Abstract
Apolipoprotein E (apoE) secreted by macrophages in the artery wall exerts an important protective effect against the development of atherosclerosis, presumably through its ability to promote lipid efflux. Previous studies have shown that increases in cellular free cholesterol levels stimulate apoE transcription in macrophages and adipocytes; however, the molecular basis for this regulation is unknown. Recently, Taylor and colleagues [Shih, S. J., Allan, C., Grehan, S., Tse, E., Moran, C. & Taylor, J. M. (2000) J. Biol. Chem. 275, 31567–31572] identified two enhancers from the human apoE gene, termed multienhancer 1 (ME.1) and multienhancer 2 (ME.2), that direct macrophage- and adipose-specific expression in transgenic mice. We demonstrate here that the nuclear receptors LXRα and LXRβ and their oxysterol ligands are key regulators of apoE expression in both macrophages and adipose tissue. We show that LXR/RXR heterodimers regulate apoE transcription directly, through interaction with a conserved LXR response element present in both ME.1 and ME.2. Moreover, we demonstrate that the ability of oxysterols and synthetic ligands to regulate apoE expression in adipose tissue and peritoneal macrophages is reduced in Lxrα−/− or Lxrβ−/− mice and abolished in double knockouts. Basal expression of apoE is not compromised in Lxr null mice, however, indicating that LXRs mediate lipid-inducible rather than tissue-specific expression of this gene. Together with our previous work, these findings support a central role for LXR signaling pathways in the control of macrophage cholesterol efflux through the coordinate regulation of apoE, ABCA1, and ABCG1 expression.
Macrophage lipid metabolism plays a central role in the pathogenesis of atherosclerosis (1, 2). Macrophages accumulate cholesteryl ester through scavenger receptor-mediated uptake of oxidatively modified low-density lipoprotein (LDL). Recent studies have shown that efflux of free cholesterol from the cell is accomplished through a pathway involving the putative cholesterol/phospholipid transporter ABCA1 (3–5). Alterations in the rates of macrophage cholesterol uptake or efflux each have the potential to influence foam cell formation and the development of the atherosclerotic lesion. An understanding of the regulatory pathways that control macrophage lipid flux may identify new opportunities for intervention in the process of atherogenesis.
Apolipoprotein E (apoE) is important for both systemic and cellular cholesterol metabolism. As a surface constituent of plasma lipoproteins and a high-affinity ligand for the LDL receptor, apoE mediates hepatic uptake of chylomicron remnants, very low density lipoprotein (VLDL), and some subpopulations of high-density lipoprotein (HDL). The critical role of apoE in these pathways is underscored by the massive accumulation of lipoproteins and lipoprotein remnants in the plasma of both humans and mice lacking functional apoE (6, 7). In contrast to the majority of apolipoproteins, which are synthesized primarily in the liver and intestine, apoE is also secreted by cells outside the enterohepatic axis, including macrophages and adipocytes (8, 9). In these cells, apoE modulates cellular cholesterol metabolism by facilitating cholesterol efflux. Human monocyte-derived macrophages have been shown to efflux cholesterol into apoE-containing particles, even in the absence of exogenously added cholesterol acceptors (10). Furthermore, macrophages derived from apoE−/− mice exhibit a diminished capacity to efflux cholesterol and other lipids to HDL3, lipid-free apoAI, or lipid-free apoE3 compared with those from wild-type mice (9, 11).
ApoE is also an important modulator of atherogenesis. ApoE null mice develop atherosclerosis on a normal chow diet, displaying foam-cell rich fatty lesions by 3 months of age (6, 7). Although impaired hepatic clearance and elevated levels of proatherogenic lipoproteins clearly contribute, these changes do not entirely explain the phenotype. Elegant in vivo studies have demonstrated that secretion of apoE by macrophages also exerts an important protective effect. Selective expression of apoE in macrophages of apoE−/− mice through bone marrow transplantation or transgenic expression decreases atherosclerosis (12, 13). Importantly, this decrease occurs even in the absence of changes in plasma lipoprotein levels. Conversely, transplantation of apoE−/− bone marrow into apoE+/+ mice confers increased susceptibility to atherosclerosis (14). Very recently, adenovirus-mediated gene transfer of apoE was shown to cause regression of established lesions (15). Effects of apoE on platelet, smooth muscle cell, and lymphocyte function have all been described; however, the protective role of macrophage apoE expression most likely results from its ability to promote lipid efflux (8, 9).
ApoE is abundantly expressed in lipid-loaded foam cells from atherosclerotic lesions (16). Moreover, increasing the cellular concentration of free cholesterol has been shown to stimulate transcription of the apoE gene in both macrophages and adipocytes (17). These observations imply the existence of a molecular sensor that regulates tissue-specific expression of the apoE gene in response to lipid loading. The mechanisms that underlie tissue-specific and lipid-inducible expression of apoE, however, are poorly understood. Recently, Taylor and coworkers (18) identified two enhancers from the human apoE flanking sequences, termed multienhancer 1 (ME.1) and multienhancer 2 (ME.2), that direct macrophage- and adipose-specific expression of a reporter gene in transgenic mice. ME.1 and ME.2 are 95% identical in sequence and presumably contain conserved cis-acting sequences required for the expression of apoE in both adipocytes and macrophages. Importantly, ME.1 and ME.2 are distinct from the previously characterized hepatic control region that facilitates expression of the apoE/C-I/C-I′/C-IV/C-II gene cluster in liver.
We demonstrate here that in addition to controlling the tissue-specific expression of apoE in macrophages and adipose tissue, the ME.1 and ME.2 enhancers are also likely to facilitate gene induction in response to cellular lipid loading. We show that the nuclear receptors LXRα and LXRβ and their oxysterol ligands are key regulators of apoE expression in both macrophages and adipocytes. LXR/RXR heterodimers regulate apoE transcription through direct interaction with a conserved LXR response element (LXRE) present in both ME.1 and ME.2. We further show that the ability of oxysterols to regulate apoE expression in both adipose tissue and peritoneal macrophages is abolished in Lxr null mice. These findings support a central role for LXR signaling pathways in the control of macrophage cholesterol efflux through the coordinate regulation of apoE, ABCA1, and ABCG1 expression.
Materials and Methods
Reagents and Plasmids.
pCMX expression plasmids for RXRα and LXRα have been described. pSG5-LXRβ and pCMX-VP16-LXRα were gifts from Steven Kliewer (Glaxo Wellcome) and Ron Evans (Salk Institute), respectively. T0901317 was provided by Bei Shan (Tularik, South San Francisco, CA) and LG268, by Rich Heyman (Ligand Pharmaceuticals, La Jolla, CA). Oxysterols (Sigma), LG268, and phorbol 12-myristate 13-acetate (Sigma) were dissolved in ethanol or DMSO before use in cell culture. The apoE proximal promoter (bp −890 to +93) was amplified by PCR from human genomic DNA and cloned into pGL3-luciferase (Promega) to create pGL-apoE(−890). ME.1 and ME.2 were cloned by PCR from human genomic DNA and subcloned into pTK-luciferase or pGL-apoE(−890).
Cell Culture, Transfections, and Stable Cell Lines.
THP-1 cells were cultured in RPMI medium 1640 containing 10% (vol/vol) FBS. NIH 3T3 and 3T3-F442A cells were grown in DMEM containing 10% (vol/vol) calf serum, and HepG2 cells were grown in MEM containing 10% (vol/vol) FBS. Peritoneal macrophages were obtained from thioglycolate-injected mice as described (19). Cells were cultured in DMEM supplemented with 10% (vol/vol) lipoprotein-deficient serum (LPDS) and receptor ligands for 42 h. Transient transfections of HepG2 cells were performed in triplicate in 48-well plates. Cells were transfected with reporter plasmid (100 ng per well), receptor plasmids (5–50 ng per well), pCMV-β-galactosidase (50 ng per well), and pTKCIII (to a total of 205 ng per well) with the use of the MBS mammalian transfection kit (Stratagene). After transfection, cells were incubated in MEM containing 10% (vol/vol) LPDS and the indicated ligands or vehicle control for 24 h. Luciferase activity was normalized to β-galactosidase activity. pBABE-Puro and pBABE-LXRα expression vectors (20) were packaged into retrovirus by transient transfection of Phoenix E cells as described (21). NIH 3T3 fibroblasts or 442A preadipocytes were infected at 50% confluence with approximately equal titers of retrovirus. Stable cell lines were selected with puromycin (2 μg/ml).
Animals and Diets.
Lxr knockout mice (ref. 22; J.-M. Lobaccaro and D.J.M., unpublished observations) were maintained on a mixed-strain (A129, Bl/6) background. Age-matched, 2.5- to 4-month-old male mice were fed a cereal-based powdered diet ad libitum (Teklad 7001; Harlan Teklad, Madison, WI) supplemented with cholesterol, LG268, or T0901317. Diets were supplemented at a level sufficient to provide the appropriate dose [mg/kg body weight] on consumption of 5 g of diet by a 25-g mouse per day. Animal experiments were approved by the Institutional Animal Care and Research Advisory Committee of the University of Texas Southwestern Medical Center.
RNA Isolation and Northern Analysis.
Total RNA was isolated by using Trizol reagent (Life Technologies, Grand Island, NY), and Northern analysis was performed as described (20). Blots were normalized with the use of probes to 36B4 or cyclophilin and quantitated by PhosphorImager analysis (Molecular Dynamics). Total RNA was prepared from mouse tissues with the use of the RNA Stat60 reagent (Tel-Test, Friendswood, TX).
Gel-Shift Assays.
In vitro translated RXRα, LXRα, and LXRβ were generated from pCMX-RXRα, pCMX-hLXRα, and pSG5-LXRβ plasmids with the TNT Quick Coupled Transcription/Translation System (Promega). Gel-shift assays were performed as described (23) with the use of in vitro translated proteins and the following oligonucleotides (only one strand shown): ME LXRE, 5′-gatcgctgccagggtcactggcggtcaaaggcag-3′; ME LXRE mutant, 5′-gatcgct gccaggaacactggcgaacaaaggcag-3′; prox LXRE, 5′-caaactcctgaccttaagtgattcgcccac-3′.
Results
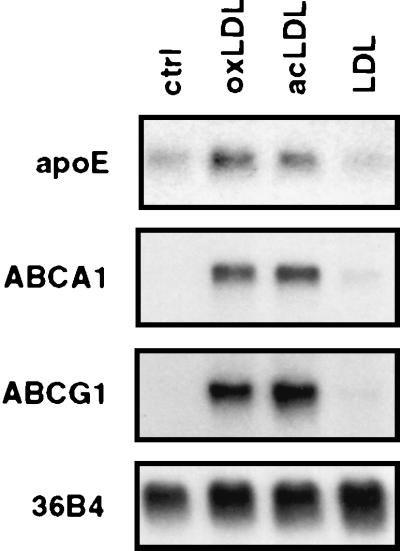
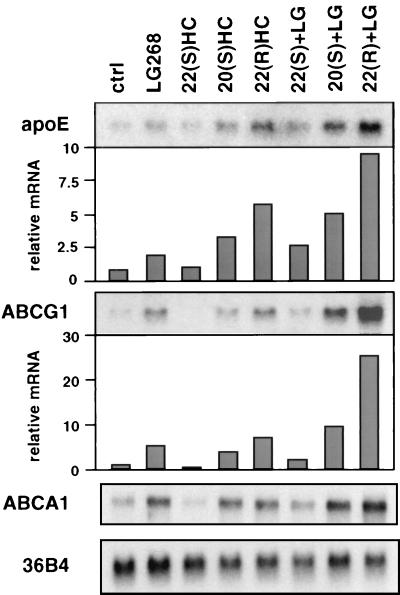
Previous work has shown that apoE gene expression in macrophages is induced in response to lipid loading. We found that treatment of phorbol 12-myristate 13-acetate-differentiated human THP-1 macrophages with either acetylated LDL or highly oxidized LDL led to a coordinate induction of apoE, ABCA1, and ABCG1 expression (Fig. 1). In contrast, native LDL had no effect on expression of these genes, suggesting that particle uptake by the cell is required for induction. We have previously described roles for the nuclear receptors LXRα and LXRβ in the regulation of macrophage gene expression by native and modified lipids. In particular, we have shown that lipid induction of ABCA1 and ABCG1 expression in macrophages depends on LXRs (19, 20, 24). To address whether LXRs might also play a role in lipid-regulated apoE expression, we examined the influence of LXR/RXR heterodimer ligands on differentiated THP-1 macrophages. As shown in Fig. 2, treatment of these cells for 48 h with the RXR ligand LG268 (50 nM) or with the LXR ligands 20(S)- or 22(R)-hydroxycholesterol (2 μg/ml) resulted in a significant induction in apoE mRNA expression. The combination of an RXR and an LXR ligand had an additive effect. In contrast, 22(S)-hydroxycholesterol, which does not activate LXR, had no effect. Interestingly, changes in apoE expression were paralleled by changes in expression of mRNAs encoding ABCA1 and ABCG1.
Figure 1.
Coordinate induction of apoE, ABCA1, and ABCG1 expression in THP-1 macrophages by modified LDL. Differentiated THP-1 macrophages were incubated for 48 h in RPMI medium 1640 containing 10% (vol/vol) LPDS with mevinolin and mevalonic acid, and 100 μg/ml (protein) LDL, highly oxidized LDL, or acetylated LDL, as indicated. Total RNA (10 μg per lane) was electrophoresed through formaldehyde-containing gels, transferred to Nylon, and hybridized to 32P-labeled cDNA probes. 36B4 was used as a control (ctrl) for loading and integrity of the RNA.
Figure 2.
Coordinate regulation of apoE, ABCG1, and ABCA1 expression in THP-1 macrophages by LXR ligands. Differentiated THP-1 macrophages were incubated for 48 h in RPMI medium 1640 containing 10% (vol/vol) LPDS with 50 nM LG268 (LG), 2.0 μg/ml 22(S)-hydroxycholesterol [22(S)HC], 2.0 μg/ml 20(S)HC, 2.0 μg/ml 22(R)HC, or vehicle control (ctrl), as indicated. Northern analysis was performed as described above. Blots were quantified by PhosphorImaging and normalized to 36B4.
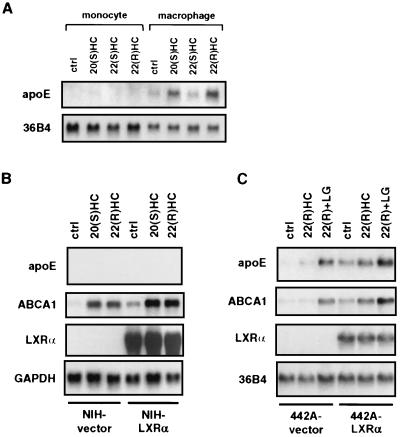
We next investigated whether the ability of LXR ligands to regulate apoE expression is limited to macrophages or whether it is also relevant to other cell types. First, because apoE expression is normally restricted to differentiated macrophages, we asked whether regulation by LXR ligands was differentiation-dependent. As shown in Fig. 3A, the ability of LXR ligands to regulate apoE expression is restricted to macrophages. This finding is in contrast to ABCA1 expression, which is responsive to LXR ligands in both differentiated and undifferentiated cells (data not shown). We also investigated apoE regulation in NIH 3T3 fibroblasts and preadipocytes. Retroviral expression of LXRα in NIH 3T3 cells facilitates expression of ABCA1 and ABCG1 (20). Surprisingly, we found that LXR ligands were unable to induce apoE expression in these cells, even in the presence of high levels of LXRα (Fig. 3B). In contrast, retroviral expression of LXRα in 3T3-F442A preadipocytes facilitated induction of apoE mRNA by LXR ligands (Fig. 3C). The basal induction of target genes by LXR ligands observed in NIH-vector and 442A-vector cells is likely to be mediated by LXRβ, which is expressed at a significant level in both cell types (20). Together, these findings suggest that induction of apoE expression by LXR ligands depends on additional tissue-specific and differentiation-dependent regulatory factors.
Figure 3.
Macrophage- and adipocyte-specific induction of ApoE expression by LXR and LXR ligands. (A) Differentiation-dependent induction of apoE expression by LXR ligands. Undifferentiated (monocyte) or differentiated (macrophage) THP-1 cells were incubated in RPMI medium 1640 containing 10% (vol/vol) LPDS and 2.0 μg/ml 22(S)HC, 2.0 μg/ml 20(S)HC, 2.0 μg/ml 22(R)HC, or vehicle control (ctrl) for 48 h. (B and C) Retroviral expression and activation of LXRα induces apoE expression in preadipocytes but not in fibroblasts. NIH 3T3 fibroblasts and 3T3-F442A preadipocytes were transduced with a retroviral vector encoding LXRα (NIH-LXRα, 442A-LXRα) or the empty vector alone (NIH-vector, 442A-vector). Stable cell lines were cultured for 48 h in DMEM containing 10% (vol/vol) LPDS in the presence of 50 nM LG268 (LG), 2.0 μg/ml 22(R)HC, or vehicle control (ctrl). Northern analysis was performed as described above.
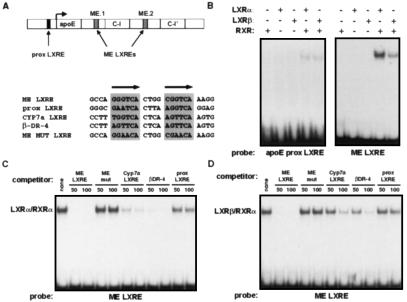
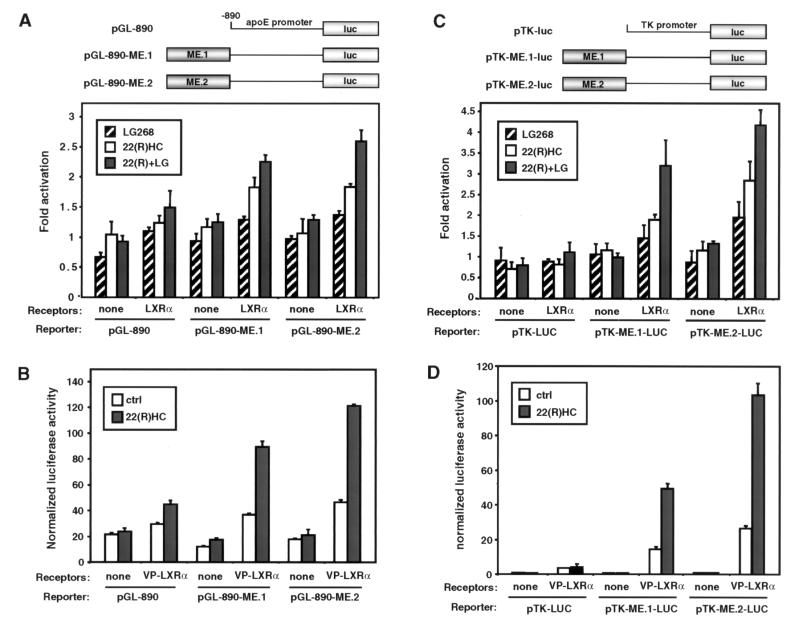
Next we asked whether the apoE gene might be a direct target for regulation by the LXR/RXR heterodimer. Recently, Taylor and coworkers (18) identified two enhancers from the apoE flanking sequences (ME.1 and ME.2) that direct macrophage- and adipose-specific expression of a reporter gene in transgenic mice. ME.1 and ME.2 are located 3′ of the apoE and apoC-I genes, respectively (Fig. 5A). We tested the ability of LXR/RXR heterodimers to regulate the apoE proximal promoter and the ME.1 and ME.2 enhancers in transient transfection assays. Luciferase reporter constructs that contained −890 to +93 of the apoE promoter alone (pGL-890) or that contained ME.1 or ME.2 upstream of this apoE promoter sequence (pGL-890-ME.1 and pGL-890-ME.2) were cotransfected into HepG2 cells along with expression vectors for LXRα and RXRα. As shown in Fig. 4A, the basal −890 promoter was very weakly activated by the LXR/RXR heterodimer in a ligand-dependent manner. The constructs that also contained one of the macrophage enhancers, however, were activated to a much greater degree. To confirm further that these luciferase reporters were targets for LXR regulation, we tested their ability to be activated by cotransfection of a VP16-LXRα expression vector. The VP16-LXRα fusion protein encoded by this vector has enhanced constitutive activity but also retains its ability to be activated by ligand. Consistent with the results obtained with wild-type LXRα, the VP16-LXRα expression vector activated all three constructs; however, the strongest activation was seen with the ME.1- and ME.2-containing reporters (Fig. 4B).
Figure 5.
The apoE ME.1 and ME.2 enhancers are direct targets for binding LXR/RXR heterodimers. (A) Genomic structure of the apoE locus and location of potential LXREs. ApoE LXRES are aligned with a known LXRE from the cholesterol-7-α-hydroxylase gene (CYP7α LXRE) and an idealized LXRE (β-DR-4). Also shown is the sequence of a mutant ME LXRE (ME MUT LXRE) used in C and D. (B) Direct binding of LXRα/RXRα and LXRβ/RXRα to a low-affinity LXRE present in the proximal promoter and a high-affinity site conserved in both ME.1 and ME.2. Gel mobility-shift assays were performed with the use of in vitro translated receptors as described in Materials and Methods. (C and D) Sequence-specific competition for LXRα/RXRα (C) and LXRβ/RXRα (D) binding to the ME LXRE.
Figure 4.
The LXR/RXR heterodimer activates the apoE ME.1 and ME.2 enhancers. (A) LXRα/RXRα activates the −890-bp apoE proximal promoter fused to either ME.1 or ME.2. HepG2 cells were transfected with pGL-890, pGL-890-ME.1, or pGL-890-ME.2 with or without CMX-mLXRα and CMX-RXRα and CMV-β-galactosidase. After transfection, cells were incubated for 24 h in MEM supplemented with 10% (vol/vol) LPDS and 22(R)HC (5.0 μg/ml), LG268 (50 nM), or vehicle control. Luciferase activity was normalized for transfection efficiency with the use of β-galactosidase activity. The data are expressed as fold activation in the presence of the indicated ligand versus in the absence of ligand and represent the average of triplicate experiments. (B) VP16-LXRα activates the apoE proximal promoter fused to either ME.1 or ME.2. Transient transfections were performed by using CMX-VP16-LXRα and CMX-RXRα as indicated. The results are shown as normalized luciferase units. (C) ME.1 and ME.2 function as LXRα/RXR-responsive enhancers when fused to a heterologous promoter. Transfections were performed with the use of pTK-Luc, pTK-ME.1-Luc, or pTK-ME.2-Luc reporters. (D) ME.1 and ME.2 are activated by VP16-LXRα. Transfections were performed with the use of pTK-Luc, pTK-ME.1-Luc, or pTK-ME.2-Luc along with CMX-VP16-LXRα and CMX-RXRα.
We also tested the ability of the ME.1 and ME.2 enhancers to be activated by LXRα when linked to a heterologous thymidine kinase (TK) promoter. As shown in Fig. 4C, both the ME.1 and ME.2 TK-luciferase reporters (pTK-ME.1-LUC and pTK-ME.2-LUC) were significantly activated by cotransfection of LXRα and RXRα expression vectors in a ligand-dependent manner. In contrast, there was no effect on the basal TK promoter (pTK-LUC). Cotransfection of the ME.1 and ME.2 TK-luciferase reporters with VP16-LXRα and RXR expression vectors resulted in a greater than 50-fold activation (Fig. 4D).
We next endeavored to identify the cis-acting sequences that mediate regulation of the apoE gene by the LXR/RXR heterodimer. The preferred binding site for LXR/RXR is a DR-4 (direct repeat with four-nucleotide spacer) hormone response element (25). Sequence analysis revealed one potential LXRE in the −890 to +93 proximal apoE promoter as well as a potential LXRE that is conserved in both the ME.1 and ME.2 enhancers (Fig. 5A). Gel-shift analysis with in vitro translated proteins confirmed that both sequences bound LXRα/RXRα and LXRβ/RXRα heterodimers, although the affinity of the ME LXRE was much greater (Fig. 5B). Binding to the ME LXRE was specific, as indicated by its ability to be competed by an excess of unlabeled ME LXRE, CYP7A1 LXRE, or consensus DR-4 oligonucleotide (Fig. 5 C and D). Consistent with its apparent affinity in the direct binding experiment (Fig. 5B), the proximal promoter LXRE was a relatively poor competitor for LXRα/RXRα and LXRβ/RXRα binding to the ME LXRE. Our identification of a low-affinity LXRE in the proximal promoter and a high-affinity LXRE in ME.1/ME.2 is entirely consistent with the degree of activation observed in the transient transfection studies (Fig. 4).
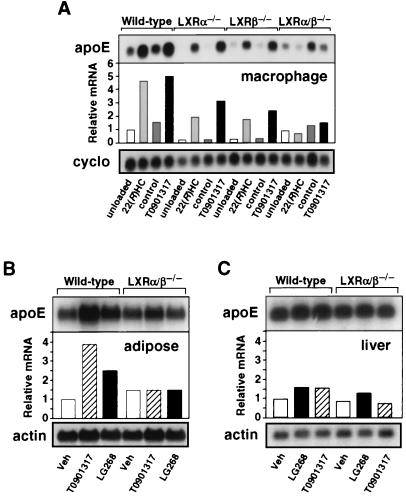
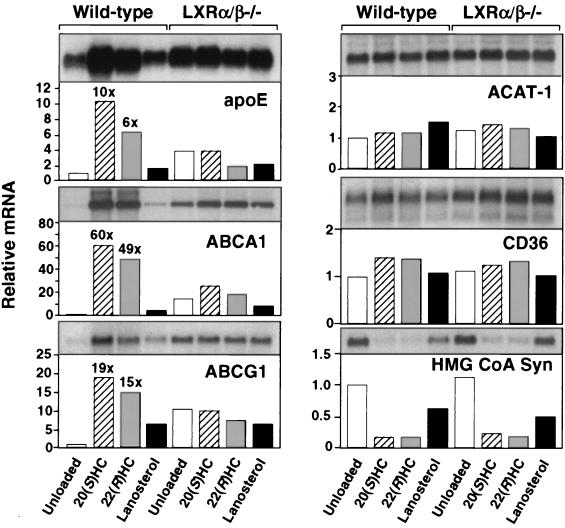
Finally, we examined the effects of LXR ligands on apoE expression in cells and tissues derived from wild-type and Lxr knockout mice. Consistent with the in vitro data presented above, treatment of wild-type mice with either LG268 or the synthetic LXR agonist T0901317 induced expression of apoE 2.5- to 4-fold in adipose tissue but had a minimal effect on expression of this gene in liver (Fig. 6 B and C). A similar induction of apoE expression was observed after treatment of peritoneal macrophages derived from wild-type mice with 22(R)-hydroxycholesterol, lanosterol, or T0901317 (Figs. 6A and 7). These observations are consistent with our identification of an LXRE in the apoE macrophage/adipocyte-specific enhancers but not in the hepatic control region. Moreover, they suggest that the low-affinity LXRE in the proximal promoter is not sufficient for LXR induction in the absence of ME.1 and ME.2 activity. As shown above for human THP-1 macrophages, treatment of murine macrophages with LXR ligands led to the coordinate induction of apoE, ABCA1, and ABCG1 expression (Fig. 7). In contrast, expression of several other genes involved in macrophage lipid metabolism, including the PPARγ target gene CD36 and ACAT-1, was not altered in response to LXR ligands. As expected, the SREBP target gene hydroxymethylglutaryl-CoA synthase was repressed by oxysterols.
Figure 6.
Tissue-specific induction of apoE expression in vivo by LXR ligands. (A) Sterol-induced expression of apoE in macrophages requires LXR. Peritoneal macrophages were isolated as described in Materials and Methods and cultured for 42 h in the presence of 100 μM mevalonic acid (control) or mevalonic acid plus 5 μM compactin (unloaded), 10 μM 22(R)-hydroxycholesterol [22(R)HC], or 10 μM T0901317. (B and C) LXR and RXR ligands induce apoE expression in an LXR-dependent manner in adipose tissue (B) but not in liver (C). Wild-type or Lxrα/β−/− mice were fed ad libitum diets containing 0.2% cholesterol and vehicle (Veh), LXR agonist (T0901317, 50 mg/kg body weight) or RXR-specific agonist (LG268, 30 mg/kg body weight) for 10 days. Northern analysis was performed as above. Blots were quantitated by PhosphorImager, standardized against actin or cyclophilin (cyclo), and mathematically adjusted to establish a unit of 1 for the wild-type group receiving vehicle.
Figure 7.
Coordinate regulation of genes involved in macrophage sterol efflux by LXRs. Peritoneal macrophages were isolated from wild-type or Lxrα/β−/− mice and incubated for 42 h with 100 μM mevalonic acid (control) or mevalonic acid plus 5 μM compactin (unloaded), 10 μM 20(S)-hydroxycholesterol [20(S)HC], 10 μM 22(R)-hydroxycholesterol [22(R)HC], or 10 μM lanosterol. Northern analysis was performed as described above.
To confirm that the observed induction of apoE was mediated by LXRs, we analyzed mice with targeted disruptions of the Lxrα gene, the Lxrβ gene, or both (19). Induction of apoE in adipose tissue by LG268 and T0901317 was abolished in Lxrα−/− Lxrβ−/− mice (Fig. 6B). A similar dependence on LXRα and LXRβ expression was observed in peritoneal macrophages derived from these mice. Induction of apoE in macrophages by LXR ligands was reduced in Lxrα−/− or Lxrβ−/− mice and completely abolished in double knockouts (Figs. 6A and 7). In contrast, loss of LXR expression had no effect on apoE expression in liver (Fig. 6C). These data indicate that both LXRα and LXRβ are functional regulators of lipid-inducible apoE expression in vivo. As expected, there was no difference in CD36, ACAT-1, or hydroxymethyl glutaryl-CoA synthase expression between wild-type and Lxr null mice. Thus, LXRs control expression of a distinct subset of genes involved in the regulation of cholesterol efflux. They do not seem to influence cholesterol esterification (ACAT-1), scavenger receptor-mediated lipid uptake (CD36), or sterol synthesis (hydroxymethyl glutaryl-CoA synthase). Interestingly, expression of apoE in Lxrα−/− or Lxrβ−/− single knockout mice was compromised to a greater degree than expression of ABCA1 or ABCG1 (ref. 19 and data not shown). This finding suggests that expression of apoE is more sensitive to the absolute level of LXR protein expression in the cell. Taken together, the above findings definitively identify the apoE gene as a direct transcriptional target of LXRα and LXRβ in macrophages and adipose tissue.
Discussion
The LXRs are a distinct pair of nuclear receptors that are emerging as key regulators of mammalian cholesterol homeostasis. LXRα is expressed primarily in liver, kidney, intestine, adipose tissue, and certain macrophages, whereas LXRβ is widely expressed (25, 26). The physiologic ligands for both receptors are now recognized to be specific oxysterols (27, 28). Targeted disruption of the Lxrα gene in mice delineated an important role for LXRα in the regulation of hepatic cholesterol metabolism. Lxrα null mice fail to induce transcription of the gene encoding cholesterol 7α-hydroxylase (CYP7A1), the rate-limiting enzyme in bile acid synthesis, in response to dietary cholesterol (22). Disruption of both Lxrα and Lxrβ in mice led to a more severe defect, indicating that these two receptors have at least partially overlapping physiologic functions (J.J.R. and D.J.M., unpublished observations).
Recent work has also defined a role for LXRs in the regulation of gene expression in response to cellular lipid loading. ABCA1 (also called ABC-1) and ABCG1 (also called ABC-8 or white), two members of the ATP-binding cassette (ABC) family of transporter proteins, are highly induced in lipid-loaded macrophages. Loss of function mutations in the ABCA1 gene result in Tangier disease, which is characterized by marked cholesterol accumulation in macrophages and other reticuloendothelial cells (3, 4). This phenotype, along with the observation that fibroblasts from Tangier patients are impaired in their ability to donate cholesterol to apoAI, suggests that ABCA1 plays a pivotal role in cellular cholesterol efflux (5). ABCG1 has also been suggested to be involved in lipid efflux, although its function is less well understood (29). We and others have recently shown that ABCA1 and ABCG1 are direct transcriptional targets of LXRs (19, 20, 24, 30). This discovery shed light on the function of LXRs in mammalian physiology. In intestine, induction of ABCA1 expression by LXR or RXR ligands dramatically reduces dietary cholesterol absorption, presumably by actively pumping cholesterol from the enterocyte into the lumen of the gut (19). In lipid-loaded macrophages, induction of ABCA1 expression by LXR ligands similarly promotes cholesterol efflux to extracellular acceptors such as apoAI (20). These results established that both dietary cholesterol absorption and the rate of cholesterol efflux in peripheral cells are controlled by a nuclear receptor signaling pathway.
In this work we have demonstrated that the apoE gene is also a direct target of LXRα and LXRβ in macrophages. We have shown that, in addition to controlling the tissue-specific expression of apoE in macrophages and adipose tissue, the ME.1 and ME.2 enhancers identified by Taylor and colleagues (18) are also likely to facilitate gene induction in response to cellular lipid loading. As basal apoE expression is preserved in Lxr mutant mice, the principal role of LXRs in apoE regulation is to mediate the lipid inducibility of the gene. The inability of LXR ligands to activate apoE expression in cell types other than macrophages and adipocytes strongly suggests that the tissue specificity of apoE expression is controlled by additional as yet undefined factors.
Together with previous work, these observations support the hypothesis that LXRs act as molecular sensors that link the process of lipid loading to specific changes in macrophage gene expression and function. Activation of LXR in macrophages leads to the coordinate induction of multiple genes potentially involved in cholesterol efflux, including ABCA1, ABCG1, and apoE. Induction of these genes in response to lipid loading may serve to limit the accumulation of lipid and thus protect against the development of fatty lesions and atherosclerosis. Our observations suggest that the LXR pathway may be a useful target for the pharmacological regulation of macrophage lipid metabolism. Given the well-characterized role of apoE as an antiatherogenic factor, our results imply the possibility that LXR ligands may protect against the development of atherosclerotic lesions. This hypothesis has yet to be tested.
Acknowledgments
We thank Steven Kliewer for pSG5-LXRβ, Rich Heyman (Ligand Pharmaceuticals) for LG268, Bei Shan (Tularik) for T0901317, Ron Evans for CMX-VP16LXRα, and Peter Edwards for a critical reading of the manuscript. P.T. is an Assistant Investigator, D.J.M. is an Associate Investigator, and B.A.L and J.J.R. are Fellows of the Howard Hughes Medical Institute. This work was supported by grants from the Robert A. Welch Foundation (to D.J.M.) and the Jonsson Comprehensive Cancer Center (to P.T.).
Abbreviations
- ABC
ATP-binding cassette
- LDL
low-density lipoprotein
- HC
hydroxycholesterol
- apoE
apolipoprotein E
- ME.1
ME.2, multienhancers 1 and 2
- LXRE
LXR response element
- LPDS
lipoprotein-deficient serum
- TK
thymidine kinase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.021488798.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.021488798
References
- 1.Ross R. Annu Rev Physiol. 1995;57:791–804. doi: 10.1146/annurev.ph.57.030195.004043. [DOI] [PubMed] [Google Scholar]
- 2.Steinberg D. J Biol Chem. 1997;272:20963–20966. doi: 10.1074/jbc.272.34.20963. [DOI] [PubMed] [Google Scholar]
- 3.Bodzioch M, Orsó E, Klucken J, Langmann T, Böttcher A, Diederich W, Drobnik W, Barlage S, Büchler C, Porsch-Özcürümez M, et al. Nat Genet. 1999;22:347–351. doi: 10.1038/11914. [DOI] [PubMed] [Google Scholar]
- 4.Brooks-Wilson A, Marcil M, Clee S M, Zhang L-H, Roomp K, van Dam M, Yu L, Brewer C, Collins J A, Molhuizen H O F, et al. Nat Genet. 1999;22:336–345. doi: 10.1038/11905. [DOI] [PubMed] [Google Scholar]
- 5.Lawn R M, Wade D P, Garvin M R, Wang X, Schwartz K, Porter J G, Seilhamer J J, Vaughan A M, Oram J F. J Clin Invest. 1999;104:R25–R31. doi: 10.1172/JCI8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plump A S, Smith J D, Hayek T, Aalto-Setala K, Walsh A, Verstuyft J G, Rubin E M, Breslow J L. Cell. 1992;71:343–353. doi: 10.1016/0092-8674(92)90362-g. [DOI] [PubMed] [Google Scholar]
- 7.Zhang S H, Reddick R L, Piedrahita J A, Maeda N. Science. 1992;258:468–471. doi: 10.1126/science.1411543. [DOI] [PubMed] [Google Scholar]
- 8.Curtiss L K, Boisvert W A. Curr Opin Lipidol. 2000;11:243–251. doi: 10.1097/00041433-200006000-00004. [DOI] [PubMed] [Google Scholar]
- 9.Mazzone T. Curr Opin Lipidol. 1996;7:303–307. doi: 10.1097/00041433-199610000-00008. [DOI] [PubMed] [Google Scholar]
- 10.Zhang W Y, Gaynor P M, Kruth H S. J Biol Chem. 1996;271:28641–28646. doi: 10.1074/jbc.271.45.28641. [DOI] [PubMed] [Google Scholar]
- 11.Langer C, Huang Y, Cullen P, Wiesenhutter B, Mahley R W, Assmann G, von Eckardstein A. J Mol Med. 2000;78:217–227. doi: 10.1007/s001090000096. [DOI] [PubMed] [Google Scholar]
- 12.Linton M F, Atkinson J B, Fazio S. Science. 1995;267:1034–1037. doi: 10.1126/science.7863332. [DOI] [PubMed] [Google Scholar]
- 13.Bellosta S, Mahley R W, Sanan D A, Murata J, Newland D L, Taylor J M, Pitas R E. J Clin Invest. 1995;96:2170–2179. doi: 10.1172/JCI118271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fazio S, Babaev V R, Murray A B, Hasty A H, Carter K J, Gleaves L A, Atkinson J B, Linton M F. Proc Natl Acad Sci USA. 1997;94:4647–4652. doi: 10.1073/pnas.94.9.4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tangirala R K, Pratico D, FitzGerald G A, Chun S, Tsukamoto K, Maugeais C, Usher D, Pure E, Rader D J. J. Biol. Chem. 2000. [DOI] [PubMed] [Google Scholar]
- 16.O'Brien K D, Deeb S S, Ferguson M, McDonald T O, Allen M D, Alpers C E, Chait A. Am J Pathol. 1994;144:538–548. [PMC free article] [PubMed] [Google Scholar]
- 17.Mazzone T, Basheeruddin K, Poulos C. J Lipid Res. 1989;30:1055–1064. [PubMed] [Google Scholar]
- 18.Shih S J, Allan C, Grehan S, Tse E, Moran C, Taylor J M. J Biol Chem. 2000;275:31567–31572. doi: 10.1074/jbc.M005468200. [DOI] [PubMed] [Google Scholar]
- 19.Repa J J, Turley S D, Lobaccaro J A, Medina J, Li L, Lustig K, Shan B, Heyman R A, Dietschy J M, Mangelsdorf D J. Science. 2000;289:1524–1529. doi: 10.1126/science.289.5484.1524. [DOI] [PubMed] [Google Scholar]
- 20.Venkateswaran A, Laffitte B A, Joseph S B, Mak P A, Wilpitz D C, Edwards P A, Tontonoz P. Proc Natl Acad Sci USA. 2000;97:12097–12102. doi: 10.1073/pnas.200367697. . (First Published October 17, 2000; 10.1073/pnas.200367697) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tontonoz P, Hu E, Spiegelman B M. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 22.Peet D J, Turley S D, Ma W, Janowski B A, Lobaccaro J M, Hammer R E, Mangelsdorf D J. Cell. 1998;93:693–704. doi: 10.1016/s0092-8674(00)81432-4. [DOI] [PubMed] [Google Scholar]
- 23.Laffitte B A, Kast H R, Nguyen C M, Zavacki A M, Moore D D, Edwards P A. J Biol Chem. 2000;275:10638–10647. doi: 10.1074/jbc.275.14.10638. [DOI] [PubMed] [Google Scholar]
- 24.Venkateswaran A, Repa J J, Lobaccaro J-M A, Bronson A, Mangelsdorf D J, Edwards P A. J Biol Chem. 2000;275:14700–14707. doi: 10.1074/jbc.275.19.14700. [DOI] [PubMed] [Google Scholar]
- 25.Willy P J, Umesono K, Ong E S, Evans R M, Heyman R A, Mangelsdorf D J. Genes Dev. 1995;9:1033–1045. doi: 10.1101/gad.9.9.1033. [DOI] [PubMed] [Google Scholar]
- 26.Peet D J, Janowski B A, Mangelsdorf D J. Curr Opin Genet Dev. 1998;8:571–575. doi: 10.1016/s0959-437x(98)80013-0. [DOI] [PubMed] [Google Scholar]
- 27.Janowski B A, Willy P J, Devi T R, Falck J R, Mangelsdorf D J. Nature (London) 1996;383:728–731. doi: 10.1038/383728a0. [DOI] [PubMed] [Google Scholar]
- 28.Janowski B A, Grogan M J, Jones S A, Wisely G B, Kliewer S A, Corey E J, Mangelsdorf D J. Proc Natl Acad Sci USA. 1999;96:266–271. doi: 10.1073/pnas.96.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klucken J, Buchler C, Orso E, Kaminski W E, Porsch-Ozcurumez M, Liebisch G, Kapinsky M, Diederich W, Drobnik W, Dean M, et al. Proc Natl Acad Sci USA. 2000;97:817–822. doi: 10.1073/pnas.97.2.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Costet P, Luo Y, Wang N, Tall A R. J Biol Chem. 2000;275:28240–28245. doi: 10.1074/jbc.M003337200. [DOI] [PubMed] [Google Scholar]