Abstract
Resistance to proteolytic enzymes and heat is thought to be a prerequisite property of food allergens. Allergens from peanut (Arachis hypogaea) are the most frequent cause of fatal food allergic reactions. The allergenic 2S albumin Ara h 2 and the homologous minor allergen Ara h 6 were studied at the molecular level with regard to allergenic potency of native and protease-treated allergen. A high-resolution solution structure of the protease-resistant core of Ara h 6 was determined by NMR spectroscopy, and homology modelling was applied to generate an Ara h 2 structure. Ara h 2 appeared to be the more potent allergen, even though the two peanut allergens share substantial cross-reactivity. Both allergens contain cores that are highly resistant to proteolytic digestion and to temperatures of up to 100 °C. Even though IgE antibody-binding capacity was reduced by protease treatment, the mediator release from a functional equivalent of a mast cell or basophil, the humanized RBL (rat basophilic leukaemia) cell, demonstrated that this reduction in IgE antibody-binding capacity does not necessarily translate into reduced allergenic potency. Native Ara h 2 and Ara h 6 have virtually identical allergenic potency as compared with the allergens that were treated with digestive enzymes. The folds of the allergenic cores are virtually identical with each other and with the fold of the corresponding regions in the undigested proteins. The extreme immunological stability of the core structures of Ara h 2 and Ara h 6 provides an explanation for the persistence of the allergenic potency even after food processing.
Keywords: 2S albumin-type peanut allergen, allergy, Ara h 2, Ara h 6, food allergy, peanut (Arachis hypogaea)
Abbreviations: 2D, two-dimensional; 3D, three-dimensional; DTT, dithiothreitol; EAST, enzyme allergosorbent test; HSQC, heteronuclear single-quantum coherence; LDS, lithium dodecyl sulphate; NOE, nuclear Overhauser effect; nsLTP, non-specific lipid-transfer protein; prAra h 2/6, protease-resistant core of Ara h 2/6; rAra h 2/6, recombinant Ara h 2/6; RBL, rat basophilic leukaemia; rmsd, root mean square deviation
INTRODUCTION
Peanuts (Arachis hypogaea) are a major cause of severe IgE-mediated food allergy in children and adults. According to studies in Great Britain and the U.S.A., approx. 0.6% of the population is affected by peanut allergy [1,2]. Compared with other food allergies, peanut allergy is characterized by particularly severe symptoms. A large fraction of all food-induced anaphylactic reactions and of all lethal reactions as a result of food allergy are caused by peanuts [3]. Although the typical peanut dose causing allergic reactions is in the range 100–1000 mg, in some sensitive patients, very low doses of approx. 100 μg of peanut protein can trigger adverse reactions [4]. In contrast with many other food allergies such as milk, soya bean (Glycine max) and wheat (Triticum aestivum) allergies, peanut allergy is rarely outgrown after childhood and often shows lifelong persistence [5]. Specific immunotherapy, which is effective for several respiratory and insect venom allergies [6], is complicated by serious side effects in the case of peanut allergy [7]. Since no treatment to cure type I food aller-gies is yet available, avoidance is the only option for peanut-sensitive persons – an approach that is complicated by the wide-spread application of peanuts as food ingredients and by the omnipresence of peanut contamination in processed food.
To date, three major allergens, Ara h 1, Ara h 2 and Ara h 3 [8–10], and four minor allergens, Ara h 4, Ara h 5, Ara h 6 and Ara h 7 [11], have been identified in peanuts. With the exception of the profilin Ara h 5, these allergens are seed storage proteins. Very recently, Ara h 8, a homologue of the major birch (Betula alba) pollen allergen, Bet v 1, was identified as a major allergen in birch-pollen-allergic patients with concomitant peanut allergy [12]. The major allergens are responsible for IgE binding in 45–90% of peanut-sensitized individuals [8,10,13]. The sequence identity between Ara h 2 and Ara h 6 is 59%, and is 35% between Ara h 2 and Ara h 7. Both Ara h 2 and Ara h 6 contain a high proportion of nitrogen- and sulphur-containing amino acids (glutamine, arginine and cysteine). Ara h 2 is synthesized as a precursor protein with an additional N-terminal 21-residue signal sequence, typical for proteins transported from the cytoplasm to target organelles. A second isoform of Ara h 2, Ara h 2.02, is largely identical with Ara h 2, but possesses a 12-residue insert in the middle of the sequence [14]. In contrast with the thoroughly studied Ara h 2, little is yet known about Ara h 6 and Ara h 7. In Ara h 2, initially isolated from peanuts [8], ten linear IgE-binding peptides were identified that are located throughout the length of the amino acid sequence [13]. Three of these epitopes (amino acid positions 27–36, 57–66 and 65–74) appeared to be immuno-dominant, because peptides containing these epitopes bound IgE antibodies of all sera tested [13]. Single amino acid changes, especially in the centre of the epitopes, can result in loss of peanut-specific IgE binding [15].
In contrast with the majority of 2S albumins, Ara h 2 is not post-translationally modified, except for disulphide formation, and the protein occurs in peanut seeds as one single polypeptide chain without subunit structure. This special feature allowed the development of an Escherichia coli-based expression and purification strategy for the large-scale production of properly folded Ara h 2 [16]. The recombinant form of Ara h 6 (rAra h 6) including a hexahistidine tag was shown to be recognized by 38% of peanut allergic patient sera [17], but no information is available about linear or structural IgE-binding epitopes.
As members of the 2S albumin family, Ara h 2, Ara h 6 and Ara h 7 belong to the prolamine protein superfamily [18] that additionally contains nsLTPs (non-specific lipid-transfer proteins), α-amylase/trypsin inhibitors and puroindolines [19]. This superfamily is characterized by the conserved cysteine pattern CXnCXn-CCXnCXCXnCXnC. 2S albumins are proteins with a molecular mass of 12–15 kDa, generally containing eight or more disulphide-bridged cysteine residues. The physiological function of these proteins is still unclear, but a role as a nitrogen and sulphur donor was suggested on the basis of their amino acid composition, their high abundance in seeds and their mobilization during germination [20,21]. The majority of 2S albumins are synthesized as long precursor proteins that undergo one or more post-translational modification steps. Napins, for example, a group of rapeseed (Brassica napus) 2S albumins, are synthesized as 20 kDa precursors and are subsequently cleaved at four positions, resulting in two subunits of 4.5 kDa and 10 kDa that are linked by disulphide bonds [22–24].
Structural data on 2S albumins are available from a low-resolution solution structure of BnIb, a napin type 2S albumin from rape seeds [25], from the solution structure of RicC3, a storage protein from castor bean (Ricinus communis) [26], and from the solution structure of SFA-8 from sunflower (Helianthus annuus) seeds [27]. Crystal or solution structures are available for members of the related class of bifunctional inhibitors and nsLTPs: RBI [ragi bifunctional inhibitor: trypsin/α-amylase inhibitor from ragi or Indian finger millet (Eleusine coracana)]; [28], α-amylase inhibitor from wheat [29], HPS (hydrophobic protein from soya bean) [30] and nsLTP from wheat [31], maize (Zea mays) [32,33], barley (Hordeum vulgare) [34], rice (Oryza sativa) [35,36] and onion (Allium cepa) [37]. The sequence identity of less than 17–20% between 2S albumins and nsLTPs renders homology modelling difficult. The known structures of the 2S albumins, the napins and the nsLTPs identified these proteins as belonging to the class of all-α proteins, differing only in the α-helix arrangement in their compact hydrophobic cores.
Despite the fact that many food allergens or stable allergen fragments have been shown to resist conditions of the gastrointestinal tract, little is known about the structures that survive digestion and thus have the potential to sensitize the immune system.
Features such as post-translational modifications or stability have turned out to be important for the severity of the allergic reaction or cross-reactivity [38–40], and the protein tertiary structure plays a prominent role for the molecular mechanisms of an allergic reaction, i.e. activation of sensitized basophils. In fact, thorough knowledge of the structure of IgE-binding epitopes seems to be key to the development of hypoallergenic variants of use as vaccines for specific immunotherapy [41]. In the present study, we solved a high-resolution structure of a protease-stable core fragment of the 2S albumin-type food allergen Ara h 6 and demonstrated that partial digestion of the allergen does not abolish the allergenic potential of either Ara h 2 or Ara h 6.
EXPERIMENTAL
Expression and purification of Ara h 6 and Ara h 2
Ara h 6 (GenBank® accession number AY871100) and Ara h 2 (GenBank® accession number AAM78596) were expressed as thioredoxin-fusion proteins in E. coli Origami cells (DE3; Novagen) and were purified as described for Ara h 2 [16]. For uniform (>95%) 15N and 13C/15N labelling, rAra h 6 was isolated and purified from E. coli cultures grown in M9 minimal medium enriched with 15NH4Cl or 15NH4Cl/[13C]glucose using the identical purification protocol. 15N-labelled leucine (Novagen) was added to the medium according to the manufacturer's instructions. Natural Ara h 2 was purified as described in [16].
Proteolytic digestion
Proteolytic reactions with trypsin and chymotrypsin were performed in 50 mM Tris/HCl, pH 8.0, over a period of 2–4 h at 37 °C using 1 μM enzyme for 50 μM protein. The reaction was stopped by adding SDS- and DTT (dithiothreitol)-containing buffers and incubation at 95 °C for 10 min. The fragments were subsequently purified using size-exclusion chromatography or reversed-phase chromatography.
CD spectroscopy
CD studies were performed on a Jasco 600 spectropolarimeter equipped with a CDF-426S Peltier temperature-control system interfaced with a Julabo F200 water bath. Far-UV CD spectra were recorded at 20 and 100 °C in a 1-mm-pathlength quartz cuvette (200 μl) (Hellma) at protein concentrations of 5–10 μM with and without 1 mM DTT. Spectra were recorded with a scanning speed of 20 nm/min, and control buffer spectra were subtracted. Thermal denaturation was monitored by recording the [θ]222 (molar ellipticity at a fixed wavelength of 222 nm) while heating or cooling at 1 °C/min. Measurements were performed in 1-cm- or 1-mm-pathlength quartz cuvettes at protein concentrations of 0.8–1.5 μM and 15–20 μM with and without 0.5 mM DTT. All samples were dissolved in 10 mM potassium phosphate, pH 7.0. The results are expressed as mean residue weight (MRW) molar ellipticity [θ]MRW=θ/(c×d×N), where θ is the observed ellipticity, c is the protein concentration, d is the optical pathlength and N is the number of amino acid residues.
Gel electrophoresis
Protein purity and apparent molecular mass were routinely monitored by SDS/PAGE using 19% polyacrylamide gels. Samples were diluted with Roti-Load 1 reducing sample buffer (Roth) and were denatured for 5–10 min at 95 °C. Commercial 10% and 12% NuPAGE® Bis-Tris polyacrylamide gels (Invitrogen) were used to analyse proteolytic products. Samples were diluted in NuPAGE® LDS (lithium dodecyl sulphate) sample buffer [10% (w/v) glycerol, 141 mM Tris base, 106 mM Tris/HCl, 2% (w/v) LDS, 0.51 mM EDTA, 0.22 mM SERVA Blue G250, 0.175 mM Phenol Red] in the presence or absence of 2.5 mg/ml DTT respectively and denatured as described.
N-terminal sequencing
Digested and undigested Ara h 2 and Ara h 6 were subjected to SDS/PAGE and transferred on to a PVDF membrane by semidry blotting. Protein bands were stained with Coomassie Brilliant Blue, excised, and N-terminally sequenced by the Edman degradation method.
NMR spectroscopy
Uniformly 15N or 15N/13C-labelled Ara h 6 samples in 50 mM potassium phosphate buffer, pH 7.0, in 1H2O/2H2O (9:1) were used for structure determination. All NMR spectra were acquired on Bruker DRX 600 MHz and DMX 750 MHz spectrometers with pulsed-field gradient capabilities at 25 °C. In addition to the triple-resonance experiments described previously [42], the following set of heteronuclear edited NOESY experiments were conducted in order to obtain distance restraints: 3D (three-dimensional) 15N-NOESY-HSQC (heteronuclear single-quantum coherence) (120 ms mixing time), 3D 13C-NOESY-HSQC (120 ms mixing time), 3D 15N-HMQC-NOESY-HSQC (150 ms mixing time), 3D 13C/13C-HMQC-NOESY-HSQC (120 ms mixing time), 3D 13C/15N-HMQC-NOESY-HSQC (120 ms mixing time), 2D (two-dimensional) [1H,1H] aromatic 13C-edited NOESY and 13C-filtered 2D NOESY. Scalar 3JHNHA coupling constants were obtained using the HNHA experiment. Slowly exchanging amide protons were identified from a series of 15N-HSQC spectra recorded after the sample freeze-dried from 1H2O had been dissolved in 2H2O. {1H}15N NOE (nuclear Overhauser effect) values were measured using a standard pulse sequence [43] with a relaxation delay of 6 s, for proton saturation a train of 120 °C high-power pulses was applied for the final 3 s of the relaxation delay. DNH residual dipolar couplings were obtained from a series of J-modulated HSQC spectra of uniformly 15N-labelled prAra h 6 (protease-resistant core of Ara h 6) weakly aligned by addition of 40 mg/ml Pf1 filamentous phage [44]; the 1JNH scalar coupling contribution as observed in reference spectra of the unaligned sample was subtracted. The NMR data were processed using in-house-written software and analysed with the program packages NMRView [45] and NDEE (SpinUp, Dortmund, Germany).
Structure calculation
On the basis of the assignment of the 1H, 13C and 15N resonances of prAra h 6, a total of 384 interresidual distance restraints could be derived from the 2D- and 3D-NOESY spectra in an iterative procedure. NOE cross-peaks were manually classified as strong, medium or weak according to their intensities and were converted into distance restraints d<2.7 Å (1 Å=0.1 nm), d<3.5 Å, and d<5.0 Å, respectively. Of the 36 measured 3JHNHA scalar coupling constants [42], 23 were smaller than 6.0 Hz, indicating backbone torsion angles between −80° and −40°. A hydrogen bond was postulated if the acceptor of a slowly exchanging amide proton could be identified from the results of preliminary structure calculations. For the 22 thus derived hydrogen bonds, the distance between the amide proton and the acceptor was restrained to d<2.3 Å, and the distance between the amide nitrogen and the acceptor to d<3.3 Å. In addition to the four canonical cystines of 2S albumins, a disulphide bond between the additional two cysteines was suggested by the results of a standard Ellman reaction. For all disulphide bonds, an S–S distance restraint of d=2.02 Å was included. These experimental restraints served as inputs for the calculation of 60–320 structures using restrained molecular dynamics with X-PLOR 3.851 (http://xplor.csb.yale.edu/xplor/). To this end, a three-stage simulated annealing protocol was carried out as described in [47], with the following modifications. For conformational space sampling 40 ps of molecular dynamics with 2 fs time steps were simulated at a temperature of 2000 K, followed by 30 ps of slow cooling to 1000 K and 15 ps of cooling to 100 K, both with a time step of 1 fs. A conformational database term for both backbone and side-chain dihedral angles [48] was included in the target function in order to improve the stereochemical properties of the structures. After simulated annealing the structures were subjected to 200 steps of Powell minimization of the full target function, followed by 1000 steps without recourse to the conformational database potential.
Of the 320 structures resulting from the final round of structure calculation, the 14 structures with the lowest overall energies and fewest experimental restraint violations were selected for refinement using additional restraints from residual dipolar couplings. Refinement comprised simulated annealing with a harmonic X-PLOR potential energy term for the dipolar couplings included in the target function of the simulated annealing protocol during the cooling stages. The axial and rhombic alignment tensor components were estimated from the distribution of the residual dipolar couplings. The force constant of the alignment tensor was increased gradually from 0.01 to 1.0 kcal·mol−1·Hz−2 (1 kcal =4.184 kJ). After refinement, the 14 structures with the lowest overall energies and restraint violations were used for further characterization. Structures were visualized with MOLMOL [49] and VMD [50]. The structure was deposited in the PDB under accession number 1W2Q.
Homology modelling
For the homology model of Ara h 2 based on the experimental structure of prAra h 6, the programs Swiss-Pdb Viewer [51] and Swiss-Model (http://www.expasy.ch/swissmod/SWISS-MODEL.html) were used. The modelled structure was energy-minimized (1000 steps, conjugate gradient) using SYBYL version 6.5 (Tripos, St. Louis, MO, U.S.A.).
Human sera
Sera were drawn from ten peanut-allergic patients who showed IgE binding to Ara h 2 on peanut extract immunoblots [52]. Serum from a non-allergic individual was used as a negative control. Sera were collected from patients after giving informed consent, at the Medical Hospital Borstel, Borstel, Germany.
EAST (enzyme allergosorbent test) and EAST inhibition
rAra h 2 and rAra h 6 were coupled to CNBr-activated paper discs (Schleicher & Schuell) at 0.5 μg of protein per disc [53]. After blocking, the discs were incubated with 50 μl of serum from peanut-allergic patients overnight. Specific IgE bound to the discs was determined with a commercial EAST (Allergopharma Spez. IgE ELISA) according to the manufacturer's instructions. IgE concentrations were expressed as units/ml. For statistical comparison of IgE antibody reactivities to wild-type or processed Ara h 2 and Ara h 6, one-way ANOVA analysis according to Tukey was performed using the SigmaStat (Systat, Erkrath, Germany) software package.
For investigation of cross-reactivity, allergen discs were incubated overnight with a serum pool from five peanut-allergic subjects and 50 μl of inhibitor (final concentration at 100 μg/ml to 0.1 ng/ml) overnight at room temperature (22 °C). Discs were incubated without inhibitor and without serum to determine TSB (total specific binding) and NSB (non-specific binding) respectively. For the preparation of the serum pools, serial dilutions of the individual sera were tested for IgE antibody reactivity to Ara h 2 and Ara h 6 respectively. Since the individual IgE antibody reactivities varied considerably, individual sera for the pools were adjusted to the same IgE antibody reactivity before the pools were prepared. After serum incubation, the discs were washed three times with TBS-T (100 mM Tris/HCl, pH 7.4, 100 mM NaCl, 2.5 mM MgCl2 plus 0.5% Tween 20), and the assay was continued as described above. Inhibition levels were calculated using the equation:
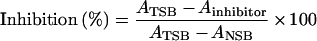 |
Mediator release from humanized RBL (rat basophilic leukaemia) cells
Activation of mast cells and basophils by allergen-specific cross-linking of IgE bound to high-affinity receptors on these cells is an essential event of the immediate-type allergic reaction. Thus mediator-release assays utilize the same principle as the skin prick test and, in contrast with IgE-binding assays, determine the biological potency of allergens. The mediator-release assays followed a well-established protocol [54]. Briefly, RBL 30/25 cells expressing the α-chain of the human FcϵRI (high-affinity IgE receptor) [55] were plated in flat-bottomed 96-well cell-culture plates (Nunc) at 1.5×105 cells per well. The cells were passively sensitized overnight with IgE-containing human sera diluted 1:10–1:40. Optimal dilutions were determined by testing dilution series (1:5–1:80) of the individual sera for maximal release using three peanut extract concentrations (1.0, 0.1 and 0.01 μg/ml) as challenge antigen. For the mediator-release experiments with Ara h 2 and Ara h 6, the sensitized RBL 30/25 cells were challenged with allergen concentrations ranging from 1 μg/ml to 0.1 ng/ml. The antigen-specific release was quantified by measuring β-hexosaminidase activity and was expressed as a percentage of the total β-hexosaminidase content that was obtained by lysing the cells with Triton X-100. For measurements of spontaneous release and possible non-specific effects, naïve RBL cells were incubated with Tyrode's buffer, cell culture medium and patients' sera respectively. Sensitization with purified polyclonal human IgE (Biodesign) and subsequent stimulation with anti-(human IgE) (Nordic Immunology Laboratories) served as a positive control.
RESULTS AND DISCUSSION
Thermal and proteolytic stability of the peanut allergens Ara h 2 and Ara h 6
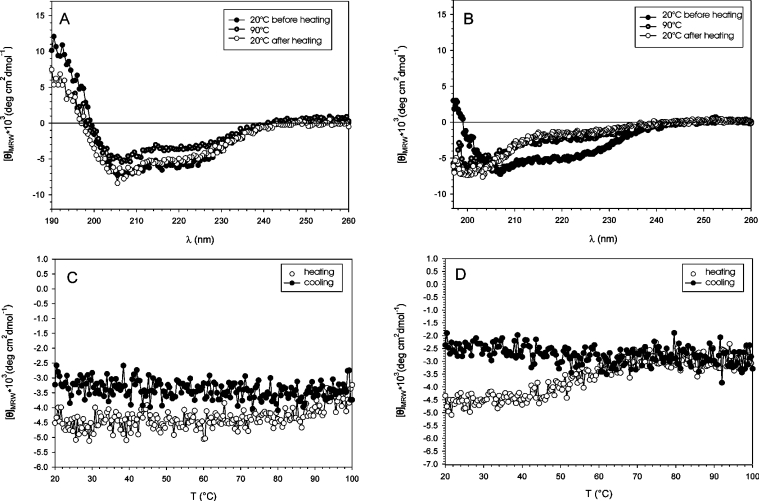
Thermal stability of rAra h 2 and rAra h 6 was investigated at the secondary-structure level using CD spectroscopy (results for Ara h 2 are shown in Figure 1). The spectral minima at 208–210 nm and 222 nm combined with the maximum at 190–193 nm (Figures 1A and 1B) indicates a largely α-helical protein. In the absence of DTT, no [θ]222 thermal transition was observed in the temperature range 20–90 °C (Figure 1C), and [θ]222 decreased only slightly and reversibly during heating. Three far-UV-CD spectra recorded at 20 °C, 90 °C and, after cooling, 20 °C were virtually identical. The 90 °C spectrum of Ara h 6 shows a slight decrease at θ208 and θ222 that reverses on cooling to 20 °C. This marginal loss of ellipticity at high temperatures is most likely to be caused by reversible melting of terminal α-helical regions. In the presence of DTT, both allergens show a co-operative unfolding transition with transition temperatures between 60 and 70 °C, typical for single-domain proteins (Figure 1D). Unfolding is irreversible as indicated by the cooling graph.
Figure 1. Thermal stability of reduced and oxidized Ara h 2.
Far-UV CD spectra of Ara h 2 (10 μM) in the absence (A) and presence (B) of 1 mM DTT at 20 °C (●), 90 °C (○) and 20 °C (○) after cooling in 10 mM potassium phosphate buffer (pH 8). A 1 mm pathlength was used with a scanning speed of 20 nm/min, 6-fold accumulation rate, corrected for buffer effects. Denaturation (○) and refolding (●) of Ara h 2 (1.5 μM) in 10 mM potassium phosphate buffer (pH 8) in the absence (C) or presence (D) of 0.5 mM DTT. Transitions were monitored with a CD signal at 222 nm with a 1 cm pathlength and heating rate 1 °C/min. deg, degrees.
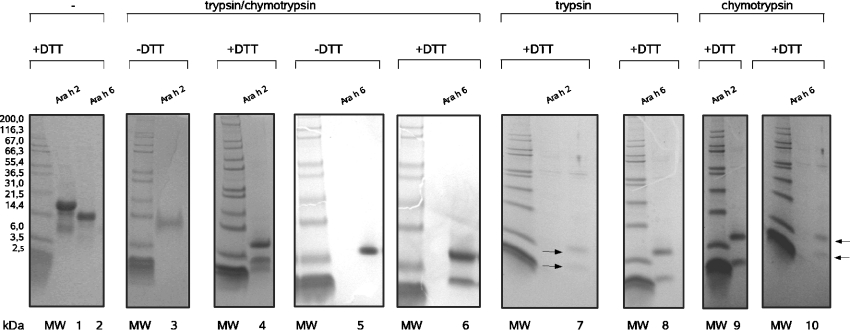
Samples of rAra h 2 and rAra h 6 were incubated with trypsin or chymotrypsin, or a mixture of both, and analysed by SDS/PAGE under reducing and non-reducing conditions (Figure 2). After DTT reduction, two distinct bands appeared in all samples after combined trypsin and chymotrypsin digestion. For rAra h 2, SDS/PAGE showed an additional band with poor intensity probably caused by contamination or by a second variant of the small fragment. The molecular masses of the two fragments were estimated as 4 and 9 kDa from comparison with molecular-mass standards. rAra h 2 and rAra h 6 fragment sizes were highly similar, and no protease-dependent variability was detected. In the absence of DTT, only one band was observed after electrophoretic separation, suggesting the formation of a proteolytic product composed of two fragments connected by disulphide bonds. These proteolytic fragments were stable in the presence of trypsin and chymotrypsin for at least 15 h.
Figure 2. Digestion products of Ara h 2 and Ara h 6.
Intact and digested protein samples were separated by SDS/PAGE under reducing (in the presence of DTT) and non-reducing conditions using 10% Coomassie Brilliant Blue-stained NuPAGE® Bis-Tris SDS/polyacrylamide gels (Invitrogen). The proteins were digested with trypsin and/or chymotrypsin for 10 min in 50 mM Tris buffer, pH 8, at 37 °C. The reaction was stopped by adding NuPAGE® SDS buffer containing 2.5 mg/ml DTT as required and by incubating at 90 °C for 5 min. Mark12 (Invitrogen) was used as molecular-mass standards (MW). Sizes are indicated in kDa. Lanes 1 and 2, undigested Ara h 2 and Ara h 6 in DTT-containing sample buffer; lanes 3 and 4, trypsin- and chymotrypsin-digested Ara h 2 in DTT-free and DTT-containing sample buffer; lanes 5 and 6, trypsin- and chymotrypsin-digested Ara h 6 in DTT-free and DTT-containing sample buffer; lanes 7 and 8, trypsin-digested Ara h 2 and Ara h 6 in DTT-containing sample buffer; lanes 9 and 10, chymotrypsin-digested Ara h 2 and Ara h 6 in DTT-containing sample buffer.
Analysis by N-terminal sequencing of products of the proteolytic cleavage of rAra h 2 yielded two major and several minor products, suggesting spurious inhomogeneity of the fragments. The sequence GAGSSQH was identified as the sequence ranging from Gly62 to His69 of the Ara h 2 large fragment (Figure 3), indicating proteolytic cleavage preceding Gly62. The sequence ELQGDF could be identified as the sequence stretching from positions Gly7 to Phe12, indicating proteolytic shortening of the N-terminus. Both sequences correspond to products expected of tryptic and chymotryptic reactions. Because of the approx. 4 kDa decrease in molecular mass due to proteolysis, it is likely that, in addition to these six N-terminal residues, other amino acids were removed in the region preceding Gly62. The existence of minor fractions suggests partially unspecific cleavage in addition to cleavage at well-defined sequence regions.
Figure 3. Alignment of Ara h 2 and Ara h 6.
Sequences of the light and heavy chains of the digestion products are indicated + and X respectively; identical amino acids have a dark-grey background; similar amino acids have a light-grey background.
In the case of rAra h 6, the sequences GRQGDSSE-E and SQDQQQR were obtained for the major fragments. The first sequence corresponded to sequence Gly8–Glu17, again indicating shortening of the N-terminus. The second sequence corresponded to proteolytic cleavage preceding Ser53 (Figure 3). The masses of the two fragments estimated by comparison with molecular-mass standards are 4 and 10 kDa respectively. The combined molecular mass of 14 kDa is only slightly lower than the mass of intact Ara h 6 of 14.9 kDa. Thus it is most likely that all cystines were intact in the proteolytic products, forming stable disulphide-bonded cores. Although Ara h 2 possesses 41 potential chymotrypsin- and trypsin-cleavage sites (Ara h 6 has 36) distributed almost evenly in the sequence, only small regions were apparently accessible to proteases, whereas the rest of Ara h 2 and Ara h 6 remained intact under chymotrypsin and trypsin treatment.
rAra h 2 and rAra h 6 exhibited significant stability against temperature and proteolytic digestion under non-reducing conditions, a prerequisite for a protein to retain its allergenic properties during food processing and digestion in the gastrointestinal tract [40]. Indeed, thermal stability and resistance to proteases of at least a core structure has been suggested as a predictive measure in the context of safety assessments of genetically modified plants, and stability in simulated gastric fluid [38] and against food-processing procedures such as heating, cooking or roasting [56] demonstrated the integrity against these influences for a number of food allergens. The significantly lower transition temperature of the reduced proteins emphasizes the contribution of the intact disulphide bonds to protein stability.
Structural characterization of the protease-resistant cores of rAra h 6 and rAra h 2
Comparison of the 1H/15N-HSQC spectra of prAra h 6 and the intact rAra h 6 showed a virtually perfect overlay of all well-resolved resonances, indicating highly similar tertiary structures of the two protein species (see the Supplementary Material at http://www.BiochemJ.org/bj/395/bj3950463add.htm). The background of unresolved broad signals in the centre of the spectrum of the intact protein is in the spectral region of random coil chemical shifts. These signals are thus attributed to amide resonances undergoing chemical exchange intermediate on the NMR time scale or reflecting the conformational heterogeneity typical for less structured regions. The absence of this ‘unstructured hump’ in prAra h 6 indicates that the cleavage affects mostly the unstructured regions of the protein, leaving the folded core intact. An analogous result was obtained for rAra h 2. The NMR spectra of prAra h 2 allowed iterative derivation of a total of 414 experimental distance restraints and 23 dihedral angle restraints based on the known sequence-specific assignments [42] from a set of multidimensional heteronuclear edited NMR experiments (see the Supplementary Material).
Restrained molecular dynamics calculations resulted in an ensemble of 14 structures with all distance violations smaller than 0.25 Å and all violations of dihedral restraints smaller than 2.2° (see the Supplementary Material). Inclusion of the 28 residual dipolar couplings in the final refinement reduces the rmsd (root mean square deviation) of the backbone heavy atoms of the region of regular secondary structure from 1.1 to 0.65 Å without significantly affecting other experimental restraints and geometric parameters (bonds and angles). Except for the first short and flexible helix, all α-helices are well defined. Heteronuclear {1H}15N NOE experiments (see the Supplementary Material) indicate significant flexibility on the picosecond to nanosecond timescale for the N-terminus as well as Gly38 to Ser53, indicating that the ill-defined structure in these regions is at least not solely caused by the lack of experimental distance restraints. These experiments further indicate slightly increased flexibility of the C-terminus in spite of its fixation by two disulphide bonds.
prAra h 6 structure
prAra h 6/rAra h 6 consists of several α-helices connected by disordered loops (Figure 4A). The light chain is split into α-helices H1 (Cys16–Val20) and H2 (Cys28–Met37), joined by a central six-residue loop at the top of an α-helix bundle formed by H4 (Met74–Glu84) and H5 (Val95–Cys109). H3 (Ser53–Met67) is followed by a short loop, and H4 is connected to the N-terminus of the light chain via cystine Cys16–Cys73 and with the C-terminus of the heavy chain via cystines Cys86–Cys126 and Cys75–Cys117. H4 and H5 are linked by a ten-residue loop with an interhelical angle of 145°. The C-terminus of H5 has 310-character. H5 is followed by a loop connected to the termini of H4 by two disulphide bonds. H1 to H5 and the connecting loops define a right-handed superstructure. The compact hydrophobic core is formed mainly by H2, H4 and H5. This core is expected to generally define the global fold of 2S albumins, and the four conserved disulphide bonds stabilize further the protein, as shown by the results of the experiments on thermal stability. The observation of 29 slowly exchanging amide protons in a 1H/15N-HSQC recorded 30 min after dissolving a sample of freeze-dried Ara h 6 in 2H2O (pH 6.4) confirms further the rigidity of the protease-resistant core.
Figure 4. Conformational features of Ara h 2 and Ara h 6.
(A) Secondary-structure elements of rAra h 6 (Asp12–Glu39; Ser50–Asp118) based on the lowest energy structure. Disulphide bonds are indicated in yellow. The proteolytically digested protein consists of two subunits connected by disulphide bonds, H1 and H2, H3, H4 and H5. The Figure was generated using VMD [50]. (B) Secondary-structure elements of an Ara h 2 model structure based on the experimental structure of prAra h 6. The Figure was generated using MOLMOL [49] (C) Superposition of the helical regions of Ara h 6 (red) with RicC3 (blue) and SFA-8 (green) [26,27]. The Figure was generated using MOLMOL [49].
Homology-based model of Ara h 2
Experimental determination of the 3D structure of Ara h 2 was seriously hampered by the low quality of the NMR spectra, with broad and overlapping signals in the random coil region indicating a high percentage of flexible areas without regular secondary structure. The α-helical regions of Ara h 6 show 75% sequence identity with the corresponding regions of Ara h 2, suggesting that the folded parts of these proteins are highly similar. On the basis of the experimental structure of prAra h 6, a 3D model of the major peanut allergen Ara h 2 could thus be calculated (Figure 4B), resulting in a backbone rmsd of 0.3 Å between Ara h 2 and Ara h 6 for the helical regions. According to this model, Ara h 2 also shows a right-handed α-helical superstructure. The extension of the loop element connecting H2 and H3 of Ara h 6 is almost doubled in Ara h 2. Ara h 2 possesses only four disulphide bonds, and the missing cystine (Cys86–Cys126 in Ara h 6) prevents anchorage of the C-terminus, which is most likely to be flexible and without any regular secondary-structure elements. The loop between H2 and H3 extended by ten additional residues in Ara h 2 as compared with Ara h 6 is probably affected by proteolytic digestion, as the corresponding two fragments of Ara h 2 and Ara h 6 are of similar size, in agreement with the flexible character of this loop.
Comparison with other 2S albumins
Superposition of the backbones of the helical regions of Ara h 6, RicC3 from R. communis [26], and SFA-8 from sunflower seeds [27] results in atomic rmsds of 2.5 and 2.6 Å respectively (Figure 4C), of the same order as the rmsd between the RicC3 and SFA-8 backbones. The three proteins indeed differ mainly in the length of the helices and in the interhelical angles, but not in their general arrangement as a right-handed superstructure.
Compared with RicC3 and SFA-8, Ara h 6 contains a fifth disulphide bond, Cys86–Cys126, linking the C-terminus to the compact fold. The region Asn110–Ser127, however, does not seem to form a regular secondary structure or pack against the stable core of Ara h 6, as suggested by the complete lack of long-range NOESY cross peaks. Therefore it is not expected that this region strongly contributes to the stability of the global fold of Ara h 6 in spite of the additional disulphide bond.
IgE-binding capacity of Ara h 2, Ara h 6 and their protease-resistant cores
Sera from ten peanut-allergic patients that had IgE binding to Ara h 2 on peanut extract immunoblots were used to determine the IgE-binding capacity of rAra h 2, rAra h 6, prAra h 2 and prAra h 6 by an EAST (Figure 5). In contrast with Western blotting, EAST keeps the allergens under native conditions during the entire procedure. Wild-type Ara h 2 had a higher IgE antibody-binding capacity than Ara h 6 (Figure 5A). prAra h 2 and prAra h 6 retained substantial portions of their IgE antibody reactivity (Figures 5B and 5C).
Figure 5. Comparison of IgE antibody reactivities to wild-type and processed rAra h 2 and rAra h 6.

The broken line represents identical IgE antibody reactivities: (A) wild-type Ara h 2 compared with wild-type Ara h 6; (B) wild-type Ara h 2 compared with processed Ara h 2; (C) wild-type Ara h 6 compared with processed Ara h 6.
Cross-reactivity of Ara h 2 and Ara h 6 by EAST inhibition analysis
Cross-inhibition experiments of IgE binding were performed with solid-phase-bound Ara h 2 and Ara h 6. Almost complete self-inhibition was obtained with rAra h 2 (Figure 6A). Ara h 6 also showed dose-dependent inhibition of the IgE antibody reactivities to Ara h 2. However, the maximal inhibition obtained was approx. 60% of that of Ara h 2, and the slope of the inhibition curve was different (Figure 6A). This indicated substantial partial cross-reactivity, but also epitope differences between the two homologous allergens.
Figure 6. Inhibition of IgE antibody reactivities to rAra h 2 (A) and rAra h 6 (B) by wild-type and processed rAra h 2 and rAra h 6 respectively.
In the reverse experiment with rAra h 6 immobilized at the solid phase, again, complete inhibition was obtained with the homologous inhibitor Ara h 6 (Figure 6B). In contrast, the inhibition by rAra h 2 reached a plateau at approx. 70%, indicating that at least one epitope of Ara h 6 is lacking on Ara h 2 and thus, again, indicating partial immunological identity of the two allergens. In conclusion, the EAST inhibition clearly demonstrated significant cross-reactivity of the two peanut allergens Ara h 2 and Ara h 6.
Biological potency of Ara h 2 and Ara h 6 assessed by mediator release from RBL 30/25 cells
The results of the mediator-release assay performed with sera from four peanut-allergic subjects showed that Ara h 2 is a more potent allergen than Ara h 6 with regard to the ability to induce mediator release. Ara h 2 always induced higher mediator releases and at lower antigen concentrations compared with those effects induced by Ara h 6. The maximal Ara h 6-induced release reached only up to 50% of the maximal Ara h 2-induced release (Figures 7A–7D).
Figure 7. Mediator release induced by peanut extract, natural Ara h 2 (nAra h 2), wild-type and processed rAra h 2 and rAra h 6.
(A–D) Peanut-allergic patients; (E) non-allergic control subject.
To assess whether rAra h 2 shared similar allergenic properties with its natural homologue, natural Ara h 2 was included in the mediator-release experiments. Comparison of the release induced by rAra h 2 and natural Ara h 2 revealed a trend towards a somewhat lower allergenic potential for rAra h 2 (Figures 7B–7D). The observed effect was most likely not due to inadequate folding of rAra h 2, but to the fact that natural Ara h 2 represents at least two different isoforms [14] that might have slightly different allergenic potential. Taken together, the results of the allergen-induced mediator-release experiments confirmed that both rAra h 2 and rAra h 6 have considerable allergenic activity, and support further the notion that Ara h 2 and Ara h 6 are highly cross-reactive allergens.
Influence of proteolytic processing on the allergenicity of rAra h 2 and rAra h 6
Both analytical methods showed that proteolytic treatment with trypsin and chymotrypsin did not influence the IgE antibody-binding capacity to any extent. The inhibition curves (Figures 6A and 6B) demonstrated that both processed Ara h 2 and Ara h 6 inhibited human IgE antibodies to the same extent, regardless of homologous (e.g. inhibition of IgE antibody reactivities to Ara h 2 by Ara h 2) or heterologous (e.g. inhibition of IgE antibody reactivities to Ara h 2 by Ara h 6) inhibition.
Wild-type and prAra h 2 were similar in their ability to induce mediator release (Figures 7A and 7D). One experiment (Figure 7C) showed a significantly higher release induced by one of six concentrations (1 ng/ml) of processed Ara h 2, whereas unprocessed Ara h 2 induced higher release in one case (Figure 7B) at higher (≥10 ng/ml) challenge concentrations. The mediator release caused by Ara h 6 (Figure 7A–7D) demonstrated that wild-type and prAra h 6 were virtually indistinguishable in their ability to induce mediator release.
Conclusion
In summary, the results of the immunological characterization of Ara h 2 and Ara h 6 provide evidence that, in addition to similar structural properties, these two peanut allergens share substantial cross-reactivity with regard to IgE antibody-binding capacity as well as in their allergenic potency, as shown by the results of the mediator-release experiments. However, the results also show that rAra h 2 did not always have the same allergenic potency as compared with its natural counterpart. And, most importantly, partial digestion yielded stable immunologically active core structures. Even though the IgE antibody-binding assay showed reduction of antibody-binding capacity, the functional assay (mediator release from a functional equivalent of mast cells or basophils, the humanized RBL cells) demonstrated that reduction in IgE antibody-binding capacity does not necessarily translate into reduced allergenic potency. Wild-type Ara h 2 and Ara h 6 have virtually identical allergenic potency as the allergens that were treated with digestive enzymes. It is tempting to speculate whether these stable core structures, prAra h 2 and prAra h 6, represent the molecular entities that are responsible for sensitization of atopic subjects to 2S albumins from peanut.
Online data
Acknowledgments
Financial support by the Deutsche Forschungsgemeinschaft is gratefully acknowledged (Be 730/3-1,4; Ro617/11-4). We thank S. Fox (Borstel, Germany), U. Herzing, and N. Herz (both Bayreuth, Germany) for expert assistance.
References
- 1.Emmett S. E., Angus F. J., Fry J. S., Lee P. N. Perceived prevalence of peanut allergy in Great Britain and its association with other atopic conditions and with peanut allergy in other household members. Allergy. 1999;54:380–385. doi: 10.1034/j.1398-9995.1999.00768.x. [DOI] [PubMed] [Google Scholar]
- 2.Sicherer S. H., Morrow E. H., Sampson H. A. Dose–response in double-blind, placebo-controlled oral food challenges in children with atopic dermatitis. J. Allergy Clin. Immunol. 2000;105:582–586. doi: 10.1067/mai.2000.104941. [DOI] [PubMed] [Google Scholar]
- 3.Sampson H. A. Update on food allergy. J. Allergy Clin. Immunol. 2004;113:805–819. doi: 10.1016/j.jaci.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 4.Wensing M., Penninks A. H., Hefle S. L., Koppelman S. J., Bruijnzeel-Koomen C. A., Knulst A. C. The distribution of individual threshold doses eliciting allergic reactions in a population with peanut allergy. J. Allergy Clin. Immunol. 2002;110:915–920. doi: 10.1067/mai.2002.129235. [DOI] [PubMed] [Google Scholar]
- 5.Carr W. W. Clinical pearls and pitfalls: Peanut allergy. Allergy Asthma Proc. 2005;26:145–147. [PubMed] [Google Scholar]
- 6.Valenta R. The future of antigen-specific immunotherapy of allergy. Nat. Rev. Immunol. 2002;2:446–453. doi: 10.1038/nri824. [DOI] [PubMed] [Google Scholar]
- 7.Nelson H. S., Lahr J., Rule R., Bock A., Leung D. Treatment of anaphylactic sensitivity to peanuts by immunotherapy with injections of aqueous peanut extract. J. Allergy Clin. Immunol. 1997;99:744–751. doi: 10.1016/s0091-6749(97)80006-1. [DOI] [PubMed] [Google Scholar]
- 8.Burks A. W., Williams L. W., Connaughton C., Cockrell G., O'Brien T. J., Helm R. M. Identification and characterization of a second major peanut allergen, Ara h II, with use of the sera of patients with atopic dermatitis and positive peanut challenge. J. Allergy Clin. Immunol. 1992;90:962–969. doi: 10.1016/0091-6749(92)90469-i. [DOI] [PubMed] [Google Scholar]
- 9.Burks A. W., Williams L. W., Helm R. M., Connaughton C., Cockrell G., O'Brien T. Identification of a major peanut allergen, Ara h I, in patients with atopic dermatitis and positive peanut challenges. J. Allergy Clin. Immunol. 1991;88:172–179. doi: 10.1016/0091-6749(91)90325-i. [DOI] [PubMed] [Google Scholar]
- 10.Rabjohn P., Helm E. M., Stanley J. S., West C. M., Sampson H. A., Burks A. W., Bannon G. A. Molecular cloning and epitope analysis of the peanut allergen Ara h 3. J. Clin. Invest. 1999;103:535–542. doi: 10.1172/JCI5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kleber-Janke T., Crameri R., Appenzeller U., Schlaak M., Becker W. M. Selective cloning of peanut allergens, including profilin and 2S albumins, by phage display technology. Int. Arch. Allergy Immunol. 1999;119:265–274. doi: 10.1159/000024203. [DOI] [PubMed] [Google Scholar]
- 12.Mittag D., Akkerdaas J., Ballmer-Weber B. K., Vogel L., Wensing M., Becker W. M., Koppelman S. J., Knulst A. C., Helbling A., Hefle S. L., et al. Ara h 8, a Bet v 1-homologous allergen from peanut, is a major allergen in patients with combined birch pollen and peanut allergy. J. Allergy Clin. Immunol. 2004;114:1410–1417. doi: 10.1016/j.jaci.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 13.Stanley J. S., King N., Burks A. W., Huang S. K., Sampson H., Cockrell G., Helm R. M., West C. M., Bannon G. A. Identification and mutational analysis of the immunodominant IgE binding epitopes of the major peanut allergen Ara h 2. Arch. Biochem. Biophys. 1997;342:244–253. doi: 10.1006/abbi.1997.9998. [DOI] [PubMed] [Google Scholar]
- 14.Chatel J. M., Bernard H., Orson F. M. Isolation and characterization of two complete Ara h 2 isoforms cDNA. Int. Arch. Allergy Immunol. 2003;131:14–18. doi: 10.1159/000070429. [DOI] [PubMed] [Google Scholar]
- 15.Burks A. W., King N., Bannon G. A. Modification of a major peanut allergen leads to loss of IgE binding. Int. Arch. Allergy Immunol. 1999;118:313–314. doi: 10.1159/000024114. [DOI] [PubMed] [Google Scholar]
- 16.Lehmann K., Hoffmann S., Neudecker P., Suhr M., Becker W. M., Rösch P. High-yield expression in Escherichia coli, purification, and characterization of properly folded major peanut allergen Ara h 2. Protein Expression Purif. 2003;31:250–259. doi: 10.1016/s1046-5928(03)00190-6. [DOI] [PubMed] [Google Scholar]
- 17.Kleber-Janke T., Becker W. M. Use of modified BL21(DE3) Escherichia coli cells for high-level expression of recombinant peanut allergens affected by poor codon usage. Protein Expression Purif. 2000;19:419–424. doi: 10.1006/prep.2000.1265. [DOI] [PubMed] [Google Scholar]
- 18.Kreis M., Forde B. G., Rahman S., Miflin B. J., Shewry P. R. Molecular evolution of the seed storage proteins of barley, rye and wheat. J. Mol. Biol. 1985;183:499–502. doi: 10.1016/0022-2836(85)90017-8. [DOI] [PubMed] [Google Scholar]
- 19.Shewry P. R., Beaudoin F., Jenkins J., Griffiths-Jones S., Mills E. N. Plant protein families and their relationships to food allergy. Biochem. Soc. Trans. 2002;30:906–910. doi: 10.1042/bst0300906. [DOI] [PubMed] [Google Scholar]
- 20.Youle R. J., Huang A. H. Albumin storage protein and allergens in cottonseeds. J. Agric. Food Chem. 1979;27:500–503. doi: 10.1021/jf60223a017. [DOI] [PubMed] [Google Scholar]
- 21.Barciszewski J., Szymanski M., Haertle T. Analysis of rape seed napin structure and potential roles of the storage protein. J. Protein Chem. 2000;19:249–254. doi: 10.1023/a:1007085627485. [DOI] [PubMed] [Google Scholar]
- 22.Gehrig P. M., Biemann K. Assignment of the disulfide bonds in napin, a seed storage protein from Brassica napus, using matrix-assisted laser desorption ionization mass spectrometry. Pept. Res. 1996;9:308–314. [PubMed] [Google Scholar]
- 23.Muren E., Rask L. Processing in vitro of pronapin, the 2S storage-protein precursor of Brassica napus produced in a baculovirus expression system. Planta. 1996;200:373–379. doi: 10.1007/BF00231392. [DOI] [PubMed] [Google Scholar]
- 24.Muntz K., Christov V., Saalbach G., Saalbach I., Waddell D., Pickardt T., Schieder O., Wustenhagen T. Genetic engineering for high methionine grain legumes. Nahrung. 1998;42:125–127. doi: 10.1002/(sici)1521-3803(199808)42:03/04<125::aid-food125>3.3.co;2-1. [DOI] [PubMed] [Google Scholar]
- 25.Rico M., Bruix M., Gonzalez C., Monsalve R. I., Rodriguez R. 1H NMR assignment and global fold of napin BnIb, a representative 2S albumin seed protein. Biochemistry. 1996;35:15672–15682. doi: 10.1021/bi961748q. [DOI] [PubMed] [Google Scholar]
- 26.Pantoja-Uceda D., Bruix M., Gimenez-Gallego G., Rico M., Santoro J. Solution structure of RicC3, a 2S albumin storage protein from Ricinus communis. Biochemistry. 2003;42:13839–13847. doi: 10.1021/bi0352217. [DOI] [PubMed] [Google Scholar]
- 27.Pantoja-Uceda D., Shewry P. R., Bruix M., Tatham A. S., Santoro J., Rico M. Solution structure of a methionine-rich 2S albumin from sunflower seeds: relationship to its allergenic and emulsifying properties. Biochemistry. 2004;43:6976–6986. doi: 10.1021/bi0496900. [DOI] [PubMed] [Google Scholar]
- 28.Gourinath S., Alam N., Srinivasan A., Betzel C., Singh T. P. Structure of the bifunctional inhibitor of trypsin and α-amylase from ragi seeds at 2.2 Å resolution. Acta Crystallogr. D Biol. Crystallogr. 2000;56:287–293. doi: 10.1107/s0907444999016601. [DOI] [PubMed] [Google Scholar]
- 29.Oda Y., Matsunaga T., Fukuyama K., Miyazaki T., Morimoto T. Tertiary and quaternary structures of 0.19 α-amylase inhibitor from wheat kernel determined by X-ray analysis at 2.06 Å resolution. Biochemistry. 1997;36:13503–13511. doi: 10.1021/bi971307m. [DOI] [PubMed] [Google Scholar]
- 30.Baud F., Pebay-Peyroula E., Cohen-Addad C., Odani S., Lehmann M. S. Crystal structure of hydrophobic protein from soybean: a member of a new cysteine-rich family. J. Mol. Biol. 1993;231:877–887. doi: 10.1006/jmbi.1993.1334. [DOI] [PubMed] [Google Scholar]
- 31.Gincel E., Simorre J. P., Caille A., Marion D., Ptak M., Vovelle F. Three-dimensional structure in solution of a wheat lipid-transfer protein from multidimensional 1H-NMR data: a new folding for lipid carriers. Eur. J. Biochem. 1994;226:413–422. doi: 10.1111/j.1432-1033.1994.tb20066.x. [DOI] [PubMed] [Google Scholar]
- 32.Shin D. H., Lee J. Y., Hwang K. Y., Kim K. K., Suh S. W. High-resolution crystal structure of the non-specific lipid-transfer protein from maize seedlings. Structure. 1995;3:189–199. doi: 10.1016/s0969-2126(01)00149-6. [DOI] [PubMed] [Google Scholar]
- 33.Gomar J., Petit M. C., Sodano P., Sy D., Marion D., Kader J. C., Vovelle F., Ptak M. Solution structure and lipid binding of a nonspecific lipid transfer protein extracted from maize seeds. Protein Sci. 1996;5:565–577. doi: 10.1002/pro.5560050402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lerche M. H., Kragelund B. B., Bech L. M., Poulsen F. M. Barley lipid-transfer protein complexed with palmitoyl CoA: the structure reveals a hydrophobic binding site that can expand to fit both large and small lipid-like ligands. Structure. 1997;5:291–306. doi: 10.1016/s0969-2126(97)00186-x. [DOI] [PubMed] [Google Scholar]
- 35.Lee J. Y., Min K., Cha H., Shin D. H., Hwang K. Y., Suh S. W. Rice non-specific lipid transfer protein: the 1.6 Å crystal structure in the unliganded state reveals a small hydrophobic cavity. J. Mol. Biol. 1998;276:437–448. doi: 10.1006/jmbi.1997.1550. [DOI] [PubMed] [Google Scholar]
- 36.Samuel D., Liu Y. J., Cheng C. S., Lyu P. C. Solution structure of plant nonspecific lipid transfer protein-2 from rice (Oryza sativa) J. Biol. Chem. 2002;277:35267–35273. doi: 10.1074/jbc.M203113200. [DOI] [PubMed] [Google Scholar]
- 37.Tassin-Moindrot S., Caille A., Douliez J. P., Marion D., Vovelle F. The wide binding properties of a wheat nonspecific lipid transfer protein: solution structure of a complex with prostaglandin B2. Eur. J. Biochem. 2000;267:1117–1124. doi: 10.1046/j.1432-1327.2000.01109.x. [DOI] [PubMed] [Google Scholar]
- 38.Astwood J. D., Leach J. N., Fuchs R. L. Stability of food allergens to digestion in vitro. Nat. Biotechnol. 1996;14:1269–1273. doi: 10.1038/nbt1096-1269. [DOI] [PubMed] [Google Scholar]
- 39.Aalberse R. C., van Ree R. Crossreactive carbohydrate determinants. Clin. Rev. Allergy Immunol. 1997;15:375–387. doi: 10.1007/BF02737733. [DOI] [PubMed] [Google Scholar]
- 40.Aalberse R. C. Structural biology of allergens. J. Allergy Clin. Immunol. 2000;106:228–238. doi: 10.1067/mai.2000.108434. [DOI] [PubMed] [Google Scholar]
- 41.Valenta R., Kraft D. From allergen structure to new forms of allergen-specific immunotherapy. Curr. Opin. Immunol. 2002;14:718–727. doi: 10.1016/s0952-7915(02)00402-8. [DOI] [PubMed] [Google Scholar]
- 42.Lehmann K., Schweimer K., Neudecker P., Rösch P. Sequence-specific 1H, 13C and 15N resonance assignments of Ara h 6, an allergenic 2S albumin from peanut. J. Biomol. NMR. 2004;29:93–94. doi: 10.1023/B:JNMR.0000019459.57344.ce. [DOI] [PubMed] [Google Scholar]
- 43.Dayie K. T., Wagner G. Relaxation-rate measurements of 15N-1H groups with pulsed-field gradients and preservation of coherence pathways. J. Magn. Reson. 1996;111A:121–126. [Google Scholar]
- 44.Hansen M. R., Mueller L., Pardi A. Tunable alignment of macromolecules by filamentous phage yields dipolar coupling interactions. Nat. Struct. Biol. 1998;5:1065–1074. doi: 10.1038/4176. [DOI] [PubMed] [Google Scholar]
- 45.Johnson B. A., Blevins R. A. NMRView: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 46. Reference deleted.
- 47.Schweimer K., Sticht H., Nerkamp J., Boehm M., Breitenbach M., Vieths S., Rösch P. NMR spectroscopy reveals common structural features of the birch pollen allergen Bet v 1 and the cherry allergen Pru a 1. Appl. Magn. Reson. 1999;17:449–456. [Google Scholar]
- 48.Kuszewski J., Schwieters C., Clore G. M. Improving the accuracy of NMR structures of DNA by means of a database potential of mean force describing base-base positional interactions. J. Am. Chem. Soc. 2001;123:3903–3918. doi: 10.1021/ja010033u. [DOI] [PubMed] [Google Scholar]
- 49.Koradi R., Billeter M., Wüthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 50.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graphics. 1996;14:33–38. 27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 51.Guex N., Peitsch M. C. SWISS-MODEL and the swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 52.Suhr M., Wicklein D., Lepp U., Becker W. M. Isolation and characterization of natural Ara h 6: evidence for a further peanut allergen with putative clinical relevance based on resistance to pepsin digestion and heat. Mol. Nutr. Food Res. 2004;48:390–399. doi: 10.1002/mnfr.200400028. [DOI] [PubMed] [Google Scholar]
- 53.Ceska M., Lundkvist U. A new and simple radioimmunoassay method for the determination of IgE. Immunochemistry. 1972;9:1021–1030. doi: 10.1016/0019-2791(72)90112-7. [DOI] [PubMed] [Google Scholar]
- 54.Ballmer-Weber B. K., Hoffmann A., Wuthrich B., Luttkopf D., Pompei C., Wangorsch A., Kastner M., Vieths S. Influence of food processing on the allergenicity of celery: DBPCFC with celery spice and cooked celery in patients with celery allergy. Allergy. 2002;57:228–235. doi: 10.1034/j.1398-9995.2002.1o3319.x. [DOI] [PubMed] [Google Scholar]
- 55.Vogel L., Luttkopf D., Hatahet L., Haustein D., Vieths S. Development of a functional in vitro assay as a novel tool for the standardization of allergen extracts in the human system. Allergy. 2005;60:1021–1028. doi: 10.1111/j.1398-9995.2005.00803.x. [DOI] [PubMed] [Google Scholar]
- 56.Kopper R. A., Odum N. J., Sen M., Helm R. M., Stanley J. S., Burks A. W. Peanut protein allergens: the effect of roasting on solubility and allergenicity. Int. Arch. Allergy Immunol. 2005;136:16–22. doi: 10.1159/000082580. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.