Abstract
Land plants underwent tremendous evolutionary change following the divergence of the ancestral lineage from algal relatives. Several important developmental innovations appeared as the embryophyte clade diversified, leading to the appearance of new organs and tissue types. To understand how these changes came about, we need to identify the fundamental genetic developmental programs that are responsible for growth, patterning, and differentiation and describe how these programs were modified and elaborated through time to produce novel morphologies. Class III homeodomain–leucine zipper (class III HD–Zip) genes, identified in the model plant Arabidopsis thaliana, provide good candidates for basic land plant patterning genes. We show that these genes may have evolved in a common ancestor of land plants and their algal sister group and that the gene family has diversified as land plant lineages have diversified. Phylogenetic analysis, expression data from nonflowering lineages, and evidence from Arabidopsis and other flowering plants indicate that class III HD–Zip genes acquired new functions in sporophyte apical growth, vascular patterning and differentiation, and leaf development. Modification of expression patterns that accompanied diversification of class III HD–Zip genes likely played an important role in the evolution of land plant form.
LAND plants (embryophytes) compose a monophyletic group that together with the charophycean green algae form the streptophyte clade (Mishler et al. 1994; Kenrick and Crane 1997; Bhattacharya et al. 1998). Recent molecular phylogenetic analyses resolve the charophycean group Charales as sister to land plants (Karol et al. 2001; Delwiche et al. 2002) (Figure 1). These phylogenetic analyses along with comparative analysis of ultrastructure and biochemistry indicate that land plants evolved from a freshwater charophycean green algal ancestor and that ancestor possessed certain developmental features that were inherited by land plants and are shared with extant charophytes (Graham 1993; Graham et al. 2000; Cook 2004).
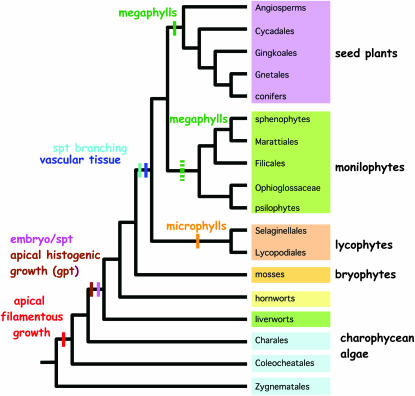
Figure 1.
Major developmental innovations in land plant evolution with relationships of green plants inferred from recent molecular and morphological phylogenies (Bremer et al. 1987; Gugerli et al. 2001; Karol et al. 2001; Pryer et al. 2001). Some nodes are controversial, such as those of the bryophytes, but the discrepancies do not affect the positions of the major innovations. The production of tissues from an apical meristem maps to the common ancestor of land plants although apical filamentous growth evolved in an algal ancestor (McCourt et al. 2004). Leaf-like organs evolved independently at least three times in the lycophytes, monilophytes, and seed plants. The simpler leaves of lycophytes are called microphylls and the more complex leaves of monilophytes and seed plants are called megaphylls. The broken green bar in the monilophyte ancestor represents the uncertainty concerning the number of origins of leaves within this clade. Roots likely had multiple origins as well, but the paleobotanical record is unclear and so we have not mapped them here.
However, the origin and diversification of embryophytes from algal ancestors also involved dramatic evolutionary changes. Several innovations allowed for the diversification of the relatively simple and diminutive ancestral land plant into a lineage of increasingly complex and diverse forms and life histories. These evolutionary innovations included the origin of the diploid sporophyte (embryo), histogenesis directly from an apical meristem, apical growth and branching in the sporophyte generation, the origin of lignified conducting and support tissues, the origin of roots, and the origin of leaves (Figure 1) (Graham et al. 2000; Niklas 2000; Sussex and Kerk 2001; Boyce and Knoll 2002; Cooke et al. 2002; Friedman et al. 2004). It has also been proposed that evolutionary changes in auxin action were essential for increasing complexity of land plant form (Cooke et al. 2002).
The key to understanding morphological evolution in multicellular organisms is determining fundamental components of the developmental patterning systems that have been modified through time to produce novel body plans (Carroll 2000). To learn how evolutionary changes in development have played a role in producing morphological diversity and complexity in plants, it is essential to understand the genetic basis of developmental evolution (Graham et al. 2000; Niklas 2000; Sussex and Kerk 2001; Boyce and Knoll 2002; Cooke et al. 2002; Friedman et al. 2004). We must look to model genetic systems and focus on developmental genes that are known to play a fundamental role in the establishment of growth and patterning throughout the plant body and throughout the life of a plant. Genes such as these would provide likely candidates for part of a developmental tool kit that has been modified through time to allow the origin of the new tissues and organs that have characterized land plant evolution.
Accumulating evidence on the developmental roles of class III homeodomain–leucine zipper (class III HD–Zip) proteins implicate this family of genes as intriguing candidates for part of a basic plant patterning tool kit as they have been shown to be involved in several key developmental processes in the sporophyte body. There are five Arabidopsis class III HD–Zip genes: AtHB8, CORONA/AtHB15 (CNA), PHABULOSA (PHB), PHAVOLUTA (PHV), and REVOLUTA (REV). Class III HD–Zip genes encode transcription factors with an N-terminal homeodomain immediately followed by a leucine zipper domain (Sessa et al. 1998). C-terminal to the HD–Zip is a START domain (Ponting and Aravind 1999; Schrick et al. 2004) followed by an extensive C-terminal region (more than half of the protein sequence) of unknown function that is highly conserved.
Four of the five genes, CNA, PHB, PHV, and REV, play a role in initiation and function of the shoot apical meristem (SAM) as well as initiation of axillary SAMs (Talbert et al. 1995; Otsuga et al. 2001; Emery et al. 2003; Green et al. 2005; Prigge et al. 2005). REV and PHB functions appear to be critical for the formation of an embryonic SAM, with rev phb phv and rev phb cna seedlings lacking a functional SAM (Emery et al. 2003; Prigge et al. 2005). Loss-of-function rev plants often fail to initiate lateral branches and floral meristems, indicating that REV regulates the initiation of axillary buds (Talbert et al. 1995; Otsuga et al. 2001). Consistent with their role in SAM establishment, PHB, REV, and CNA are initially expressed throughout the globular proembryo but their expression later becomes restricted to a central apical position (McConnell et al. 2001; Emery et al. 2003; Prigge et al. 2005).
PHB, PHV, and REV are also involved in establishment of adaxial identity and growth of leaves and other leaf-derived lateral organs. All three genes are expressed in leaf anlagen (primordia prior to emergence), with expression becoming restricted to adaxial domains and provascular tissue of primordia and young leaves (McConnell et al. 2001; Otsuga et al. 2001; Emery et al. 2003; Prigge et al. 2005). This expression pattern is evident during embryogenesis with expression of PHB, PHV, and REV in the adaxial domains of the cotyledons from their inception and also centrally in the developing hypocotyl. CNA is also transiently expressed adaxially in cotyledons and centrally in torpedo-stage embryos. Dominant gain-of-function mutations of PHB and PHV result in an abaxial-to-adaxial conversion of tissue types in cotyledons and leaves and in loss of blade outgrowth (McConnell and Barton 1998; McConnell et al. 2001; Zhong and Ye 2004). Conversely, phb phv rev seedlings often have only a single radial structure, which has been interpreted as representing an abaxialized cotyledon (Emery et al. 2003).
All Arabidopsis class III HD–Zip genes have been implicated in patterning and differentiation of vascular tissues of the shoot system. For the genes that have been examined in detail, vascular expression commences in the provascular cells, possibly defining them, and subsequently becomes restricted to the residual procambium and differentiating xylem (Baima et al. 1995; Scarpella et al. 2000; Kang and Dengler 2001; Kang et al. 2003; Prigge et al. 2005). Loss-of-function rev plants are reported to produce less secondary vascular tissue than wild-type plants (Zhong and Ye 1999) and to have altered patterns of interfascicular fiber differentiation (Zhong and Ye 1999; Prigge et al. 2005; our unpublished data). Dominant gain-of-function rev mutations result in a radialization of vascular bundles in stems with xylem surrounding phloem as well as an altered arrangement of bundles (Zhong and Ye 2001, 2004; Emery et al. 2003). A similar phenotype has been reported for phb phv cna loss-of-function plants, which suggests that, in stem vascular patterning, REV function may be antagonized by the action of PHB, PHV, and CNA (Prigge et al. 2005). Orthologs of ATHB8, CNA, and REV have all been shown to be expressed and play a role in vascular differentiation in Zinnia, with specific roles in xylem differentiation (Ohashi-Ito et al. 2002, 2003, 2005). Furthermore, orthologs of ATHB8, ATHB15/CNA, and PHV are expressed in the vascular cambium and immature secondary xylem of poplar, suggesting a role in secondary vascular development (Hertzberg et al. 2001; Schrader et al. 2004; Ko et al. 2006). On the basis of analogous spatial relationships, Emery et al. (2003) speculated that a genetic system consisting of class III HD–Zip and KANADI genes that pattern vascular tissues was co-opted to pattern leaves in seed plants.
In summary, class III HD–Zip genes in flowering plants are involved with fundamental developmental processes that represent most of the key innovations of land plants, including formation and function of shoot apical and axillary meristems, patterning of three-dimensional tissues, differentiation of lignified conducting and support tissues, and leaf development. Furthermore, some class III HD–Zip genes have been implicated in auxin response and polar transport (Baima et al. 2001; Zhong and Ye 2001; Mattsson et al. 2003) and in root development (Zhong and Ye 2001; Hawker and Bowman 2004) in Arabidopsis, extending the scope of their potential roles in development of the land plant body.
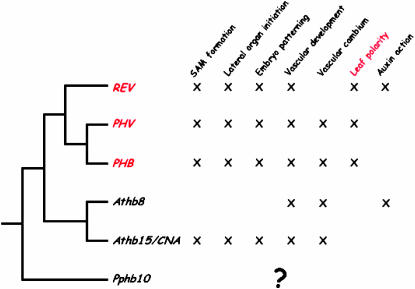
There exists a complex pattern of functional overlap and redundancy among class III HD–Zip genes in Arabidopsis. There is also evidence of antagonistic or complementary function and what appears to be a restriction of some functions, such as adaxial patterning, to particular genes. Phylogenetic analysis of the five Arabidopsis genes indicates that PHB and PHV are sister to each other and with REV form a clade (the “REV clade”) that is sister to ATHB8 plus CNA (the “CNA clade”) (Emery et al. 2003) (Figure 2). Functions such as vascular patterning and differentiation, embryo patterning, and SAM function occur in genes from both REV and CNA and clades. However, adaxial patterning is restricted to genes of the REV clade.
Figure 2.
Phylogenetic relationships and functions of Arabidopsis class III HD–Zip genes.
One explanation for the distribution of functions among Arabidopsis class III HD–Zip genes is that apical meristem function and vascular patterning are more ancient developmental roles for class III HD–Zip genes and that leaf polarity was a new function derived in the common ancestor of the REV clade genes (Emery et al. 2003). Alternatively, it is possible that adaxial polarity was a function of a gene ancestral to all five Arabidopsis class III HD–Zips but was lost in the CNA clade ancestor following divergence from the REV clade ancestor. Phylogenetic interpretation of fossil and anatomical evidence indicates that sporophyte apical meristems and branching evolved prior to the evolution of vascular tissues (Edwards 1986; Kenrick and Crane 1991, 1997) (Figure 1) and that tracheids (water-conducting cells) evolved once in the common ancestor of all tracheophytes (Kenrick and Crane 1997; Cook and Friedman 1998). Leaves evolved later, and independently, in several vascular plant lineages, including lycophytes, monilophytes, and seed plants (Donoghue et al. 1989; Gensel 1992; Kenrick and Crane 1997; Doyle 1998) (Figure 1). Thus the phylogenetic distribution of class III HD–Zip functions in Arabidopsis suggests a scenario in which class III HD–Zip genes were associated with basic growth and patterning in ancient land plants and diversified and acquired new functions that allowed the modification of land plant development and the origin of new tissues and organs such as vascular tissue and leaves.
Class III HD–Zip homologs have been identified in representatives of all land plant lineages (Aso et al. 1999; Sakakibara et al. 2001; Floyd and Bowman 2004), indicating that the gene family was an early component of the land plant developmental program. To further evaluate the hypothesis that the evolution of class III HD–Zip genes was associated with the evolution of plant form, it is essential to determine the phylogenetic relationships of land plant class III HD–Zip genes and to attempt to assess the functions of class III HD–Zip genes in taxa representing the major lineages and diverse body plans of the embryophytes. Furthermore, it is not known if class III HD–Zip genes evolved prior to the origin of land plants and were thus a part of the ancestral land plant genome or represent a gene family that evolved uniquely within the land plant lineage. In the absence of knowledge of the phylogenetic relationships of class III HD–Zips and their functions throughout the land plant clade, we cannot distinguish between alternative hypotheses for the distribution of functions of class III HD–Zip genes in Arabidopsis or do more than speculate that these genes have played a role in the evolution of land plant form.
To further investigate the possibility that class III HD–Zip genes constitute part of an ancient land plant developmental tool kit, we undertook a study to identify homologs in all major land plant lineages including bryophytes, lycophytes, ferns, gymnosperms, and charophycean algae. Here we present the phylogenetic distribution of the class III HD–Zip gene family and explore the evolutionary relationships of these genes. We also present expression data for genes from three nonflowering vascular plants. Finally, we discuss the implications of class III HD–Zip expression in flowering and nonflowering plants in the context of gene phylogeny and organismic phylogeny to formulate a more informed hypothesis about the functional evolution of class III HD–Zip genes.
MATERIALS AND METHODS
Taxon selection:
Taxa representing all major land plant clades as well as the two closest lineages of charophycean algae, Charales and Coleochaetales, were sampled. These taxa include the seed plants Ginkgo biloba, Pseudotsuga menziesii, and Taxus globosa; the monilophytes Psilotum nudum and Ceratopteris richardii; the lycophytes Selaginella kraussiana and S. moellendorffii; the moss Physcomitrella patens; the hornwort Phaeoceros carolinianus; the liverwort Marchantia polymorpha; and the algae Chara corallina and Coleochaete scutata.
RNA and DNA extraction and cDNA synthesis:
For Physcomitrella, leafy shoots (gametophores) of the haploid gametophyte generation were collected. For the thalloid bryophytes Marchantia and Phaeoceros and the alga Chara, total RNA was extracted from marginal and apical gametophytic tissue, respectively. Entire Coleochaete thalli were used for RNA and DNA extraction. Phaeoceros sporophytes were cut above the level of emergence from the gametophyte thallus and RNA was extracted separately. Trizol reagent (Invitrogen, San Diego) was used to extract RNA from developing male and female cones of T. globosa. RNA was extracted from sporophyte shoot apices (including the SAM and young leaves) for all other vascular plant taxa, including Ceratopteris, Psilotum, Ginkgo, and Pseudotsuga. RNA was also extracted from whole haploid gametophytes of Ceratopteris. Total RNA was extracted from Physcomitrella and Selaginella using the RNeasy kit (QIAGEN, Chatsworth, CA). The RNA extraction protocol of Kiefer et al. (2000) was used for all other taxa (except Taxus). Extraction of RNA from Psilotum, Chara, and Ginkgo required two phenol extractions prior to the final chloroform extraction and subsequent cleaning with the RNeasy kit. 5′-RACE ready cDNA was generated from total RNA following the protocol of the SMART RACE kit (CLONTECH, Palo Alto, CA). Genomic DNA was extracted from all taxa using the Nucleon Phytopure DNA extraction kit (Amersham Biosciences). For Chara, Phaeoceros, and Pseudotsuga, additional purifications using QIAGEN columns were needed before we were able to amplify sequence from the DNA.
Primer design and PCR:
Degenerate primers were designed on the basis of aligned amino acid sequences in conserved regions of the coding regions of all five Arabidopsis family members as well as PpHB10 (Sakakibara et al. 2001) (supplemental Figure S1 at http://www.genetics.org/supplemental/). Two forward and two reverse degenerate primers were used in nested PCR reactions using cDNA as template. Bands matching the expected size were excised from agarose gels, extracted with the QIAquick gel extraction kit (QIAGEN), and cloned directly into the pCRII vector (TOPO TA cloning kit, Invitrogen). A total of 18–54 colonies were grown in TB culture and plasmid DNA was isolated. A preliminary screen for different gene family members was done by a series of restriction digests (AluI, Sau3AI, HaeIII) on PCR-amplified (using M13 primers) cloned fragments. Clones representing different banding patterns were sequenced by the College of Biological Sciences automated sequencing facility, University of California at Davis, using an ABI PRISM 3100 Genetic Analyzer.
For isolation of full-length cDNA clones, primers specific to each partially cloned sequence were designed and used in 5′- and 3′-RACE PCR reactions using the SMART RACE kit (CLONTECH). Two primers were designed for each RACE and nested RACE PCRs were necessary. End primers were designed on the basis of sequenced 5′- and 3′-ends. The complete cDNA sequences were then amplified using PCR, TA cloned, and sequenced. We also designed 3′-RACE primers to amplify the full-length cDNA of the Ceratopteris gene, CrHB1, for which only the HD–Zip encoding region was previously published (GenBank AB013791). Primers based on the cDNA sequences were used to amplify the corresponding genomic sequences for at least one sequence from each land plant species to compare intron positions and length.
Identification of homologs:
One hundred clones resulting from the TA cloning of degenerate PCR products served as templates in PCR reactions using gene-specific primers for all of the sequences already identified for a given taxon. Any clones not positive for known sequences were screened using a series of restriction digests. One representative clone from each pattern group was sequenced. We tested this method for detecting Arabidopsis class III HD–Zip genes. The same degenerate primers were used in a PCR reaction with Arabidopsis thaliana Landsberg erecta seedling cDNA and the products were TA cloned. All five Arabidopsis class III HD–Zip genes were present in 100 randomly chosen clones.
Discontiguous megaBLAST similarity searches of the NCBI trace archives of whole-genome sequencing of P. patens and Selaginella moellendorffii (Department of Energy Joint Genome Institute) were performed. Sequence traces were assembled into contigs in Sequencher 4.2 for Macintosh (Gene Codes, Ann Arbor, MI). Messenger RNA (mRNA) sequences were inferred from genomic sequences on the basis of comparisons with aligned amino acid translations of class III HD–Zip genes from all other land plants. Trace archive accession numbers used to assemble these sequences are available in supplemental Table S2 at http://www.genetics.org/supplemental/ and inferred genomic and mRNA sequences are available in FASTA format also as supplemental material at http://www.genetics.org/supplemental/. In addition, BLAST similarity searches using amino acid sequences of REVOLUTA, CORONA, and Athb8 were performed on The Institute for Genomic Research Rice Genome Annotation Database and four class III HD–Zip genes from Zinnia elegans were obtained from GenBank. Inferred amino acid sequences for Oryza and Zinnia sequences were included in the alignment and phylogenetic analyses.
Sequence analysis, alignment, and phylogenetic analysis:
Sequence contigs were assembled using Sequencher 4.2 for Macintosh (Gene Codes). Translated amino acid sequences were initially aligned in ClustalX. Coding nucleotide sequences were entered into Se-Al v2.0a11 for Macintosh (Rambaut 1996) and manually aligned using amino acid translations. Ambiguously aligned regions were excised and the remaining alignment, including 757 amino acid characters, was exported as both Nexus and Phylip files for further analysis.
Bayesian phylogenetic analysis was performed using Mr. Bayes 3.1 (Huelsenbeck and Ronquist 2001). This version of the software conducts two independent analyses simultaneously. The mixed model option (aamodelpr=mixed) was used to estimate the appropriate amino acid fixed-rate model. The analysis was run for 500,000 generations, which was sufficient for the standard deviation of the split frequencies to drop below 0.01. To allow for the burn-in phase, the first 500 trees (10% of the total number of saved trees) were discarded.
Maximum-likelihood analysis of amino acid sequences was performed using Phylip 3.2 (Felsenstein 1989). We used the Jones–Taylor–Thornton probability model with constant rates. To estimate clade support, a bootstrap analysis was performed with 1000 replicates.
Histology:
Shoot apices of Ginkgo, Pseudotsuga, and Selaginella were fixed in a solution of 1.5% glutaraldehyde, 1% paraformaldehyde, and 4% acrolein in PIPES buffer (84 mm PIPES, 8.4 mm EGTA, and 1.6 mm MgSO4) at pH 6.8. Specimens were left in fixative a minimum of 24 hr and then rinsed in PIPES buffer and dehydrated through an ethanol series to 95% ethanol.
Specimens were then infiltrated with catalyzed monomer A of the JB-4 embedding kit (Polysciences, Warrington, PA) and embedded in an oxygen-free environment following the basic protocol provided with the kit. Blocks were serially sectioned at 5 μm on an HM 355S (Microm) rotary microtome using glass knives. Slides were stained in 0.1% toluidine blue, examined, and photographed on a Zeiss Axioskop microscope equipped with a Zeiss Axiocam digital camera using bright-field microscopy.
In situ hybridization:
Tissues were fixed in formalin acetic acid overnight and then dehydrated through an ethanol series to 100% ethanol. The ethanol was gradually replaced with Histo-Clear II (National Diagnostics, Atlanta) The Histo-Clear was gradually replaced with Paraplast X-tra (Fisher Scientific) at 56°. The Paraplast was replaced twice daily for 3 days after which the tissues were embedded. The embedded specimens were sectioned at 10–12 μm, mounted on ProbeOn Plus slides (Fisher Scientific), and dried overnight at 37°.
Digoxigenin (DIG)-labeled antisense and sense RNA probes were prepared from full-length or partial cDNA clones of PmC3HDZ1, PmC3HDZ2, GbC3HDZ1, GbC3HDZ2, GbC3HDZ3, SkC3HDZ1, and SkC3HDZ2 using DIG labeling mix (Roche Diagnostics). Our prehybridization, hybridization, and posthybridization procedures were based on those of Vielle-Calzada et al. (1999) with some modifications. A detailed protocol is available from the authors upon request. After staining was stopped, the slides were dehydrated, dried, and permanently covered using Cytoseal (Richard-Allan Scientific). Slides were examined and photographed on a Zeiss Axioskop microscope equipped with a Zeiss Axiocam digital camera using either bright-field or differential interference contrast microscopy.
RESULTS
Class III HD–Zip genes in nonflowering plants:
Class III HD–Zip mRNA sequences were amplified and cloned from the growing-tip regions of all taxa except Coleochaete (Table 1). We detected a single, unique expressed class III HD–Zip gene in each of three taxa: Chara (CcC3HDZ1), Phaeoceros (PcC3HDZ1), and Marchantia (MpC3HDZ1). The same transcript was amplified in RT–PCR from Phaeoceros sporophytic and gametophytic tissue. From gametophores of the moss Physcomitrella, we identified two expressed genes (PpC3HDZ1 and PpC3HDZ2) in addition to the previously published gene PpHB10 (Sakakibara et al. 2001). Similarity searches of the P. patens genomic trace archives revealed the presence of two additional genes (PpC3HDZ3 and PpC3HDZ4; supplemental Figure S2 at http://www.genetics.org/supplemental/) for a total of five. Using RT–PCR, we also detected mRNA sequences for these two sequences in gametophores.
TABLE 1.
New class III HD-Zip sequences cloned in this study
| Species | Sequence name | Sequence type | GenBank accession no. |
|---|---|---|---|
| C. corallina | CcC3HDZ1 | cDNA | DQ385513 |
| M. polymorpha | MpC3HDZ1 | cDNA, genomic | DQ385514, DQ385532 |
| P. carolinianus | PcC3HDZ1 | cDNA | DQ385515 |
| P. patens | PpC3HDZ1 | cDNA, genomic | DQ385516, DQ385533 |
| P. patens | PpC3HDZ2 | cDNA, genomic | DQ385517DQ385534 |
| P. patens | PpHB10 | cDNA, genomic | DQ385518, DQ385535 |
| S. kraussiana | SkC3HDZ1 | cDNA, genomic | DQ385519, DQ385536 |
| S. kraussiana | SkC3HDZ2 | cDNA | DQ385520 |
| P. nudum | PnC3HDZ1 | cDNA, genomic | DQ385521, DQ385537 |
| P. nudum | PnC3HDZ2 | cDNA | DQ385522 |
| P. nudum | PnC3HDZ3 | cDNA | DQ385523 |
| C. richardii | CrC3HDZ1 | cDNA | DQ385524 |
| G. biloba | GbC3HDZ1 | cDNA, genomic | DQ385525, DQ385538 |
| G. biloba | GbC3HDZ2 | cDNA | DQ385526 |
| G. biloba | GbC3HDZ3 | cDNA | DQ385527 |
| P. menziesii | PmC3HDZ1 | cDNA, genomic | DQ385528, DQ385539 |
| P. menziesii | PmC3HDZ2 | cDNA, genomic | DQ385529, DQ385540 |
| T. globosa | TgC3HDZ1 | cDNA | DQ385530 |
| T. globosa | TgC3HDZ2 | cDNA | DQ385531 |
In all vascular plant taxa we detected multiple gene family members (Table 1). Two class III HD–Zip sequences were cloned from S. kraussiana (SkC3HDZ1 and SkC3HDZ2). A similarity search of S. moellendorffii genomic trace archives also revealed two class III HD–Zip genes (SmC3HDZ1 and SmC3HDZ2; supplemental Figure S2 at http://www.genetics.org/supplemental/). From C. richardii we cloned one new sequence, CrC3HDZ1, in addition to the full-length cDNA and genomic sequences of CrHB1 (Aso et al. 1999). We found three expressed genes in Ginkgo (GbC3HDZ1, GbC3HDZ2, and GbC3HDZ3) and two each from Taxus (TgC3HDZ1 and TgC3HDZ2) and Pseudotsuga (PmC3HDZ1 and PmC3HDZ2).
Genomic sequences for the coding regions of MpC3HDZ, PpHB10, PpC3HDZ1, PpC3HDZ2, SkC3HD1, CrHB1, PnC3HDZ, PmC3HDZ1, PmC3HDZ2, GbC3HDZ1, and GbC3HDZ2 were amplified and sequenced (Table 1). Partial genomic sequence for CcC3HDZ1 was amplified and sequenced.
Inferred amino acid sequences of all land plant and Chara class III HD–Zip genes are highly conserved and easily alignable for most of their length (supplemental Figure S1 at http://www.genetics.org/supplemental/). The exception is the region between the HD–Zip domain and the START domain (supplemental Figure S1 at http://www.genetics.org/supplemental/). In that part of the alignment, some sequences are conspicuously longer than others. Of these, the Marchantia and Phaeoceros sequences are similar, the Physcomitrella sequences are alignable to each other, and the Selaginella sequences as well as two monilophyte sequences (CrC3HDZ1 and PnC3HDZ3) seem to have alignable elements. Beyond that, it is not clear that any of these groups of sequences align with each other or with the Chara sequence (supplemental Figure S1 at http://www.genetics.org/supplemental/). The average pairwise identity of all identified class III HD–Zip protein sequences is 59.6% with the low being 41% (between the Chara sequence and three different vascular plant sequences) and the highest identity being 91% (between Ginkgo GbC3HDZ2 and Taxus TgC3HDZ2). The coding sequences range in length from 2457 nt (Selaginella SkC3HDZ1) to 2733 nt (Chara CcC3HDZ1), encoding amino acid sequences of 818–910 aa. Most length differences in the coding sequence are due to variable regions flanking the HD–Zip domain (supplemental Figure S1 at http://www.genetics.org/supplemental/). All class III HD–Zip sequences encode an HD–Zip domain, a START domain, and a conserved C terminus (Figure 3; supplemental Figure S1 at http://www.genetics.org/supplemental/) with one exception. The exception is the C. richardii gene CrHB1. We cloned the full-length cDNA on the basis of the previously published HD–Zip encoding sequence and found that this mRNA encodes an ORF for the HD–Zip region, followed by 1375 nucleotides with no ORFs, and terminated with a poly(A) tail. BLAST searches of the C-terminal sequence of the CrHB1 cDNA revealed no significant similarity to any known Arabidopsis genes.
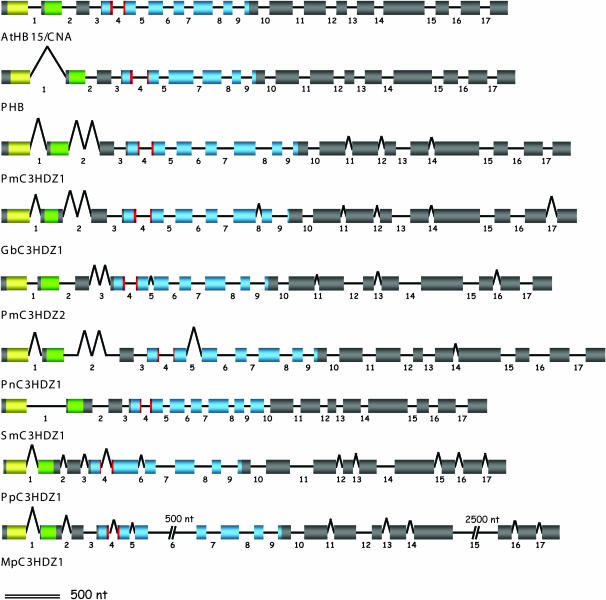
Figure 3.
Intron–exon structure of class III HD–Zip coding regions. Introns within the coding region are numbered 1–17 according to the maximum number present in Arabidopsis genes. Exons are represented by wider, shaded bars; introns by black lines. Lines and bars are proportional in length and represent total sequence length except for MpC3HDZ1 in which segments of two long introns are omitted as indicated by double breaks. Length of missing segments are indicated above. Yellow, homeodomain; green, leucine zipper; blue, START domain; red, miR165/166 binding site; gray, carboxy terminus.
Comparison of genomic sequences indicates that introns and splice sites are largely conserved (Figure 3; supplemental Figure S1 at http://www.genetics.org/supplemental/). There are 17 internal introns (within the coding region) in three of the five Arabidopsis class III HD–Zip genes. PHB lacks internal intron 6 and ATHB8 lacks intron 15. Most genomic sequences from other land plants also have 17 introns and the splice sites map to within or between the same codons in most cases (supplemental Figure S1 at http://www.genetics.org/supplemental/). However, the splice sites in two of the Physcomitrella sequences (PpHB10 and PpC3HDZ2) occur within a sequence that cannot be aligned with other taxa (supplemental Figure S1 at http://www.genetics.org/supplemental/). Intron 5 is missing in all of the Physcomitrella sequences. One of the Selaginella sequences, SkC3HDZ1, is missing intron 15 although the intron appears to be present in the S. moellendorffii ortholog. No additional introns were identified in genomic sequences from nonflowering plants.
Evidence of microRNA regulation:
Comparison of the aligned nucleotide sequences of streptophyte class III HD–Zip sequences at the miR165/166 binding site indicates nearly complete conservation at the nucleotide level for all land plant sequences (Floyd and Bowman 2004; Figure 4). The nucleotide sequence of Chara CcC3HDZ1 differs at five nucleotide positions from all land plant sequences, resulting in five additional mismatches to miR165/166 (Figure 4).
Figure 4.
Alignment of the nucleotides composing the miR165/166 binding site in land plant class III HD–Zip mRNAs. x, a conserved position; –, a nucleotide position that varies: at the top of the alignment these symbols indicate conserved and variable positions in land plant sequences and at the bottom of the alignment indicate conserved and variable positions with the Chara sequence included in the alignment. Vertical lines indicate complementarity of the mRNA sequence to miR165/166. Arrows show five additional mismatches of the Chara sequence to miR166.
Phylogeny of class III HD–Zip genes:
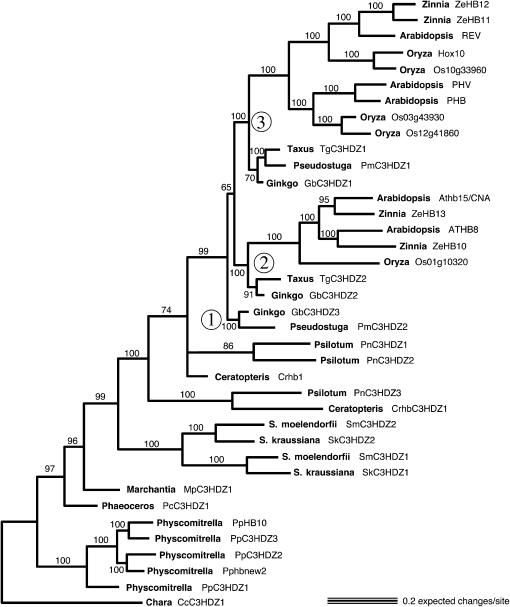
Bayesian analysis of amino acid sequences produced a tree with good resolution and mostly well-supported clades (Figure 5). The tree was rooted with the Chara sequence (CcC3HDZ1). A clade including all five moss sequences is resolved as a sister to all other land plant sequences. Within the Physcomitrella clade, PpC3HDZ1 is sister to the other four genes and these relationships are strongly supported (100%). The single Phaeoceros gene (PcC3HDZ1) is sister to the sequences of Marchantia plus vascular plants and the single Marchantia sequence (MpC3HDZ1) is resolved as sister to a clade including all vascular plant sequences. Both of these relationships have high clade credibility. Within the vascular plant clade, Selaginella sequences were identified as a monophyletic sister group to all other vascular plant genes and this relationship is highly supported (99%). Within this lineage there are two clades, each including one S. kraussiana sequence and one S. moellendorffii sequence (Figure 5). Sister to the Selaginella clade is a group including monilophyte (Psilotum and Ceratopteris) and seed plant sequences with 100% credibility. Within this clade, three of the monilophyte sequences, CrHB1 and PnC3HDZ1 plus PnC3HDZ2, reside in an unresolved polytomy with the seed plant sequences, but this relationship does not have high clade credibility. The seed plant clade is highly supported (99%).
Figure 5.
Bayesian phylogram of class III HD–Zip genes. Numbers above the branches indicate posterior probability values. Circled numbers 1, 2, and 3 indicate the three highly supported seed plant clades.
Seed plant class III HD-Zip sequences are resolved into two clades that include both gymnosperm and angiosperm sequences (both with 100% credibility values) (circled numbers 2 and 3 in Figure 5) and a third comprising one Ginkgo (GbC3HDZ1) and one Pseudotsuga (PmC3HDZ2) sequence also with 100% clade credibility (circled number 1 in Figure 5). The two clades including angiosperm sequences are resolved as sister groups, but this relationship is not well supported. In each of the two clades including angiosperm and gymnosperm sequences, gymnosperm sequences form a sister group to angiosperm sequences, but neither gymnosperm clade has >95% clade credibility. One seed plant clade includes Ginkgo GbC3HDZ1, Taxus TgC3HDZ1, Pseudotsuga PmC3HDZ1, and Arabidopsis REV, PHB, and PHV and the other includes Ginkgo GbC3HDZ2, Taxus TgC3HDZ2, and Arabidopsis ATHB8 and CNA. REV is sister to two Zinnia sequences (ZeHB11 and ZeHB12) and REV plus Zinnia sequences are sister to two Oryza sequences (Hox10 and Os10g33960). All these relationships are highly supported. CNA and ATHB8 are each most closely related to a Zinnia sequence (ZeHB13 and ZeHB10, respectively). Together, the Zinnia and Arabidopsis sequences are sister to a single predicted rice gene (Os01g10320) and all of these relationships are highly supported.
Maximum-likelihood analysis produced a tree nearly identical to the Bayesian tree (not shown). Bootstrapping indicated similar levels of support, as did the Bayesian posterior probabilities. The exception was that there was <50% bootstrap support for the resolution of the branching order of Marchantia, Phaeoceros, the moss sequence clade, and the Selaginella sequence clade relative to each other and the monilophytes.
Pseudotsuga class III HD–Zip expression:
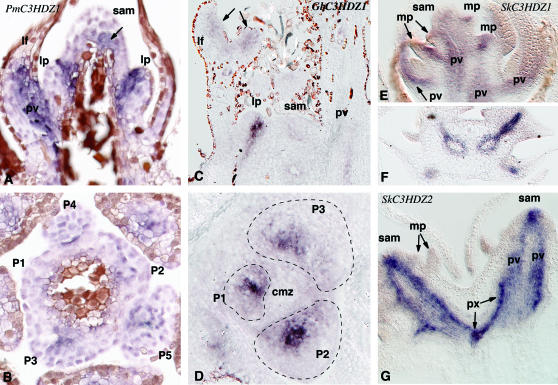
Signal from the PmC3HDZ1 probe is evident as light staining throughout the apical meristem initial zone (above the point at which leaf primordia form) (Figure 6A). Signal is more intense in the peripheral zone and throughout the youngest leaf primordia (Figure 6, A and B). In older leaf primordia, the staining is strongest in the adaxial region (compare P4 and P5 in Figure 6B), although the production of tannins in maturing leaf cells obscures staining outside of the developing vascular tissue. The darkest staining occurs in the region where the provascular strand is differentiating (Figure 6B). In longitudinal view, continuous expression of PmC3HDZ1 is evident from the adaxial region of primordia into the stem corresponding to the position of the provascular strand (Figure 6A). We were unable to detect a signal from the PmC3HDZ2 probe.
Figure 6.
In situ hybridization of class III HD–Zip genes in Pseudotsuga, Ginkgo, and Selaginella. (A) Nearly median longitudinal section through shoot apex of Pseudotsuga hybridized to PmC3HDZ1. The brown coloration is due to the presence of tannins. (B) Transverse section through shoot apex of Pseudotsuga at the level of emergence of leaf primordia, hybridized to PmC3HDZ1. Primordia are labeled P1–P5 from youngest to oldest. (C) Nearly median longitudinal section through shoot apex of Ginkgo hybridized to GbC3HDZ1. (D) Transverse section through shoot apex of Ginkgo at the level of emergence of leaf primordia, hybridized to GbC3HDZ1. Primordia are labeled P1–P3 from youngest to oldest. (E) Median longitudinal section through shoot apex of Selaginella hybridized to SkC3HDZ1. (F) Transverse section through shoot of Selaginella several nodes below the SAM hybridized to SkC3HDZ1. (G) Median longitudinal section through shoot apex of Selaginella hybridized to SkC3HDZ2. cmz, central mother cell zone; lf, leaf; lp, leaf primordium; mi, microphyll; mp, microphyll primordium; pv, provascular tissue; px, protoxylem; SAM, shoot apical meristem; x, xylem.
Ginkgo class III HD–Zip expression:
GbC3HDZ1 mRNA was detected as weak signal in the apical meristem (Figure 6, C and D). A distinct ring of light staining is evident in the peripheral zone (Figure 6D). Staining was also evident throughout the youngest leaf primordia but was restricted to adaxial domains of older primordia (Figure 6D). Very strong staining was observed in the location where leaf provascular tissue would later differentiate (Figure 6D). In longitudinal section, a continuous path of staining was observed leading from the strong signal in the young primordia into the ground tissue, defining the path of the leaf provascular connection from the tip of the primordium to the stem provasculature (Figure 6C). No signal was detected for the probes for GbC3HDZ2 and GbC3HDZ3.
Selaginella class III HD–Zip expression:
Signal from the SkC3HDZ1 antisense probe is weak in the apex at the level of the initial cells (Figure 6E). Strong signal is evident in a localized manner on the adaxial side of expanding leaves (microphylls) (Figure 6E). This position corresponds to where the ligule begins to grow on the adaxial side of the microphyll and where tracheary tissue first differentiates in the microphyll. In lower nodes this focus of expression extends both outward into expanded microphylls and inward toward the nearest stem provascular strand, clearly defining where the microphyll vascular trace will differentiate (Figure 6E). Expression remains restricted to the provascular strand and does not extend into the lamina as the lamina expands. Weak staining from the SkC3HDZ1 probe is visible in the stem provasculature more distally from the apex (Figure 6, E and F). The signal for this probe in stem and leaf provasculature is limited to the outer cell layers of the provascular strand that differentiate into phloem and pericycle (Figure 6F).
Expression for the second gene, SkC3HDZ2, is strong in the apical cells and diverges just below the position of the apical cells into two bands that define the location of the two provascular strands (Figure 6G). At the point where SkC3HDZ1 expression is evident in the stem provasculature, SkC3HDZ2 signal is restricted to the center of the provascular strand that will differentiate into xylem (Figure 6G). In older tissues, SkC3HDZ2 signal diminishes in the provascular strand except for highly localized points of strong signal that correspond to the first-maturing tracheary elements (protoxylem) in both stem and microphyll (Figure 6G).
DISCUSSION
Evolution of class III HD–Zip genes:
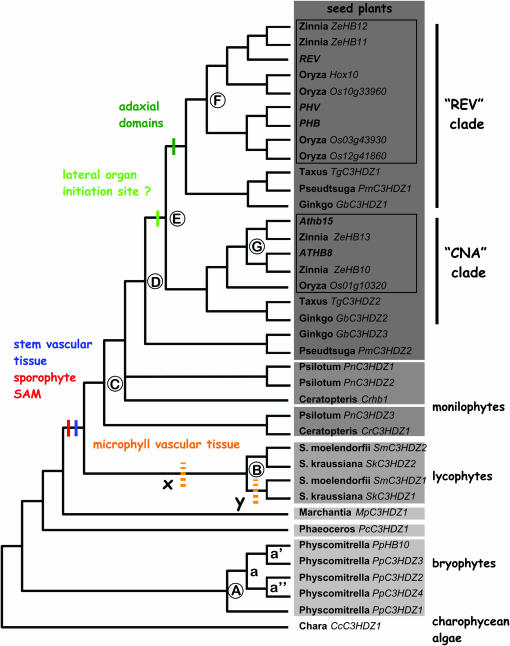
The inferred phylogenetic tree of class III HD–Zip genes is largely consistent with that of streptophytes (Figures 1 and 5). Chara has been recently resolved as more closely related to the land plants than to Coleochaete and we detected a class III HD–Zip gene only in Chara. Bryophyte sequences diverge from the basal nodes of the tree as three distinct lineages corresponding to liverworts, hornworts, and mosses. Vascular plant sequences form a monophyletic group with lycophytes sister to euphyllophytes. The ancestors of liverwort, hornwort, moss, lycophyte, and vascular plant lineages can be inferred to have inherited a single class III HD–Zip gene, inherited from an algal ancestor. Four duplications must be inferred within Physcomitrella, giving rise to five genes (A, a, a′, a″ in Figure 7). Similarly, a single ancestral gene was duplicated within the lycophyte lineage, giving rise to two genes in Selaginella (B in Figure 7). These two genes have orthologs in two different Selaginella species that diverged from the common ancestor of all but two extant Selaginella species according to recent phylogenetic analysis (Korall et al. 1999), suggesting an ancient duplication event. Whether this occurred within the Selaginellaceae or earlier in lycophyte evolution is not known. The analysis of class III HD–Zip gene diversity in additional lycophyte genera will resolve this issue.
Figure 7.
Hypothesis for functional evolution of class III HD–Zip genes in vascular plants. Putative origin of new functions are mapped onto the Bayesian gene cladogram based on the phylogram in Figure 5. Clades of angiosperm sequences in the REV and CNA clades are boxed. Circled letters A–F indicate inferred duplications described in the discussion. The broken bars labeled “x” and “y” in the Selaginella clade indicate uncertainty about whether specialization for microphyll vascularization evolved before or after duplication B.
Monilophyte (Psilotum and Ceratopteris) sequences were resolved into two clades, each including both Ceratopteris and Psilotum sequences. However, there was not significant support for placement of PnC3HDZ1, PnC3HDZ2, and CrHB1 in a clade with seed plant sequences. Collapsing the nodes would result in a polytomy of one highly supported clade (PnC3HDZ1 plus CrC3HDZ1), a seed plant clade, and the remaining Ceratopteris and Psilotum sequences. The analysis seems to indicate that there was a duplication of a monilophyte class III HD–Zip sequence prior to divergence of the ancestors of Psilotum and Ceratopteris (C in Figure 7), but we cannot determine if this duplication occurred before or after the divergence of monilophyte and seed plant ancestors. CrHB1 is a truncated sequence, encoding only a HD–Zip domain, which reduces the number of characters from this sequence and hence may make phylogenetic placement of this sequence difficult.
Seed plant class III HD–Zip sequences were resolved into three highly supported monophyletic groups (circled 1, 2, and 3 in Figure 5). Two of these include sequences from both gymnosperms and angiosperms and correspond to the REV and CNA clades of Arabidopsis (Figures 2 and 7). The third clade (circled 3 in Figure 5) includes only gymnosperm sequences. All three lineages include a Ginkgo sequence and the monophyly of the three seed plant clades is well supported (Figure 5). This indicates that three genes were present in the common ancestor of the seed plants included in the analysis (angiosperms and gymnosperms). Thus two duplications (D and E in Figure 7) must have occurred in a seed plant ancestor prior to the divergence of extant taxa, giving rise to three class III HD–Zip genes, one of which was lost in the angiosperm ancestor. The remaining two genes diversified further within angiosperms (F and G in Figure 7), giving rise to the REV and CNA clades, but not within gymnosperm lineages. We identified only two class III HD–Zip genes in both Taxus and Pseudotsuga. This could be explained by our failure to detect a third gene in these taxa, by the loss of one gene in each of these two lineages, or by a third gene that is not expressed in shoot apices.
Orthologs of both ATHB8 and CNA exist in Zinnia, an asterid, but only a single gene sister to both Arabidopsis and Zinnia sequences was found in Oryza, suggesting that the duplication giving rise to those two eudicot genes occurred after the divergence of monocots and eudicots but before the divergence of asterid and rosid ancestors. In this CNA clade, the gymnosperm sequences were resolved as the sister group to the angiosperm sequences, but there was not significant support for gymnosperm monophyly.
In the REV clade, the gymnosperm sequences formed a sister group to the angiosperm sequences, but, again, gymnosperm monophyly did not have significant support. The angiosperm sequences were resolved into two clades, each of which included both monocot and eudicot sequences. Thus a duplication of the REV clade ancestral gene must have occurred early in angiosperm evolution, giving rise to REV and a PHB/PHV ancestor. There are two rice sequences that together are orthologous to both PHB and PHV, indicating that there have been independent duplications in monocot and eudicot lineages of the PHB/PHV ancestral gene.
Conservation of class III HD–Zip sequences:
The time since divergence of the common ancestor of Chara and land plants is estimated to be >450 million years (Graham 1993). The algal sequence (CcC3HDZ1) is no less than 41% identical to amino acid sequences from other lineages, including flowering plants (range 41–91% and mean 59%). For comparison, average pairwise identity of a sample of class I KNOX proteins for which orthologs have been identified for most land plants was found to range between 26 and 76% with a mean of 40%. A sample of MIKCc MADS-box amino acid sequences, including Chara as well as land plants, are on average 42.6% identical, ranging between 31 and 60%. Thus, relative to other ancient groups of plant transcription factors, class III HD–Zip proteins are remarkably conserved across time and broad phylogenetic distance.
Sequences from all nonflowering lineages have introns in the same positions as Arabidopsis class III HD–Zip genes (supplemental Figure S1 at http://www.genetics.org/supplemental/). Thus it appears that genomic structure of the coding region has also largely been conserved, at least in land plants (Figure 3). Although we have not yet amplified the complete genomic sequence of Chara C3HDZIP1, the sequence obtained thus far shows that Chara shares introns 4, 5, and 15 with land plants and that these introns are relatively long. Tanabe et al. (2005) found similar results for the Chara MADS-box gene CgMADS1. Intron–exon structure was largely conserved between the Chara gene and the land plant MADS-box genes, but the Chara introns were much longer than the homologous land plant introns. MADS-box genes from two other charophycean algal species were also found to have longer introns than land plant homologs. This may indicate that charophycean algal genomes are characterized by more intronic sequence than land plants.
Developmental interpretation of class III HD–Zip expression:
The expression patterns of the two Selaginella genes are distinctly different. SkC3HDZ2 is expressed in and may therefore have a role in apical meristem function and in the origin of the stem vascular tissue (Figure 6, E–G). As tissues differentiate within the provascular strands of stems and leaves, expression patterns suggest that SkC3HDZ2 is involved with the differentiation of xylem. In contrast, SkC3HDZ1 is not expressed strongly until microphyll expansion is evident. This gene is associated initially with microphyll vascularization, which begins at the base of the microphyll subadjacent to the ligule. It appears that expression then extends outward in the provascular tissue as the microphyll expands and inward toward the nearest stem provascular strand. In later stages, SkC3HDZ1 expression is limited to the tissues that will become pericycle and phloem in both microphylls and stems. In situ expression patterns suggest that the two genes may have complementary roles in patterning the shoot apex and stem vascular tissues and that two different genes may initiate stem and microphyll vascularization. Expression data do not indicate that Selaginella class III HD–Zip genes are involved with microphyll initiation nor was either gene expressed in a way that suggests a role in adaxial/abaxial leaf polarity as REV clade genes are in Arabidopsis.
The expression of Ginkgo GbC3HDZ1 and Pseudotsuga PmC3HDZ1 are similar to each other and similar to Arabidopsis REV clade genes (Figure 6, A–D). In contrast to Selaginella, foci of expression in the apex occur in the meristem peripheral zone where leaf primordia form. The strongest expression occurs where the leaf provascular strand develops and expression continues from the tip of the primordium into the stem, as does the provascular strand. Expression data and functional data from Arabidopsis indicate that REV clade genes regulate apical meristem formation and growth, adaxial patterning/identity in leaves, and provascular patterning and differentiation. Expression of GbC3HDZ1 and PmC3HDZ1 indicates that these genes are involved in the same developmental processes in Ginkgo and Pseudotsuga as REV clade genes are in Arabidopsis. In contrast to Selaginella, in Ginkgo and Pseudotsuga, a primary gene is involved in all of these processes. The lack of signal for non-REV clade genes in Ginkgo and Pseudotsuga indicates that these genes may be expressed at lower levels or be limited to fewer tissues than REV clade genes in the shoot apex. In Arabidopsis, the CNA clade genes ATHB8 and CNA are not expressed in leaf adaxial domains and either are not detectable or are weakly detected in the SAM (Prigge et al. 2005).
Conservation of functions in vascular plants:
Phylogenetic analysis of class III HD–Zip genes indicates that a single class III HD–Zip gene diversified within the seed plant lineage, giving rise to all seed plant class III HD-Zip genes. Likewise, a single gene diversified within the lycophyte lineage, resulting in two Selaginella genes. This means that all seed plant class III HD–Zip genes are equally related to both Selaginella class III HD–Zip genes. The single ancestral gene in each lineage must have provided all essential developmental activity associated with class III HD–Zip genes. Thus we should expect class III HD–Zip genes in seed plants and lycophytes to be involved in similar developmental processes that evolved prior to the divergence from their last common ancestor into lycophyte and euphyllophyte lineages.
Gene expression patterns in gymnosperms and lycophytes and functional analyses of class III HD–Zip function in flowering plants indicate shared developmental roles. Expression patterns of class III HD–Zip genes from the lycophyte Selaginella (Figure 6, E–G) indicate that both SkC3HDZ1 and SkC3HDZ2 are involved in vascular patterning and that SkC3HDZ2 may be particularly important to apical meristem function, stem vascular patterning, and xylem differentiation. In situ hybridization results for two gymnosperm genes, GbC3HDZ1 (Figure 6, A and B) and PmC3HDZ1 (Figure 6, C and D), also indicate that they function in the apical meristem and in vascular patterning and differentiation. These are some of the demonstrated functions of class III HD–Zip genes in angiosperms (Figure 2). Sporophyte apical meristems and vascular tissue evolved prior to the divergence of lycophytes and euphyllophytes (Figure 1). Thus we can trace these shared functions to the last common ancestor of extant vascular plants (Figure 7). Since the common ancestor of all vascular plants was most likely leafless (Stewart and Rothwell 1993; Kenrick and Crane 1997), we can infer that two roles of the single ancestral vascular plant class III HD–Zip gene were apical meristem function and axial vascular patterning. The apparent role of all five Arabidopsis class III HD–Zips as well as Ginkgo GbC3HDZ1 and Pseudotsuga PmC3HDZ1 in stem vascular patterning and differentiation reflects the conservation of an ancient function. The roles of seed plant REV clade genes, and perhaps of CNA, in apical meristem function (Figure 2) also reflects conservation of an ancestral function. The inferred role of SkC3HDZ2, in both apical meristem function and stem vascular patterning, represents retention of both ancestral functions.
Origin of novel functions and the evolution of form:
Phylogenetic analysis and the fossil record indicate that the lycophyte and euphyllophyte (all other vascular plants) common ancestors diverged >400 million years ago (Kenrick and Crane 1997). Our analysis of gene expression in Selaginella and seed plants revealed many differences that likely reflect this long evolutionary separation and appear to be associated with morphological differences in the two lineages.
Leaves:
In the lycophyte Selaginella, SkC3HDZ2 is expressed in the apical cells, stem provasculature, and differentiating xylem, which we have shown to be likely ancestral patterns. SkC3HDZ1 is associated with the onset of microphyll vascularization and phloem/pericycle differentiation. Since microphylls evolved within the lycophyte lineage (Gensel 1992; Kenrick and Crane 1997), any role in microphyll vascularization must represent recruitment within that lineage for the development of a novel organ. Whether this occurred before or after the duplication of the single ancestral gene, we cannot say. Analysis of class III HD–Zip diversity and expression patterns in other lycophyte genera would provide insight into the antiquity of the duplication evident in the lycophyte clade and may help discern among possible scenarios of functional evolution.
In the seed plant apex, the strongest expression of REV clade genes occurs in the peripheral zone and is associated with the origin of leaf primordia and the leaf provascular strand. Later, adaxial restriction in emerging primordia is associated with polarity and laminar outgrowth. Only genes in the REV clade (REV, PHB, PHV, GbC3HDZ1, PmC3HDZ1) have been shown to become limited to adaxial domains during primordium development. The role of conferring adaxial identity may be a function that can be mapped to the REV clade ancestor and may represent a new function acquired by that gene following class III HD–Zip duplication early in seed plant history.
Thus class III HD–Zip genes that were the products of independent, lineage-specific diversifications were recruited independently within lycophtyes and seed plants for new roles in leaf development. These findings have implications for understanding the underlying developmental differences of microphylls and megaphylls and the SAMs that produce them.
Evolution of vascular tissue:
We know that class III HD–Zip homologs are expressed in growing regions of the haploid generations of Chara, in all bryophytes, and in the sporophyte of Phaeoceros. It has been demonstrated that class III HD–Zip genes play a role in vascular development and differentiation in Arabidopsis where they are expressed. Expression analysis further implicates class III HD–Zip genes with vascular development in other flowering plants, gymnosperms, and lycophytes. Since it is clear that vascular tissues evolved after bryophyte lineages diverged, class III HD–Zip gene involvement in the development of vasculature must also represent a derived function. The association of class III HD–Zip genes with vascular development maps to the last common ancestor of extant lycophytes and seed plants. Thus it is possible that class III HD–Zip genes played a role in the origin of vascular tissues by acquiring novel functions directing cell differentiation. There are fossil plants that in most ways resemble ancient vascular plants (leafless, dichotomizing axes with central conducting strands) except that they lack conducting cells with differentially thickened secondary walls (Edwards 1986; Kenrick and Crane 1991, 1997). These fossils (protracheophytes) indicate that the developmental patterning of stem tissues was in place prior to the origin of tracheary elements with differential secondary-wall thickenings. In vascular plants, class III HD–Zip genes are associated with both the patterning of conducting tissues and the differentiation of tracheary elements. Thus it is possible that class III HD–Zip transcription factors played a role in the patterning of conducting tissues in protracheophytes and that their role in tracheary element differentiation was acquired later. The conducting tissues of mosses may be either analogous or homologous to those of vascular plants (Ligrone et al. 2000), and investigation of class III HD–Zip gene function in mosses may clarify this issue.
Ancient land plant functions:
We have demonstrated that class III HD–Zip genes predate the origin of vascular plants and even embryophytes. Given that bryophyte sporophytes lack leaves, roots, and vascular tissues and the alga Chara lacks a sporophyte altogether, the question arises as to what the ancestral role of class III HD–Zip genes might be? Class III HD–Zip genes are expressed during development of the haploid bodies of Chara, Marchantia, Phaeoceros, and Physcomitrella and in the hornwort (Phaeoceros) sporophyte. It is not yet known if class III HD–Zip genes are expressed in the sporophytes of other bryophytes. This suggests that class III HD–Zip genes initially played a role in gametophytic development and were co-opted to roles in sporophytic development early in land plant evolution. Class III HD–Zip genes appear to be involved in apical growth in all vascular plants. Apical growth is a feature that land plants share with Chara and perhaps Coleochaete, but does not characterize earlier-diverging lineages of Charophycean green algae (Graham et al. 2000; McCourt et al. 2004). Thus, apical growth is a likely candidate for an ancestral function. However, additional investigations into the expression patterns of class III HD–Zip genes in nonvascular plants are needed and functional analyses would provide for more robust conclusions about class III HD–Zip gene activity in all taxa.
Evolution of miRNA regulation:
Finally, while we have shown evidence that class III HD–Zip genes in land plants are regulated by miR165/166 (Floyd and Bowman 2004), the nucleotide sequence of the miR165/166 binding site is not conserved in the algal gene CcC3HDZ1 (Figure 4). Regulation of class III HD–Zip mRNAs by miR165/166 appears to be restricted to embryophytes. It is possible that microRNA (miRNA) regulation of the Chara class III HD–Zip mRNA was lost in that lineage. Another possibility is that CcC3HDZ1 is regulated by a different miRNA. However, it is also possible, and more parsimonious, to hypothesize that regulation by miR165/166 evolved once in the common ancestor of embryophytes.
In recent years numerous miRNAs have been identified in metazoans and land plants, but in no other eukaryotic lineage (reviewed in Lau et al. 2001; Pasquinelli et al. 2003; Bartel 2004; Bartel and Chen 2004; Floyd and Bowman 2005), suggesting that miRNA regulation in plants and metazoans evolved independently (Bartel 2004; Floyd and Bowman 2005). In both of these major groups of multicellular organisms, miRNAs are involved in numerous processes essential for normal development. This has led to the speculation that the origin of miRNA regulation may have played important roles in facilitating developmental patterning that evolved in both groups (Reinhart et al. 2002; Bartel 2004). Multicellularity evolved in the charophycean algal lineages prior to the divergence of the charalean/land plant ancestral lineage. However, land plants differ from their algal relatives in having both haploid and diploid generations that are composed of truly three-dimensional tissues (Graham et al. 2000). Our finding that the otherwise highly conserved algal class III HD–Zip homolog in Chara is likely not regulated by the same miRNA as all land plant homologs may be evidence that the origin of miRNA regulation of these genes was important for the evolution of three-dimensional developmental patterning in land plants. Continued investigation of the functions of class III HD–Zip genes and their regulation by miR165/166 in Chara and bryophyte lineages is likely to provide additional insight into the significance of miRNA regulation in body-plan evolution.
Conclusions:
Our data suggest that the developmental roles of the class III HD–Zip gene family most certainly expanded as land plants became more complex through gene duplications, neofunctionalization, and subfunctionalization. We can associate class III HD–Zip genes with apical meristem function and vascular patterning in all vascular plants. We have also shown evidence that class III HD–Zip genes played a role in the independent evolution of microphylls in lycophytes and of megaphylls in seed plants. Furthermore, our data suggest that regulation of class III HD–Zip genes by miR165/166 is restricted to land plants. Class III HD–Zip genes in flowering plants are associated with many other aspects of development critical to vascular plant evolution that are beyond the scope of this article, including branching, root development, and secondary growth. As we continue to investigate the phylogenetic distribution and functions of class III HD–Zip genes in streptophytes we will learn more about their ancestral and derived functions. It is clear that class III HD–Zip genes have evolved and diversified in parallel with land plants and are associated with the development of major body-plan innovations such as apical meristems, vascular tissue, and leaves. So far evidence suggests that these genes are part of an ancient developmental patterning tool kit that was modified through time to produce a diversity of land plant forms.
Acknowledgments
We thank Judith Lucas, Randi Puckett, Amy Short, Kristy Fournier, and Jarae Ng for assistance with probe preparation, PCR, plasmid preps, and restriction digests. Harriet Oldenhoff completed RACE PCR and cloning of cDNA and genomic sequences of PpC3HDZ1 and PpC3HDZ2 from Physcomitrella. We thank Ernesto Sandoval, Tim Metcalf, and Doug Walker of the University of California at Davis Conservatory, Andrew Groover and Dave Johnson of the United States Department of Agriculture Institute of Forest Genetics (Placerville, CA), and Sue Nichol for assistance with obtaining and cultivating plants used in this study. Francisco Vergara-Silva provided RNA samples of Taxus. Present and past members of the Bowman lab, including John Alvarez, Stuart Baum, John Emery, Yuval Eshed, Nathaniel Hawker, and Anat Izhaki, provided stimulating discussion and technical advice that contributed to this work. Two anonymous reviewers provided thoughtful comments that assisted in the improvement of the manuscript. We are grateful for funding in the form of a University of California, Davis, Katherine Esau Post-Doctoral Fellowship awarded to S.K.F., a University of California at Davis undergraduate research grant (“P.U.F.”) awarded to Judith Lucas, and National Science Foundation grant IBN0332556 awarded to J.L.B. and grant IOB0515435 awarded to J.L.B. and S.K.F.
References
- Aso, K., M. Kato, J. A. Banks and M. Hasebe, 1999. Characterization of homeodomain-leucine zipper genes in the fern Ceratopteris richardii and the evolution of the homeodomain-leucine zipper gene family in vascular plants. Mol. Biol. Evol. 16: 544–552. [DOI] [PubMed] [Google Scholar]
- Baima, S., F. Nobili, G. Sessa, S. Lucchetti, I. Ruberti et al., 1995. The expression of the Athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development 121: 4171–4182. [DOI] [PubMed] [Google Scholar]
- Baima, S., M. Possenti, A. Matteucci, E. Wisman, M. M. Altamura et al., 2001. The Arabidopsis ATHB-8 HD-Zip protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol. 126: 643–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, D. P., 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Bartel, D. P., and C.-Z. Chen, 2004. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 5: 396–400. [DOI] [PubMed] [Google Scholar]
- Bhattacharya, D., K. Weber, S. S. An and W. Berning-Koch, 1998. Actin phylogeny identifies Mesostigma viride as a flagellate ancestor of the land plants. J. Mol. Evol. 47: 544–550. [DOI] [PubMed] [Google Scholar]
- Boyce, C. K., and A. H. Knoll, 2002. Evolution of developmental potential and the multiple independent origins of leaves in Paleozoic vascular plants. Paleobiology 28: 70–100. [Google Scholar]
- Bremer, K., C. J. Humphries, B. D. Mishler and S. P. Churchill, 1987. On cladistic relationships in green plants. Taxon 36: 339–349. [Google Scholar]
- Carroll, S. B., 2000. Endless forms: the evolution of gene regulation and morphological diversity. Cell 101: 577–580. [DOI] [PubMed] [Google Scholar]
- Cook, M. E., 2004. Cytokinesis in Coleochaete orbicularis (Charophyceae): an ancestral mechanism inherited by plants. Am. J. Bot. 91: 313–320. [DOI] [PubMed] [Google Scholar]
- Cook, M. E., and W. E. Friedman, 1998. Tracheid structure in a primitive extant plant provides an evolutionary link to earliest fossil tracheids. Int. J. Plant Sci. 159: 881–890. [Google Scholar]
- Cooke, T. J., D. B. Poli, A. E. Sztein and J. D. Cohen, 2002. Evolutionary patterns in the auxin action. Plant Mol. Biol. 49: 319–338. [PubMed] [Google Scholar]
- Delwiche, C. F., K. Karol, M. T. Cimino and K. J. Sytsma, 2002. Phylogeny of the genus Coleochaete (Coleochaetales, Charophyta) and related taxa inferred by analysis of the chloroplast gene rbcL. J. Phycol. 38: 394–403. [Google Scholar]
- Donoghue, M. J., J. A. Doyle, J. Gautier, A. G. Kluge and T. Rowe, 1989. The importance of fossils in phylogeny reconstruction. Annu. Rev. Ecol. Syst. 20: 431–460. [Google Scholar]
- Doyle, J. A., 1998. Phylogeny of vascular plants. Annu. Rev. Ecol. Syst. 29: 567–599. [Google Scholar]
- Edwards, D. S., 1986. Aglaophyton major, a non-vascular land-plant from the Devonian Rhynie Chert. Bot. J. Linn. Soc. 98: 173–204. [Google Scholar]
- Emery, J. F., S. K. Floyd, J. Alvarez, Y. Eshed, N. P. Hawker et al., 2003. Radial patterning of Arabidopsis shoots by class III HD-Zip and KANADI genes. Curr. Biol. 13: 1768–1774. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J., 1989. PHYLIP: phylogeny inference package (version 3.2). Cladistics 5: 164–166. [Google Scholar]
- Floyd, S. K., and J. L. Bowman, 2004. Ancient microRNA target sequences in plants. Nature 428: 485–486. [DOI] [PubMed] [Google Scholar]
- Floyd, S. K., and J. L. Bowman, 2005. Micromanaging the plant genome, pp. 244–278 in Plant Epigenetics, edited by P. Meyer. Blackwell Scientific, London.
- Friedman, W. E., R. C. Moore and m. d. Purugganan, 2004. The evolution of plant development. Am. J. Bot. 91: 1726–1741. [DOI] [PubMed] [Google Scholar]
- Gensel, P. G., 1992. Phylogenetic relationships of the zosterophylls and lycopsids: evidence from morphology, paleoecology, and cladistic methods of inference. Ann. Mo. Bot. Gard. 79: 450–473. [Google Scholar]
- Graham, L. E., 1993. Origin of Land Plants. John Wiley & Sons, New York.
- Graham, L. E., M. E. Cook and J. S. Busse, 2000. The origin of plants: body plan changes contributing to a major evolutionary radiation. Proc. Natl. Acad. Sci. USA 97: 4535–4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, K. A., M. J. Prigge, R. B. Katzman and S. E. Clark, 2005. CORONA, a member of the class III homeodomain leucine zipper family of Arabidopsis, regulates stem cell specification and organogenesis. Plant Cell 17: 691–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gugerli, F., C. Sperisen, U. Buchler, I. Brunner, S. Brodbeck et al., 2001. The evolutionary split of Pinaceae from other conifers: evidence from an intron loss and a multigene phylogeny. Mol. Phylogenet. Evol. 21: 167–175. [DOI] [PubMed] [Google Scholar]
- Hawker, N. P., and J. L. Bowman, 2004. Underground polarity: roles for class III HD-Zip and KANADI genes in Arabidopsis root development. Plant Physiol. 135: 2261–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzberg, M., H. Aspeborg, J. Schrader, A. Andersson, R. Erlandsson et al., 2001. A transcriptional roadmap to wood formation. Proc. Natl. Acad. Sci. USA 98: 14732–14737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck, J., and F. Ronquist, 2001. MrBayes: Bayesian inference of phylogeny. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- Kang, J., and N. Dengler, 2001. Cell cycling frequency and expression of the homeobox gene ATHB8 during leaf vein development in Arabidopsis. Planta 216: 212–219. [DOI] [PubMed] [Google Scholar]
- Kang, J., J. Tang, P. Donnelly and N. Dengler, 2003. Primary vascular pattern and expression of ATHB-8 in shoots of Arabidopsis. New Phytol. 158: 443–454. [DOI] [PubMed] [Google Scholar]
- Karol, K. G., R. M. McCourt, M. T. Cimino and C. F. Delwiche, 2001. The closest living relatives of land plants. Science 294: 2351–2353. [DOI] [PubMed] [Google Scholar]
- Kenrick, P., and P. R. Crane, 1991. Water-conducting cells in early fossil land plants: implications for the early evolution of tracheophytes. Bot. Gaz. 152: 335–356. [Google Scholar]
- Kenrick, P., and P. R. Crane, 1997. The Origin and Early Evolution of Land Plants: A Cladistic Study. Smithsonian Institution Press, Washington, DC.
- Kiefer, E., W. Heller and D. Ernst, 2000. A simple and efficient protocol for isolation of functional RNA from plant tissues rich in secondary metabolites. Plant Mol. Biol. Rep. 18: 33–39. [Google Scholar]
- Ko, J.-H., C. Prassinos and K.-H. Han, 2006. Developmental and seasonal expression of PtaHB1, a Populus gene encoding a class III HD-Zip protein, is closely associated with secondary growth and inversely correlated with the level of microRNA (miR166). New Phytol. 169: 469–478 [DOI] [PubMed] [Google Scholar]
- Korall, P., P. Kenrick and J. P. Therrien, 1999. Phylogeny of Selaginellaceae: evaluation of generic/subgeneric relationships based on rbcL gene sequences. Int. J. Plant Sci. 160: 585–594. [Google Scholar]
- Lau, M. C., L. P. Lim, E. G. Weinstein and D. P. Bartel, 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862. [DOI] [PubMed] [Google Scholar]
- Ligrone, R., J. G. Duckett and K. S. Renzaglia, 2000. Conducting cells and the phyletic relationships of bryophytes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355: 795–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattsson, J., W. Ckurshumova and T. Berleth, 2003. Auxin signaling in Arabidopsis leaf vascular development. Plant Physiol. 131: 1327–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell, J. R., and M. K. Barton, 1998. Leaf polarity and meristem formation in Arabidopsis. Development 125: 2935–2942. [DOI] [PubMed] [Google Scholar]
- McConnell, J., J. Emery, Y. Eshed, N. Bao, J. Bowman et al., 2001. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411: 709–713. [DOI] [PubMed] [Google Scholar]
- McCourt, R. M., C. F. Delwiche and K. G. Karol, 2004. Charophyte algae and land plant origins. Trends Ecol. Evol. 19: 661–666. [DOI] [PubMed] [Google Scholar]
- Mishler, B. D., L. A. Lewis, M. A. Buchheim, K. S. Renzaglia, D. J. Garbary et al., 1994. Phylogenetic relationships of the “green algae” and “bryophytes.” Ann. Mo. Bot. Gard. 81: 451–483. [Google Scholar]
- Niklas, K. J., 2000. The evolution of plant body plans—a biomechanical perspective. Ann. Bot. 85: 411–438. [Google Scholar]
- Ohashi-Ito, K., and H. Fukuda, 2003. HD-Zip homeobox genes that include a novel member, ZeHB-13 (Zinnia)/ATHB-15 (Arabidopsis), are involved in procambium and xylem cell differentiation. Plant Cell Physiol. 44: 1350–1358. [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito, K., T. Demura and H. Fukuda, 2002. Promotion of transcript accumulation of novel Zinnia immature xylem specific HD-Zip homeobox genes by brassinosteroids. Plant Cell Physiol. 43: 1146–1153. [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito, K., M. Kubo, T. Demura and H. Fukuda, 2005. Class III homeodomain leucine-zipper proteins regulate xylem cell differentiation. Plant Cell Physiol. 46: 1646–1656. [DOI] [PubMed] [Google Scholar]
- Otsuga, D., B. DeGuzman, M. J. Prigge, G. N. Drews and S. E. Clark, 2001. REVOLUTA regulates meristem initiation at lateral positions. Plant J. 25: 223–236. [DOI] [PubMed] [Google Scholar]
- Pasquinelli, A. E., A. McCoy, E. Jiménez, E. Saló, G. Ruvkun et al., 2003. Expression of the 22 nucleotide let-7 heterochronic RNA throughout the Metazoa: A role in life history evolution? Evol. Dev. 5: 372–378. [DOI] [PubMed] [Google Scholar]
- Ponting, C. P., and L. Aravind, 1999. START: a lipid-binding domain in StAR, HD-ZIP and signalling proteins. Trends Biochem. Sci. 24: 130–132. [DOI] [PubMed] [Google Scholar]
- Prigge, M. J., D. Otsuga, J. M. Alonso, J. R. Ecker, G. N. Drews et al., 2005. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17: 61–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryer, K. M., H. Schneider, A. R. Smith, R. Cranfill, P. G. Wolf et al., 2001. Horsetails and ferns are a monophyletic group and the closest living relatives to seed plants. Nature 409: 618–622. [DOI] [PubMed] [Google Scholar]
- Rambaut, A., 1996. Se-Al: Sequence Alignment Editor. http://evolve.zoo.ox.ac.uk/
- Reinhart, B. J., E. G. Weinstein, M. W. Rhoades, B. Bartel and D. P. Bartel, 2002. MicroRNAs in plants. Genes Dev. 16: 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara, K., T. Nishiyama, M. Kato and M. Hasebe, 2001. Isolation of homeodomain-leucine zipper genes from the moss Physcomitrella patens and the evolution of homeodomain-leucine zipper genes in land plants. Mol. Biol. Evol. 18: 491–502. [DOI] [PubMed] [Google Scholar]
- Scarpella, E., S. Rueb, K. Boot, J. Hoge and A. Meijer, 2000. A role for the rice homeobox gene Oshox1 in provascular cell fate commitment. Development 127: 3655–3669. [DOI] [PubMed] [Google Scholar]
- Schrader, J., J. Nilsson, E. Mellerowicz, A. Berglund, P. Nilsson et al., 2004. A high-resolution transcript profile across the wood-forming meristem of poplar identifies potential regulators of cambial stem cell identity. Plant Cell 16: 2278–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrick, K., D. Nguyen, W. M. Karlowski and K. F. X. Mayer, 2004. START lipid/sterol-binding domains are amplified in plants and are predominantly associated with homeodomain transcription factors. Genome Biol. 5: R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa, G., C. Steindler, G. Morelli and I. Ruberti, 1998. The Arabidopsis Athb-8, -9 and -14 genes are members of a small gene family coding for highly related HD-ZIP proteins. Plant Mol. Biol. 38: 609–622. [DOI] [PubMed] [Google Scholar]
- Stewart, W. N., and G. W. Rothwell, 1993. Paleobotany and the Evolution of Plants. Cambridge University Press, New York.
- Sussex, I. M., and N. M. Kerk, 2001. The evolution of plant architecture. Curr. Opin. Plant Biol. 4: 33–37. [DOI] [PubMed] [Google Scholar]
- Talbert, P. B., H. T. Adler, D. W. Parks and L. Comai, 1995. The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana. Development 121: 2723–2735. [DOI] [PubMed] [Google Scholar]
- Tanabe, Y., M. Hasebe, H. Sekimoto, T. Nishiyama, M. Kitani et al., 2005. Characterization of MADS-box genes in charophycean green algae and its implication for the evolution of MADS-box genes. Proc. Natl. Acad. Sci USA 102: 2436–2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielle-Calzada, J. P., J. Thomas, C. Spillane, A. Coluccio, M. A. Hoeppner et al., 1999. Maintenance of genomic imprinting at the Arabidopsis medea locus requires zygotic DDM1 activity. Genes Dev. 13: 2971–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., and Z.-H. Ye, 1999. IFL1, a gene regulating interfascicular fiber differentiation in Arabidopsis, encodes a homeobox-leucine zipper protein. Plant Cell 11: 2139–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., and Z.-H. Ye, 2001. Alteration of auxin polar transport in the Arabidopsis ifl1 mutants. Plant Physiol. 126: 549–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R. Q., and Z. H. Ye, 2004. Amphivasal vascular bundle 1, a gain-of-function mutation of the IFL1/REV gene, is associated with alterations in the polarity of leaves, stems and carpels. Plant Cell Physiol. 45: 369–385. [DOI] [PubMed] [Google Scholar]