Abstract
Several constitutional chromosomal rearrangements occur on human chromosome 17. Patients who carry constitutional deletions of 17q21.3–q24 exhibit distinct phenotypic features. Within the deletion interval, there is a genomic segment that is bounded by the myeloperoxidase and homeobox B1 genes. This genomic segment is syntenically conserved on mouse chromosome 11 and is bounded by the mouse homologs of the same genes (Mpo and HoxB1). To attain functional information about this syntenic segment in mice, we have generated a 6.9-Mb deletion [Df(11)18], the reciprocal duplication [Dp(11)18] between Mpo and Chad (the chondroadherin gene), and a 1.8-Mb deletion between Chad and HoxB1. Phenotypic analyses of the mutant mouse lines showed that the Dp(11)18/Dp(11)18 genotype was responsible for embryonic or adolescent lethality, whereas the Df(11)18/+ genotype was responsible for heart defects. The cardiovascular phenotype of the Df(11)18/+ fetuses was similar to those of patients who carried the deletions of 17q21.3–q24. Since heart defects were not detectable in Df(11)18/Dp(11)18 mice, the haplo-insufficiency of one or more genes located between Mpo and Chad may be responsible for the abnormal cardiovascular phenotype. Therefore, we have identified a new dosage-sensitive genomic region that may be critical for normal heart development in both mice and humans.
THE most overt differences between the genomes of two mammalian species are the numbers and arrangement of their chromosomes. Structural alterations in the mammalian genome, particularly duplications and inversions, provide the raw material for the forces of evolution. Duplications enable genetic variants to be tested in one copy of a gene, enabling new gene functions to emerge, while inversions can lock sets of allelic variants into large haplotype blocks, enabling these to diverge as a group without genetic assortment until the inversion increases in frequency in the population.
Recently, it has been recognized that large genomic alterations involving loss or gain of millions of base pairs are common polymorphisms in the human and mouse populations (Sebat et al. 2004; Adams et al. 2005). Most of these copy number polymorphisms (CNPs) do not have any developmental or physiological consequences to the individual with the CNP. However, a subset of these alterations are not neutral and are responsible for many disease processes. Chromosomal abnormalities in somatic cells play a major role in many types of cancer (Rabbitts 1994). Constitutional chromosomal abnormalities are important causes of human genetic diseases (Shaffer and Lupski 2000). Some chromosomal rearrangements such as the deletions associated with DiGeorge, Prader–Willi/Angelman, Williams, and Smith–Magenis syndromes are generated de novo at a relatively high rate in the human population. Many other disease-associated chromosomal rearrangements have been described, but they are comparatively rare and/or their associated phenotypes are quite variable so that they have yet to be classified as “syndromes.” Until recently, constitutional deletions have been identified using conventional cytogenetic techniques, restricting the detection limit of disease-associated deletions to several million base pairs. Recently, the use of high-resolution BAC arrays has begun to identify many more disease-associated deletions previously undetected because of the low resolution of cytogenetics. Characterization of these chromosomal rearrangements offers an opportunity to identify the causative genes for many disease phenotypes (Riccardi et al. 1978; Varesco et al. 1989; Millar et al. 2000).
The many conserved linkage groups between the genomes of humans and mice makes it possible to model the chromosomal rearrangements involved in human diseases by using chromosome engineering (Ramirez-Solis et al. 1995; Yu and Bradley 2001). Mouse models that carry engineered chromosomal deletions have been successfully used to model the human chromosomal deletions that are responsible for DiGeorge syndrome (Lindsay et al. 1999, 2001; Merscher et al. 2001), Prader–Willi syndrome (Tsai et al. 1999), and Smith–Magenis syndrome (Walz et al. 2003). Deletion syndromes are very difficult to analyze in humans because one must rely on rare deletions to subclassify the phenotype. In contrast, specific subdeletions can be generated in mice, enabling specific associations to be drawn between aspects of the phenotype and genes in the deleted region. Indeed, this approach was instrumental in the identification of the causative gene for the principal cardiovascular defect in DiGeorge syndrome (Jerome and Papaioannou 2001; Lindsay et al. 2001; Merscher et al. 2001).
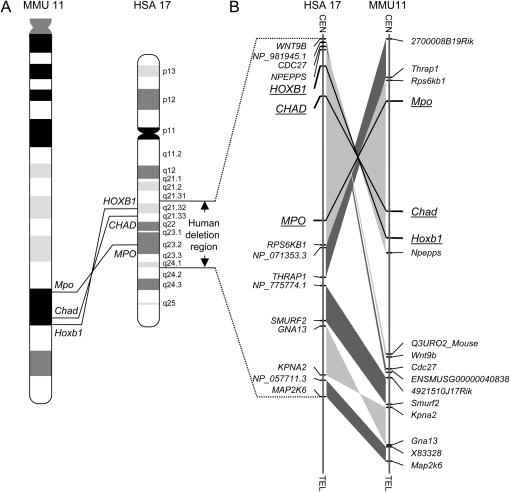
Many disease-associated chromosomal rearrangements have been reported on human chromosome 17 (Shaffer and Lupski 2000; Schinzel 2001). Mouse models for some of these disorders have been developed by using targeted manipulation of mouse chromosome 11 (Hirotsune et al. 1998; Toyo-oka et al. 2003; Walz et al. 2003). However, mouse models have not been developed for the constitutional deletions in the human chromosome region 17q21.3–q24 (Park et al. 1992; Dallapiccola et al. 1993; Khalifa et al. 1993; Levin et al. 1995; Thomas et al. 1996; Mickelson et al. 1997; Marsh et al. 2000). These de novo deletions occur at a low frequency. Children with the deletions have a distinct phenotype with the clinical features of heart defects, esophageal atresia, and hand abnormalities. The genomic region associated with the human deletions spans ∼19 Mb (Thomas et al. 1996). The syntenic region in the mouse genome is distributed between seven segments in the distal region of mouse chromosome 11 (Figure 1). In this study, we have engineered two deletions and one duplication in the largest of these syntenic regions. We characterized the phenotypic consequences of gene dosage imbalance in the rearranged regions and found that mice with the deletion between Mpo and Chad have developmental heart defects that mirror those seen in the human patients who carry the deletions of 17q21.3–q24.
Figure 1.
(A) Schematic of the genomic regions bounded by the myeloperoxidase and homeobox B1 genes in the human and mouse genomes. The deletion region associated with human 17q21.3–q24 is indicated. (B) Schematic of the syntenic segments in the deletion-associated human 17q21.3–q24 region and in the distal region of mouse chromosome 11. The genes at the ends of syntenic segments are shown. Genomic segments that show linkage conservation (i.e., identical gene order) in humans and mice are connected by dark shading if the gene orders are in the same direction relative to their respective centromeres. If the gene orders in the syntenic segments are in opposite orientations, they are connected by light shading. The endpoints of the engineered chromosomal rearrangements in mice and their human homologs are underlined.
MATERIALS AND METHODS
The targeting vectors for generating the rearrangements between Mpo and Chad:
The targeting vectors were isolated from either the 5′- or the 3′-Hprt vector genomic libraries (Zheng et al. 1999). The vectors from both libraries contain inserts of DNA that were generated by partial Sau3AI digestion of genomic DNA from the 129S5 mouse strain. A Chad-specific probe was amplified from mouse genomic DNA with primers 5′-GCC TGG TCG GGG CTG TCT AGG-3′ (forward) and 5′-GGA TTG AAG GCG TGC GCT ACC-3′ (reverse) and used to screen the 5′-Hprt library. Clone 4B8F, which contained a 7.3-kb genomic insert, was identified. Three internal NheI fragments, of 0.5, 0.8, and 1.5 kb, were deleted from the genomic insert of clone 4B8F to generate the targeting vector pTVChad2. The orientation of the deleted insert of pTVChad2 was reversed to generate the targeting vector pTVChad3. An Mpo-specific probe was amplified from mouse genomic DNA with primers TGG CAG TTT GGG GAT AGG ATT G-3′ (forward) and TAG AAG GGA AGG GAG GTG CAA G-3′ (reverse) and used to screen the 3′-Hprt library. Clone 2C10C, which contained a 10-kb insert, was identified. A 4.0-kb AflII fragment was deleted from the insert of clone 2C10C to generate the targeting vector pTVMpo2. The orientation of the deleted insert of pTVMpo2 was reversed to generate the targeting vector pTVMpo3.
The targeting vectors for the deletion between Chad and HoxB1:
The targeting vector for the HoxB1 gene pTVHoxB1 has been described previously (Medina-Martinez et al. 2000). To construct pTVChad14, an insertional targeting vector for the Chad locus with the 3′-Hprt vector backbone, a PmeI site was inserted into the NheI site within the genomic insert in pTVChad2 and this modified genomic insert was cloned into the AscI site of the 3′-Hprt vector backbone.
Gene targeting in embryonic stem cells:
Culture of the AB2.2 line of embryonic stem (ES) cells (Bradley et al. 1998) and the method for gene targeting have been described previously (Ramirez-Solis et al. 1993). For generating the rearrangements between Mpo and Chad, pTVChad2 and pTVChad3 were linearized by digestion with NheI whereas pTVMpo2 and pTVMpo3 were linearized by digestion with AflII prior to transfection. For generating Df(11)19, pTVChad14 and pTVHoxBI were linearized by digestion with PmeI and SalI, respectively. Using electroporation, the linearized targeting vectors were transfected into ES cells, which were selected in G418 or puromycin. Positive clones were identified by Southern blot analysis with one of the following probes: a 1.5-kb NheI fragment from clone 4B8F <94,386,239–94,387,768> for the Chad locus, a 0.7-kb NdeI–AflII <87,527,542–87,528,230> fragment from clone 2C10C for the Mpo locus, or a 0.7-kb EcoRI <96,182,387–96,183,050> fragment external to the 5′ homologous region of the targeting vector for the HoxB1 locus.
Generation of chromosomal rearrangements in ES cells and mice:
The pOG231 cre-expression vector (O'Gorman et al. 1997) was electroporated into double-targeted clones, and ES cell clones with recombined products were selected in hypoxanthine, aminopterin, and thymidine (HAT) medium as described previously (Ramirez-Solis et al. 1995; Liu et al. 1998). Clones of ES cells that carried the desired deletion between Chad and Mpo generated by trans recombination were identified by hybridizing Southern blots of NdeI-digested genomic DNA from HAT-resistant ES cell clones to the 0.7-kb NdeI–AflII fragment from clone 2C10C for the Mpo locus (Figure 3). The reciprocal duplication was confirmed by hybridizing the same Southern blots to the 1.5-kb NheI fragment from clone 4B8F from the Chad locus (Figure 3). The deletion was designated del(11)(Mpo-Chad)Brd, abbreviated as Df(11)18. The duplication was designated dup(11)(Mpo-Chad)Brd, abbreviated as Dp(11)18. Clones of ES cells that carried the desired deletion between Chad and HoxBI were identified by hybridizing Southern blots of NdeI-digested genomic DNA of the ES cell clones to a radiolabeled 1.5-kb NheI fragment that was isolated from clone 4B8F from the Chad locus (Figure 6). This deletion was designated del(11)(Chad-HoxB1)Brd, abbreviated as Df(11)19. Germline-transmitting chimeras were generated from ES cell lines carrying the engineered chromosomes by microinjection of blastocysts isolated from albino C57B6/J-Tyrc-Brd females as described previously (Bradley 1987).
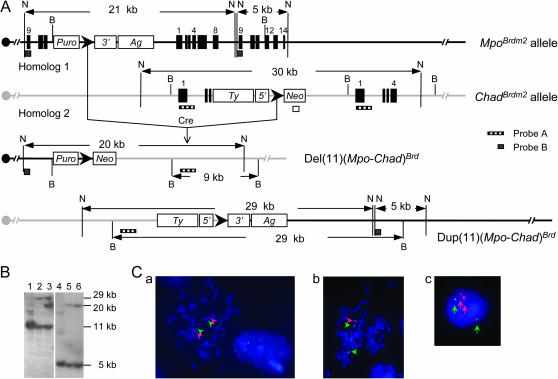
Figure 3.
Generation of a Mpo-Chad deletion and the reciprocal duplication via trans recombination. (A) Strategy to generate deletion and duplication. (B) Southern blot analysis of samples of ES cell DNA that were digested with NdeI. Wild-type AB2.2 ES cell DNA (lanes 1 and 4), double-targeted ES cell DNA (lanes 2 and 5), and ES cell DNA that contained the Mpo-Chad deletion and the reciprocal duplication (lanes 3 and 6) were hybridized to probe A (lanes 1–3) and probe B (lanes 4–6). N, NdeI; B, BamHI; 3′, 3′-Hprt; 5′, 5′-Hprt; Ag, K14-Aguoti gene; Ty, tyrosinase minigene; Puro, puromycin-resistance gene; Neo, neomycin-resistance gene; ▸, loxP site. (C) FISH analysis of chromosomal rearrangements between Mpo and Chad. The RP23-276G11 BAC probe (green) hybridized to the D11Mit171 marker, which is located outside the region, whereas the RP23-257C13 BAC probe (red) hybridized to the D11Mit179 marker located between Mpo and Chad. (a) Metaphase chromosomes from wild-type ES cells. (b) Metaphase chromosomes from ES cells that contain the Mpo-Chad deletion and the reciprocal duplication. (c) Interphase nucleus of an ES cell that contains the Mpo-Chad deletion and the reciprocal duplication.
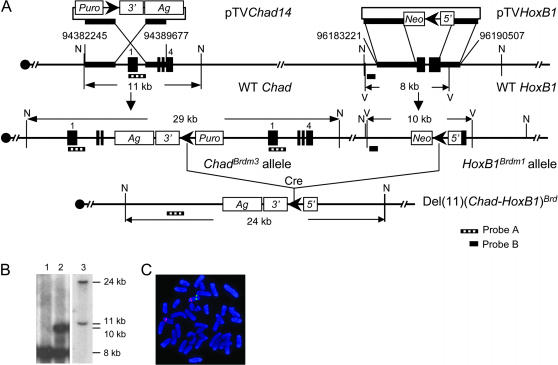
Figure 6.
Generation of the deletion between Chad and HoxB1 via cis recombination. (A) Strategy to generate the deletion. (B) Southern blot analysis of samples of DNA that were digested with NdeI and hybridized to probe B (lanes 1 and 2) or probe A (lane 3). The parental AB2.2 ES cell DNA (lane 1), HoxB1-targeted ES cell DNA (lane 2), and ES cell DNA that contained the Chad-HoxB1 deletion (lane 3). N, NdeI; V, EcoRV. For the other abbreviations, see Figure 2. (C) FISH analysis of the chromosomal deletion between Chad and HoxB1. Metaphase chromosomes from a 7G9F ES cell that contains the Chad-HoxB1 deletion. The BAC probe RP23-276G11 (red) hybridized to the D11Mit171 locus, which was located outside the rearranged region whereas the RP23-374F6 probe (green) hybridized to the Ngfr locus, which was located inside the deletion interval.
Genotyping by simple sequence length polymorphism markers:
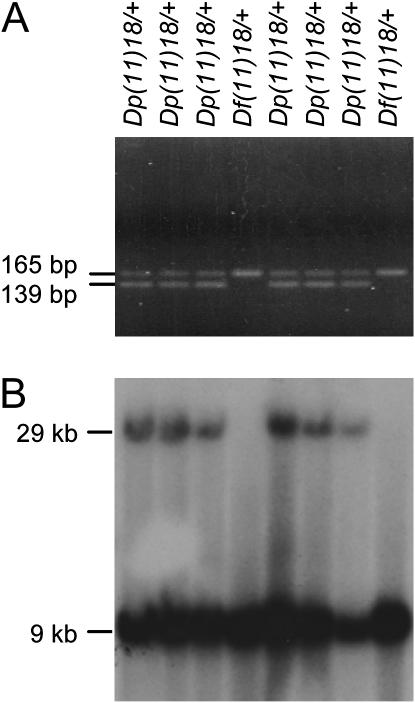
The simple sequence length polymorphism (SSLP) marker at D11Mit179 was used to distinguish chromosomes originating from the AB2.2 ES cell line (129S7) and from C57B6/J-Tyrc-Brd mice. The allele size at D11Mit179 is 139 bp in 129S7 and 165 bp in strain C57B6/J-Tyrc-Brd (H. Su and A. Bradley, unpublished results).
Fluorescent in situ hybridization:
Chromosome spreads of ES cells were prepared as described previously (Robertson 1987). BAC clones were used as probes for fluorescent in situ hybridization (FISH) (Baldini and Lindsay 1994). For the analysis of the genomic rearrangements between Mpo and Chad, BAC clone RP23-276G11 was labeled with biotin and detected with fluorescin isothiocyanate–avidin. BAC clones RP23-257C13 and RP23-351G6 were labeled with digoxigenin, and they were detected with antidigoxigenin–rhodamine antibody. RP23-351G6, which contains the Tbx2 gene, was confirmed by PCR with primers 5′-GCG CCG CTG GTG GTG CAG ACA-3′ (forward) and 5′-CCG GGG CCC ATG GCG AAT TGT-3′ (reverse). For the analysis of the deletion between Chad and HoxB1, BAC clone RP23-374F6 was labeled with biotin, and it was detected with fluorescin isothiocyanate–avidin. BAC clone RP23-276G11 was labeled with digoxigenin, and it was detected with antidigoxigenin–rhodamine antibody. Chromosomes were counterstained with 4′,6′-diamidino-2-phenylindole.
Phenotyping esophageal atresia:
E18.5 fetuses were euthanized by asphyxiation with carbon dioxide. A 0.4-ml sample of diluted India ink was orally administrated to the euthanized fetuses with a 1-ml syringe. The degree of blockage in the esophagus was determined by quantifying the amount of India ink in the stomach. A stereomicroscope equipped with an eyepiece reticule was used to measure the sizes of ink-stained areas of the stomach.
RESULTS
Generation of a chromosomal deletion and duplication between Mpo and Chad:
Human chromosome 17q21.32–q23.2 contains 99 known and predicted genes between the MPO and HOXB1 genes (NCBI build 35). The homologs for 92 of these are present on mouse chromosome 11 between the Mpo and HoxB1 genes with a conserved gene order (NCBI build 34). The human MPO–CHAD region has 51 known and predicted genes. The mouse homologs for 47 of these are also located on mouse chromosome 11 between the Mpo and Chad genes (Figure 1; supplementary Tables S1 and S2 at http://www.genetics.org/supplemental/). To examine the relevance of these regions in the constitutional deletion disorders on human chromosome 17, two deletions and a reciprocal duplication of the syntenic region between Mpo and HoxB1 were generated using a long-range Cre/loxP-mediated recombination (Ramirez-Solis et al. 1995; Liu et al. 1998; Yu and Bradley 2001; Zheng et al. 2001). When this study was initiated, the orientation of both Mpo and Chad relative to the centromere was not known. Therefore, two targeting vectors with different orientations of the 5′- and 3′-Hprt mini-gene cassettes were utilized.
AB2.2 ES cells were sequentially targeted with pTVChad3 and pTVMpo2 (Figures 2 and 3). Six double-targeted clones were isolated and they were transiently transfected with the cre-expression vector. ES cell clones that had undergone loxP recombination, which activated the Hprt selection cassette, were selected in HAT medium. Two of the six double-targeted clones did not generate any HAT-resistant (HATr) clones. This result suggested that this combination of vectors resulted in targeting of the 5′- and 3′-Hprt-loxP selection cassettes in opposite orientations on chromosome 11.
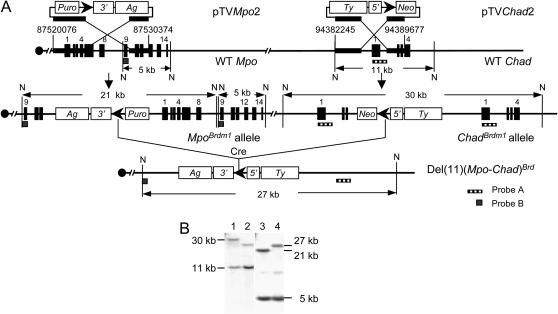
Figure 2.
Generation of the Mpo-Chad deletion via cis recombination. (A) Strategy to generate the deletion. (B) Southern blot analysis of samples of DNA that were digested with NdeI and hybridized to probe A (lanes 1 and 2) and probe B (lanes 3 and 4). Double-targeted ES cell DNA (lanes 1 and 3) and ES cell DNA that contained the Mpo-Chad deletion (lanes 2 and 4). N, NdeI; 3′, 3′-Hprt; 5′, 5′-Hprt; Ag, K14-Aguoti gene; Ty, tyrosinase minigene; Puro, puromycin-resistance gene; Neo, neomycin-resistance gene; WT, wild type; ▸, loxP site.
To generate ES cells with the deletion, double-targeted AB2.2 ES cell clones were generated with pTVMpo2 and pTVChad2. Eight double-targeted clones were isolated and they were transiently transfected with the cre-expression vector. HAT-resistant clones were generated at a frequency of 0.03% from two of the clones and 10% for six of the clones. All of the HAT-resistant clones derived from double-targeted clones with a high recombination efficiency were sensitive to both G418 and puromycin. These results suggested that an Mpo-Chad deletion was generated by cis recombination in these clones (Figure 2A). The presence of the deletion in these HAT-resistant clones was confirmed by Southern blot analysis (Figure 2B).
To generate ES cell clones with both the deletion and the reciprocal duplication, ES cells were targeted with pTVChad3 and pTVMpo3. Four clones designated 1C, 2B, 3G, and 7H were isolated and transiently transfected with the cre-expression plasmid. The efficiency of Cre/loxP-mediated recombination measured by the generation of HAT-resistant clones was 0.05% for clone 1C and 10% for the clones 2B, 3G, and 7H. The lower efficiency of loxP recombination in clone 1C suggested that the loxP sites were targeted in trans (Figure 3A). Six HAT-resistant clones were isolated from 1C cells, designated 1C1–1C6. Sib selection indicated that clone 1C6 was G418 sensitive and puromycin resistant whereas clones 1C1–1C5 were resistant to both G418 and puromycin. These results suggested that 1C6 carried a duplication whereas clones 1C1–1C5 carried both the deletion and the duplication (Yu and Bradley 2001; Zheng et al. 2001), which was confirmed by Southern blot analysis (Figure 3B) and by FISH on metaphase and interphase chromosomes (Figure 3C).
1C1 and 1C2 clones carrying both Df(11)18 and Dp(11)18 were used to generate germline chimeras. Segregation of the deletion and the duplication alleles was observed in the first generation (Figure 4). The 129S7 allele of the D11Mit179 SSLP marker (which maps within the deletion interval) was absent in 25–30% of agouti (ES-cell-derived) F1 progeny, which confirmed the presence of the deletion allele (Figure 4A), although at lower-than-expected frequency. Df(11)18/Dp(11)18 mice were established by crossing Dp(11)18/+ females with their chimeric fathers. These genetically balanced mice were viable and fertile and used to maintain both Df(11)18 and Dp(11)18 chromosomes. The tyrosinase minigene present in the Chad targeting vector cosegregated with Dp(11)18 (Figure 3), and it was phenotypically expressed as a light-brown coat color in an albino background (supplementary Figure S1 at http:/www.genetics.org/supplemental/).
Figure 4.
Genotyping F1 agouti mice. (A) SSLP analysis of DNA from eight F1 agouti mouse tails by PCR with D11Mit179 primers. The PCR product for the allele of strain 129S7 was 139 bp whereas the PCR product for the allele of strain C57B6/J-Tyrc-Brd was 165 bp. The absence of the 139-bp fragment indicated that there was a deletion in the allele of strain 129S7. (B) Southern blot analysis of BamHI-digested DNA from F1 agouti mouse tails with probe A (see Figure 3). The 29-kb fragment indicates the duplication.
Df(11)18/Df(11)18 and Dp(11)18/Dp(11)18 mice:
The Df(11)18 and Dp(11)18 chromosomes were maintained in the mixed genetic background of 129S7 and C57BL/6-Tyrc-Brd. Intercrosses between Df(11)18/Dp(11)18 mice were performed. The litters produced from these crosses were smaller than normal (mean = 4.1, n = 8, P < 0.05). Genotype analysis of these litters at 2 weeks of age revealed that 94% of mice were of the balanced genotype Df(11)18/Dp(11)18 while only 6% of mice were Dp(11)18/Dp(11)18 (Table 1). The surviving Dp(11)18/Dp(11)18 mice were smaller than their litter mates and none survived past 4 weeks of age. Timed matings were established and fetuses were isolated and genotyped at E18.5. As expected, no Df(11)18/Df(11)18 fetuses were identified since the homozygous deletion of Sfrs1 located within the deletion interval would cause embryonic lethality prior to E7.5 (Xu et al. 2005). Undoubtedly, homozygous loss of other genes in the interval would also contribute to early embryonic lethality. Just 8% of the E18.5 fetuses were Dp(11)18/Dp(11)18, indicating that this genotype also affected normal embryonic development. While the specific mechanism underlying the lethality of Dp(11)18/Dp(11)18 mice is unknown, the phenotype is apparently caused by increased gene dosage in the duplicated region. These results demonstrated that the genomic region bounded by Mpo and Chad is a gene-dosage-sensitive region.
TABLE 1.
Impact of Df(11)18, Dp(11)18, and Df(11)19 on fetal and postnatal viability
| Genotype of progeny:
|
Df(11)18/Dp(11)18 × Df(11)18/Dp(11)18
|
Df(11)18/Dp(11)18 × +/+
|
Df(11)19/+ × +/+
|
|||||
|---|---|---|---|---|---|---|---|---|
| Analysis time | Df(11)18/Dp(11)18 N (%) | Df(11)18/Df(11)18 N (%) | Dp(11)18/Dp(11)18 N (%) | Df(11)18/+ N (%) | Dp(11)18/+ N (%) | Df(11)19/+ N (%) | +/+ N (%) | |
| E18.5 | 35* (92) | 0* (0) | 3* (8) | 24 (40) | 36 (60) | 17 (47) | 19 (53) | |
| p14 | 33* (94) | 0* (0) | 2* (6) | 15* (32) | 32* (68) | 18 (46) | 21 (54) | |
| p28 | 31* (100) | 0* (0) | 0* (0) | 14* (31) | 31* (69) | 18 (46) | 21 (54) | |
Significant difference from expected Mendelian ratio (P < 0.05) based on χ2 test.
Df(11)18/+ and Dp(11)18/+ mice:
Df(11)18/+ mice were not observed at the expected 50% frequency in the ES-cell-derived progeny transmitted by chimeras derived from the injection of ES cells with the balanced Df(11)18/Dp(11)18 genotype. Test crosses between Df(11)18/Dp(11)18 and +/+ (C57B6/J-Tyrc-Brd) confirmed that just 31% progeny genotyped at 4 weeks of age carried the Df(11)18/+ genotype (Table 1). Df(11)18/+ mice identified at 4 weeks of age were overtly normal and exhibited normal mortality. During an observation period of 12 months, however, Df(11)18/+ females (n = 14) were infertile. We observed no discernible abnormality in the histology sections of ovaries of Df(11)18/+ females (n = 14). The physiological cause of the female infertility is unknown.
To identify why the Df(11)18/+ mice were underrepresented, progeny from Df(11)18/Dp(11)18 × +/+ crosses were genotyped during the neonatal period and just prior to birth (Table 1). At 2 weeks of age, 32% of mice had the Df(11)18/+ genotype; thus mortality was occurring before this time. At E18.5, Df(11)18/+ fetuses were observed at a frequency of 40%, indicating that the majority survived to term. Observation of littering females revealed that while the majority of Df(11)18/+ progeny were born alive, ∼15% died within several hours of birth.
Humans with a deletion of the conserved linkage region on human chromosome 17 exhibit ventricular septal defects, overriding of the aorta, and pulmonary stenosis (Park et al. 1992; Dallapiccola et al. 1993; Khalifa et al. 1993; Levin et al. 1995; Thomas et al. 1996; Mickelson et al. 1997; Marsh et al. 2000). In addition, these patients may exhibit other clinical features such as esophageal atresia and hand anomalies. We therefore set up Df(11)18/Dp(11)18 × +/+ timed matings and examined Df(11)18/+ and Dp(11)18/+ fetuses at E18.5 to determine if the poor survival of Df(11)18/+ mice after birth was caused by cardiac defects.
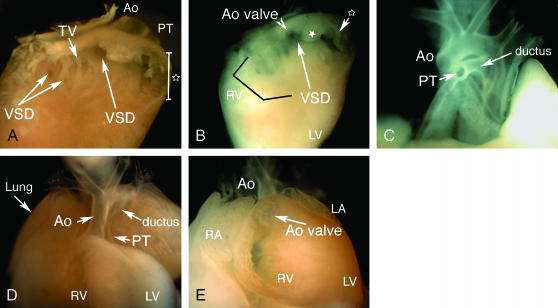
This analysis revealed that 34% of Df(11)18/+ fetuses (n = 38) exhibited structural heart defects (Table 2). Five of these exhibited two or more abnormalities that are characteristics of the tetralogy of Fallot (TOF) complex, including perimembranous ventricular septal defects (Figure 5, A and B), overriding of the aorta (Figure 5B), and hypoplastic pulmonary trunks (Figure 5C). Four of these fetuses also showed the characteristic deviation of the parietal muscle band (Figure 5B) considered pathognomonic of TOF. In addition, 8 fetuses had inlet septal defects (Figure 5A) and 8 fetuses showed transposition of the great arteries (TGA) (Figure 5, D and E). Heart defects were not detectable in Dp(11)18/+ fetuses from the same cross (n = 42) or in 32 +/+ fetuses from other control matings. Dp(11)18/+ mice appear normal and have normal life expectancy.
TABLE 2.
Cardiovascular anomalies in Df(11)18/+ fetuses at E18.5
| Cardiovascular anomalies | No. of fetuses |
|---|---|
| Inlet VSD | 8 |
| Perimembranous/conal VSD | 7 |
| Overriding of the aorta | 5 |
| Deviation of the parietal muscle band | 4 |
| Hypoplastic pulmonary trunk | 5 |
| TOF | 5 |
| TOF and inlet VSD | 4 |
| TGA (I) | 2 |
| TGA (II) | 2 |
| TGA (III) | 4 |
TGA (I), TGA without VSD (intact septum); TGA (II), TGA with perimembranous/conal VSD; TGA (III), TGA with inlet VSD. Thirty-eight Df(11)18/+ fetuses were examined and 34% of them showed cardiovascular anomalies.
Figure 5.
Cardiovascular anomalies in 2 Df(11)18/+ fetuses at E18.5. A–C show a fetus with a TOF-like group of defects. (A) An intracardiac view (from the right side) of the right ventricle after removal of the free wall reveals the presence of two distinct types of ventricular septal defect (VSDs), a malalignment type in the infundibular region (single arrow, an outflow tract anomaly) and an inlet type (two arrows on the left) (an inflow-septal anomaly). The open star indicates the subpulmonary infundibular chamber. (B) A more caudal view of the same heart shows an abnormal deviation of the parietal band (asterisk), which causes subpulmonary stenosis. Note that because of the overriding of the aorta, more than half of the aortic (Ao) valve is visible through the VSD. (C) A frontal view of the great arteries after removal of the heart shows a hypoplastic pulmonary trunk (compared to the size of the aorta). D and E show a fetus with transposition of the great arteries. (D) A frontal view shows a right-sided, anteriorly positioned aorta, while the pulmonary trunk was located behind the aorta. (E) An intracardiac view of the right ventricle shows the aortic valve located on the right ventricle. The pulmonary valve in this fetus is located on the left ventricle (not shown). Ao, aorta; LA, left atrium; LV, left ventricle; PT, pulmonary trunk; RA, right atrium; RV, right ventricle; TV, tricuspid valve.
Forty Df(11)18/+ mice were also examined for esophageal atresia and anomalies in limb and digit development at E18.5. None of the embryos exhibited blockage of the esophagus and no gross anomalies of the limbs and digits were observed under a stereomicroscope.
Intercrosses of Df(11)18/Dp(11)18 mice had reduced litter sizes, which we primarily attributed to loss of Df(11)18/Df(11)18 and Dp(11)18/Dp(11)18 embryos. However, these losses make it hard to assess if the balanced Df/Dp mice are fully viable. The heart defects observed in Df(11)18/+ mice might be caused by haplo-insufficiency of deleted gene(s) or by an effect (upregulation) of the deletion on neighboring genes. Therefore we examined Df(11)18/Dp(11)18 mice fetuses at E18.5 for heart defects. None were detectable (n = 35). These results indicate that heart defects were most likely due to haplo-insufficiency of one or more genes within or close to the deleted genomic region flanked by Mpo and Chad.
We examined the mouse genome assembly in the Df(11)18 region for genes that might cause heart defects. The Tbx2 gene (85.4 Mb from centromere) is located close to Mpo (87.4 Mb), but is outside the Df(11)18 deleted region in the C57BL/6 assembly. A homozygous mutation of Tbx2 causes developmental heart defects, including abnormalities in atrioventricular morphology and outflow tract septation (Harrelson et al. 2004). Because the gene order can vary between inbred mouse strains, primarily as a result of polymorphic inversions between the 129S7 and C57BL/6 genomes, it is possible that the Tbx2 gene was deleted in Df(11)18/+ mice. Therefore, we examined Df(11)18/+ metaphase chromosomes for the copy number of the Tbx2 gene by FISH analysis using a BAC probe that contained the Tbx2 gene (data not shown). The Tbx2 gene was detected on both the deleted and the nondeleted chromosomes in Df(11)18/+ mice, confirming that deletion of the structural gene was not causing the observed heart defects. Although it is possible that a regulatory element(s) of Tbx2 may reside within the deletion interval, it is unlikely that Df(11)18 caused the heart defects by affecting Tbx2 expression because heterozygous Tbx2 mutant mice were reported to be normal (Harrelson et al. 2004) and our whole-mount in situ hybridization analysis detected no significant alteration of the level or pattern of Tbx2 expression in E9.5 Df(11)18/+ embryos (data not shown).
Generation of a chromosomal deletion between Chad and HoxB1:
The syntenic region of the mouse chromosome 11 genomic segment bounded by Chad and HoxB1 was also deleted in human 17q21.3–q24 deletion disorder. To examine the phenotypic consequence of the deletion in this syntenic region, we generated a deletion, Df(11)19, in mice using Chad and HoxB1 as the endpoints. This effort was aided by the known orientation of HoxB1 relative to the centromere, which was first revealed during the process of generating the genomic rearrangements among HoxB1, HoxB9, and Hsd17b1 (Ramirez-Solis et al. 1995; Liu et al. 1998; Medina-Martinez et al. 2000). To generate Df(11)19 by cis recombination, AB2.2 ES cells were first targeted with pTVHoxB1 (Figure 6A). Four targeted clones were isolated and were tested for germline transmission competency. After confirmation that clone 7G could be reliably transmitted into the germline, this clone was electroporated with pTVChad14, a 3′-Hprt targeting vector (Figure 6A), and the ES cells were then selected in puromycin. After 9 days of the selection, the cells from the resistant clones were pooled, electroporated with pOG231, and subsequently selected in HAT medium. Sib selection of the HATr clones was performed with G418 and puromycin, 55% were G418 and puromycin sensitive. This sib selection result suggested that these clones carried the desired deletion, which was confirmed by Southern blot analysis (Figure 6B) and by FISH on metaphase chromosomes (Figure 6C). Df(11)19 was established in mice using standard procedures.
Heterozygous Df(11)19/+ mice appear normal and males were fertile. During an observation period of 12 months, however, Df(11)19/+ females (n = 10) were infertile. We observed no discernible abnormality in the histology sections of ovaries of Df(11)19/+ females (n = 10). The physiological cause of the female infertility is unknown.
Heterozygous E18.5 fetuses and postnatal mice from mating heterozygous Df(11)19/+ males to wild-type females were present at normal Mendelian ratios, suggesting that no heterozygous mutants are lost due to haplo-insufficiency (Table 1). A total of 43 E18.5 fetuses carrying Df(11)19/+ were examined and no heart defect was detectable.
DISCUSSION
We have generated two multi-megabase deficiencies between Mpo and HoxB1 in mice. This mouse genomic region is syntenic to a segment on human 17q21.3–q24, which is associated with human constitutional deletion disorders (Figure 1) (Park et al. 1992; Dallapiccola et al. 1993; Khalifa et al. 1993; Levin et al. 1995; Thomas et al. 1996; Mickelson et al. 1997; Marsh et al. 2000). Children with this disorder have a distinct phenotype, which includes heart defects, esophageal atresia, and hand abnormalities. Specific defects in heart development are atrial and ventricular septal defects, overriding aorta, pulmonary stenosis, patent ductus arteriosus, and dilated left atrium and ventricle (Park et al. 1992; Dallapiccola et al. 1993; Khalifa et al. 1993; Levin et al. 1995; Thomas et al. 1996; Mickelson et al. 1997; Marsh et al. 2000). While Df(11)19/+ fetuses were normal, Df(11)18/+ fetuses had defects in heart development reminiscent of the human disorder, but there were some differences. Df(11)18/+ fetuses did not develop patent ductus arteriosus and dilated left atrium and ventricle. Furthermore, Df(11)18/+ fetuses did not develop esophageal atresia or digit anomalies. Similarities in the disease profiles of human patients and Df(11)18/+ fetuses suggests that deletion of one or more genes between the myeloperoxidase and chondroadherin genes may be responsible for some defects in heart development for both organisms. Differences in the disease profiles may be due to genes that were deleted in the 17q23.2–q24 region since some of this region is located outside of the region bounded by the myeloperoxidase and chondroadherin genes. It is possible that deletion of genes on 17q23.2–q24 was responsible for the development of patent ductus arteriosus, dilated left atrium and ventricle, esophageal atresia, and hand anomalies. In addition, TGA was detected in Df(11)18/+ mouse fetuses. The apparent absence of TGA in a low number of patients may be due to reduced penetrance of the haplo-insufficient phenotype in humans. These results demonstrate that the Df(11)18/+ mouse line is a good but incomplete model for deletions on human 17q21.3–q24. To recapitulate noncardiac phenotypes of this human chromosomal disorder, deficiencies will also need to be constructed in the mouse syntenic regions of human 17q21.3–q24 located outside of the Mpo–HoxB1 segment (Figure 1).
The ventricular septal defect, overriding aorta, and pulmonary stenosis are the key structural components of TOF. TOF and TGA are among the most common congenital heart defects found in human infants. However, gene haplo-insufficiency mouse models that accurately recapitulate the TOF and TGA of haplo-insufficiency syndromes are not available. In mice, a mutation in connexin 40 is the only heterozygous mutation that has been shown to cause TOF (Gu et al. 2003). Furthermore, mice with heterozygous mutations that cause TGA are not available. Therefore, Df(11)18/+ mice represent a rare haplo-insufficiency model of TGA and TOF. TOF and TGA are morphogenetic abnormalities of the outflow tract and both phenotypic features may be simultaneously induced by mutations of single genes in humans or mice (Icardo and Sanchez de Vega 1991; Frank et al. 2002; McElhinney et al. 2003). Therefore, it is possible that a single haplo-insufficieny gene may be responsible for TOF and TGA in Df(11)18/+ fetuses.
The discovery of a haplo-insufficient region of the mouse genome that is responsible for TOF and TGA will greatly facilitate the effort to identify specific genes that cause these two diseases. Candidate genes may be evaluated by mutation analysis. Within the deleted interval of Df(11)18, eight genes, Mpo, Sfrs1, Coil, Nog, Pctp, Hlf, Tob1, and Cacna1g, have been mutagenized by using a gene-targeting approach (supplementary Table S2 at http://www.genetics.org/supplemental/) (Brunet et al. 1998; McMahon et al. 1998; Aratani et al. 1999; van Helvoort et al. 1999; Peng et al. 2000; Yoshida et al. 2000; Brennan et al. 2001; Tucker et al. 2001; Gachon et al. 2004; Lee et al. 2004; Xu et al. 2005) but none of these targeted mutations have been reported to give a heterozygous cardiac phenotype. Targeted disruptions of all of the genes in the region one by one might eventually identify the causal gene, but it is more efficient to generate and analyze subdeletions of the region bounded by Mpo and Chad. This would reduce the number of candidate genes to a few genes. The generation of subdeletions has been made simpler by the newly established Mouse Insertional and Chromosome Engineering Resource, which contains ∼100,000 chromosome-engineering vectors distributed throughout the mouse genome (Adams et al. 2004). If a haplo-insufficient gene that is responsible for heart defects can be located in a subdeletion of ∼1 Mb, a complementary strategy of BAC transgenics could be used for binning of the gene to a single BAC (Antoch et al. 1997; Kibar et al. 2003). Finally, targeted disruptions of the candidate gene may be carried out in ES cells to identify the causative gene for the TOF/TGA phenotype.
Both Df(11)18/+ and Df(11)19/+ caused infertility in female mice, suggesting that haplo-insufficiencies of at least two genes in the region are associated with impairment of female reproduction.
Modeling human chromosomal deletions in mice has become an essential approach for the study of deletion disorders of humans. This approach has provided an important alternative strategy for isolating the genes that are responsible for the clinical features associated with the human deletions. The Df(11)18/+ mouse line is a model for human cardiovascular defects associated with deletions on human chromosome 17. This model should serve as a powerful tool for the eventual identification of the haplo-insufficient gene or genes that are responsible for TOF and TGA.
Acknowledgments
We thank H. Zhang, L. Vein, G. Schuster, S. Rivera, J. Wesley, and Z. Li for technical assistance; H. Su for the information regarding 129/C57B6 SSLP polymorphisms on mouse chromosome 11; and S. Matsui for assistance on FISH analysis of Df(11)19. This work was supported by grants from the National Institutes of Health and the Wellcome Trust (A.B.) and from Roswell Park Alliance Foundation (Y.Y.).
References
- Adams, D. J., P. J. Biggs, T. Cox, R. Davies, L. van der Weyden et al., 2004. Mutagenic insertion and chromosome engineering resource (MICER). Nat. Genet. 36: 867–871. [DOI] [PubMed] [Google Scholar]
- Adams, D. J., E. T. Dermitzakis, T. Cox, J. Smith, R. Davies et al., 2005. Complex haplotypes, copy number polymorphisms and coding variation in two recently divergent mouse strains. Nat. Genet. 37: 532–536. [DOI] [PubMed] [Google Scholar]
- Antoch, M. P., E. J. Song, A. M. Chang, M. H. Vitaterna, Y. Zhao et al., 1997. Functional identification of the mouse circadian Clock gene by transgenic BAC rescue. Cell 89: 655–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aratani, Y., H. Koyama, S. Nyui, K. Suzuki, F. Kura et al., 1999. Severe impairment in early host defense against Candida albicans in mice deficient in myeloperoxidase. Infect. Immun. 67: 1828–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldini, A., and E. A. Lindsay, 1994. Mapping human YAC clones by fluorescence in situ hybridization using Alu-PCR from single yeast colonies. Methods Mol. Biol. 33: 75–84. [DOI] [PubMed] [Google Scholar]
- Bradley, A., 1987. Production and analysis of chimaeric mice, pp. 113–151 in Teratocarcinomas and Embryonic Stem Cells: A Practical Approach, edited by E. Robertson. IRL Press, Oxford/Washington, DC.
- Bradley, A., B. Zheng and P. Liu, 1998. Thirteen years of manipulating the mouse genome: a personal history. Int. J. Dev. Biol. 42: 943–950. [PubMed] [Google Scholar]
- Brennan, M. L., M. M. Anderson, D. M. Shih, X. D. Qu, X. Wang et al., 2001. Increased atherosclerosis in myeloperoxidase-deficient mice. J. Clin. Invest. 107: 419–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet, L. J., J. A. McMahon, A. P. McMahon and R. M. Harland, 1998. Noggin, cartilage morphogenesis, and joint formation in the mammalian skeleton. Science 280: 1455–1457. [DOI] [PubMed] [Google Scholar]
- Dallapiccola, B., R. Mingarelli, C. Digilio, M. G. Obregon and A. Giannotti, 1993. Interstitial deletion del(17) (q21.3q23 or 24.2) syndrome. Clin. Genet. 43: 54–55. [DOI] [PubMed] [Google Scholar]
- Frank, D. U., L. K. Fotheringham, J. A. Brewer, L. J. Muglia, M. Tristani-Firouzi et al., 2002. An Fgf8 mouse mutant phenocopies human 22q11 deletion syndrome. Development 129: 4591–4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachon, F., P. Fonjallaz, F. Damiola, P. Gos, T. Kodama et al., 2004. The loss of circadian PAR bZip transcription factors results in epilepsy. Genes Dev. 18: 1397–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, H., F. C. Smith, S. M. Taffet and M. Delmar, 2003. High incidence of cardiac malformations in connexin40-deficient mice. Circ. Res. 93: 201–206. [DOI] [PubMed] [Google Scholar]
- Harrelson, Z., R. G. Kelly, S. N. Goldin, J. J. Gibson-Brown, R. J. Bollag et al., 2004. Tbx2 is essential for patterning the atrioventricular canal and for morphogenesis of the outflow tract during heart development. Development 131: 5041–5052. [DOI] [PubMed] [Google Scholar]
- Hirotsune, S., M. W. Fleck, M. J. Gambello, G. J. Bix, A. Chen et al., 1998. Graded reduction of Pafah1b1 (Lis1) activity results in neuronal migration defects and early embryonic lethality. Nat. Genet. 19: 333–339. [DOI] [PubMed] [Google Scholar]
- Icardo, J. M., and M. J. Sanchez de Vega, 1991. Spectrum of heart malformations in mice with situs solitus, situs inversus, and associated visceral heterotaxy. Circulation 84: 2547–2558. [DOI] [PubMed] [Google Scholar]
- Jerome, L. A., and V. E. Papaioannou, 2001. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat. Genet. 27: 286–291. [DOI] [PubMed] [Google Scholar]
- Khalifa, M. M., P. M. MacLeod and A. M. Duncan, 1993. Additional case of de novo interstitial deletion del(17)(q21.3q23) and expansion of the phenotype. Clin. Genet. 44: 258–261. [DOI] [PubMed] [Google Scholar]
- Kibar, Z., S. Gauthier, S. H. Lee, S. Vidal and P. Gros, 2003. Rescue of the neural tube defect of loop-tail mice by a BAC clone containing the Ltap gene. Genomics 82: 397–400. [DOI] [PubMed] [Google Scholar]
- Lee, J., D. Kim and H. S. Shin, 2004. Lack of delta waves and sleep disturbances during non-rapid eye movement sleep in mice lacking alpha1G-subunit of T-type calcium channels. Proc. Natl. Acad. Sci. USA 101: 18195–18199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin, M. L., L. G. Shaffer, R. Lewis, M. V. Gresik and J. R. Lupski, 1995. Unique de novo interstitial deletion of chromosome 17, del(17) (q23.2q24.3) in a female newborn with multiple congenital anomalies. Am. J. Med. Genet. 55: 30–32. [DOI] [PubMed] [Google Scholar]
- Lindsay, E. A., A. Botta, V. Jurecic, S. Carattini-Rivera, Y. C. Cheah et al., 1999. Congenital heart disease in mice deficient for the DiGeorge syndrome region. Nature 401: 379–383. [DOI] [PubMed] [Google Scholar]
- Lindsay, E. A., F. Vitelli, H. Su, M. Morishima, T. Huynh et al., 2001. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature 410: 97–101. [DOI] [PubMed] [Google Scholar]
- Liu, P., H. Zhang, A. McLellan, H. Vogel and A. Bradley, 1998. Embryonic lethality and tumorigenesis caused by segmental aneuploidy on mouse chromosome 11. Genetics 150: 1155–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh, A. J., D. Wellesley, D. Burge, M. Ashton, C. Browne et al., 2000. Interstitial deletion of chromosome 17 (del(17)(q22q23.3)) confirms a link with oesophageal atresia. J. Med. Genet. 37: 701–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElhinney, D. B., E. Geiger, J. Blinder, D. W. Benson and E. Goldmuntz, 2003. NKX2.5 mutations in patients with congenital heart disease. J. Am. Coll. Cardiol. 42: 1650–1655. [DOI] [PubMed] [Google Scholar]
- McMahon, J. A., S. Takada, L. B. Zimmerman, C. M. Fan, R. M. Harland et al., 1998. Noggin-mediated antagonism of BMP signaling is required for growth and patterning of the neural tube and somite. Genes Dev. 12: 1438–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Martinez, O., A. Bradley and R. Ramirez-Solis, 2000. A large targeted deletion of Hoxb1-Hoxb9 produces a series of single-segment anterior homeotic transformations. Dev. Biol. 222: 71–83. [DOI] [PubMed] [Google Scholar]
- Merscher, S., B. Funke, J. A. Epstein, J. Heyer, A. Puech et al., 2001. TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell 104: 619–629. [DOI] [PubMed] [Google Scholar]
- Mickelson, E. C., W. P. Robinson, M. A. Hrynchak and M. E. Lewis, 1997. Novel case of del(17)(q23.1q23.3) further highlights a recognizable phenotype involving deletions of chromosome (17)(q21q24). Am. J. Med. Genet. 71: 275–279. [DOI] [PubMed] [Google Scholar]
- Millar, J. K., J. C. Wilson-Annan, S. Anderson, S. Christie, M. S. Taylor et al., 2000. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum. Mol. Genet. 9: 1415–1423. [DOI] [PubMed] [Google Scholar]
- O'Gorman, S., N. A. Dagenais, M. Qian and Y. Marchuk, 1997. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc. Natl. Acad. Sci. USA 94: 14602–14607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J. P., J. B. Moeschler, S. Z. Berg, R. M. Bauer and D. H. Wurster-Hill, 1992. A unique de novo interstitial deletion del(17)(q21.3q23) in a phenotypically abnormal infant. Clin. Genet. 41: 54–56. [DOI] [PubMed] [Google Scholar]
- Peng, J., L. Zhang, L. Drysdale and G. H. Fong, 2000. The transcription factor EPAS-1/hypoxia-inducible factor 2alpha plays an important role in vascular remodeling. Proc. Natl. Acad. Sci. USA 97: 8386–8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbitts, T. H., 1994. Chromosomal translocations in human cancer. Nature 372: 143–149. [DOI] [PubMed] [Google Scholar]
- Ramirez-Solis, R., A. C. Davis and A. Bradley, 1993. Gene targeting in embryonic stem cells. Methods Enzymol. 225: 855–878. [DOI] [PubMed] [Google Scholar]
- Ramirez-Solis, R., P. Liu and A. Bradley, 1995. Chromosome engineering in mice. Nature 378: 720–724. [DOI] [PubMed] [Google Scholar]
- Riccardi, V. M., E. Sujansky, A. C. Smith and U. Francke, 1978. Chromosomal imbalance in the Aniridia-Wilms' tumor association: 11p interstitial deletion. Pediatrics 61: 604–610. [PubMed] [Google Scholar]
- Robertson, E., 1987. Embryo-derived stem cell lines, pp. 77–112 in Teratocarcinomas and Embryonic Stem Cells: A Practical Approach, edited by E. Robertson. IRL Press, Oxford/Washington, DC.
- Schinzel, A., 2001. Catalogue of Unbalanced Chromosome Aberrations in Man. Walter de Gruyter, Berlin.
- Sebat, J., B. Lakshmi, J. Troge, J. Alexander, J. Young et al., 2004. Large-scale copy number polymorphism in the human genome. Science 305: 525–528. [DOI] [PubMed] [Google Scholar]
- Shaffer, L. G., and J. R. Lupski, 2000. Molecular mechanisms for constitutional chromosomal rearrangements in humans. Annu. Rev. Genet. 34: 297–329. [DOI] [PubMed] [Google Scholar]
- Thomas, J. A., D. K. Manchester, K. E. Prescott, R. Milner, L. McGavran et al., 1996. Hunter-McAlpine craniosynostosis phenotype associated with skeletal anomalies and interstitial deletion of chromosome 17q. Am. J. Med. Genet. 62: 372–375. [DOI] [PubMed] [Google Scholar]
- Toyo-oka, K., A. Shionoya, M. J. Gambello, C. Cardoso, R. Leventer et al., 2003. 14–3-3epsilon is important for neuronal migration by binding to NUDEL: a molecular explanation for Miller-Dieker syndrome. Nat. Genet. 34: 274–285. [DOI] [PubMed] [Google Scholar]
- Tsai, T. F., Y. H. Jiang, J. Bressler, D. Armstrong and A. L. Beaudet, 1999. Paternal deletion from Snrpn to Ube3a in the mouse causes hypotonia, growth retardation and partial lethality and provides evidence for a gene contributing to Prader-Willi syndrome. Hum. Mol. Genet. 8: 1357–1364. [DOI] [PubMed] [Google Scholar]
- Tucker, K. E., M. T. Berciano, E. Y. Jacobs, D. F. LePage, K. B. Shpargel et al., 2001. Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product. J. Cell Biol. 154: 293–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Helvoort, A., A. de Brouwer, R. Ottenhoff, J. F. Brouwers, J. Wijnholds et al., 1999. Mice without phosphatidylcholine transfer protein have no defects in the secretion of phosphatidylcholine into bile or into lung airspaces. Proc. Natl. Acad. Sci. USA 96: 11501–11506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varesco, L., H. J. Thomas, S. Cottrell, V. Murday, S. J. Fennell et al., 1989. CpG island clones from a deletion encompassing the gene for adenomatous polyposis coli. Proc. Natl. Acad. Sci. USA 86: 10018–10022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walz, K., S. Caratini-Rivera, W. Bi, P. Fonseca, D. L. Mansouri et al., 2003. Modeling del(17)(p11.2p11.2) and dup(17)(p11.2p11.2) contiguous gene syndromes by chromosome engineering in mice: phenotypic consequences of gene dosage imbalance. Mol. Cell. Biol. 23: 3646–3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X., D. Yang, J. H. Ding, W. Wang, P. H. Chu et al., 2005. ASF/SF2-regulated CaMKIIdelta alternative splicing temporally reprograms excitation-contraction coupling in cardiac muscle. Cell 120: 59–72. [DOI] [PubMed] [Google Scholar]
- Yoshida, Y., S. Tanaka, H. Umemori, O. Minowa, M. Usui et al., 2000. Negative regulation of BMP/Smad signaling by Tob in osteoblasts. Cell 103: 1085–1097. [DOI] [PubMed] [Google Scholar]
- Yu, Y., and A. Bradley, 2001. Engineering chromosomal rearrangements in mice. Nat. Rev. Genet. 2: 780–790. [DOI] [PubMed] [Google Scholar]
- Zheng, B., A. A. Mills and A. Bradley, 1999. A system for rapid generation of coat color-tagged knockouts and defined chromosomal rearrangements in mice. Nucleic Acids Res. 27: 2354–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, B., A. A. Mills and A. Bradley, 2001. Introducing defined chromosomal rearrangements into the mouse genome. Methods 24: 81–94. [DOI] [PubMed] [Google Scholar]