Abstract
The proteasome is the primary protease used by cells for degrading proteins and generating peptide ligands for class I molecules of the major histocompatibility complex. Based on the properties of cells adapted to grow in the presence of the proteasome inhibitor 4-hydroxy-5-iodo-3-nitrophenylacetyl-Leu-Leu-leucinal-vinyl sulfone (NLVS), it was proposed that proteasomes can be replaced by alternative proteolytic systems, particularly a large proteolytic complex with a tripeptidyl peptidase II activity. Here we show that NLVS-adapted cells retain sensitivity to a number of highly specific proteasome inhibitors with regard to antigenic peptide generation, accumulation of polyubiquitinated proteins, degradation of p53, and cell viability. In addition, we show that in the same assays (with a single minor exception), NLVS-adapted cells are about as sensitive as nonselected cells to Ala-Ala-Phe-chloromethylketone, a specific inhibitor of tripeptidyl peptidase II activity. Based on these findings, we conclude that proteasomes still have essential proteolytic functions in adapted cells that are not replaced by Ala-Ala-Phe-chloromethylketone-sensitive proteases.
Proteasomes are complex multisubunit proteases that are abundant and ubiquitous in eukaryotic cells (1, 2). Proteasomes degrade proteins marked for destruction by the addition of multiple ubiquitin molecules (3). This system is used for modulating the levels of specific proteins and as a general method of disposal for misfolded or damaged proteins (4). Oligopeptides generated by proteasomes are a major source of the peptide ligands of MHC class I molecules (5).
The proteolytic activity of proteasomes occurs in a barrel-shaped core structure known as the 20S proteasome. 20S proteasomes consist of 14 different proteins in four rings arrayed in an α7β7β7α7 manner. All of the proteolytic activity is thought to reside in three of the seven β subunits. The substrate specificity of 20S proteasomes has been defined largely with the use of fluorogenic oligopeptidyl substrates. This has revealed trypsin-like, chymotrypsin-like, and postglutamyl peptide hydrolyzing activities. 20S proteasomes do not recognize polyubiquitinated (polyUb) proteins and are able to degrade proteins only if they are first denatured (6). In cells, 20S proteasomes are thought to be active only in association with regulatory structures that function to locate substrates and translocate them into the 20S barrel (7).
Low-molecular-weight inhibitors of the proteasome have proved to be invaluable for studying the proteasome–ubiquitin system in mammalian cells. These include relatively nonspecific compounds such as cbz-Leu-Leu-leucinal (zLLL) (also a potent inhibitor of calpains) (8) and highly specific inhibitors such as 4-hydroxy-5-iodo-3-nitrophenylacetyl-Leu-Leu-leucinal-VS (NLVS) (9), the microbial products lactacystin (10) and epoxomicin (11), and boronic acid inhibitors boro-LLL (MG262) (12) and PS-341 (13). Although the effects of these compounds on the defined catalytic activity of purified 20S proteasomes have been extensively characterized with the use of small fluorogenic substrates, their effects on proteasomes in viable cells are somewhat less well defined.
Inasmuch as proteasomes are essential for the viability of Saccharomyces cerevisiae (14), it was surprisingly reported that propagation of mammalian cells in the presence of NLVS results in the selection of cells lacking active proteasomes (15). Notably, the absence of proteasome function in NLVS-selected cells was largely inferred from experiments using 20S proteasomes isolated from the cells. The viability of these cells was attributed to the induction of an alternative protease that substitutes for the essential functions of the proteasome. More recent studies have demonstrated an increase in levels of tripeptidyl peptidase II (TPP II) in selected cells, implicating this proteolytic complex in the survival of cells in NLVS (16, 17).
In the present study we have used a panel of proteasome inhibitors to examine the function of proteasomes in NLVS-selected cells. Our findings indicate that the residual proteolytic activity of proteasomes in these cells remains essential for the degradation of ubiquitinated proteins, antigen presentation, and cell viability.
Materials and Methods
Cell Lines.
Mouse cell lines EL4 (H-2b) and NLVS-adapted EL4 cells (EL4ad) were maintained in RPMI medium 1640 supplemented with 10% (vol/vol) FBS (RP10) at 37°C in an air/CO2 (94%/6%) atmosphere. Murine TCD8+ cell lines specific for H-2Kb complexed to a peptide corresponding to residues 366–374 from influenza nucleoprotein (NP) were generated as described (18).
The inhibitors zLLL and Ala-Ala-Phe-chloromethylketone (AAF-cmk) were purchased from Sigma or Bachem. Lactacystin was purchased from E. J. Corey (Harvard University, Cambridge, MA). NLVS was the kind gift of Hidde Ploegh (Harvard Medical School, Boston) and also was purchased from Calbiochem-Novabiochem. MG262 was purchased from Affinity (Nottingham, U.K.). Epoxomicin and YU101 were synthesized as described (19, 20). All inhibitors were dissolved in 100% DMSO.
Viral Infections and Intracellular Staining.
For intracellular staining, EL4 and EL4ad cells were incubated for 1 h at 37°C in RP10 containing the appropriate concentration of inhibitor followed by infection for 1 h with PR8 influenza virus in Autopow MEM (Life Technologies, Rockville, MD) adjusted to pH 6.8 and containing inhibitor. Peptides were added to a concentration of 0.1 μM or as indicated. After infection, EL4 or EL4ad cells and TCD8+ cells were added in 200 μl RP10 containing the appropriate inhibitor to each well of a 96-well plate. After 4 h of incubation at 37°C, brefeldin A was added, and cells were incubated for an additional 4 h to accumulate IFN-γ in the endoplasmic reticulum of activated cells.
Cytofluorography.
The expression of influenza virus nucleoprotein levels was measured by fixing cells in PBS containing 1% (wt/vol) paraformaldehyde for 30 min at room temperature followed by a 30-min incubation on ice with the H16-L10 mAb (21), used as the tissue culture supernatant. Cells were washed twice with cold balanced salt solution with 0.1% BSA (BSS/BSA) and then incubated on ice for 30 min with fluorescein-conjugated rabbit anti-mouse Ig (Dako) diluted 1:20 in BSS/BSA. Cells were washed three times in BSS/BSA and analyzed directly by cytofluorography. For intracellular cytokine staining, cells were incubated for 30 min on ice with cychrome-conjugated mouse anti-CD8 (PharMingen) diluted 1:100 in BSS/BSA. Cells were washed once in BSS/BSA and then fixed with 1% paraformaldehyde in PBS at room temperature for 20 min. Cells were washed twice and then incubated with fluorescein-conjugated mouse anti-IFN-γ (PharMingen) diluted 1:150 in PBS containing 0.2% (wt/vol) saponin. Cells were analyzed with the use of either FACScalibur or FACScan cytofluorographs (Becton Dickinson) and cellquest (Becton Dickinson) and flowjo (Tree Star, San Carlos, CA) software.
Immunoblotting.
EL4 and EL4ad cells (1 × 106) were lysed in 0.2 ml lysis buffer [50 mM Tris⋅HCl, pH 7.5/5 mM EDTA/150 mM NaCl/0.5% (wt/vol) {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}/0.2% (wt/vol) deoxycholate]. Lysates were mixed with hot (95°C) sample buffer (2% SDS/1% β-mercaptoethanol/1% glycerol/65 mM Tris⋅HCl, pH 6.5) and boiled for 10 min. Samples were separated in an 8% SDS-polyacrylamide gel according to the method of Laemmli and transferred to Immobilon P membranes (Millipore). Membranes were blocked overnight in PBS supplemented with 5% BSA and 0.3% Tween 20 and probed for polyUb by incubation with the FK2 mAb (22) (Nippon Bio-Test Laboratories, Tokyo, Japan) or for p53 with polyclonal goat anti-p53 antibody (Santa Cruz Biotechnology), followed by horseradish peroxidase-coupled rabbit anti-mouse (Dako) or rabbit anti-goat (Dako) antibody, respectively. Immunoblots were developed with the use of a chemiluminescent peroxide substrate (Pierce), and luminescence was recorded on Biomax MR film (Eastman Kodak). Images were digitized on a flatbed scanner and analyzed with imagequant software (Molecular Dynamics).
Fluorometric Assay for TPP II Activity.
EL4ad cells were incubated in RP10 containing 10 μM AAF-cmk, 10 μM lactacystin, or no inhibitor for 90 min at 37°C. Cells were washed twice in PBS and then resuspended in 50 mM Tris (pH 7.4) containing 5 mM MgCl2 and 0.2 μg/ml digitonin. Cells were transferred to black 96-well flat-bottom plates at a final concentration of 2 × 104 cells in 100 μl per well. The fluorogenic AAF-amc substrate was added to wells for a final concentration of 100 μM, and fluorescence was measured at 380-nm excitation and 465-nm emission on a Victor model 1420 multilabel counter (Wallac, Gaithersburg, MD).
Results
Effects of Proteasome Inhibitors on NLVS-Adapted Cells: Cellular Proliferation.
We reproduced the findings of Glas et al. (15) regarding the selection of EL4 cells propagated in the presence of 10 μM NLVS. As originally described, most cells die within a few days of exposure to NLVS, and some survivors begin to proliferate after ≈2 weeks in culture and do so indefinitely in the presence of the inhibitor. We were similarly able to select NLVS-resistant mouse P815 cells with the use of 10 μM NLVS. We compared the sensitivities of NLVS-selected and nonselected EL4 cells to the effects of a panel of proteasome inhibitors (lactacystin, zLLL, epoxomicin) on antigen presentation, accumulation of polyUb proteins, and cell viability, as will be described below. We failed to detect substantial differences associated with NLVS adaptation (data not shown).
Glas et al. were able to adapt EL4 cells to grow in 50 μM NLVS. These cells could differ considerably from 10 μM NLVS-adapted cells. Although we were unable to adapt cells to 50 μM NLVS, we were able to serially propagate cells obtained from Hidde Ploegh (termed EL4ad) in 50 μM NLVS as originally described.
We first examined the effects of proteasome inhibitors and the TPP II inhibitor AAF-cmk on the proliferation of EL4 and EL4ad cells. The inhibitor concentration resulting in an 80% reduction in cell number relative to control cultures after 3 days was determined (Table 1). For EL4ad cells, this analysis was performed in the absence and presence of 50 μM NLVS (the normal culture conditions for these cells). EL4 and EL4ad cells were similarly sensitive to the cytostatic effects of the proteasome inhibitors lactacystin, zLLL, and PS-341. EL4ad cells were 12.5- to 50-fold less sensitive to the cytostatic effects of the proteasome inhibitors boro-LLL and epoxomicin. Despite the structural similarity of YU101 to epoxomicin, EL4ad cells demonstrated only a slight resistance to this inhibitor relative to EL4 cells. Importantly, we failed to detect a significant difference in sensitivities of EL4 and EL4ad cells to the TPP II inhibitor AAF-cmk obtained from two different commercial sources.
Table 1.
Inhibitor concentrations resulting in 80% growth inhibition of EL4 and EL4ad cells
| Inhibitor | EL4, μM | Fold difference inhibitor
concentration
|
|
|---|---|---|---|
| EL4ad + 50 μM NLVS | EL4ad | ||
| Lactacystin | 4 | 0.75 × | 1.25 × |
| zLLL | 0.4 | 1 × | 3.75 × |
| NLVS | 8 | >6 × | >6 × |
| Epoxomicin | 0.03 | 23 × | 50 × |
| Boro-LLL | 0.04 | 12.5 × | >25 × |
| PS-341 | 0.02 | 1 × | 2 × |
| YU101 | 0.25 | 4 × | 8 × |
| AAF-cmk (Sigma) | 20 | 1 × | 1.25 × |
| AAF-cmk (Bachem) | 20 | 1.5 × | 1.5 × |
Cells were cultured in RP10 with varying concentrations of the indicated inhibitors, and the number of viable cells was determined by trypan blue exclusion every 24 h. After 72 h in culture, the concentration of inhibitor necessary to cause an 80% reduction in the number of viable cells relative to control cultures was determined. The fold difference in inhibitor concentration for EL4ad relative to EL4 cells is indicated.
Effects of Proteasome Inhibitors on NLVS-Adapted Cells: Antigen Presentation.
To compare the antigen-processing capacities of EL4 and EL4ad cells, we generated a TCD8+ cell line specific for H-2Db complexed to a peptide corresponding to residues 366–374 from NP. The presentation of this determinant is known to be proteasome-dependent (23). Cells were infected with influenza virus for 4 h before the addition of brefeldin A to block further export of class I peptide complexes from the endoplasmic reticulum, preventing peptide class I complexes from reaching saturating levels. The expression of peptide class I complexes was determined by measuring TCD8+ activation as detected by intracellular staining for IFN-γ in TCD8+ cells identified by binding a directly conjugated anti-CD8 mAb.
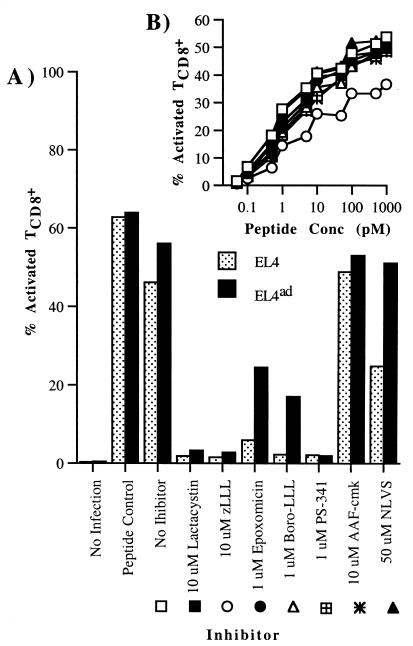
As seen in Fig. 1, EL4ad cells continuously incubated with 50 μM NLVS were able to activate Db-NP366–374-specific TCD8+ to an extent similar to that of the activation by EL4 cells or EL4ad cells in the absence of inhibitor. The generation of NP366–374 by EL4ad cells could be due to either the activity of proteasomes or, as suggested (15, 17), an alternative protease capable of producing the peptide or a suitable precursor. To distinguish between these possibilities, cells were incubated continuously from 1 h before infection with inhibitors specific for the proteasome or TPP II. Proteasome inhibitor concentrations were used at concentrations that block proteasome function without reducing protein synthesis. AAF-cmk was used at 10 μM, a concentration greater than that used by Glas et al. to completely inhibit the proliferation of EL4ad cells (15).
Figure 1.
Effect of proteasome inhibitors on antigen processing in EL4 and EL4ad cells. Cells were treated with proteasome inhibitors and then infected with PR8 influenza virus. TCD8+ cells specific for influenza NP366–374 were added 1 h after infection. Brefeldin A was added 5 h after infection. After an additional 4 h, cells were harvested, stained for CD8 and intracellular IFN-γ, and analyzed by cytofluorography. (A) The percentage of CD8+ cells positive for intracellular IFN-γ is represented graphically. (B) To ensure that proteasome inhibitors were not inhibiting IFN-γ production in TCD8+ cells, the assay described in A was performed with the use of EL4 cells titrated with limiting quantities of NP366–374 peptide as antigen-presenting cells and TCD8+ cells exposed to the same inhibitor concentrations as in A. Legend symbols are defined in A.
EL4 and EL4ad cells demonstrated similar sensitivities to lactacystin, zLLL, and PS-341, which completely blocked antigen presentation (Fig. 1A). This blockage of antigen presentation is not due to inhibitor effects on TCD8+ activation, as inhibitors had no effects on the activation of these TCD8+ cells exposed to the synthetic NP366–374 peptide over an entire dose–response curve (Fig. 1B). Nor could this blockage of antigen presentation be attributed to the effects of the inhibitors on viral gene expression, as determined by cytofluorographic detection of NP in fixed and permeabilized cells by indirect staining with the use of an anti-NP mAb (data not shown). NLVS (50 μM) had only a partial effect on Db-NP366–374 complex generation in EL4 cells, providing our initial evidence that NLVS at the concentrations used for selection has only a partial effect on proteasome activity. EL4ad cells were partially resistant to the effects of epoxomicin and boro-LLL. The TPP II inhibitor AAF-cmk had no significant effect on antigen presentation by either EL4 or EL4ad cells.
It is notable that the effects of the inhibitors on antigen processing in EL4 and EL4ad cells closely parallel their effects on cellular proliferation.
Effects of Proteasome Inhibitors on NLVS-Adapted Cells: Accumulation of PolyUb Proteins and p53.
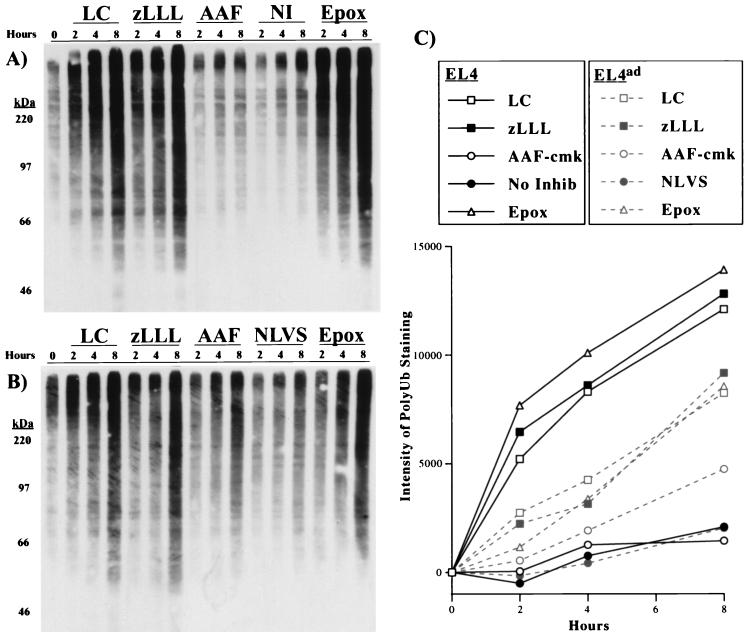
We next examined the effects of proteasome inhibitors on the levels of polyUb proteins in EL4 and EL4ad cells cultured for 2, 4, and 8 h with lactacystin, zLLL, epoxomicin, or AAF-cmk. For EL4ad cells, the media also included 50 μM NLVS. PolyUb proteins were detected by Western blotting with the FK2 mAb (22) (Fig. 2A). As expected, EL4 cells demonstrate an increase in the level of polyUb proteins in response to the proteasome inhibitors. EL4ad cells also demonstrated a marked increase in polyUb proteins, although the magnitude of the effect was approximately half of that demonstrated by EL4 cells. For this experiment, equivalent numbers of cells were loaded per lane. EL4ad cells are smaller than EL4 cells, and this size difference translates to a ≈1/3 reduction in the amount of protein per cell. Thus, nonmanipulated EL4ad cells actually possess more polyUb proteins as a percentage of total cell protein, which suggests that the adaptation process does not completely compensate for the inhibition in degradation of polyUb proteins. Accounting for the difference in cell protein, the magnitude of the accumulation of polyUb proteins is similar in EL4ad and EL4 cells. Notably, polyUb levels increased in EL4ad but not in EL4 cells treated with AAF-cmk, although this increase was minor compared with that observed with proteasome inhibitors. As will be discussed, this difference in polyUb levels does not necessarily indicate that TPP II degrades polyUb proteins.
Figure 2.
Effect of proteasome inhibitors on the accumulation of polyUb proteins in EL4 and EL4ad cells. (A and B) EL4 (A) and EL4ad (B) cells treated with inhibitors for 2, 4, and 8 h were analyzed by Western blotting with the use of the FK2 mAb, the binding of which was visualized by chemiluminescence. The inhibitors used were 10 μM lactacystin (LC), 10 μM zLLL (zLLL), 10 μM AAF-cmk (AAF), no inhibitor (NI), 50 μM NLVS (NLVS), and 1 μM epoxomicin (Epox). (C) The increase in FK2 staining was quantitated and is represented graphically.
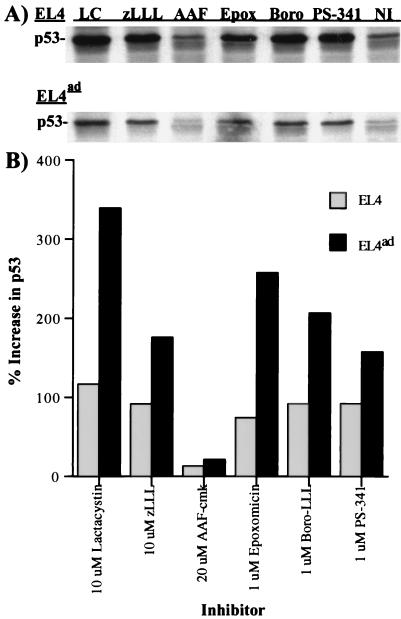
The same extracts were used to determine the effects of proteasome inhibitors on a defined cellular proteasome substrate, p53, which was detected via Western blotting with the use of a p53-specific antiserum (Fig. 3A). Less p53 was detected in EL4ad cells, again, probably because of the size difference in the cells. To account for this difference, the data are represented as the percentage increase in signal. For all proteasome inhibitors tested, the fold increase in p53 was higher in EL4ad than in EL4 cells (Fig. 3B). In contrast, AAF-cmk had only a very slight enhancing effect on p53 levels in either EL4 or EL4ad cells.
Figure 3.
Effect of proteasome inhibitors on the accumulation of p53 in EL4 and EL4ad cells. Aliquots from cells analyzed in Fig. 2 were Western blotted with the use of polyclonal p53-specific antibodies. The inhibitors used were 10 μM lactacystin (LC), 10 μM zLLL (zLLL), 10 μM AAF-cmk (AAF), 1 μM epoxomicin (Epox), 1 μM boro-LLL (Boro), 1 μM PS-341 (PS-341), and no inhibitor (NI). (A) Antibody binding was visualized by chemiluminescence. (B) The increase in p53 staining was quantitated and is represented graphically.
Taken together, these data indicate that the proteasome retains an essential role in the degradation of polyUb proteins in EL4ad cells.
Confirmation of the Effect of AAF-cmk on the Degradation of a Fluorogenic Peptide Substrate.
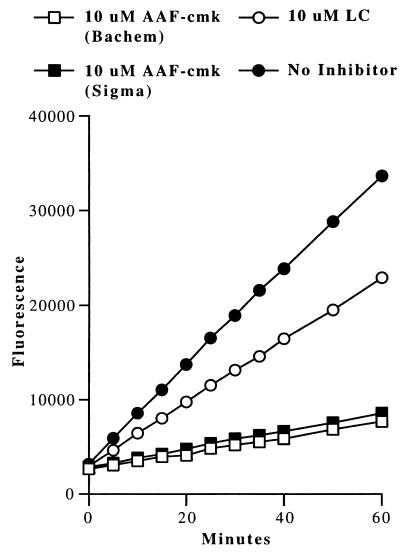
Because all of the assays performed to this point showed no significant inhibitory effect of AAF-cmk on EL4ad cells, it was important to demonstrate the activity of the AAF-cmk preparations we used. First, the chemical integrity of each AAF-cmk preparation source was confirmed by mass spectroscopy (data not shown). Next, we treated EL4ad cells with 10 μM AAF-cmk for 90 min and then incubated digitonin-permeabilized cells with the TPP II fluorogenic substrate AAF-amc. As shown in Fig. 4, AAF-amc was rapidly degraded in the absence of inhibitor by EL4ad cells. Both preparations of AAF-cmk blocked AAF-cmk degradation nearly completely, confirming their biological activity against TPP II when used to treat cultured cells. In contrast, 10 μM lactacystin had little effect on TPP II activity.
Figure 4.
Effect of AAF-cmk on AAF-amc hydrolysis in EL4ad cells. EL4ad cells were treated with 10 μM AAF-cmk or 10 μM lactacystin for 90 min at 37°C and then incubated with 100 μM AAF-amc. Hydrolysis of AAF-amc was determined by measuring fluorescence at 5–10-min intervals.
Discussion
It was previously reported that NLVS-selected EL4 cells grown in the presence of NLVS do not possess active proteasomes (15). This conclusion was inferred from the following:
(i) the inability of radioiodinated NLVS to modify proteasomes from adapted cells,
(ii) the absence of enzymatically active proteasomes in gel filtration fractions derived from adapted cells,
(iii) the increased activity of a nonproteasomal AAF-amc hydrolyzing activity recovered from adapted cells,
(iv) the enhanced sensitivity of cells to inhibition of proliferation by AAF-cmk.
These findings led to the proposal that an AAF-cmk-sensitive enzyme, possibly TPP II, can substitute for the functions of the proteasome, including the degradation of ubiquitinated proteins and the generation of antigenic peptides. These findings were further supported by the observation that cells transfected with cDNA encoding TPP II were resistant to toxic doses of NLVS without the need to first adapt cells for growth in this inhibitor (17).
In the present study, we show that NLVS-adapted cells remain fully sensitive to the effects of several proteasome inhibitors on proliferation, antigen processing, and degradation of polyUb proteins. A critical issue is whether the effects of these inhibitors are due to the inhibition of proteasomes and not other proteases. The specificity of inhibitors can never be assumed to be absolute. Lactacystin, for example, originally touted as a specific proteasome inhibitor, is not absolutely specific for the proteasome, as it has been reported to inhibit a cathepsin A activity from platelets and even purified TPP II (16). However, inhibition of TPP II required high concentrations of lactacystin; at the 10 μM concentration we used, purified TPP II was inhibited by less than 50% (16), and, furthermore, we directly demonstrate that 10 μM lactacystin has a small effect on the ability of cells to hydrolyze the TPP II substrate AAF-amc (Fig. 4). Moreover, Greier et al. (16) reported that cells adapted to grow in 6 μM lactacystin exhibit increased levels of TPP II activity, making it extremely unlikely that the effects we observe with 10 μM are due to inhibition of TPP II activity. In addition, we found that incubation of EL4ad cells with 1 μM PS-341, another highly specific inhibitor of the proteasome, also resulted in inhibition of cell growth and antigen processing as well as increases in polyUb protein and p53 levels.
NLVS was originally reported to modify covalently all of the catalytically active proteasome subunits and to block all of the defined in vitro activities of 20S proteasomes (9). More recent studies, however, indicate that EL4ad cells retain residual proteasome activity, predominantly due to the tryptic and caspase-like catalytic activities of the Z and Y proteasome subunits, respectively (17). This conclusion is in complete agreement with our finding that EL4ad cells demonstrate resistance to expoxomicin and boro-LLL, the most highly specific inhibitors in our panel for the chymotryptic activity of the proteasome.
Our findings demonstrate that EL4ad cells require proteasome function for the degradation of polyUb proteins, the processing of a model class I ligand, and proliferation. From this requirement we infer that adaptation to NLVS has not induced an alternative protease able to replace the role of proteasomes in recognition and degradation of polyUb proteins. What, then, is the role of TPP II in the adapted cells? In contrast to Glas et al., we were unable to detect any increased sensitivity of EL4ad to AAF-cmk. We did detect a slight effect of AAF-cmk on the degradation of polyUb proteins (but not p53). Although this effect could reflect the involvement of TPP II in the degradation of polyUb proteins, it is equally or more plausible that this effect reflects a less direct effect of TPP II on cellular metabolism that increases the work load of proteasomes, which are already compromised by the severe reduction of their chymotryptic activity. For example, proteasomes might serve as an alternative protease for substrates that can be handled by TPP II. In this case, blocking TPP II in EL4ad cells would increase the substrate load of proteasomes to the point where the rate of degradation of polyUb substrates is reduced to a value less than their rate of generation, resulting in the observed increase in polyUb proteins. Such a shared substrate pool between proteasomes and TPP II could account for the clear effect that TPP II overexpression has in enabling the survival of EL4 cells exposed to NLVS (17).
Acknowledgments
We thank Dr. Sandy Hayes and Beth Buschling for excellent technical assistance. We also thank Gustav Russ for critical reading of the manuscript. We are particularly grateful to Dr. Hidde Ploegh for his extreme generosity in providing both EL4ad adapted cells and sufficient quantities of NLVS to perform extensive experiments with the cells.
Abbreviations
- AAF-amc
H-Ala-Ala-Phe-7-amido-4-methylcoumarin
- AAF-cmk
Ala-Ala-Phe-chloromethylketone
- NLVS
4-hydroxy-5-iodo-3-nitrophenylacetyl-Leu-Leu-leucinal-vinyl sulfone
- EL4ad
NLVS-adapted EL4 cells
- NP
influenza nucleoprotein
- polyUb
polyubiquitinated
- zLLL
cbz-Leu-Leu-leucinal
- BSS
balanced salt solution
- TPP II
tripeptidyl peptidase II
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.021132398.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.021132398
References
- 1.Baumeister W, Walz J, Zuhl F, Seemuller E. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 2.DeMartino G N, Slaughter C A. J Biol Chem. 1999;274:22123–22126. doi: 10.1074/jbc.274.32.22123. [DOI] [PubMed] [Google Scholar]
- 3.Deveraux Q, Ustrell V, Pickart C, Rechsteiner M. J Biol Chem. 1994;269:7059–7061. [PubMed] [Google Scholar]
- 4.Yewdell J W, Antón L C, Bennink J R. J Immunol. 1996;157:1823–1826. [PubMed] [Google Scholar]
- 5.Rock K L, Goldberg A L. Annu Rev Immunol. 1999;17:739–779. doi: 10.1146/annurev.immunol.17.1.739. [DOI] [PubMed] [Google Scholar]
- 6.Kisselev A F, Akopian T N, Woo K M, Goldberg A L. J Biol Chem. 1999;274:3363–3371. doi: 10.1074/jbc.274.6.3363. [DOI] [PubMed] [Google Scholar]
- 7.Voges D, Zwickl P, Baumeister W. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 8.Rock K L, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg A L. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 9.Bogyo M, McMaster J S, Gaczynska M, Tortorella D, Goldberg A L, Ploegh H. Proc Natl Acad Sci USA. 1997;94:6629–6634. doi: 10.1073/pnas.94.13.6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Craiu A, Gaczynska M, Akopian T, Gramm C F, Fenteany G, Goldberg A L, Rock K L. J Biol Chem. 1997;272:13437–13445. doi: 10.1074/jbc.272.20.13437. [DOI] [PubMed] [Google Scholar]
- 11.Meng L, Mohan R, Kwok B H, Elofsson M, Sin N, Crews C M. Proc Natl Acad Sci USA. 1999;96:10403–10408. doi: 10.1073/pnas.96.18.10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCormack T, Baumeister W, Grenier L, Moomaw C, Plamondon L, Pramanik B, Slaughter C, Soucy F, Stein R, Zuhl F, et al. J Biol Chem. 1997;272:26103–26109. doi: 10.1074/jbc.272.42.26103. [DOI] [PubMed] [Google Scholar]
- 13.Palombella V J, Conner E M, Fuseler J W, Destree A, Davis J M, Laroux F S, Wolf R E, Huang J, Brand S, Elliott P J, et al. Proc Natl Acad Sci USA. 1998;95:15671–15676. doi: 10.1073/pnas.95.26.15671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heinemeyer W, Kleinschmidt J A, Saidowsky J, Escher C, Wolf D H. EMBO J. 1991;10:555–562. doi: 10.1002/j.1460-2075.1991.tb07982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glas R, Bogyo M, McMaster J S, Gaczynska M, Ploegh H L. Nature (London) 1998;392:618–622. doi: 10.1038/33443. [DOI] [PubMed] [Google Scholar]
- 16.Geier E, Pfeifer G, Wilm M, Lucchiari-Hartz M, Baumeister W, Eichmann K, Niedermann G. Science. 1999;283:978–981. doi: 10.1126/science.283.5404.978. [DOI] [PubMed] [Google Scholar]
- 17.Wang E W, Kessler B M, Borodovsky A, Cravatt B F, Bogyo M, Ploegh H L, Glas R. Proc Natl Acad Sci USA. 2000;97:9990–9995. doi: 10.1073/pnas.180328897. . (First Published August 22, 2000; 10.1073/pnas.180328897) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen W, Anton L C, Bennink J R, Yewdell J W. Immunity. 2000;12:83–93. doi: 10.1016/s1074-7613(00)80161-2. [DOI] [PubMed] [Google Scholar]
- 19.Yewdell J W, Frank E, Gerhard W. J Immunol. 1981;126:1814–1819. [PubMed] [Google Scholar]
- 20.Elofsson M, Splittgerber U, Myung J, Mohan R, Crews C M. Chem Biol. 1999;6:811–822. doi: 10.1016/s1074-5521(99)80128-8. [DOI] [PubMed] [Google Scholar]
- 21.Yewdell J W, Frank E, Gerhard W. J Immunol. 1981;126:1814–1819. [PubMed] [Google Scholar]
- 22.Fujimuro M, Sawada H, Yokosawa H. FEBS Lett. 1994;349:173–180. doi: 10.1016/0014-5793(94)00647-4. [DOI] [PubMed] [Google Scholar]
- 23.Cerundolo V, Benham A, Braud V, Mukherjee S, Gould K, Macino B, Neefjes J, Townsend A. Eur J Immunol. 1997;27:336–341. doi: 10.1002/eji.1830270148. [DOI] [PubMed] [Google Scholar]