Abstract
Hydrogen bonds occurring in the catalytic triad (Asp32, His64 and Ser221) and the oxyanion hole (Asn155) are very important to the catalysis of peptide bond hydrolysis by serine proteases. For the subtilisin NK (nattokinase), a bacterial serine protease, construction and analysis of a three-dimensional structural model suggested that several hydrogen bonds formed by four residues function to stabilize the transition state of the hydrolysis reaction. These four residues are Ser33, Asp60, Ser62 and Thr220. In order to remove the effect of these hydrogen bonds, four mutants (Ser33→Ala33, Asp60→Ala60, Ser62→Ala62, and Thr220→Ala220) were constructed by site-directed mutagenesis. The results of enzyme kinetics indicated that removal of these hydrogen bonds increases the free-energy of the transition state (ΔΔGT). We concluded that these hydrogen bonds are more important for catalysis than for binding the substrate, because removal of these bonds mainly affects the kcat but not the Km values. A substrate, SUB1 (succinyl-Ala-Ala-Pro-Phe-p-nitroanilide), was used during enzyme kinetics experiments. In the present study we have also shown the results of FEP (free-energy perturbation) calculations with regard to the binding and catalysis reactions for these mutant subtilisins. The calculated difference in FEP also suggested that these four residues are more important for catalysis than binding of the substrate, and the simulated values compared well with the experimental values from enzyme kinetics. The results of MD (molecular dynamics) simulations further demonstrated that removal of these hydrogen bonds partially releases Asp32, His64 and Asn155 so that the stability of the transition state decreases. Another substrate, SUB2 (H-D-Val-Leu-Lys-p-nitroanilide), was used for FEP calculations and MD simulations.
Keywords: catalytic mechanism, enzyme kinetics, hydrogen bond, molecular dynamics simulation, site-directed mutagenesis, subtilisin NAT
Abbreviations: E, enzyme; S, substrate; E–S, enzyme–substrate non-covalent complex; ETS, enzyme-transition state complex; FEP, free-energy perturbation; MD, molecular dynamics; NK, nattokinase; SUB1, substrate 1 (succinyl-Ala-Ala-Pro-Phe-p-nitroanilide); SUB2, substrate 2 (H-D-Val-Leu-Lys-p-nitroanilide)
INTRODUCTION
NK (nattokinase) is a potent fibrinolytic enzyme from Bacillus natto [1]. Functionally similar to plasminogen on the basis of substrate specificity, NK dissolves fibrin directly. In addition, it also enhances the body's production of both plasmin and other clot-dissolving agents, including urokinase. In some aspects, NK is actually superior to conventional clot-dissolving drugs [2], which have many benefits including convenient oral administration, known efficacy, prolonged effects, cost effectiveness and prevention of clot formation. NK has been demonstrated to have pH and temperature stability and so can be found in the gastrointestinal tract [3].
As a member of the subtilisin family of serine proteases, NK has the same conserved catalytic triad (Asp32, His64 and Ser221) and oxyanion hole (Asn155) [4]. The serine protease subtilisin is an important industrial enzyme, as well as a model for understanding the relationship between enzyme structure and function [5]. Subtilisin has many advantages, such as rapid gene sequencing and cloning, widespread expression, ease of purification and the availability of abundant crystal structures. For these reasons, subtilisin became a model system for protein engineering studies in the 1980s. Twenty years later, mutations in well over 50% of the 275 amino acids in subtilisin have been reported in the scientific literature. Most subtilisin molecular engineering has involved the catalytic amino acids, substrate-binding regions and investigation of stabilizing mutations [6]. Temperature and pH stability is the property of subtilisin that is most amenable to enhancement, yet perhaps the least well understood.
Wells and Carter [7] have reported on the catalytic mechanism of subtilisin. A large body of literature indicates that these enzymes catalyse reactions by binding the substrate in the transition state more tightly than the reactants or products [8]. Enzymes often accomplish this using weak binding forces such as hydrogen bonds, electrostatic and hydrophobic interactions [9]. Based on the Wells and Carter [7] mechanism, we aimed to determine the relationship between structure and function in NK. In order to investigate the importance of hydrogen bonds in the catalytic triad and oxyanion hole of NK, the four residues Ser33, Asp60, Ser62 and Thr220 were chosen and each mutated into alanine. Both enzyme kinetics and FEP (free-energy perturbation) calculations were used to evaluate the catalytic function of these four residues in NK. MD (molecular dynamics) simulations were used to further investigate how these four residues affected the stability of the transition state of NK.
Materials
Bacillus subtilis var. natto strain AS 1.107 was purchased from the Institute of Microbiology, Chinese Academy of Sciences (Beijing, China). Escherichia coli BL21(DE3) and the vector pET-28a were from Novagen. Bovine fibrinogen, thrombin and urokinase were purchased from Sigma (St. Louis, MO, U.S.A.). SUB1 (substrate 1; succinyl-Ala-Ala-Pro-Phe-p-nitroanilide) was purchased from Sigma. Ampicillin and kanamycin were from Takara (Dalian, China). Restriction enzymes and DNA-modifying enzymes were purchased from Promega. Sephadex G-200 was purchased from Pharmacia Biotech (Uppsala, Sweden).
Construction and purification of NK mutants
B. subtilus var. natto was used as the source of genomic DNA. E. coli BL21 (DE3) was the host bacterial strain for the pET-28a vector expression system. The NK mature peptide gene [aprN(MP)] was cloned into the plasmid pET-28a, and each of the four mutations (Ser33→Ala33, Asp60→Ala60, Ser62→Ala62, and Thr220→Ala220) was introduced into the aprN(MP) gene by sited-directed mutagenesis. Mutagenesis was performed by the overlap extension method [10,11]. As shown in Table 1, oligonucleotide sequences for the wild-type NK were used for PCR. Restriction enzymes BamHI and SacI were used to modify the aprN(MP) gene and pET-28a. The mutant phagemids were verified by dideoxy-mediated sequencing [12,13]. Subtilisin mutants were cultured at 37 °C in Luria–Bertani medium and purified on a Sephadex G-200 column as described by Zhang et al. [14].
Table 1. Synthetic oligonucleotides used for site-directed mutagenesis.
Underlined sequences denote the sites indicated; mut, mutated.
| Subsitution | Oligonucleotide sequences |
|---|---|
| Wild-type | Forward: 5′-TGTGGATCCGCGCAATCTGTTCCTTATGGC-3′, BamHI site |
| Reverse: 5′-CTGGAGCTCTTGTGCAGCTGCTTGTACGTT-3′, SacI site | |
| Mutant | |
| S33A | Forward: 5′-CGTAAAAGTAGCTGTTATCGACGCAGGAATTGACTCTTCTCATCC-3′, mut site |
| Reverse: 5′-GGATGAGAAGAGTCAATTCCTGCGTCGATAACAGCTACTTTTACG-3′, mut site | |
| D60A | Forward: 5′-GAAACAAACCCATACCAGGCAGGCAGTTCTCACGGTA-3′, mut site |
| Reverse: 5′-TACCGTGAGAACTGCCTGCCTGGTATGGGTTTGTTTC-3′, mut site | |
| S62A | Forward: 5′-CCATACCAGGACGGCGCTTCTCACGGTACGCAT-3′, mut site |
| Reverse: 5′-GGTATGGTCCTGCCGCGAAGAGTGCCATGCGTA-3′, mut site | |
| T220A | Forward: 5′-CGGCGCTTATAACGGAGCATCCATGGCGACTCC-3′, mut site |
| Reverse: 5′-GGAGTCGCCATGGATGCTCCGTTATAAGCGCCG-3′, mut site |
Assay of enzyme activity and kinetic studies
Quantitative analysis of fibrinolytic activity was conducted by the standard fibrin plate method [15].
Kinetic parameters were assayed with the substrate SUB1 in 1 ml of 100 mM Tris/HCl (pH 8.60), 4% (v/v) DMSO at (25±0.2) °C as described by Carter and Wells [7]. Enzyme concentrations were determined spectrophotometrically at 280 nm. Subtilisin mutants were assayed at concentrations of about 5 nM for the most active mutants and 1 μM for the least active mutants. Initial rates of hydrolysis were measured at substrate concentrations in the range 0.2–5×Km.
MD simulations and FEP calculations
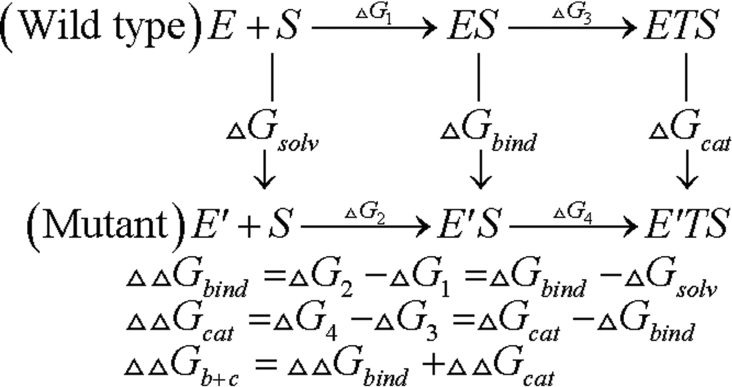
A refined three-dimensional model of NK was constructed by the method of Zheng et al. [16]. Based on this model, the simulated mutations (Ser33→Ala33, Asp60→Ala60, Ser62→Ala62, and Thr220→Ala220) were performed by the TRITON program [17,18]. Three simulations of MD were carried out: (i) on the enzyme itself (E), (ii) on the enzyme–substrate non-covalent complex (E–S), and (iii) on the model of the enzyme-transition state complex (ETS) for acylation catalysis by the serine protease (Figure 1). In each case, we began with the three-dimensional model of NK. For the E–S, we docked SUB2 (substrate 2; H-D-Val-Leu-Lys-p-nitroanilide) with neutral N- and C-terminal residues into the active site of NK and orientated it so that its scissile bond was in position to be attacked by Ser221. For the ETS, we used a structure in which His64 was protonated and the Oγ of Ser221 formed a covalent bond with the carbonyl carbon of the substrate, which was sp3 hybridized.
Figure 1. Thermodynamic cycle for substrate binding and catalysis.
In all three modelled structures E, E—S and ETS, WATBOX216 waters were added to the system to fill the space within 18 Å (1 Å=0.1 nm) of the Oγ (Ser221) in the XLEaP module of AMBER7 [2]. The system energy was minimized with a harmonic constraint of 100 kJ/mol per Å2 for 5000 steps, including 2500 steps of steepest descent, followed by 2500 steps of conjugated gradient. After that, the system was equilibrated using MD at constant temperature (300 K) and pressure with SHAKE [19] for 50 ps. A constraint of 5 kJ/mol per Å2 was used. Next, the free-energy calculations were initiated and continued for 100 ps. In all of the theoretical simulations, an 8 Å nonbonded cutoff was used. A PARM99 force field was used for the enzyme and a GAFF force field was used for the ligand.
The free-energy simulations using the GIBBS module of the AMBER program were carried out on these systems using the method of dynamically modified windows [20]. The free-energies for the forward (λ=1→0) direction were obtained in each system. In all calculations, a duration of 40 ps was used for this simulation with timed steps of 0.002 fs. The nonbonded cutoff was 15 Å.
RESULTS AND DISCUSSION
Mutant design and preparation
Based on the three-dimensional structure of NK [16], the hydrogen bonds in the active centre of NK are discussed below. As shown in Table 2, there are 16 important hydrogen bonds interacting with the catalytic triad (Asp32, His64 and Ser221) and the oxyanion hole (Asn155). Four hydrogen bonds, including hydrogen bond-1, hydrogen bond-4, hydrogen bond-13 and hydrogen bond-14, are formed between the HN atom and the O atom, which are both backbone atoms. Since these four hydrogen bonds are not altered by the site-directed mutagenesis of the catalytic residues, we have disregarded these four hydrogen bonds. The other 12 hydrogen bonds are more important because they are formed not only between backbone atoms and residue atoms but also between residue atoms. It is evident that the mutation of any of these residues will disrupt the hydrogen bonds.
Table 2. Hydrogen bonds around the catalytic triad (Asp32, His64 and Ser221) and oxyanion hole (Asn155) of NK.
These hydrogen bonds were determined using DeepView 3.7.; bond length was determined by MD simulation.
| Proton-supplying group | Proton-accepting group | ||||
|---|---|---|---|---|---|
| H-bonds | Residue | Atom | Residue | Atom | Bond length (Å) |
| 1 | Asp32 | HN | Val93 | O | 2.2±0.2 |
| 2 | Ser33 | HN | Asp32 | Oδ1 | 1.8±0.1 |
| 3 | Ser33 | HOγ | Asp60 | Oδ1 | 1.7±0.1 |
| 4 | Gly61 | HN | Ser33 | O | 2.0±0.3 |
| 5 | Ser62 | HOγ | Ser33 | Oγ | 2.0±0.3 |
| 6 | Ser62 | HN | Asp60 | Oδ1 | 2.7±0.7 |
| 7 | Ser63 | HN | Asp60 | Oδ1 | 2.7±0.7 |
| 8 | His64 | HNe1 | Asp32 | Oδ1 | 1.8±0.1 |
| 9 | His64 | HNe1 | Asp32 | Oδ2 | 2.7±0.5 |
| 10 | His64 | HN | Ser62 | Oγ | 2.3±0.5 |
| 11 | Thr66 | HN | Asp60 | Oδ2 | 1.9±0.2 |
| 12 | Thr66 | HOγ | Asp60 | Oδ2 | 1.8±0.2 |
| 13 | Val68 | HN | His64 | O | 2.2±0.3 |
| 14 | Val95 | HN | Asp32 | O | 2.0±0.2 |
| 15 | Thr220 | HOγ | Asn155 | Oδ | 2.4±0.5 |
| 16 | Thr220 | HN | Asn155 | Oδ | 2.0±0.2 |
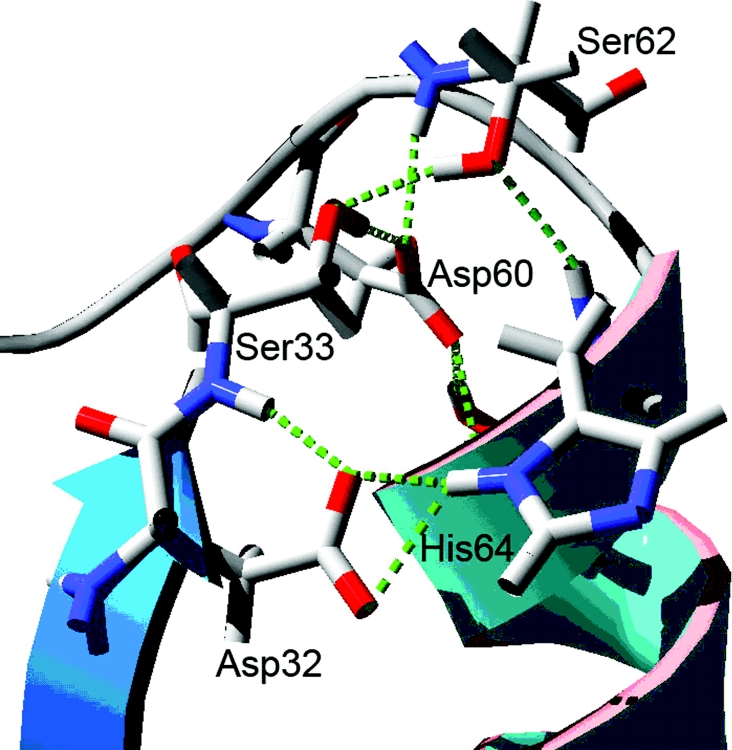
We further determined that four residues have an important function for the stability of the NK active site. These four residues are Ser33, Asp60, Ser62 and Thr220. As shown in Figure 2, the loop which connects the α-helix and His64 is very important. Several hydrogen bonds formed by Ser33 link the β-sheet, including Asp32, with the loop. It is obvious that the loop's movement may directly affect the stability of the Asp32 and His64 residues. The loop includes Asp60 and Ser62 residues. As shown in Figure 3, the hydrogen bonds formed by the residues Asp60 and Ser62 play an important role in the stability of the His64 residue. The hydrogen bonds formed by Ser33 have a great effect on the stability of the Asp32 residue. As shown, the removal of these hydrogen bonds will partially release the Asp32 and His64 backbone atoms so that the Asp32 and His64 residues can move freely. Therefore we first chose these three residues for site-directed mutagenesis. As demonstrated by Braxton and Wells [21] and Mizushima et al. [22], the Thr220 residue can stabilize the transition state in subtilisin BPN′ and we have shown a similar finding for the hydrogen bonds in NK as shown in Figure 3(D). Therefore we examined these hydrogen bonds in NK, to identify the residues that account for stability in NK.
Figure 2. The loop structure of NK including Ser33, Asp60 and Ser62.
The backbone atoms of NK are indicated by secondary structure colours. Other residues are in conventional scale colours, and labelled by their residue names. This Figure was rendered using DeepView 3.7.
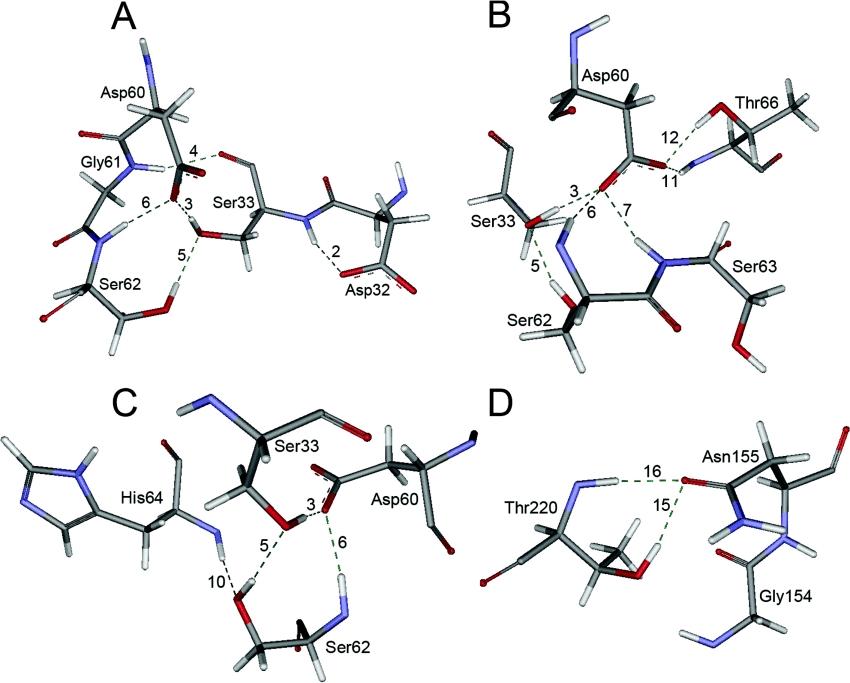
Figure 3. The hydrogen bonding interactions of Ser33, Asp60, Ser62 and Thr220 residues.
(A) Hydrogen bond interactions of Ser33. (B) Hydrogen bond interactions of Asp60. (C) Hydrogen bond interactions of Ser62. (D) Hydrogen bond interactions of Thr220. The residues from NK are in standard atomic colours, labelled by residue name, and shown with the line rendering. This picture was created using Weblab Viewlite 5.0. Dashed lines are hydrogen bonds.
Kinetic analysis and FEP calculations of variant subtilisins
A series of hydrogen bonds were removed by site-directed mutagenesis to investigate the roles of Ser33, Asp60, Ser62 and Thr220 in enzyme catalysis. Four mutants (Ser33→Ala33, Asp60→Ala60, Ser62→Ala62, and Thr220→Ala220) were constructed and used for kinetic analysis. In the present study, alanine was used to minimize structural disruption and alternative hydrogen-bonding effects from site-directed mutagenesis. Kinetic analysis was carried out according to the method of Carter and Wells [7]. As shown in Table 3, removal of each of these four residues resulted in a greater reduction (<3–14-fold) in the kcat value, and a smaller increase (<3-fold) in the Km. The effects of this removal on catalytic efficiency were also quantified in terms of the incremental free-energy of transition state stabilization of the mutants compared with the wild-type enyzme. The values are given in Table 3. The ΔΔGT values of the four mutants are all above 0. Therefore we concluded that the hydrogen bonds formed by the residues Ser33, Asp60, Ser62 and Thr220 function predominantly in the catalytic step to stabilize the catalytic triad and the oxyanion hole.
Table 3. Kinetic constants for the wild-type and mutant NK reactions with SUB1, and the results of three different types of FEP calculation.
Another substrate, SUB2, was used in FEP calculations. −∞, the results of FEP calculations trend to negative infinity; NA, not available.
| Enzyme | kcat (s−1) | Km (μM) | kcat/Km (s−1·mM−1) | ΔΔGT (kcal/mol) | ΔGsolv (kcal/mol) | ΔGbind (kcal/mol) | ΔGcat (kcal/mol) | ΔΔGbind (kcal/mol) | ΔΔGcat (kcal/mol) | ΔΔGb+c (kcal/mol) |
|---|---|---|---|---|---|---|---|---|---|---|
| Wild-type | 52.1 | 196 | 265.3 | NA | NA | NA | NA | NA | NA | NA |
| Mutant | ||||||||||
| S33A | 16.5 | 358 | 46.1 | 1.1 | 7.1 | 7.4 | 8.7 | 0.3 | 1.3 | 1.6 |
| D60A | 3.7 | 527 | 7.0 | 2.2 | −∞ | −∞ | −∞ | NA | NA | NA |
| S62A | 8.2 | 386 | 21.2 | 1.6 | 6.4 | 7.1 | 8.1 | 0.7 | 1.0 | 1.7 |
| T220A | 6.5 | 432 | 15.0 | 1.8 | 9.1 | 9.8 | 10.9 | 0.7 | 1.1 | 1.8 |
By comparison of the NK and subtilisin BPN′ sequences we found the same specificity at the P1 position in NK as in subtilisin BPN′. They both possess the same Gly166 at the S1 subsite which is more sensitive to binding phenylalanine than binding lysine or arginine at the P1 position [23]. Moreover, Fujita et al. [24] have proved that SUB1 is more suitable for enzyme kinetics analysis of NK than is SUB2. As a result SUB1 was chosen for enzyme kinetics analysis. However, SUB1 does not display a similar clot-dissolving property with NK [1]. In order to accurately display the mechanism of action of NK, SUB2 was chosen for FEP calculations and MD simulations.
The results of FEP calculations also indicate the function Ser33, Asp60, Ser62 and Thr220 residues. As shown in Table 3, mutating Ser33 into Ala33 with water, Michaelis complex or tetrahedral intermediates involves a free-energy change of 7.1 kcal/mol (1 kcal=4.184 kJ), 7.3506 kcal/mol or 8.7 kcal/mol respectively. This demonstrates that the solvation of the Ala33 mutant is unfavoured compared with the native enzyme (containing Ser33) in all three cases; the enzyme itself, the E–S and the tetrahedral intermediate. The mutants Ser62→Ala62 and Thr220→Ala220 are similar to the Ser33→Ala33 mutant, but the Asp60→Ala60 mutant is different. Mutating Asp60 to alanine involves a free-energy change of −∞, this value is variable and does not indicate the advantage gained in mutating Asp60 to alanine. Although this result deviates from known enzyme kinetics data, it does suggest the possibility that the solvation of the Ala60 mutant is enhanced compared with the native enzyme (containing Asp60) in all three cases. The calculated ΔΔGbind and ΔΔGcat of the Ser33→Ala33 mutant are 0.3 kcal/mol and 1.3 kcal/mol respectively. The ΔΔGbind is 6-fold less than the ΔΔGcat, which further indicates that the hydrogen bonds formed by Ser33 function predominantly in the catalytic step and not in the binding step. The ΔΔGb+c of the Ser33→Ala33 mutant is 1.6 kcal/mol. This value is similar to the experimental value of ΔΔGT (1.1 kcal/mol) although the substrates for the ΔΔGb+c and ΔΔGT are different. As shown in Table 3, the mutants Ser62→Ala62 and Thr220→Ala220 have similar properties in this respect compared with the Ser33→Ala33 mutant.
MD simulations
Based on the catalytic mechanism of subtilisin [7], the MD simulations focused on the structural alternations in the catalytic triad and the oxyanion hole of NK. Our investigation of the crystal structures of subtilisins with or without their proteinaceous inhibitors revealed a number of interesting findings about the catalytic triad. In all subtilisins, either with or without inhibitors, the Oδ1(Asp32)–e1(His64) distance is 3.0–3.2 Å, and the Oδ2(Asp32)–e1(His64) distance is 2.5–2.7 Å. These statistical data are in accordance with our MD results. As shown in Table 2, the HNe1(His64)–Oδ1(Asp32) distance remains approx. 1.8 Å, which is the distance trajectory of hydrogen bond-8, and the HNe1(His64)–Oδ2(Asp32) distance remains approx. 2.7 Å, which is the distance trajectory of hydrogen bond-9. With the aid of hydrogen bond-8 and hydrogen bond-9, the carboxyl terminal of Asp32 can act on HNe1(His64) strengthening the negative charge on the imidazole of His64 [25]. Therefore it was clear that these two hydrogen bonds are strong and stable enough to exist in all three states and in the binding and catalytic steps.
In the crystal structures of subtilisins without inhibitors, the Ser221 residues can move freely. The Ne2(His64)–Oγ(Ser221) distance remains approx. 2.5–4.0 Å. For example, the Ne2(His64)–Oγ(Ser221) distance in the complex of subtilisin BPN′ with chymotrypsin inhibitor 2 M59k (PDB accession number 1LW6) is 2.64 Å, and that of the Bacillus lentus subtilisin (PDB 1C9J) is 3.93 Å. However, in these crystal structures of serine proteases with inhibitors, the Ser221 residue cannot move freely. The Ne2(His64)–Oγ(Ser221) distance is restrained to 2.5–2.7 Å, which is the hydrogen bond distance. These statistical data suggest that substrates or inhibitors induce the inter-approach of Ne2(His64) and HOγ(Ser221) in enzyme–substrate interactions. This inductive process is in accordance with the catalytic mechanism of Carter and Wells [7]. Therefore the Michaelis complex structure that was modelled, which has a starting distance of 4.0 Å for Ne2(His64)–HOγ(Ser221), was chosen as our initial co-ordinate.
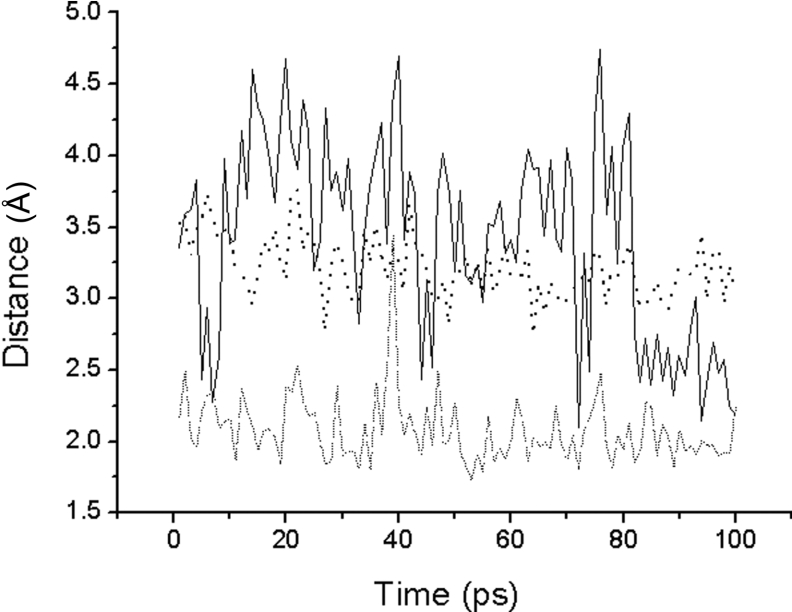
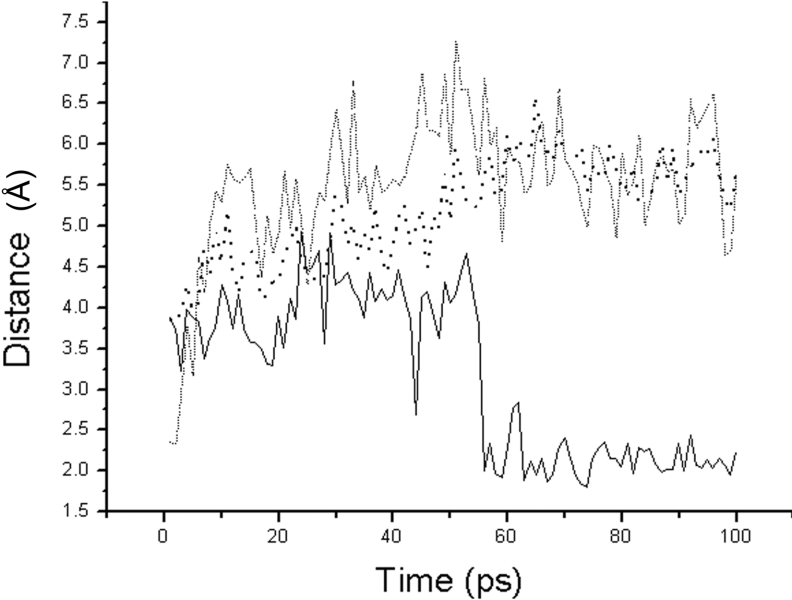
As shown in Figure 4, the 100 ps of MD simulation also supports the inductive interaction of substrates. As can be seen, the Oγ(Ser221)–C(Lys278) distance gradually decreases from 3.6 to 3.0 Å during the 100 ps of MD simulation, which suggests that Oγ(Ser221) is very stable and able to attack C(Lys278). The hydrogen bond interaction between HNδ2(Asn155) and O(Lys278) is also stable at 2.0 Å during the 100 ps of MD simulation. The His64 residue; however, does not remain stable during the first 80 ps of MD simulation. The Ne2(His64)–Oγ(Ser221) distance is increased by 2.5 to 4.5 Å, which is wider and stronger than that of the native enzyme without the bound substrate. After 80 ps, the Ne2(His64)–Oγ(Ser221) distance is stable at 2.5 Å, which is the strong hydrogen bond distance. However, the interaction between Ne2(His64) and Oγ(Ser221) is weaker in the native enzyme without bound substrates and the distance gradually decreases from 4.0 to 3.0 Å during the 100 ps of MD simulation. We have therefore demonstrated that the substrate induces interaction between His64 and Ser221 residues when binding in the catalytic triad.
Figure 4. The distance trajectories for the native enzyme bound with substrate.
The solid line is the distance between Ne2(His64) and HOγ(Thr220). The dashed line is the distance between HNδ2(Asn155) and O(Lys278). The dotted line is the distance between Oγ(Ser221) and C(Lys278).
Effects of the hydrogen bonds from Ser33 on the catalytic efficiency of NK
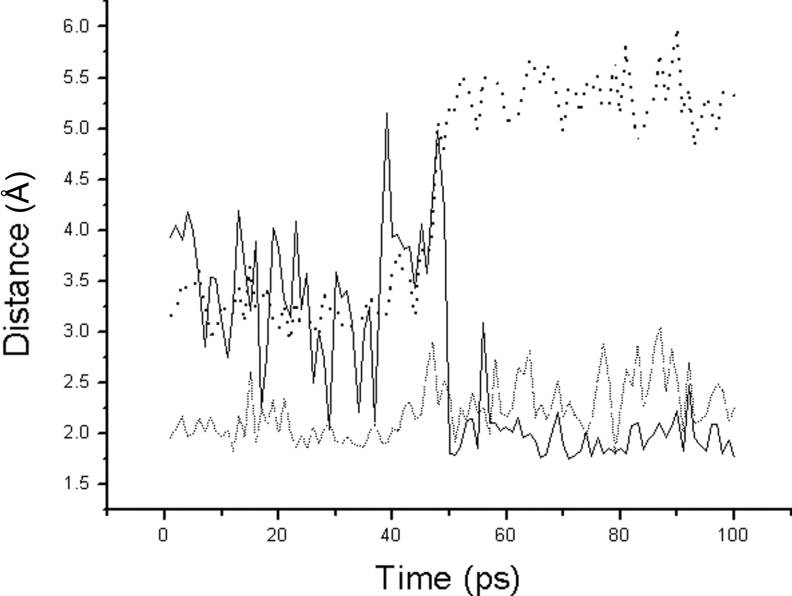
Substituting Ala33 for Ser33 breaks hydrogen bond-3 and hydrogen bond-5 formed by the hydroxyl terminal of Ser33 as shown in Figure 3(A). The removal of these two hydrogen bonds directly results in decreasing catalytic efficiency of NK, which has been demonstrated by enzyme kinemics and FEP calculation as shown in Table 3. But why does catalytic efficiency decrease, and what effects does this have on the catalytic triad in the Ser33→Ala33 mutant? During the 100 ps MD simulation without substrate, the Ne2(His64)–HOγ(Ser221) distance in the Ser33→Ala33 mutant consistently remains approx. 4.0 Å. This distance trajectory does not trend to descend as in the native enzyme. It is evident that the Ala33 substitution does not contribute to the stability of Ser221 when there is no bound substrate. However, in the presence of a substrate, the Ne2(His64)–HOγ(Ser221) distance rapidly decreases from 4.0 to 2.0 Å after 50 ps, as shown in Figure 5. This suggests that the strong hydrogen bond interaction restrains the movement of Ser221. However, the distances for HNδ2(Asn155)–O(Lys278) and Oγ(Ser221)–C(Lys278) increase to greater than 5.0 Å, which are both greater than the interaction distance. Therefore we concluded that the removal of hydrogen bond-3 and hydrogen bond-5 partially releases the Asp32 backbone atoms and allows Asp32 to move freely. When there is no substrate, Asp32 is found at the same starting co-ordinates as in the native enzyme. When the substrate is in complex with the enzyme, Asp32 moves closer to His64, which increases the alkalinity of Ne2(His64). At the same time, with the aid of the substrate's inductive interaction, the strong attraction of Ne2(His64) for the hydroxyl group of Ser221 causes it to rapidly depart from the correct position for nucleophilic attack. Without the co-operation of Oγ(Ser221), HNδ2(Asn155) does not remain stable and dissociates from the substrate.
Figure 5. The distance trajectories for the Ser33→Ala33 mutant bound with substrate.
The solid line is the distance between Ne2(His64) and HOγ(Thr220). The dashed line is the distance between HNδ2(Asn155) and O(Lys278). The dotted line is the distance between Oγ(Ser221) and C(Lys278).
Threonine has the same function as serine at position 33. Comparing the sequence identity of the subtilisin family, we found that residue 33 is a conserved serine or threonine residue. Moreover, Thr33 can form the same hydrogen bonds as Ser33, which are revealed in the crystal structure of subtilisin Carlsberg and subtilisin Savinase.
Effects of the Ser62 hydrogen bonds on the catalytic efficiency of NK
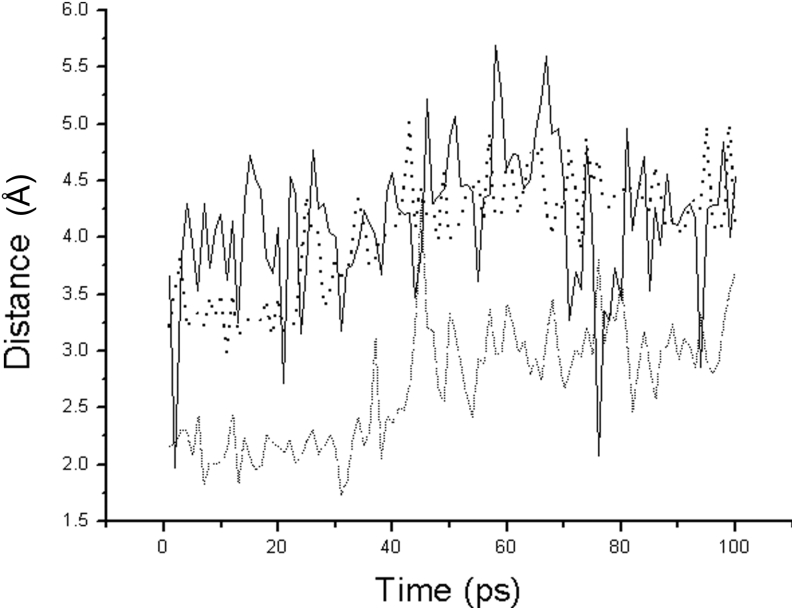
Substituting Ala62 for Ser62 breaks hydrogen bond-5 and hydrogen bond-10 formed by the hydroxyl group of Ser62 as shown in Figure 3(C). The removal of hydrogen bond-5 and hydrogen bond-10 partially releases the His64 backbone atoms. As a consequence, His64 can move freely. During the 100 ps MD simulation without substrate, the Ne2(H His64)–HOγ(Ser221) distance in the Ser62→Ala62 mutant ranges between 2.0 and 4.5 Å. However, when the substrate is binding to enzyme, the unreleased residue Asp32 will restrain the movement of the His64 residue, even attracting it away from the correct position to interact with HOγ(Ser221). As shown in Figure 7, during the 100 ps MD simulation, the Ne2(His64)–HOγ(Ser221) distance remains approx. 4.0 Å, which is greater than the functional interaction distance. The Oγ(Ser221)–C(Lys278) distance increases from 3.0 to 3.4 Å, which indicates that Oγ(Ser221) is dissociating from C(Lys278) during the 100 ps MD simulation. However the removal of hydrogen bond-5 and hydrogen bond-10 doesn't affect the stability of the oxyanion hole (Asn155). The distance HNδ2(Asn155)–O(Lys278) is consistently approx. 2.0 Å during the 100 ps MD simulation.
Figure 7. The distance trajectories for the Ser62→Ala62 mutant bound with substrate.
The solid line is the distance between Ne2(His64) and HOγ(Thr220). The dashed line is the distance between HNδ2(Asn155) and O(Lys278). The dotted line is the distance between Oγ(Ser221) and C(Lys278).
Similar to residue 33, residue 62 is either a conserved serine or aspartic acid residue. Thus Asn62 has the same role as Ser62 in subtilisins.
Effects of the Asp60 hydrogen bonds on catalytic efficiency of NK
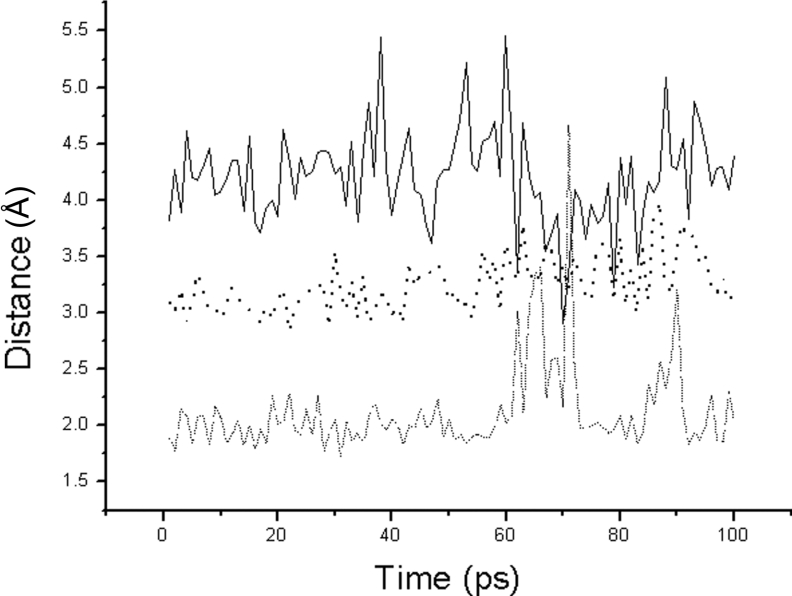
Substituting Ala60 for Asp60 breaks five hydrogen bonds (hydrogen bond−3, −6, −7, −11 and −13) formed by the carboxyl group of Asp60 as shown in Figure 3(B). The removal of these five hydrogen bonds releases the Asp32 and His64 backbone atoms, which is similar to mutating Ser33 and Ser62 at the same time. When there is no bound substrate, without the restraint of Asp32, Ne2(His64) rotates into close proximity to HOγ(Ser221) and moves around HOγ(Ser221) into a functional interaction distance. During the first 50 ps of MD simulation without substrate, the Ne2(His64)–HOγ(Ser221) distance of the Asp60→Ala60 mutant gradually decreases from 4.0 to 3.0 Å, and then fluctuates between 2.0 and 3.5 Å after 50 ps. When the substrate is binding to the enzyme, the catalytic activity increases during the first 50 ps of MD simulation. As shown in Figure 6, the HNδ2(Asn155)–O(Lys278) distance is stable at 2.0 Å, which is the functional hydrogen bond interaction distance. The Oγ(Ser221)–C(Lys278) distance is also stable at 3.0 Å which is the functional interaction distance for nucleophilic attack. In addition, the Ne2(His64)–HOγ(Ser221) distance fluctuates between 2.0 and 5.0 Å as in the native enzyme. Therefore it was clear that the MD results during the first 50 ps are in accordance with the results of the FEP calculations shown in Table 3.
Figure 6. The distance trajectories for the Asp60→Ala60 mutant bound with substrate.
The solid line is the distance between Ne2(His64) and HOγ(Thr220). The dashed line is the distance between HNδ2(Asn155) and O(Lys278). The dotted line is the distance between Oγ(Ser221) and C(Lys278).
However, after 50 ps, the alkalinity of the imidazole of His64 is rapidly increased in the interaction with the released Asp32, which strengthens the interaction between Ne2(His64) and HOγ(Ser221). At the same time, with the help of the substrate's inductive interaction, Ne2(His64) attracts the hydroxyl group of Ser221 to move away from the correct position for nucleophilic attack. As shown in Figure 6, the Ne2(His64)–HOγ(Ser221) distance decreases to 2.0 Å, and the Oγ(Ser221)–C(Lys278) distance rapidly increases to 5.5 Å. As shown, the catalytic activity rapidly decreases after a 50 ps MD simulation.
Effects of the Thr220 hydrogen bonds on catalytic efficiency of NK
Substituting Ala220 for Thr220 breaks hydrogen bond-15 formed by the hydroxyl group of Thr220 as shown in Figure 3(D). As shown, the stability of Asn155 relies on two hydrogen bonds, hydrogen bond-15 and hydrogen bond-16. The removal of hydrogen bond-15 partially releases the Asn155 residue, and forces HNδ2(Asn155) to move freely [22]. As shown in Figure 8, the HNδ2(Asn155)–O(Lys278) distance gradually decreases from 2.0 to 3.0 Å during the 100 ps MD simulation [21]. The instability of HNδ2(Asn155) directly results in the instability of Oγ(Ser221). The Oγ(Ser221)–C(Lys278) distance increases to 4.5 Å, as shown in Figure 8. However, the instability of HNδ2(Asn155) doesn't affect the interaction between Ne2(His64) and HOγ(Ser221). The Ne2(His64)–HOγ(Ser221) distance always increases from 2.0 to 4.5 Å as in the native enzyme.
Figure 8. The distance trajectories for the Thr220→Ala220 mutant bound with substrate.
The solid line is the distance between Ne2(His64) and HOγ(Thr220). The dashed line is the distance between HNδ2(Asn155) and O(Lys278). The dotted line is the distance between Oγ(Ser221) and C(Lys278).
Braxton and Wells [21] have demonstrated that the stabilizing effects of Thr220 are mediated either via a short-range charedipolar interaction or by dynamic fluctuations in the protein structure, which permits direct hydrogen bonding with oxyanion. The results calculated by Mizushima et al. [22] further demonstrated that -OH(Thr220) is important for catalysis but not to as great an extent as a hydrogen-bonding group, such as Asn155, positioned directly in the oxyanion hole.
Acknowledgments
This work was mostly supported by the National Natural Scientific Foundation (grant numbers 30370366 and 30470111) and supported in part by a grant (number 92-2313-B-007-002) from the NSC (National Science Council Taiwan). The AMBER studies were conducted at the National Centre for High Performance Computing, Taiwan. We are grateful to Dr Keng-chang Tsai for his fruitful discussions concerning the results of enzyme kinetics and MD simulation.
References
- 1.Sumi H. A novel fibrinolytic enzyme in the vegetable cheese natto: a typical and popular soybean food in the Japanese diet. Experientia. 1987;43:1110–1111. doi: 10.1007/BF01956052. [DOI] [PubMed] [Google Scholar]
- 2.Jovin I. S., Müller B. G. Inter-relationships between the fibrinolytic system and lipoproteins in the pathogenesis of coronary atherosclerosis. Atherosclerosis. 2004;174:225–233. doi: 10.1016/j.atherosclerosis.2004.01.026. [DOI] [PubMed] [Google Scholar]
- 3.Ko J. H., Yan J. P., Zhu L., Qi Y. P. Identification of two novel fibrinolytic enzymes from Bacillus subtilis QK02. Comp. Biochem. Physiol. 2004;137:65–74. doi: 10.1016/j.cca.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 4.Bryan P. N. Protein engineering of subtilisin. Biochim. Biophys. Acta. 2000;1543:203–222. doi: 10.1016/s0167-4838(00)00235-1. [DOI] [PubMed] [Google Scholar]
- 5.Takagi H., Koga M., Katsurada S., Yabuta Y., Shinde U., Inouye M. Functional analysis of the propeptides of subtilisin E and aqualysin I as intramolecular chaperones. FEBS Lett. 2001;508:210–214. doi: 10.1016/s0014-5793(01)03053-8. [DOI] [PubMed] [Google Scholar]
- 6.Yang Y. H., Jiang L., Zhu L. Q., Wu Y. J., Yang S. L. Thermal stable and oxidation-resistant variant of subtilisin E. J. Biotech. 2000;81:113–118. doi: 10.1016/s0168-1656(00)00272-8. [DOI] [PubMed] [Google Scholar]
- 7.Carter P., Wells J. A. Dissecting the catalytic triad of a serine protease. Nature (London) 1988;332:564–568. doi: 10.1038/332564a0. [DOI] [PubMed] [Google Scholar]
- 8.Baker D., Sali A. Protein structure, prediction and structural genomics. Science (Washington, DC) 2001;294:93–96. doi: 10.1126/science.1065659. [DOI] [PubMed] [Google Scholar]
- 9.Skolnick J., Fetrow J. S. From genes to protein structure and function: novel applications of computational approaches in the genomic era. Trends Biotechnol. 2000;18:34–39. doi: 10.1016/s0167-7799(99)01398-0. [DOI] [PubMed] [Google Scholar]
- 10.Higuchi R., Krummel B., Saiki R. K. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 12.Brow M. A. D. Sequencing with Taq DNA polymerase. In: Innis M. A., Gelfand D. H., Sninsky J. J., White T. J., editors. PCR protocols: A guide to Methods and Applications. San Diego, CA, U.S.A.: Academic Press; 1990. pp. 189–196. [Google Scholar]
- 13.Innis M. A., Myambo K. B., Gelfand D. H., Brow M. A. DNA sequencing with Thermus aquaticus DNA polymerase and direct sequencing of polymerase chain reaction-amplified DNA. Proc. Natl. Acad. Sci. U.S.A. 1988;85:9436–9440. doi: 10.1073/pnas.85.24.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng Y., Huang Q., Zhang R. H., Zhang Y. Z. Purification and characterization of a fibrinolytic enzyme produced by Bacillus amyloliquefaciens DC-4 screened from douchi, a traditional Chinese soybean food. Comp. Biochem. Physiol. 2003;134:45–52. doi: 10.1016/s1096-4959(02)00183-5. [DOI] [PubMed] [Google Scholar]
- 15.Astrup T., Mullertz S. The fibrin plate method for estimating fibrinolytic activity. Arch. Biochem. Biophys. 1952;40:346–351. doi: 10.1016/0003-9861(52)90121-5. [DOI] [PubMed] [Google Scholar]
- 16.Zheng Z. L., Zou G. L., Liu Z. G., Tsai K. C., Lie A. F., Zou G. L. Construction of a 3D model of nattokinase, a novel fibrinolytic enzyme from Bacillus natto. A novel nucleophilic catalytic mechanism for nattokinase. J. Mol. Graph. Mod. 2005;23:373–380. doi: 10.1016/j.jmgm.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 17.Prokop M., Damborsky J., Koca J. TRITON: in silico construction of protein mutants and prediction of their activities. Bioinformatics. 2000;16:845–846. doi: 10.1093/bioinformatics/16.9.845. [DOI] [PubMed] [Google Scholar]
- 18.Damborsky J., Prokop M., Koca J. TRITON: graphic software for rational engineering of enzymes. Trends Biochem. Sci. 2001;26:71–73. doi: 10.1016/s0968-0004(00)01708-4. [DOI] [PubMed] [Google Scholar]
- 19.Van Gunsteren W. F., Berendsen H. J. C. Algorithms for macromolecular dynamics and constraint dynamics. Mol. Phys. 1977;34:1311–1327. [Google Scholar]
- 20.Pearlman D. A., Kollman P. A. A new method for carrying out free energy perturbation calculation: dynamically modified windows. J. Chem. Phys. 1989;90:2460–2470. [Google Scholar]
- 21.Braxton S., Wells J. A. The importance of a distal hydrogen bonding group in stabilizing the transition state in subtilisin BPN′. J. Biol. Chem. 1991;266:11797–11800. [PubMed] [Google Scholar]
- 22.Mizushima N., Hirono S., Pearlman D., Kollman P. A. Free energy perturbation calculation on binding and catalysis after mutating threonine 220 in subtilisin. J. Biol. Chem. 1991;18:11801–11809. [PubMed] [Google Scholar]
- 23.Ballinger M. D., Tom J., Wells J. A. Designing subtilisin BPN′ to cleave substrates containing dibasic residues. Biochemistry. 1995;34:13312–13319. doi: 10.1021/bi00041a006. [DOI] [PubMed] [Google Scholar]
- 24.Fujita M., Nomura K., Hong K., Ito Y., Asada A., Nishimuro S. Purification and characterization of a strong fibrinolytic enzyme (Nattokinase) in the vegetable cheese Natto, a popular soybean fermented food in Japan. Biochem. Biophys. Res. Commun. 1993;30:1340–1347. doi: 10.1006/bbrc.1993.2624. [DOI] [PubMed] [Google Scholar]
- 25.Thomas P. G., Russell A. J., Fersht A. R. Tailoring the pH dependence of enzyme catalysis using protein engineering. Nature (London) 1985;318:375–376. [Google Scholar]