Abstract
The G12 subfamily of heterotrimeric G proteins, comprised of the α-subunits Gα12 and Gα13, has been implicated as a signaling component in cellular processes ranging from cytoskeletal changes to cell growth and oncogenesis. In an attempt to elucidate specific roles of this subfamily in cell regulation, we sought to identify molecular targets of Gα12. Here we show a specific interaction between the G12 subfamily and the cytoplasmic tails of several members of the cadherin family of cell-surface adhesion proteins. Gα12 or Gα13 binding causes dissociation of the transcriptional activator β-catenin from cadherins. Furthermore, in cells lacking the adenomatous polyposis coli protein required for β-catenin degradation, expression of mutationally activated Gα12 or Gα13 causes an increase in β-catenin-mediated transcriptional activation. These findings provide a potential molecular mechanism for the previously reported cellular transforming ability of the G12 subfamily and reveal a link between heterotrimeric G proteins and cellular processes controlling growth and differentiation.
G proteins transmit a wide variety of extracellular signals to effector molecules within the cell (1, 2). G proteins consist of two functional units, a guanine nucleotide-binding α-subunit and a β-subunit/γ-subunit dimer, and are classified according to their α-subunit into four subfamilies: Gs, Gi, Gq, and G12. The G12 subfamily, which is comprised of the α-subunits Gα12 and Gα13, has been implicated in such cellular processes as Rho-dependent cytoskeletal shape changes, activation of c-Jun N-terminal kinase, and stimulation of Na+/H+ exchange (3). In addition, there is substantial evidence that G12 proteins mediate signaling pathways involved in cell growth and tumorigenesis (4–6). Whereas G12 proteins have been directly linked to regulatory molecules such as the Rho-directed guanine nucleotide exchange factor p115 (7, 8) and the GTPase-activating protein RasGAP1 (9), the significance of these interactions in relation to the developmental and oncogenic roles of G12 proteins has not been clearly defined.
Classical cadherins are plasma membrane glycoproteins that mediate a variety of cell–cell processes through binding to cadherins of adjacent cells (10, 11). Cadherins contain a single plasma membrane-spanning region near the C terminus; the majority of the protein extends out of the cell whereas a short C-terminal region extends into the cytoplasm. Although the extracellular region is the “business end” of the protein with regard to cell–cell adhesion, it has become clear that the cytoplasmic domain is also critical for the function of the protein. Cadherin-mediated cell adhesion depends on this region binding to key cytoplasmic proteins that link the cadherin to the actin cytoskeleton. Among these proteins, β-catenin has been revealed as an important multifunctional signaling component in cells (12, 13). Indeed, interaction with β-catenin is required for most, if not all, of the effects of cadherin on cell biology (10). Through mechanisms not well understood, β-catenin can dissociate from cadherins and migrate from the plasma membrane through the cytoplasm and to the nucleus, where it participates in transcriptional regulation.
Cytoplasmic β-catenin is bound by a protein complex that includes the adenomatous polyposis coli (APC) protein, and this association results in degradation of β-catenin via the ubiquitin/proteasome pathway. The extracellular Wingless/Wnt ligand can trigger a signaling pathway that blocks this degradation, causing accumulation of β-catenin that leads to its nuclear entry, where it complexes with lymphoid enhancer-binding factor to activate key developmental patterning and growth regulatory genes (14, 15). Mutations in APC also can result in stabilization and nuclear accumulation of β-catenin, as can mutations at key Ser and Thr residues in β-catenin itself (12, 16), thus leading to activation of transforming genes (11). Most colorectal carcinomas and adenomas harbor mutations in APC, and the resulting β-catenin accumulation is thought to be the primary transforming agent in these tumors (16).
In this paper, we describe an activation-dependent interaction between G12 proteins and the cytoplasmic region of cadherins. We present evidence that this interaction results in release of β-catenin from cadherins, causing it to translocate to the cytoplasm and nucleus. We further show that, in cells lacking APC, β-catenin-mediated transcriptional activation is up-regulated by expression of activated Gα12 or Gα13. These findings provide a molecular mechanism for the transforming ability of G12 proteins and demonstrate a link between heterotrimeric G proteins and signaling pathways that mediate cell growth and differentiation.
Materials and Methods
Materials.
Saccharomyces cerevisiae strain Y190 and pAS2 plasmid were from Stephen Elledge (Baylor College of Medicine, Houston, TX). The cDNAs for Gα12, Gα12Q229L, Gα13Q226L, and GαzQ205L were from Henry Bourne (University of California, San Francisco), and cDNAs for Gαi2QL and GαsQL were from Patrick Burgon (Harvard University, Boston). The cDNA for epithelial cadherin (E-cadherin) was from David Rimm (Yale University, New Haven, CT). Anti-Gα12 antibodies were from Tom Gettys (Medical University of South Carolina, Charleston) and Tohru Kozasa (University of Illinois, Chicago), anti-Gα13 antibody was purchased from Santa Cruz Biotechnology, and anti-E-cadherin and anti β-catenin antibodies were purchased from Zymed. Gα12 baculovirus construct was provided by Alfred Gilman (University of Texas Southwestern Medical Center, Dallas).
Yeast Two-Hybrid Screening.
The Gα12 two-hybrid bait was constructed by subcloning the cDNA for Gα12QL into pAS2. General procedures for yeast transformation and two-hybrid screening methodology have been described (17, 18). A human brain library in the vector pACT2 (CLONTECH) was screened, plates were incubated at 24°C for 4–7 days, and colonies of >1-mm diameter were assayed for β-galactosidase activity. Plasmids were isolated from positive-scoring colonies and sequenced.
Production of G Proteins.
Gα12 and Gαz were produced in Sf9 cells as described (19, 20). Gαs was produced in Escherichia coli (21) and Gβγ was purified from bovine brain (20). In vitro transcription/translation of Gα13QL and Gαi2QL was performed by using a coupled reticulocyte lysate system (Promega) according to the manufacturer's instructions.
In Vitro Protein-Binding Studies.
C-terminal domains of cadherin cDNAs were amplified by PCR, subcloned into pGEX-5X-1 (Amersham Pharmacia), and verified by sequencing. Fusion proteins were produced in E. coli strain BL21-DE3 and isolated from cell lysates by using glutathione Sepharose 4B (Amersham Pharmacia). Glutathione Sepharose was washed with 10 mM Hepes (pH 8.0)/1 mM DTT/150 mM NaCl and stored in the same buffer.
Approximately 60 pmol of purified Gα12 (see above) was incubated with either 10 μM GTPγS or 10 μM GDP for 2 h at 30°C in 50 mM Hepes (pH 8.0)/1 mM EDTA/3 mM DTT/0.05% polyoxyethylene 10 lauryl ether containing 10 mM MgSO4. Reactions were divided and then incubated with glutathione Sepharose-bound glutathione-S-transferase (GST)-cadherins for 2 h at 4°C with gentle agitation. Glutathione Sepharose was pelleted, washed with 50 mM Hepes, pH 8.0/1 mM EDTA/3 mM DTT/0.05% polyoxyethylene 10 lauryl ether/10 mM MgSO4, and immunoblotted for Gα12. For in vitro transcribed/translated proteins, equal amounts of radiolabeled proteins were incubated with GST-cadherins in the same manner as above, and precipitated material was separated by SDS/PAGE and analyzed by autoradiography.
For studies using radioactive guanine nucleotides, Gα12 was incubated with either 10 μM [35S]GTPγS or 10 μM [32P]GDP (specific activity 10,000–20,000 cpm/pmol) for 2 h at 30°C in 50 mM Hepes, pH 8.0/1 mM EDTA/3 mM DTT/0.05%polyoxyethylene 10 lauryl ether/5 mM MgSO4 and was subsequently purified by gel filtration through G-50 Sephadex equilibrated in the same buffer. Radiolabeled G protein (0.5–2 pmol) was incubated with glutathione Sepharose-bound GST-cadherin fusion proteins for 2 h at 4°C in the same buffer. Glutathione Sepharose was pelleted, washed three times, and then resuspended in the same buffer, and this suspension was assayed by scintillation counting along with the supernatant and washes. Assays with Gαs and Gαz were performed in the same fashion, except that Gαs was incubated with nucleotide for 20 min, and Gαz was incubated with nucleotide in 50 mM Hepes, pH 8.0/5 mM EDTA/3 mM DTT/0.05% polyoxyethylene 10 lauryl ether/0.75 mM MgSO4. [32P]GDP was prepared from α-[32P]GTP (NEN; specific activity 3,000 Ci/mmol) as described (21).
Cell Culture and Indirect Immunofluorescence.
HEK293 cells were maintained in DMEM containing 10% FBS, and SW480 cells were maintained in Leibovitz's L-15 medium containing 10% FBS. SW480 cells stably expressing E-cadherin were selected in the same medium supplemented with 200 μg/ml hygromycin B, and clonal populations were isolated and maintained in the same medium containing 100 μg/ml hygromycin B. Transfections were performed by using Lipofectamine (Life Technologies, Grand Island, NY) according to the manufacturer's instructions.
For immunofluorescence, cells grown on glass cover slips were washed in PBS, fixed in 4% paraformaldehyde for 10 min, washed twice in PBS, and then incubated 15 min in blocking buffer (PBS containing 10% calf serum/0.2% saponin). Primary antibodies were diluted 1:100 in blocking buffer and cover slips were incubated in this solution for 1 h before washing three times in PBS containing 10% calf serum. Fluorophore-conjugated secondary antibody (Molecular Probes) was diluted 1:100 in blocking buffer and cover slips were incubated in this solution for 1 h before washing three times in PBS containing 10% calf serum and once in PBS. Cover slips were mounted on SlowFade reagent (Molecular Probes) and visualized with an Axioskop fluorescence microscope (Zeiss).
Transcriptional Activation Assays.
Cells were transfected with TOPFLASH or FOPFLASH (Upstate Biotechnology, Lake Placid, NY) and pRL-TK (Promega), along with additional plasmids as indicated. After 48 h, cells were washed with PBS, lysed in passive lysis buffer (Promega), cleared by centrifugation, and assayed for protein. Lysates were analyzed by using a dual luciferase assay system (Promega), in which lysates were first assayed for firefly (TOPFLASH or FOPFLASH) luciferase activity, and then the reactions were quenched and immediately reassayed for Renilla (pRL-TK) luciferase activity. Luminometry readings were performed by using a Lumat LB-9501 luminometer (Berthold, Nashua, NH).
Results
Activated G12 Proteins Bind to the Cytoplasmic Region of Cadherins.
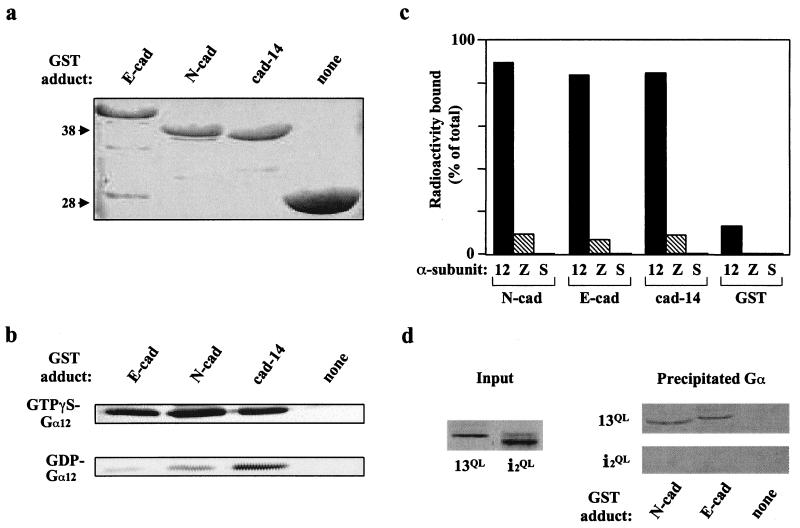
The yeast two-hybrid system was used to screen a human brain cDNA library for proteins interacting with a mutationally activated variant (Q229L) of Gα12 (22). Attaining levels of Gα12QL “bait” expression in yeast that were adequate for this screen required use of a lowered growth temperature throughout the procedure (17). From 5 × 106 independent clones in the library, an average of 4.3 × 106 transformants were screened in three separate rounds. These three screens yielded a total of 14 potentially positive Gα12 interactors, one of which was identified as the cytoplasmic C-terminal 98 residues of cadherin-14. The Gα12–cadherin interaction was characterized in a cell-free system by using fusions of the C-terminal 98 residues of cadherin-14, as well as the corresponding C-terminal regions of E-cadherin and neural cadherin (N-cadherin), to GST (Fig. 1a). Purified Gα12 was loaded with either GTPγS or GDP and then applied to immobilized GST-cadherin proteins. Whereas the cadherin-14 tail bound similar amounts of GTPγS-bound and GDP-bound Gα12, the E-cadherin and N-cadherin tails bound Gα12 in an activation-specific manner, with both proteins precipitating a much greater percentage of GTPγS-loaded Gα12 compared with GDP-loaded Gα12 (Fig. 1b).
Figure 1.
Interaction between G12 proteins and the cytoplasmic region of cadherins. (a) Production of GST-fusion proteins of the C-terminal 98 amino acids of cadherins E, N, and 14, as well as GST alone. Coomassie blue staining of bacterially produced proteins is shown; molecular mass standards (in kDa) are indicated at left. (b) Immunoblot of GTPγS-loaded Gα12 and GDP-loaded Gα12 that was precipitated by the indicated GST-cadherin fusion proteins. (c) Quantitation of 35S-GTPγS-labeled Gα12, Gαz, and Gαs that was precipitated by the indicated GST-cadherin fusion proteins. Precipitated radioactivity is presented as a percentage of the total radioactivity for each sample. For each Gα protein, total radioactivity was typically 8,000–10,000 cpm, with a specific activity of roughly 10,000 cpm/pmol. (d) Autoradiographs of mutationally activated Gα proteins produced by in vitro transcription/translation (Input), and of these proteins after precipitation with the indicated GST-cadherin fusion proteins (Precipitated Gα).
Because GTPγS-binding stoichiometry for Gα12 is typically less than 30% (ref. 19; data not shown), the GTPγS-liganded Gα12 in the above experiments presumably contained a significant amount of the GDP-bound form. Therefore, to more accurately gauge the activation dependence of the Gα12–cadherin interaction, we loaded Gα12 with radiolabeled GTPγS and repeated the GST “pull-downs” with results quantitated by the amount of precipitated radioactivity. All three GST-cadherin fusion proteins precipitated GTPγS-liganded Gα12, but none precipitated GTPγS-liganded Gαz or GTPγS-liganded Gαs (Fig. 1c). When Gα12 was loaded with radioactive GDP, a modest affinity for GST-cadherin proteins was observed; however, preincubation of GDP-liganded Gα12 with purified Gβγ decreased this affinity to nearly background levels (data not shown), suggesting that the heterotrimeric form of Gα12 does not bind to the cadherin cytoplasmic domain.
To determine whether the specific affinity for cadherin cytoplasmic regions seen for Gα12 is shared by Gα13, we used the cDNA for mutationally activated (Q226L) Gα13 to produce the radiolabeled protein in an in vitro transcription/translation system, and applied this protein to immobilized GST-cadherin fusion proteins. Gα13QL was precipitated by fusions of E- or N-cadherin, but not by GST alone (Fig. 1d). In contrast, mutationally activated Gαi2 produced in the same fashion (Gαi2QL) was not precipitated by cadherins (Fig. 1d).
Expression of Activated G12 Proteins Causes Relocalization of β-Catenin.
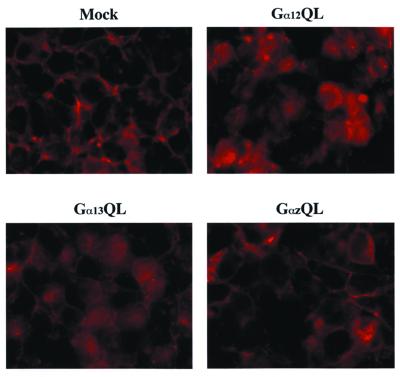
An intriguing possibility arising from the observed G12–cadherin interaction was that G12 proteins could displace β-catenin from cadherins, thereby leading to activation of β-catenin-mediated transcriptional responses. To address this hypothesis, we examined whether expression of mutationally activated G12 proteins altered the subcellular distribution of β-catenin. In mock-transfected HEK293 cells, β-catenin was localized almost exclusively at the plasma membrane and appeared particularly concentrated at areas of cell–cell contact (Fig. 2). After transfection with either Gα12QL or Gα13QL, a marked shift from this peripheral staining to a more diffuse, uniform staining was observed in many cells. This effect was specific to Gα12QL and Gα13QL, because expression of mutationally activated Gαz (GαzQL) did not trigger this shift in β-catenin distribution (Fig. 2).
Figure 2.
Gα12 and Gα13 induce β-catenin release from a plasma membrane localization. HEK293 cells were transfected with either empty vector or constructs encoding the indicated mutationally activated Gα proteins. β-catenin subcellular distribution was analyzed by indirect immunofluorescence 48 h posttransfection.
Expression of G12 Proteins Up-Regulates β-Catenin-Mediated Transcriptional Activation.
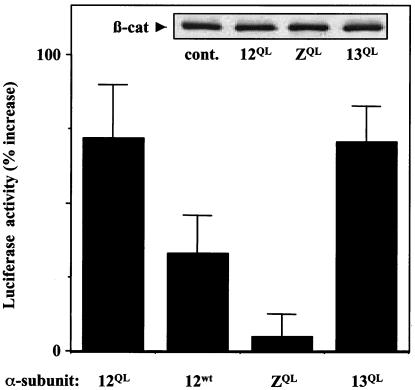
SW480 human colon carcinoma cells lack functional APC, and thus β-catenin degradation is impaired in these cells (23). We reasoned that in SW480 cells, G12-induced release of β-catenin from the plasma membrane might lead to up-regulation of β-catenin/lymphoid enhancer-binding factor-mediated transcriptional activation. To test this, we used the plasmid TOPFLASH, which contains a luciferase reporter gene positioned downstream of several lymphoid enhancer-binding factor-binding sites. Expression of Gα12QL in SW480 cells caused a substantial (72 ± 18%) increase in nuclear β-catenin activity; this effect was mirrored by Gα13QL and to a lesser degree by wild-type Gα12, but not by GαzQL (Fig. 3). Immunoblot analysis showed that total β-catenin levels in SW480 cells were unchanged by expression of these mutationally activated G proteins (Fig. 3 Inset), and that GαzQL was expressed at levels equal to or greater than Gα12QL in these cells (data not shown). Furthermore, expression of neither Gαi2QL nor GαsQL caused increased nuclear β-catenin activity in SW480 cells (data not shown). The plasmid FOPFLASH, in which TOPFLASH has been mutated to abolish lymphoid enhancer-binding factor/β-catenin binding, produced a signal only 3–5% of the TOPFLASH-generated signal in all plasmid combinations tested here (data not shown).
Figure 3.
Gα12 and Gα13 up-regulate β-catenin-mediated transcriptional activation. SW480 cells were transfected with TOPFLASH, pRL-TK, and the indicated mutationally activated (QL) or wild-type (wt) Gα protein. Empty vector was used to adjust samples to an equal amount of total DNA for all transfections. Cell lysates were normalized for protein and were analyzed for firefly luciferase (TOPFLASH) activity and Renilla luciferase (pRL-TK) activity as an internal standard for transfection efficiency. TOPFLASH activity was normalized for pRL-TK activity in each sample and is shown here as a percent increase above normalized TOPFLASH activity in cells not transfected with Gα protein. (Inset) For transfections with mutationally activated Gα proteins (QL) and with no Gα protein (cont.), whole-cell lysates were normalized for protein and then analyzed by immunoblot for endogenous β-catenin (β-cat) levels.
We also tested cells harboring functional APC for G12-induced up-regulation of β-catenin-mediated transcriptional activation. Rat-1 cells showed a very slight increase in TOPFLASH luciferase activity in response to expression of Gα12QL or Gα13QL, whereas HEK293 cells showed only negligible changes in activity under the same conditions (data not shown). As expected of cells competent in β-catenin degradation, basal levels of β-catenin-responsive reporter activity in these cells were at least 10-fold lower than the levels observed in SW480 cells. These results suggest that the pathway for β-catenin proteolysis must be compromised in cells (e.g., loss of APC function) for G12 proteins to trigger an increase in nuclear β-catenin levels.
G12 Proteins Destabilize β-Catenin Binding to Cadherins.
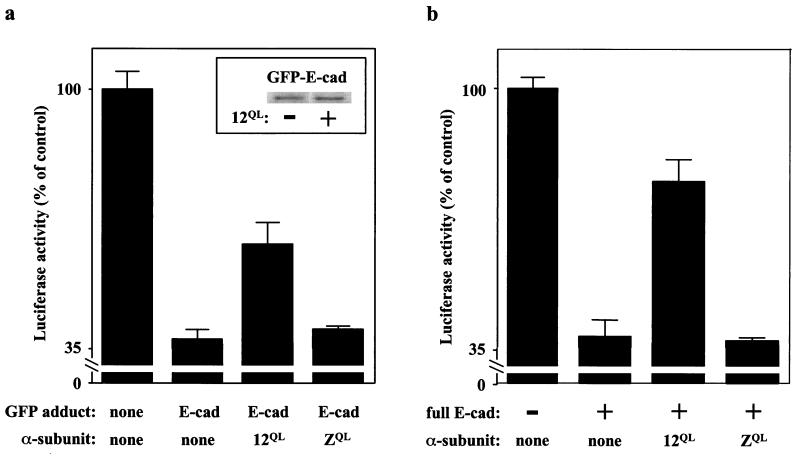
To provide further evidence that G12 proteins can affect β-catenin/cadherin association in vivo, we took advantage of the fact that β-catenin-mediated transcriptional activation can be attenuated by expression of the C-terminal domain of cadherin, which can bind β-catenin and block its interaction with regulatory molecules (24). The cytoplasmic C-terminal 98 residues of E-cadherin were fused to the C terminus of green fluorescent protein (GFP), and this construct was transfected into SW480 cells along with the TOPFLASH reporter plasmid. As expected, expression of GFP-E-cadherin caused a substantial decrease in β-catenin-mediated nuclear activity (Fig. 4a). To test whether Gα12 could free β-catenin from this cadherin-bound state, Gα12QL was coexpressed along with GFP-E-cadherin. Expression of Gα12QL markedly attenuated the cadherin-induced inhibition of β-catenin-mediated nuclear activity, whereas expression of GαzQL was without effect (Fig. 4a). Importantly, expression of Gα12QL did not hinder expression of GFP-E-cadherin (Fig. 4a Inset). We also produced stable transfectants of full-length E-cadherin in SW480 cells. Mirroring the effect observed in SW480 cells transiently expressing the E-cadherin tail (Fig. 4a), β-catenin-mediated nuclear activity was significantly lower in SW480 cells stably expressing E-cadherin compared with SW480 cells, and expression of Gα12QL partially reversed this inhibition; expression of GαzQL in these cells was without effect (Fig. 4b).
Figure 4.
Gα12 disrupts cadherin/β-catenin association in cells. (a) Effect in transiently transfected cells. SW480 cells were transfected with TOPFLASH, pRL-TK, and the indicated combinations of GFP fusion protein and mutationally activated (QL) G protein. Normalized TOPFLASH activity was calculated for each sample (Fig. 3) and is presented as a percent of normalized TOPFLASH activity in cells transfected with GFP alone. (Inset) Levels of GFP-E-cadherin in SW480 cells, with and without coexpressed Gα12QL, were determined by immunoblot analysis. (b) Effect in stably transfected cells. SW480 cells stably expressing full-length E-cadherin were transfected with TOPFLASH, pRL-TK, and the indicated mutationally activated (QL) Gα proteins. Normalized TOPFLASH activity for each sample is presented as a percent of normalized TOPFLASH activity in SW480 cells not transfected with Gα protein.
Discussion
Dissociation of β-catenin from the adherens junction is not a well-understood process. Tyrosine phosphorylation of β-catenin has been implicated in this process (25, 26), as has a protein termed IQGAP1 in a related process of α-catenin dissociation (27). However, a direct mechanism for release of β-catenin from cadherins has not been demonstrated. Our finding that Gα12 and Gα13 interact with cadherins in a fashion that triggers release of β-catenin provides such a mechanism and reveals a link between heterotrimeric G proteins and cellular processes controlling growth and development. In addition, these findings provide a possible molecular mechanism for the previously reported transforming ability of G12 proteins.
The interaction of G12 proteins with E-cadherin may be particularly significant with regard to oncogenesis. Many gastric and breast cancers harbor E-cadherin mutations (11), and most human epithelial cancers lose E-cadherin function at some stage of their development (28). Also, introduction of E-cadherin into tumor cells was found to restrict cell proliferation and block the progression from adenoma to invasive carcinoma in a mouse model system (29). A protein such as Gα12 or Gα13 that destabilizes the E-cadherin–β-catenin interaction could potentially effect a cancerous phenotype by (i) disrupting E-cadherin linkage to the cytoskeleton, and thus disrupting E-cadherin-dependent cell–cell adhesion, or (ii) increasing the pool of free β-catenin in the cell, allowing up-regulation of β-catenin-mediated transcriptional events. Thus, if E-cadherin acts as a tumor suppressor by sequestering β-catenin at the plasma membrane, a mutationally activated G12 protein could act as an oncogene by constitutively triggering β-catenin release from E-cadherin. According to this model, it is possible that mutationally activated Gα12 or Gα13 could act in concert with a mutation that hinders β-catenin degradation (e.g., an APC mutation) to trigger cellular transformation. Indeed, our observation that G12 proteins up-regulate β-catenin-mediated transcriptional activation in an APC−/− cell line, but not appreciably so in cells harboring the functional β-catenin proteolytic machinery, suggests that G12 proteins require additional mutations in cells to participate in oncogenesis.
A noteworthy observation in our study was the differing ability of the type I cadherins (N-cadherin and E-cadherin) and type II cadherin (cadherin-14) in discriminating between the active and inactive forms of Gα12. Unlike type I cadherins, type II cadherins have not been well characterized, although initial studies suggest that type II cadherins mediate cell–cell binding interactions similar to the type I cadherins (30). Within a 72-aa region of the cytoplasmic tail, cadherin-14 is only 57% identical to N-cadherin and 54% identical to E-cadherin; by comparison, N-cadherin is 74% identical to E-cadherin within a contiguous 89-aa region of the cytoplasmic tail. Mutagenesis studies based on differences between these type I cadherins and cadherin-14 may reveal key determinants of the activation dependence of Gα12 interaction with N- and E-cadherin.
Although results of disruption of Gα12 in mice are not yet available, mice lacking Gα13 are impaired in angiogenesis and die at day 10.5 of embryogenesis (31). Interestingly, mice homozygous for a truncation in the cytoplasmic region of vascular endothelial-cadherin also exhibit angiogenesis defects (32). An intriguing possibility is that interactions between G12 proteins and cadherins are involved in critical processes of early development. Perhaps also noteworthy in this regard is that mice lacking β-catenin are defective in gastrulation (33), whereas Drosophila lacking the Gα12/Gα13 orthologue Concertina also fail to undergo gastrulation (34). Further elucidation of the relationship between the G12 proteins and the cadherin–β-catenin complex may provide a more clear understanding of key events in early development.
Acknowledgments
We thank Henry Bourne, Tom Gettys, David Rimm, Al Gilman, Tohru Kozasa, Patrick Burgon, and Stephen Elledge for providing reagents; Bill Tschantz, Jim Otto, Dan Kaplan, and Pat Fields for helpful discussions; and Ying Chen, Michael Datto, and Chandra Theesfeld for advice on technical aspects. This work was supported by grants from the National Institutes of Health (GM 55717) and the Pardee Foundation (to P.J.C.), and the GlaxoWellcome-Duke University Collaborative Research Program in Cancer. T.E.M. was supported by a National Institutes of Health postdoctoral fellowship.
Abbreviations
- APC
adenomatous polyposis coli
- E-cadherin
epithelial cadherin
- N-cadherin
neural cadherin
- GST
glutathione-S-transferase
- GFP
green fluorescent protein
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.021350998.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.021350998
References
- 1.Hamm H E. J Biol Chem. 1998;273:669–672. doi: 10.1074/jbc.273.2.669. [DOI] [PubMed] [Google Scholar]
- 2.Iiri T, Farfel Z, Bourne H R. Nature (London) 1998;394:35–38. doi: 10.1038/27831. [DOI] [PubMed] [Google Scholar]
- 3.Dhanasekaran N, Dermott J M. Cell Signalling. 1996;8:235–245. doi: 10.1016/0898-6568(96)00048-4. [DOI] [PubMed] [Google Scholar]
- 4.Chan A M, Fleming T P, McGovern E S, Chedid M, Miki T, Aaronson S A. Mol Cell Biol. 1993;13:762–768. doi: 10.1128/mcb.13.2.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang H, Wu D, Simon M I. FEBS Lett. 1993;3:319–322. doi: 10.1016/0014-5793(93)80896-3. [DOI] [PubMed] [Google Scholar]
- 6.Jones T L, Gutkind J S. Biochemistry. 1998;37:3196–3202. doi: 10.1021/bi972253j. [DOI] [PubMed] [Google Scholar]
- 7.Kozasa T, Jiang X, Hart M J, Sternweis P M, Singer W D, Gilman A G, Bollag G, Sternweis P C. Science. 1998;280:2109–2111. doi: 10.1126/science.280.5372.2109. [DOI] [PubMed] [Google Scholar]
- 8.Hart M J, Jiang X, Kozasa T, Roscoe W, Singer W D, Gilman A G, Sternweis P C, Bollag G. Science. 1998;280:2112–2114. doi: 10.1126/science.280.5372.2112. [DOI] [PubMed] [Google Scholar]
- 9.Jiang Y, Ma W, Wan Y, Kozasa T, Hattori S, Huang X-Y. Nature (London) 1998;395:808–813. doi: 10.1038/27454. [DOI] [PubMed] [Google Scholar]
- 10.Barth A I M, Nathke I S, Nelson W J. Curr Opin Cell Biol. 1997;9:683–690. doi: 10.1016/s0955-0674(97)80122-6. [DOI] [PubMed] [Google Scholar]
- 11.Behrens J. Cancer Metastasis Rev. 1999;18:15–30. doi: 10.1023/a:1006200102166. [DOI] [PubMed] [Google Scholar]
- 12.Willert K, Nusse R. Curr Opin Genet Dev. 1998;8:95–102. doi: 10.1016/s0959-437x(98)80068-3. [DOI] [PubMed] [Google Scholar]
- 13.Bullions L C, Levine A J. Curr Opin Oncol. 1998;10:81–87. doi: 10.1097/00001622-199801000-00013. [DOI] [PubMed] [Google Scholar]
- 14.Eastman Q, Grosschedl R. Curr Opin Cell Biol. 1999;11:233–240. doi: 10.1016/s0955-0674(99)80031-3. [DOI] [PubMed] [Google Scholar]
- 15.Peifer M, Polakis P. Science. 2000;287:1606–1609. doi: 10.1126/science.287.5458.1606. [DOI] [PubMed] [Google Scholar]
- 16.Morin P J, Sparks A B, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler K W. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 17.Glick J L, Meigs T E, Casey P J. In: G Proteins: Techniques of Analysis. Manning D R, editor. Boca Raton, FL: CRC; 1999. pp. 59–75. [Google Scholar]
- 18.Bai C, Elledge S J. Methods Enzymol. 1997;283:141–156. doi: 10.1016/s0076-6879(97)83013-3. [DOI] [PubMed] [Google Scholar]
- 19.Kozasa T, Gilman A G. J Biol Chem. 1995;270:1734–1741. doi: 10.1074/jbc.270.4.1734. [DOI] [PubMed] [Google Scholar]
- 20.Fields T A, Casey P J. J Biol Chem. 1995;270:23119–23125. doi: 10.1074/jbc.270.39.23119. [DOI] [PubMed] [Google Scholar]
- 21.Graziano M P, Freissmuth M, Gilman A G. J Biol Chem. 1989;264:409–418. [PubMed] [Google Scholar]
- 22.Voyno-Yasenetskaya T, Conklin B R, Gilbert R L, Hooley R, Bourne H R, Barber D L. J Biol Chem. 1994;269:4721–4724. [PubMed] [Google Scholar]
- 23.Munemitsu S, Albert I, Souza B, Rubinfeld B, Polakis P. Proc Natl Acad Sci USA. 1995;92:3046–3050. doi: 10.1073/pnas.92.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sadot E, Simcha I, Shtutman M, Ben-Ze'ev A, Geiger B. Proc Natl Acad Sci USA. 1998;95:15339–15344. doi: 10.1073/pnas.95.26.15339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoschuetzky H, Aberle H, Kemler R. J Cell Biol. 1994;127:1375–1380. doi: 10.1083/jcb.127.5.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muller T, Choidas A, Reichmann E, Ullrich A. J Biol Chem. 1999;274:10173–10183. doi: 10.1074/jbc.274.15.10173. [DOI] [PubMed] [Google Scholar]
- 27.Kuroda S, Fukata M, Nakagawa M, Fujii K, Nakamura T, Ookubo T, Izawa I, Nagase T, Nomura N, Tani H, et al. Science. 1998;281:832–835. doi: 10.1126/science.281.5378.832. [DOI] [PubMed] [Google Scholar]
- 28.Christofori G, Semb H. Trends Biochem Sci. 1999;24:73–76. doi: 10.1016/s0968-0004(98)01343-7. [DOI] [PubMed] [Google Scholar]
- 29.Perl A-K, Wilgenbus P, Dahl U, Semb H, Christofori G. Nature (London) 1998;392:190–193. doi: 10.1038/32433. [DOI] [PubMed] [Google Scholar]
- 30.Shimoyama Y, Takeda H, Yoshihara S, Kitajima M, Hirohashi S. J Biol Chem. 1999;274:11987–11994. doi: 10.1074/jbc.274.17.11987. [DOI] [PubMed] [Google Scholar]
- 31.Offermanns S, Mancino V, Revel J-P, Simon M I. Science. 1997;275:533–536. doi: 10.1126/science.275.5299.533. [DOI] [PubMed] [Google Scholar]
- 32.Carmeliet P, Lampugnani M G, Moons L, Breviario F, Compernolle V, Bono F, Balconi G, Spagnuolo R, Oostuyse B, Dewerchin M, et al. Cell. 1999;98:147–157. doi: 10.1016/s0092-8674(00)81010-7. [DOI] [PubMed] [Google Scholar]
- 33.Haegel H, Larue L, Ohsugi M, Fedorov L, Herrenknecht K, Kemler R. Development (Cambridge, UK) 1995;121:3529–3537. doi: 10.1242/dev.121.11.3529. [DOI] [PubMed] [Google Scholar]
- 34.Parks S, Wieschaus E. Cell. 1991;64:447–458. doi: 10.1016/0092-8674(91)90652-f. [DOI] [PubMed] [Google Scholar]