosome 3 QTL affecting reduced male hybrid fertility in the D. santomea backcross males. Is it possible that the coincidence of QTL affecting postzygotic reproductive isolation and morphological variation is a byproduct of adaptive fixation of genes affecting loss of black pigmentation in D. santomea, as predicted from the Dobzhansky-Muller model of evolution of hybrid incompatibility (Dobzhansky 1937; Muller 1940)? If so, one expects that the same genes have pleiotropic effects on both traits, an hypothesis that can be tested by simultaneous high-resolution mapping of pigmentation and fertility in male interspecific hybrids.
QTL affecting hybrid fertility in other Drosophila species pairs:
Our data fully recapitulate the most prominent feature of the genetic architecture of hybrid male sterility observed in other Drosophila species pairs, namely, a disproportionately large effect of the X chromosome (reviewed by Coyne and Orr 2004). In addition, the low amount of QTL overlap seen here when comparing reciprocal hybridizations has also been observed in reciprocal hybridizations of D. simulans/D. mauritiana (Perez et al. 1993; Palopoli and Wu 1994). The breadth of the QTL peaks observed in this study, spanning several markers, also suggests that high-resolution mapping will reveal multiple closely linked QTL affecting male fertility in D. santomea/D. yakuba hybrids, as observed for D. simulans/D. mauritiana hybrids (Cabot et al. 1994; Palopoli and Wu 1994; Perez and Wu 1995; Davis and Wu 1996; True et al. 1996; Tao et al. 2003b). While we must be reserved in speculating about candidate genes given the rather coarse resolution of our mapping, we note that Ods (at 16D) is in the X-linked QTL for all three fertility-scoring methods in males resulting from a backcross to D. santomea, but is only within the QTL for the presence of spermatids in males from the backcross to D. yakuba.
Acknowledgments
We thank Justin Cassidy and Sarah Powers for technical assistance and Mary Anna Carbone and Ted Morgan for helpful discussions. This work was funded by National Institutes of Health research grants to J.A.C. (GM 58260) and T.F.C.M. (GM45344, GM 58260). This is a publication of the W. M. Keck Center for Behavioral Biology.
References
- Basten, C. J., B. S. Weir and Z-B. Zeng, 1999. QTL Cartographer: Version 1.13. Department of Statistics, North Carolina State University, Raleigh, NC.
- Cabot, E. L., A. W. Davis, N. A. Johnson and C.-I Wu, 1994. Genetics of reproductive isolation in the Drosophila simulans clade: complex epistasis underlying hybrid male sterility. Genetics 137: 175–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone, M. A., A. Llopart, M. DeAngelis, J. Coyne and T. F. C. Mackay, 2005. Quantitative trait loci affecting the difference in pigmentation between Drosophila yakuba and D. santomea. Genetics 171: 211–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cariou, M.-L., J.-F. Silvain, V. Daubin, J.-L. Dalage and D. Lachaise, 2001. Divergence between Drosophila santomea and allopatric or sympatric populations of D. yakuba using paralogous amylase genes and migration scenarios along the Cameroon volcanic line. Mol. Ecol. 10: 649–660. [DOI] [PubMed] [Google Scholar]
- Chang, A. S., 2004. Conspecific sperm precedence in sister species of Drosophila with overlapping ranges. Evolution 58: 781–789. [DOI] [PubMed] [Google Scholar]
- Churchill, G. A., and R. W. Doerge, 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne, J. A., 1984. Genetic basis of male sterility in hybrids between two closely related species of Drosophila. Proc. Natl. Acad. Sci. USA 81: 4444–4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne, J. A., and A. Berry, 1994. Effects of the fourth chromosome on the sterility of hybrids between Drosophila simulans and its relatives. Heredity 85: 224–227. [DOI] [PubMed] [Google Scholar]
- Coyne, J. A., and B. Charlesworth, 1986. Location of an X-linked factor causing sterility in male hybrids of Drosophila simulans and D. mauritiana. Heredity 57: 243–246. [DOI] [PubMed] [Google Scholar]
- Coyne, J. A., and B. Charlesworth, 1989. Genetic analysis of X-linked sterility in hybrids between three sibling species of Drosophila. Heredity 62: 97–106. [DOI] [PubMed] [Google Scholar]
- Coyne, J. A., and H. A. Orr, 2004. Speciation. Sinauer Associates, Sunderland, MA.
- Coyne, J. A., J. Rux and J. R. David, 1991. Genetics of morphological differences and hybrid sterility between Drosophila sechellia and its relatives. Genet. Res. 57: 113–122. [DOI] [PubMed] [Google Scholar]
- Coyne, J. A., S. Elwyn, S. Y. Kim and A. Llopart, 2004. Genetic studies of two sister species in the Drosophila melanogaster subgroup, D. yakuba and D. santomea. Genet. Res. 84: 11–26. [DOI] [PubMed] [Google Scholar]
- Coyne, J. A., S. Elwyn and E. Rolán-Alvarez, 2005. Impact of experimental design on Drosophila sexual isolation studies: direct effects and comparison to field hybridization data. Evolution 59: 2588–2601. [PubMed] [Google Scholar]
- Davis, A. W., and C.-I Wu, 1996. The broom of the sorcerer's apprentice: the fine structure of a chromosomal region causing reproductive isolation between two sibling species of Drosophila. Genetics 143: 1287–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermitzakis, E. T., J. P. Masly, H. M. Waldrip and A. G. Clark, 2000. Non-Mendelian segregation of sex chromosomes in heterospecific Drosophila males. Genetics 154: 687–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky, T., 1937. Genetics and the Origin of Species. Columbia University Press, New York.
- Doerge, R. W., and G. A. Churchill, 1996. Permutation tests for multiple loci affecting a quantitative character. Genetics 142: 285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drysdale, R. A., M. A. Crosby and The FlyBase Consortium, 2005. FlyBase: genes and gene models. Nucleic Acids Res. 33: D390–D395 (http://flybase.org/). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer, D. S., and T. F. C. Mackay, 1996. Introduction to Quantitative Genetics, Ed. 4. Longman, London. [DOI] [PMC free article] [PubMed]
- Haldane, J. B. S., 1922. Sex ratio and unisexual sterility in animal hybrids. J. Genet. 12: 101–109. [Google Scholar]
- Hollocher, H., and C.-I Wu, 1996. The genetics of reproductive isolation in the Drosophila simulans clade: X vs. autosomal effects and male vs. female effects. Genetics 143: 1243–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, N. A., H. Hollocher, E. Noonburg and C.-I Wu, 1993. The effects of interspecific Y chromosome replacements on hybrid sterility within the Drosophila simulans clade. Genetics 135: 443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly, D., C. Bazin, L. W. Zeng and R. S. Singh, 1997. Genetic basis of sperm and testis length differences and epistatic effect on hybrid inviability and sperm motility between Drosophila simulans and D. sechellia. Heredity 78: 354–362. [DOI] [PubMed] [Google Scholar]
- Lachaise, D., M. L. Cariou, J. R. David, F. Lemeunier, L. Tsacas et al., 1988. Historical biogeography of the Drosophila melanogaster species subgroup. Evol. Biol. 22: 159–225. [Google Scholar]
- Lachaise, D., M. Harry, M. Solignac, F. Lemeunier, V. Bénassi et al., 2000. Evolutionary novelties in islands: Drosophila santomea, a new melanogaster sister species from São Tomé. Proc. R. Soc. Lond. Ser. B 267: 1487–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie, C. C., 1997. The weaker sex is heterogametic: 75 years of Haldane's rule. Genetics 147: 937–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehring, A. J., J. Li, M. D. Schug, S. Smith, M. DeAngelis et al., 2004. Quantitative trait loci for sexual isolation between Drosophila simulans and D. mauritiana. Genetics 167: 1265–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehring, A. J., A. Llopart, S. Elwyn, J. A. Coyne and T. F. C. Mackay, 2006. The genetic basis of prezygotic reproductive isolation between Drosophila santomea and D. yakuba due to mating preference. Genetics 173: 215–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, H. J., 1940. Bearing of the Drosophila work on systematics, pp. 185–268 in The New Systematics, edited by J. Huxley. Clarendon Press, Oxford.
- Palopoli, M. F., and C.-I. Wu, 1994. Genetics of hybrid male sterility between Drosophila sibling species: a complex web of epistasis is revealed in interspecific studies. Genetics 138: 329–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez, D. E., and C.-I Wu, 1995. Further characterizations of the Odysseus locus of hybrid sterility in Drosophila: one gene is not enough. Genetics 140: 201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez, D. E., C.-I Wu, N. A. Johnson and M.-L. Wu, 1993. Genetics of reproductive isolation in the Drosophila simulans clade: DNA marker-assisted mapping and characterization of a hybrid-male sterilty gene, Odysseus (Ods). Genetics 143: 261–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, Y., S. Chen, D. L. Hartl and C. C. Laurie, 2003. a Genetic dissection of hybrid incompatibilities between Drosophila simulans and D. mauritiana. I. Differential accumulation of hybrid male sterility effects on the X and autosomes. Genetics 164: 1383–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, Y., Z-B. Zeng, J. Li, D. L. Hartl and C. C. Laurie, 2003. b Genetic dissection of hybrid incompatibilities between Drosophila simulans and D. mauritiana. II. Mapping hybrid male sterility loci on the third chromosome. Genetics 164: 1399–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting, C.-T., S.-C. Tsaur, M.-L. Wu and C.-I Wu, 1998. A rapidly evolving homeobox at the site of a hybrid sterility gene. Science 282: 1501–1504. [DOI] [PubMed] [Google Scholar]
- True, J. R., B. S. Weir and C. C. Laurie, 1996. A genome-wide survey of hybrid incompatibility factors by the introgression of marked segments of Drosophila mauritiana chromosomes into Drosophila simulans. Genetics 142: 819–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, S., and W. R. Atchley, 1996. Mapping quantitative trait loci for complex binary diseases using line crosses. Genetics 143: 1417–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Z-B., 1994. Precision mapping of quantitative trait loci. Genetics 136: 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]