Abstract
The novel family of SPOC domain proteins is composed of broadly conserved nuclear factors that fall into two subclasses, termed large and small, based on protein size. Members of the large subgroup, which includes Drosophila SPEN and human SHARP, have been characterized as transcriptional corepressors acting downstream of a variety of essential cell signaling pathways, while those of the small subclass have remained largely unstudied. Since SPEN has been implicated in Drosophila eye development, and the small SPOC protein NITO is also expressed in the developing eye, we have used this context to perform a structure–function analysis of NITO and to examine the relationship between the two SPOC family subclasses. Our results demonstrate that the phenotypes obtained from overexpressing NITO share striking similarity to those associated with loss of spen. Dosage-sensitive genetic interactions further support a model of functional antagonism between NITO and SPEN during Drosophila eye development. These results suggest that large and small SPOC family proteins may have opposing functions in certain developmental contexts.
CONSERVED signaling pathways are used reiteratively throughout development to specify the cell and tissue types composing an adult organism. Since these pathways do not function independently of each other, cells must receive and respond to multiple interconnected signals. One critical strategy for information integration occurs at the level of the nuclear effectors of these signal transduction cascades that act in a concerted fashion to regulate expression of target genes required for proper development. While in most cases the underlying molecular mechanisms remain poorly understood, the use of large-scale, unbiased genetic screens in model systems such as Drosophila has proven to be a powerful approach to identify and dissect these conserved nuclear circuitries.
One potential mediator of nuclear signal integration identified by such screens is split ends (spen). A role for spen as a nuclear effector was first revealed in several independent genetic screens designed to isolate new downstream players in the Drosophila receptor tyrosine kinase (RTK) signaling pathway (Dickson et al. 1996; Rebay et al. 2000; Therrien et al. 2000). From these and subsequent investigations, spen has been positioned as a positive regulator and/or effector of RTK-mediated signaling events in multiple developmental contexts, including the eye and the embryonic central nervous system (Chen and Rebay 2000; Rebay et al. 2000). Additional studies have implicated spen in a diverse spectrum of cellular processes including neuronal cell fate specification and survival, axon guidance, cell cycle, hox gene regulation, and cell positioning (Kolodziej et al. 1995; Gellon et al. 1997; Staehling-Hampton et al. 1999; Wiellette et al. 1999; Chen and Rebay 2000; Kuang et al. 2000; Lane et al. 2000; Rebay et al. 2000; Brumby et al. 2004; Mace and Tugores 2004; Mutsuddi et al. 2004).
Importantly, spen appears to operate downstream of multiple signaling pathways. In addition to its role in RTK-mediated signaling events, spen functions as a context-specific positive regulator of Wingless signaling and as a likely regulator of Notch signaling (Oswald et al. 2002; Schreiber et al. 2002; Kuroda et al. 2003; Lin et al. 2003). Together these results suggest a complex role for SPEN as a nuclear effector and potential integrator of multiple signaling pathways.
spen encodes the founding member of a family of proteins characterized by three N-terminal RNA recognition motifs (RRMs) and a novel C-terminal domain, called the SPEN paralog ortholog conserved domain, or SPOC domain (Figure 1A)(Wiellette et al. 1999; Kuang et al. 2000; Rebay et al. 2000). SPEN orthologs have been identified in worms, flies, mosquitos, mice, humans, and other vertebrates, and more recent studies have identified proteins in plants and yeast carrying the SPOC domain in conjunction with other functional motifs (Sanchez-Pulido et al. 2004). The RRMs suggest a role for SPOC family proteins in RNA or DNA binding and in the case of SPEN are necessary for nuclear localization (I. Rebay, unpublished data), while the SPOC domain of SPEN and its human and mouse orthologs SHARP (SMRT/HDAC1 associated repressor protein) and MINT (Msx2-interacting nuclear target protein) have been implicated in transcriptional regulation and repression (Shi et al. 2001; Oswald et al. 2002; Kuroda et al. 2003; Yang et al. 2005).
Figure 1.
Alignment of conserved SPOC domain across multiple species. Residues with * are divergent between large and small SPOC proteins. Alignments and similarity/identity comparisons were performed with MacVector's ClustalW program. Accession numbers for GenBank sequences are as follows: Dm NITO, NP_610339; Dm SPEN, NP_722616; Hs SHARP, Q96T58; HsOTT/RBM15, CAC38861.1; Mm MINT, Q62504; Mm RBM15, AAH57038; Ce SPEN, E88320; CeNITO, AAC1912. Abbreviations for species are as follows: Dm, Drosophila melanogaster; Hs, Homo sapiens; Mm, Mus musculus; and Ce, Caenorhabditis elegans. Numbers refer to amino acid residues.
SPOC family proteins can be further divided into two subclasses on the basis of their size. We will refer to members of these subclasses as “large” SPOC family proteins, which include SPEN, MINT, and SHARP, and “small” SPOC family proteins, which are also well conserved from worms to humans. In contrast to large SPOC family proteins, almost nothing is known about the functions of small SPOC proteins. Thus far, only the human small SPOC family member one twenty two (OTT)/RNA-binding motif protein-15 (RBM15) has been studied. Specifically, chromosomal translocations identified in cases of acute megakaryocytic leukemia revealed a fusion with MAL (megakaryocytic acute leukemia)/MKL1 (megakaryoblastic leukemia-1) that results in a chimeric protein that includes almost the entire coding region of both genes, with RBM15/OTT at the N terminus and MAL/MKL1 at the C terminus (Ma et al. 2001; Mercher et al. 2001). Recent evidence suggests that the RBM15–MKL1 fusion may contribute to leukemogenesis through an increased ability to activate serum response factor (SRF) target genes (Cen et al. 2003).
As the Drosophila genome encodes both large and small SPOC family proteins, SPEN and SPENITO (NITO), respectively, this provides an opportunity for comparing the two SPOC subfamilies in a genetically tractable system. To explore the relationship between small and large SPOC family proteins we have examined the effects of genetically manipulating NITO levels in the eye, a tissue in which spen loss of function results in developmental defects (Dickson et al. 1996; Rebay et al. 2000). We found that overexpression of nito perturbs eye development, resulting in phenotypes similar to those observed in spen mutants and suggesting the possibility of functional antagonism between NITO and SPEN in this context. Dosage-sensitive genetic interactions between spen and nito further support an antagonistic relationship between these two genes during eye development.
MATERIALS AND METHODS
Drosophila stocks and transgenic lines:
The UAS-nito full-length construct was generated by fusing the GH11110 cDNA sequence, derived from a Berkeley Drosophila Genome Project clone, in frame to a 5′-Myc tag downstream of a UAS promoter (pUAST). UAS-nitoΔC was generated by fusing the 5′-fragment of nito, corresponding to amino acids 2–593, to a 5′-Myc tag downstream of UAS promoter (pUAST). UAS-nitoΔN was made by fusing two 3′-nito fragments, corresponding to amino acids 471–793 and generated from GH11100 cDNA by PCR, to a 5′-Myc tag downstream of a UAS promoter (pUAST). The 3′ fragment contains an SV40 NLS inserted by PCR with specific primers.
nito-RNAi was generated against the 5′ end of the nito coding region using previously described methodology (Kalidas and Smith 2002). A 606-bp fragment of nito genomic DNA was amplified using primers 5′-RI Dm44A G (5′-CAGAATTCGAGTAGTCATCGAGACGGAGCCGG-3′) and 3′-H3 Dm44A G (5′-CTTAAGCTTCTGCAAAGCATCTTAGATTAGCCAAGG-3′), and a 548-bp cDNA fragment, corresponding to the reverse complement of the genomic sequence but lacking the internal intron, was amplified from full length nito cDNA using primers 5′-H3 Dm44A cDNA (5′-CTTAAGCTTCTATATTCCGGTCTGGTTGTGG-3′) and 3′-KpnI Dm44A cDNA (5′- CTGGTACCGAGTAGTCATCGAGACGGAGC-3′). The genomic fragment was cut with EcoRI and HindIII and the cDNA fragment cut with HindIII and KpnI and both fragments were ligated into a pUAST vector digested with EcoRI and KpnI. UAS-myc-nito-FL, UAS-myc-nitoΔC, UAS-myc-nitoΔN, and UAS-nito RNAi were used to generate transgenic lines as previously described (Rebay et al. 1993).
The following fly stocks were used: w1118; sevmed-Gal4, UAS-spenDN (Lin et al. 2003); sev-Gal4 (weak and medium generated by I. Rebay and strong from Bloomington Stock Center); spenxfm911/Cyo (Rebay et al. 2000).
Western blots:
Western blot to examine expression of UAS-nito transgenes was carried out by crossing UAS lines to hsp70Gal4, heat-shocking adults for 1 hr, followed by a 1.5 hr recovery. A total of 25 fly heads were collected for each sample, homogenized in 2× SDS buffer, and run on a gel. Protein levels were examined using mouse anti-MYC mAb 9E10 (1:500; a gift from R. Fehon).
Immunohistochemistry:
Larval eye discs were dissected in Drosophila S2 cell media, fixed in 4% paraformaldehyde in 1× PBS for 10 min at room temperature, washed 3 times in PT (1× PBS + 0.1% Triton X-100), blocked 1 hr in PNT (1× PBS + 0.1% Triton X-100 + 1% NGS). Primary antibodies were incubated overnight at 4° in PNT on a rotator. Mouse anti-elav (Developmental Studies Hybridoma Bank) was used at 1:50. Samples were then washed in PT and incubated with goat anti-mouse HRP (1:500; Jackson ImmunoResearch, West Grove, PA) in PNT for 2 hr, washed, and developed. For acridine orange staining, larval eye discs were dissected in S2 media, incubated with 1 mm acridine orange diluted 1:500 in S2 media for 10 min, rinsed, and mounted in 1× PBS for immediate viewing.
RT–PCR analysis:
Sixty pairs of eye discs were dissected from control (w: GMR-GAL4) and GMR-GAL4> nito-RNAi third instar larvae. Total RNA was isolated using Trizol Reagent (Invitrogen) according to the manufacturer's instructions and cDNA was synthesized using random primers (Promega) from 1 μg of total RNA. PCR amplification was performed with nito primers 5′-AGGTTCCTCTTCTTCAGTTCCCCC-3′ and 5′-TTGGTGTCGTTTGTGGACCCTG-3′ and Rps17 primers 5′-CGAACCAAGACGGTG AAGAAG-3′ and 5′-CCTGCAACTTGATGGAGATACC-3′ to compare expression levels. NIH Image was used to quantitate expression levels of nito relative to Rps17. Each experiment was performed twice.
RESULTS AND DISCUSSION
Sequence conservation defines two distinct SPOC family subclasses:
SPOC family proteins fall into two apparent subclasses based on their size. To determine whether such a distinction might be functionally significant, sequence alignments of the conserved C-terminal SPOC motif were performed to compare the level of sequence conservation in the SPOC family in general and subclass members in particular (Figure 1). Analysis revealed only 27% identity and 50% overall similarity between the SPOC domains of SPEN and NITO, the Drosophila representatives of the large and small subfamilies, respectively; however, upon comparison of the SPOC domains of these proteins with those of their respective subclass family members, a higher level of conservation was revealed. Drosophila SPEN and human SHARP exhibit 58% sequence identity and 79% overall sequence similarity, while Drosophila NITO and human RBM15/OTT share 47% sequence identity and 62% overall sequence similarity. Comparable results were obtained by comparing the RRM motifs (data not shown). These results reveal a higher level of sequence conservation within SPOC family subclasses relative to the family in general, raising the possibility that subclasses may have adopted divergent functions.
Overexpression of nito perturbs adult eye morphology:
To better understand the relationship between large and small SPOC proteins, we were interested in determining if spen and nito function synergistically or antagonistically in vivo. Because the large SPOC family member spen is required for Drosophila eye development (Dickson et al. 1996; Rebay et al. 2000; Lin et al. 2003) and the fly eye provides a uniquely powerful system in which to explore functional relationships between signaling molecules (Zipursky and Rubin 1994), we focused our analyses on this tissue. RT–PCR confirmed that nito, like spen, is expressed in the developing eye disc (Figure 4 and data not shown), further validating the approach.
Figure 4.
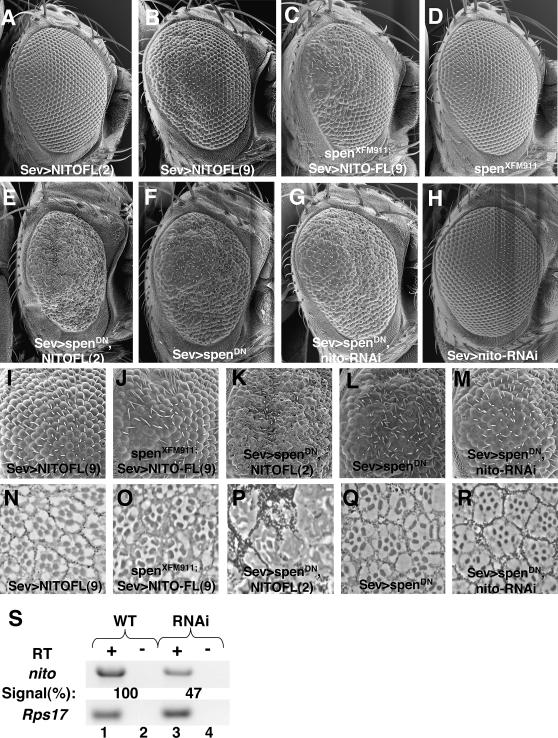
Nito and spen function antagonistically. (A–H) Scanning electron micrograph (SEM) of adult eyes, oriented posterior to the left. (I–M) Higher magnification view of the posterior region of eyes shown in C–G. (N–R) Eye sections of genotypes indicated in I–M. (A) UAS-nito-FL(2)/+; sevweak-GAL4/+. (B, I, and N) UAS-nito-FL(9)/+; sevweak-GAL4/+. (C, J, and O) spenXFM911/UAS-nito-FL(9); sevweak-GAL4/+. (D) spenXFM911/CyO. (E, K, and P) UAS-nito-FL(2)/+; sevweak-GAL4, UAS-spenDN/TM6B. (F, L, and Q) sevweak-GAL4, UAS-spenDN/TM6B. (G, M, and R) sevweak-GAL4, UAS-spenDN/UAS-nito-RNAi. (H) sevweak-GAL4/UAS-nito-RNAi. Numbers in parentheses [(2) and (9)] refer to independent transgenic insertions of UAS-nito-FL. (S) RNAi efficiently knocks down nito expression. RT–PCR with nito (top) and Rps17 primers (bottom) with RT (+; lanes 1 and 3) and without RT control (−; lanes 2 and 4). The percentage of nito expression in GMR-GAL4>RNAi discs relative to wild-type eye discs is normalized to expression levels for the Rps17 control.
Because no nito mutants are currently available, an in vivo structure–function analysis was undertaken to investigate nito function during eye development. While the phenotypes resulting from overexpression of a gene must be interpreted with caution, such overexpression models frequently result in sensitized genetic systems that can provide powerful tools for investigating in vivo relationships between signaling molecules. Myc-tagged full-length NITO (NITO-FL), NITO lacking the N terminus (NITOΔN; an exogenous nuclear localization sequence was added to ensure proper nuclear targeting), and NITO lacking the C terminus (NITOΔC) were cloned downstream of a UAS promoter and the transgenes were expressed in flies using eye-specific GAL4 drivers (Figure 2A). Three different sevenless-Gal4 (sev-Gal4) drivers, which promote expression in photoreceptors R1, R3, R4, R6, R7, the cone cells, and the “mystery” cells, which are poorly understood interommatidial cells that are never recruited to the ommatidia and ultimately apoptose, were utilized in this study: sevstrong couples the sev enhancer to the hsp70 promoter, resulting in the highest levels of expression; sevmedium contains both the sev enhancer and sev promoter and expresses at an intermediate level; sevweak contains the same regulator sequences as sevmedium but expresses at lower levels, presumably as a consequence of position effect of the transgene. To avoid unnecessary confusion, we will refer to these collectively as sev-Gal4 and will identify the specific driver in the figure legends as appropriate.
Figure 2.
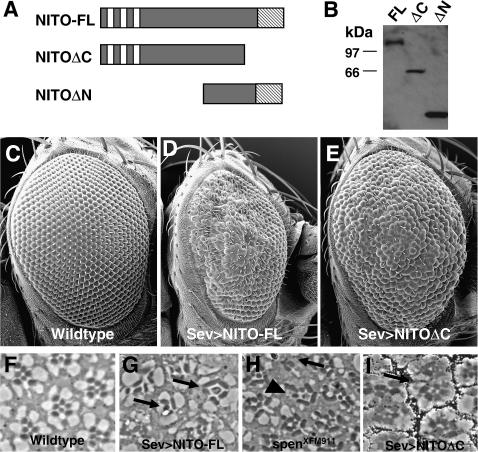
Overexpression of nito results in rough eye phenotypes. (A) Overexpression of nito constructs. (B) Western blot with anti-myc antibody using head lysates from hsp70Gal4;UAS-nito flies. (C–E) SEM of adult eyes. (C) Wild-type (w1118) control. (D) sevmedium-GAL4/UAS-nito-FL. (E) sevstrong-GAL4/UAS-nitoΔC. (F–H) Sections of adult eyes. (F) Wild-type (w1118) control. (G) sevmedium-GAL4/UAS-nito-FL. Arrows indicate ommatidia missing photoreceptors. (H) spenXFM911/spenXFM911 mutant eye clones. Arrow indicates ommatidium missing photoreceptors. Arrowhead indicates disorganized ommatidium. (I) sevstrong-GAL4/UAS-nitoΔC. Arrow indicates ommatidial fusion.
Sev-Gal4-driven overexpression of NITO-FL and NITOΔC yielded dosage-dependent adult rough eye phenotypes (Figure 2, C–E), while overexpression of NITOΔN was indistinguishable from wild type (data not shown). Western blots confirmed the expression of all transgenes (Figure 2B), and immunohistochemistry showed nuclear localization of NITO in all cases (data not shown), indicating that the lack of a NITOΔN phenotype is not due to the absence or mislocalization of protein.
While overexpression of NITO-FL and NITOΔC both perturb eye development, the resulting phenotypes are distinct. These data are not unexpected given previous results for the large SPOC family protein, SPEN, in which overexpression of SPENΔC functions as a dominant negative with respect to spen (Chen and Rebay 2000). We therefore speculate that NITOΔC functions analogously as a dominant negative relative to nito, whereas NITO-FL expression simply augments the pool of full-length NITO. Specifically, we observe that overexpression of NITO-FL results in roughening of the posterior part of the eye and an overall decrease in eye size (Figure 2D), whereas overexpression of NITOΔC more uniformly perturbs the external morphology of the eye (Figure 2E).
To distinguish the NITO-FL and NITOΔC rough eye phenotypes at the cellular level, adult eyes were sectioned and examined for defects. In wild-type ommatidia, photoreceptors are arranged in a trapezoidal array with seven of the eight photoreceptors visible in one plane of view (Figure 2F). The regular trapezoidal arrangement of photoreceptors is disturbed in both overexpression systems (Figure 2, G–H). When NITO-FL is overexpressed we see a decrease in the number of photoreceptors per ommatidia, elongated rhabdomeres, as well as a general disorganization of the ommatidia (Figure 2G). These observations suggest that the rough eye phenotype is due to a loss of photoreceptors and possible defects in the accessory cells, which normally provide support for the rhabdomeres in the ommatidia. This phenotype is strikingly reminiscent of that seen in sections of spen mutant eye clones (Figure 2H)(Dickson et al. 1996), raising the possibility that overexpressed nito may function antagonistically with respect to spen in the developing eye.
Eyes overexpressing NITOΔC also appear disorganized compared to wild type, although in contrast to NITO-FL ommatidia, photoreceptor number is not strongly affected. Rather, the most prevalent defect appears to be ommatidial fusions (Figure 2I) suggesting that cone and pigment cells, rather than photoreceptors, are most affected. Given that the Gal4 driver used for these experiments is expressed primarily in a subset of photoreceptors, the cone cells and interommatidial mystery cells, the accessory cell defects we observe upon nito overexpression may be due in part to indirect effects on pigment cells. Thus, NITO-FL ommatidia have defects in photoreceptor number and ommatidial morphology, while NITOΔC ommatidia have defects in accessory cells required for the spacing of ommatidia.
nito overexpression impairs cell survival in the developing eye:
To further investigate the defects caused by overexpressing nito, we examined the effects of increasing nito expression in early eye development. First, we examined recruitment of the photoreceptor neurons into ommatidia by looking at expression of the pan-neural marker ELAV in the larval precursor to the eye, the eye imaginal disc (Figure 3, A–C). Consistent with the differences observed in the adult phenotypes, the larval phenotypes associated with sev-Gal4-driven expression of NITO-FL and NITOΔC are also distinct.
Figure 3.
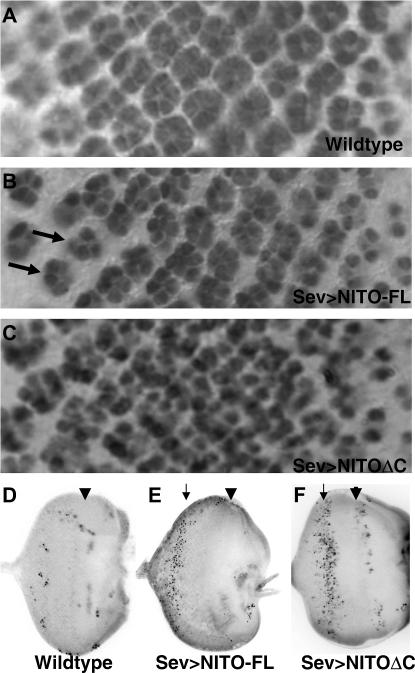
Developmental defects associated with nito overexpression. Eye imaginal discs are oriented posterior to the left. (A–C) Elav staining of third instar larval eye imaginal discs. (A) Wild-type (w1118) control. (B) sevstrong-GAL4/UAS-nito-FL. Arrows indicate ommatidial clusters missing photoreceptors. (C) sevstrong-GAL4/UAS-nitoΔC. (D–F) Acridine orange staining of third instar larval eye discs. Location of cell death is indicated with arrow; arrowhead indicates the morphogenetic furrow. (D) Wild-type (w1118) control. (E) sevstrong-GAL4/UAS-nito-FL. (F) sevstrong-GAL4/UAS-nitoΔC.
In eye discs overexpressing NITO-FL, initial recruitment of photoreceptors appears normal (Figure 3B). However, approximately seven rows posterior to the furrow there is a decrease in the number of photoreceptors per ommatidium. Thus while NITO-FL expression does not perturb initial photoreceptor recruitment, subsequent development and/or survival are compromised, resulting in the reduced number of photoreceptors observed in the adult eye. The loss of photoreceptors upon overexpression of NITO-FL is also similar to spen mutant clones, which have reduced numbers of photoreceptors in mutant ommatidia in the developing imaginal disc (D. Doroquez and I. Rebay, unpublished data), consistent with the observations made in adult eye sections (Figure 2). In contrast to NITO-FL and spen mutant clones, and consistent with the ommatidial fusions observed in adult eye sections, overexpression of NITOΔC causes loss of spacing between ommatidia in the larval eye disc, while recruitment of photoreceptors is not affected (Figure 3C).
To examine the possibility that the phenotypes associated with overexpression of NITO-FL and NITOΔC were due primarily to cell death, we stained eye discs with the apoptotic marker acridine orange (Figure 3, D–F). In the wild-type eye disc very little cell death is observed (Figure 3D). In NITO-FL eye discs a stripe of cell death occurs in the posterior part of the differentiating eye disc (Figure 3E), consistent with the loss of photoreceptors observed in the ELAV-probed eye disc (Figure 3B) and similar to the elevated cell death phenotype observed in spen mutant clones (D. Doroquez and I. Rebay, unpublished data). However, coexpression of the apoptotic inhibitor p35 or introduction of the H99 deficiency that removes the proapoptotic genes hid, reaper, and grim did not suppress the NITO-FL rough eye phenotypes (data not shown), suggesting that increased apoptotic cell death is unlikely to be the primary factor contributing to the NITO-FL-associated eye defects. In discs overexpressing NITOΔC, increased cell death is observed more anteriorly relative to that for NITO-FL (Figure 3F vs. 3E), consistent with the ommatidial spacing defects observed in the ELAV-probed disc (Figure 3C).
SPEN and NITO act antagonistically:
The potential for functional antagonism between SPEN and NITO was suggested by the similarity of phenotypes observed in adult eye sections overexpressing NITO-FL and in spen mutant eye clones. To further investigate this potential antagonism, we performed a series of dose-sensitive genetic interactions between spen and nito.
First, we examined the effects of reducing spen levels in the NITO-FL overexpression background. If NITO-FL antagonizes SPEN function, as suggested by our phenotypic analysis, further reducing spen should exacerbate the NITO-FL overexpression phenotype. An important requirement for such an experiment is the need for dose-sensitive NITO-FL phenotypes. Two observations suggest NITO-FL provides a dose-sensitive phenotype ideal for studying genetic interactions: first, expression of independent transgenic lines with the same sev-Gal4 driver results in a range of phenotypes (Figure 4, A and B); and second, expression of a given NITO-FL transgene with sevweak results in a mild rough eye phenotype (Figure 4B), whereas expression of the same line at a higher level using the sevmedium produces a more severe phenotype (Figure 2D). Consistent with our hypothesis of an antagonistic relationship between spen and nito, we found that heterozygosity for a null spen allele enhanced the rough eye phenotype associated with NITO-FL expression, as demonstrated by an increased number of ommatidia lacking photoreceptors (Figure 4, B–D, I, J, N, and O).
Next, we investigated the consequences of increasing or decreasing nito levels in the background of a dominant negative spen transgene (spenDN), which encodes the C-terminal 936 amino acids of spen (Lin et al. 2003) and also produces dose-sensitive phenotypes (D. Doroquez and I. Rebay, unpublished results). Because both transgenes are capable of perturbing eye development on their own, to distinguish between additive and synergistic interactions we used a NITO-FL transgenic line that when expressed with sevweak exhibits only very mild perturbations of the adult eye (Figure 4A). As expected given the NITO structure–function analysis, NITO-FL causes an enhancement of the spenDN rough eye phenotype, an increase in necroses in the eye, and a complete loss of organization (Figure 4, A, E, F, K, L, P, and Q). Thus, overexpression of nito and overexpression of spenDN appear to act in the same direction, suggesting opposing functions for NITO and SPEN.
As loss-of-function mutations in nito have not been isolated, we generated a nito transgenic dsRNA construct to investigate the consequences of reducing endogenous nito expression levels with respect to spen function. RT–PCR from Drosophila eye discs confirmed that this construct mediates partial knockdown of nito expression (Figure 4S). In vivo, while dsRNA-mediated knockdown of nito expression does not perturb eye morphology on its own, nito-RNAi partially rescues the rough eye phenotype resulting from overexpression of spenDN (Figure 4, G, H, L, M, Q, and R), again suggesting antagonism between nito and spen. Eye sections show fewer missing ommatidia in nito-RNAi, spenDN adult eyes relative to those overexpressing spenDN alone, as well as fewer missing photoreceptors in ommatidia lacking the full complement of photoreceptors and more normal rhabdomere morphology (Figure 4, Q and R). Together, these dose-sensitive genetic interactions argue for mutual antagonism between the large SPOC family member spen and the small SPOC family representative nito during Drosophila eye development.
It remains to be determined if the antagonistic relationship between nito and spen is maintained in developmental contexts outside of the eye. Previous work examining the role of SPEN in Wingless signaling suggested the presence of a redundant partner for SPEN (Lin et al. 2003), for which NITO would be a good candidate, given their sequence conservation. In situ hybridization for nito and spen suggests that both are also ubiquitously expressed throughout embryonic development (data not shown), and considering the broad range of embryonic phenotypes attributed to spen mutants (Kolodziej et al. 1995; Chen and Rebay 2000; Kuang et al. 2000), exploration of context-specific interactions between spen and nito in the embryo will likely improve our understanding of the relationships between these two related proteins. We predict that certain developmental events will require synergism between nito and spen, whereas others, as we demonstrate in the eye, will require antagonism.
At the cellular level, spen is implicated as a positive component of Wingless and RTK/RAS signaling (Chen and Rebay 2000; Rebay et al. 2000; Lin et al. 2003), and large SPOC family proteins SHARP and MINT are implicated as negative regulators of Notch signaling (Oswald et al. 2002; Kuroda et al. 2003). Given the ability of nito to antagonize spen function in the developing eye, it seems reasonable to speculate that NITO also acts as a downstream regulator/effector of some or all of these pathways. Furthermore, the antagonism between nito and spen may provide a mechanism for differential regulation of output from these pathways.
Mechanistically, how might one envision the mutual antagonism between SPEN and NITO? Large SPOC proteins have been previously shown to serve as transcriptional corepressors (Shi et al. 2001; Oswald et al. 2002; Kuroda et al. 2003; Yang et al. 2005). Thus one attractive possibility is that small SPOC proteins might serve as transcriptional activators. In this model, by virtue of their conserved RRM and SPOC motifs, small and large SPOC proteins might compete for access to common binding partners. The resulting complexes, depending on whether they contain SPEN or NITO, would then either repress or activate transcription. In a slight variation of the model, one could propose that SPOC proteins might be able to either repress or activate transcription and so depending on context would act either synergistically or antagonistically. Unfortunately, we have found that Drosophila cultured cells do not provide an appropriate environment in which to assay the activity of SPOC proteins (J. Jemc, D. Doroquez and I. Rebay, unpublished results) so we have not been able to test this model with respect to SPEN and NITO. However, using mammalian COS cells, we have observed that while the SPOC motif of SHARP represses transcription as previously published (Shi et al. 2001; Oswald et al. 2002), the SPOC motif of RBM15, the human NITO ortholog, strongly activates transcription (J. Jemc and I. Rebay, unpublished data). Thus, perhaps the antagonistic relationship between SPEN and NITO that we report here in the context of Drosophila eye development reflects a conserved antagonistic relationship between large and small SPOC proteins that is manifested at the level of transcriptional output.
In conclusion, we have demonstrated an antagonistic relationship between the large and small SPOC family proteins in the developmental context of the Drosophila eye. The finding that SPOC family proteins function as downstream effectors of a variety of signaling pathways suggests they may act to fine tune transcriptional output downstream of these cascades. Thus, it will be extremely interesting to determine whether the antagonistic relationship we have observed between NITO and SPEN in the eye is a general property of small and large SPOC proteins, or if it is unique to Drosophila eye development. Determination of transcriptional targets and cofactors will be required to understand how SPOC family proteins function to regulate and integrate information from these signaling pathways.
Acknowledgments
We thank R. Fehon for anti-MYC, the Developmental Studies Hybridoma Bank for anti-ELAV and anti-Cut, and the Bloomington Stock Center for fly stocks. We thank J. T. Littleton, D. Doroquez, and M. Rosenzweig for critical reading of this manuscript and all members of the Rebay lab for advice during the course of this work. Scanning electron microscopy was performed at the W. M. Keck Facility for Biological Imaging. This work was supported by NIH grant RO1 EY-012549 to I.R.
References
- Brumby, A., J. Secombe, J. Horsfield, M. Coombe, N. Amin et al., 2004. A genetic screen for dominant modifiers of a cyclin E hypomorphic mutation identifies novel regulators of S-phase entry in Drosophila. Genetics 168: 227–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cen, B., A. Selvaraj, R. C. Burgess, J. K. Hitzler, Z. Ma et al., 2003. Megakaryoblastic leukemia 1, a potent transcriptional coactivator for serum response factor (SRF), is required for serum induction of SRF target genes. Mol. Cell. Biol. 23: 6597–6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, F., and I. Rebay, 2000. split ends, a new component of the Drosophila EGF receptor pathway, regulates development of midline glial cells. Curr. Biol. 10: 943–946. [DOI] [PubMed] [Google Scholar]
- Dickson, B. J., A. van der Straten, M. Dominguez and E. Hafen, 1996. Mutations modulating Raf signaling in Drosophila eye development. Genetics 142: 163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellon, G., K. W. Harding, N. McGinnis, M. M. Martin and W. McGinnis, 1997. A genetic screen for modifiers of Deformed homeotic function identifies novel genes required for head development. Development 124: 3321–3331. [DOI] [PubMed] [Google Scholar]
- Kalidas, S., and D. P. Smith, 2002. Novel genomic cDNA hybrids produce effective RNA interference in adult Drosophila. Neuron 33: 177–184. [DOI] [PubMed] [Google Scholar]
- Kolodziej, P. A., L. Y. Jan and Y. N. Jan, 1995. Mutations that affect the length, fasciculation, or ventral orientation of specific sensory axons in the Drosophila embryo. Neuron 15: 273–286. [DOI] [PubMed] [Google Scholar]
- Kuang, B., S. C. Wu, Y. Shin, L. Luo and P. Kolodziej, 2000. split ends encodes large nuclear proteins that regulate neuronal cell fate and axon extension in the Drosophila embryo. Development 127: 1517–1529. [DOI] [PubMed] [Google Scholar]
- Kuroda, K., H. Han, S. Tani, K. Tanigaki, T. Tun et al., 2003. Regulation of marginal zone B cell development by MINT, a suppressor of Notch/RBP-J signaling pathway. Immunity 18: 301–312. [DOI] [PubMed] [Google Scholar]
- Lane, M. E., M. Elend, D. Heidmann, A. Herr, S. Marzodko et al., 2000. A screen for modifiers of cyclin E function in Drosophila melanogaster identifies Cdk2 mutations, revealing the insignificance of putative phosphorylation sites in Cdk2. Genetics 155: 233–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, H. V., D. B. Doroquez, S. Cho, F. Chen, I. Rebay et al., 2003. Splits ends is a tissue/promoter specific regulator of Wingless signaling. Development 130: 3125–3135. [DOI] [PubMed] [Google Scholar]
- Ma, Z., S. W. Morris, V. Valentine, M. Li, J. A. Herbrick et al., 2001. Fusion of two novel genes, RBM15 and MKL1, in the t(1;22)(p13;q13) of acute megakaryoblastic leukemia. Nat. Genet. 28: 220–221. [DOI] [PubMed] [Google Scholar]
- Mace, K. A., and A. Tugores, 2004. The product of the split ends gene is required for the maintenance of positional information during Drosophila development. BMC Dev. Biol. 4: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercher, T., M. B. Coniat, R. Monni, M. Mauchauffe, F. N. Khac et al., 2001. Involvement of a human gene related to the Drosophila spen gene in the recurrent t(1;22) translocation of acute megakaryocytic leukemia. Proc. Natl. Acad. Sci. USA 98: 5776–5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutsuddi, M., C. M. Marshall, K. A. Benzow, M. D. Koob and I. Rebay, 2004. The spinocerebellar ataxia 8 noncoding RNA causes neurodegeneration and associates with staufen in Drosophila. Curr. Biol. 14: 302–308. [DOI] [PubMed] [Google Scholar]
- Oswald, F., U. Kostezka, K. Astrahantseff, S. Bourteele, K. Dillinger et al., 2002. SHARP is a novel component of the Notch/RBP-Jkappa signalling pathway. EMBO J. 21: 5417–5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebay, I., R. G. Fehon and S. Artavanis-Tsakonas, 1993. Specific truncations of Drosophila Notch define dominant activated and dominant negative forms of the receptor. Cell 74: 319–329. [DOI] [PubMed] [Google Scholar]
- Rebay, I., F. Chen, F. Hsiao, P. A. Kolodziej, B. H. Kuang et al., 2000. A genetic screen for novel components of the Ras/mitogen-activated protein kinase signaling pathway that interact with the yan gene of Drosophila identifies split ends, a new RNA recognition motif-containing protein. Genetics 154: 695–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Pulido, L., A. M. Rojas, K. H. van Wely, A. C. Martinez and A. Valencia, 2004. SPOC: a widely distributed domain associated with cancer, apoptosis and transcription. BMC Bioinformatics 5: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber, S. L., A. Preiss, A. C. Nagel, I. Wech and D. Maier, 2002. Genetic screen for modifiers of the rough eye phenotype resulting from overexpression of the notch antagonist hairless in drosophila. Genesis 33: 141–152. [DOI] [PubMed] [Google Scholar]
- Shi, Y., M. Downes, W. Xie, H. Y. Kao, P. Ordentlich et al., 2001. Sharp, an inducible cofactor that integrates nuclear receptor repression and activation. Genes Dev. 15: 1140–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staehling-Hampton, K., P. J. Ciampa, A. Brook and N. Dyson, 1999. A genetic screen for modifiers of E2F in Drosophila melanogaster. Genetics 153: 275–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therrien, M., D. K. Morrison, A. M. Wong and G. M. Rubin, 2000. A genetic screen for modifiers of a kinase suppressor of Ras-dependent rough eye phenotype in Drosophila. Genetics 156: 1231–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiellette, E. L., K. W. Harding, K. A. Mace, M. R. Ronshaugen, F. Y. Wang et al., 1999. spen encodes an RNP motif protein that interacts with Hox pathways to repress the development of head-like sclerites in the Drosophila trunk. Development 126: 5373–5385. [DOI] [PubMed] [Google Scholar]
- Yang, X., J. Li, H. Qin, H. Yang, P. Zhou et al., 2005. Mint represses transactivation of the type II collagen gene enhancer through interaction with alpha A-crystallin-binding protein 1. J. Biol. Chem. 280: 18710–18716. [DOI] [PubMed] [Google Scholar]
- Zipursky, S. L., and G. M. Rubin, 1994. Determination of neuronal cell fate: lessons from the R7 neuron of Drosophila. Annu. Rev. Neurosci. 17: 373–397. [DOI] [PubMed] [Google Scholar]