Abstract
The cAMP-dependent pathway up-regulates MITF (microphthalmia-associated transcription factor), important for key melanogenic proteins such as tyrosinase, TRP-1 (tyrosinase-related protein 1) and TRP-2. We asked whether MITF is also a key transcription factor for PKC-β (protein kinase C-β), required to phosphorylate otherwise inactive tyrosinase. When paired cultures of human melanocytes were treated with isobutylmethylxanthine, known to increase intracellular cAMP, both protein and mRNA levels of PKC-β were induced by 24 h. To determine whether MITF modulates PKC-β expression, paired cultures of human melanocytes were transfected with dn-MITF (dominant-negative MITF) or empty control vector. By immunoblotting, PKC-β protein was reduced by 63±3.7% within 48 h. Co-transfection of an expression vector for MITF-M, the MITF isoform specific for pigment cells, or empty control vector with a full-length PKC-β promoter–CAT (chloramphenicol acetyltransferase) reporter construct (PKC-β/CAT) into Cos-7 cells showed >60-fold increase in CAT activity. Melanocytes abundantly also expressed MITF-A, as well as the MITF-B and MITF-H isoforms. However, in contrast with MITF-M, MITF-A failed to transactivate co-expressed PKC-β/CAT or CAT constructs under the control of a full-length tyrosinase promoter. Together, these results demonstrate that MITF, specifically MITF-M, is a key transcription factor for PKC-β, linking the PKC- and cAMP-dependent pathways in regulation of melanogenesis.
Keywords: human melanocyte, microphthalmia-associated transcription factor (MITF), pigmentation, protein kinase C-β (PKC-β), transcription, tyrosinase
Abbreviations: BPE, bovine pituitary extract; CAT, chloramphenicol acetyltransferase; CRE, cAMP-response element; DAG, diacylglycerol; dbcAMP, dibutyryl cAMP; DMEM, Dulbecco's modified essential medium; IBMX, isobutylmethylxanthine; α-MSH, α-melanocyte-stimulating hormone; MITF, microphthalmia-associated transcription factor; dn-MITF, dominant-negative MITF; PKC-β, protein kinase C-β; RT, reverse transcriptase; TRP-1, tyrosinase-related protein 1
INTRODUCTION
Agents that elevate the intracellular level of cAMP, such as α-MSH (α-melanocyte-stimulating hormone), dbcAMP (dibutyryl cAMP) or IBMX (isobutylmethylxanthine), have been shown to induce melanogenesis in both murine and human pigment cells [1–7]. After α-MSH binds to its cell surface, MC1-R (melanocortin-1 receptor), a seven-transmembrane G-protein-coupled receptor that activates adenylate cyclase [8,9], the intracellular level of cAMP is elevated [8,10]. The expression of melanogenic proteins such as tyrosinase and TRP-1 (tyrosinase-related protein 1) and TRP-2 was also shown to be induced by cAMP. Most of the biological effects of cAMP have been shown to be mediated through cAMP-dependent PKA (protein kinase A) [11]. PKA, a serine/threonine kinase, phosphorylates the CREB [CRE (cAMP-response element)-binding protein], which then interacts with the CRE to transactivate MITF (microphthalmia-associated transcription factor) [12–14]. The major melanogenic proteins such as tyrosinase, TRP-1 and TRP-2 do not have CRE in their promoter region, excluding the possibility that the cAMP pathway directly affects transcription of these genes. It has therefore been proposed that regulation of these melanogenic genes by cAMP occurs indirectly through MITF [15–17].
MITF is a member of the b-HLH-zip (basic/helix–loop–helix/leucine zipper) transcription factors and was shown to bind to one or both of two conserved consensus elements, the M-box (AGT-CATGTGCT) or E-box (CATGTG), in the promoter of each regulated gene [18]. Promoters of tyrosinase, TRP-1 and TRP-2 were all shown to contain at least one M-box consensus sequence [19]. Ectopic expression of MITF in fibroblasts leads to phenotypes associated with melanocytic cells [20], and mutations in MITF are found in the pigmentary disorder Waardenburg syndrome type IIA [21]. MITF was subsequently shown to be a family of isoforms [13,22], with MITF-M specifically expressed in melanocytic cells [23]. The promoter region of MITF contains the DNA consensus sequences for CRE [12–14]; thus the expression of MITF is up-regulated by cAMP, and α-MSH increases the expression of MITF through a cAMP-dependent pathway [15,24]. The function of MITF has been shown to be pivotal for melanogenesis, in that dn-MITF (dominant-negative MITF) blocked cAMP-induced increases in tyrosinase expression and pigmentation [14,15,25].
The PKC (protein kinase C)-dependent pathway has emerged as a second intracellular signalling pathway regulating melanogenesis. It was first observed that addition of DAG (diacylglycerol), the endogenous activator of PKC, to cultured human melanocytes caused a rapid 3–4-fold increase in total melanin content [26], and this increase was blocked by a PKC inhibitor [26]. Moreover, topical application of active DAG species to guinea-pig skin increased melanogenesis, while a DAG species not capable of activating PKC failed to do so [27]. Conversely, topical application of a selective PKC inhibitor reduces pigmentation and blocks UV-induced tanning in guinea-pig skin [28]. In cultured pigment cells, depletion of PKC reduced the melanin content dramatically [29–31] and completely blocked α-MSH-induced melanogenesis [31]. Further work established that the β-isoform of PKC was specifically involved in melanogenesis and that the specific role of PKC-β was to phosphorylate (activate) tyrosinase [32]. The cytoplasmic domain of tyrosinase contains two serine residues at amino acid positions 505 and 509 [33], and both serine residues are phosphorylated by PKC-β, resulting in activation of tyrosinase [34].
In the present paper, we examine whether cAMP regulates the expression of PKC-β, as seen with other melanogenic proteins. Moreover, because the promoter region of PKC-β contains at least two E-boxes [35], the possible role of MITF-M as a transcription factor for PKC-β has been explored.
MATERIALS AND METHODS
Materials
DMEM (Dulbecco's modified essential medium), L15, non-essential amino acids, glutamine, M199 medium and trypsin were purchased from Gibco BRL. Recombinant basic fibroblast growth factor was purchased from Amgen, tri-iodothyronine from Collaborative Research, BPE (bovine pituitary extract) from Clonetics and cortisol from Calbiochem. Insulin, transferrin and dbcAMP were from Sigma. Nylon membranes were from Amersham. PKC antibodies were from Transduction Laboratories. Bovine calf serum and fetal calf serum were from HyClone Laboratories. All radiolabelled compounds were purchased from New England Nuclear. PVDF membranes were from Bio-Rad Laboratories. FuGene6 was purchased from Roche Diagnostics (Indianapolis, IN, U.S.A.). TRIzol® reagent was from Gibco BRL Life Technologies. CAT Enzyme Assay and Dual-Luciferase Reporter Assay systems were from Promega.
Cells and media
Neonatal foreskins were used to culture human melanocytes as previously described [32]. In brief, the epidermis was separated from the dermis after overnight incubation in 0.25% trypsin at 4 °C. Primary cultures were then established in M199 medium containing 1.8 mM Ca2+ and supplemented with 10−9 M tri-iodothyronine, 10 μg/ml insulin, 1.4×10−6 M cortisol, BPE (35 μg/ml), 80 μM dbcAMP, 10 ng/ml basic fibroblast growth factor and 5–10% (v/v) fetal bovine serum. All post-primary cultures were maintained in a low-calcium (0.03 mM) version of this medium known to selectively support melanocyte growth [36]. Cells at first to third passages were routinely used.
Immunoblot analysis
Immunoblot analysis was performed as described previously [32,34]. Cells were harvested in RIPA buffer [0.25 M Tris, pH 7.5, 0.5 M NaCl, 2.5% (w/v) SDS and 0.1% Triton X-100] containing protease inhibitors (100 μM okadaic acid, 100 μM PMSF, 100 μg/ml aprotinin, 2 μg/ml leupeptin and 100 μM sodium orthovanadate). Protein samples were subjected to SDS/PAGE (7.5% gel) and transferred to a nitrocellulose membrane or PVDF membrane electrophoretically. The membrane was pre-incubated in 100% Blotto (5 g of non-fat dry milk in 100 ml of PBS) for 3 h at room temperature (25–30 °C) with shaking, followed by an overnight incubation with antiserum [0.5–1 μg/ml in 10% (w/v) Blotto] at 4 °C. At the end of the incubation, the membrane was washed extensively with PBS containing 0.5% Tween 20, and processed using the ECL® kit. The membrane was then exposed to Kodak X-OMAT film.
Tyrosinase activity
Tyrosinase activity was measured by the method of Pomerantz [37]. In brief, 5×105 cells were briefly sonicated in 80 mM phosphate buffer (pH 6.8) containing 1% Triton X-100, and tyrosinase was extracted for 60 min at 4 °C. Cellular protein (10–50 μg) was incubated with 250 nM L-tyrosine, 25 nM L-dihydroxyphenylalanine and 5 μCi of L-[3,5-3H]tyrosine (40–60 Ci/mmol) for 30–60 min at 37 °C. The reaction was stopped by addition of 500 μl of 10% (w/v) trichloroacetic acid containing 0.2% BSA. Trichloroacetic acid-soluble material was reacted with Norit A, and released 3H2O was measured using a scintillation counter. The activity was expressed as c.p.m. of 3H2O released·(μg of protein)−1·h−1 minus the non-specific incorporation of radioactivity, determined by using lysate boiled for 30 min (background). Background was generally less than 5–10% of the sample.
Northern-blot analysis
Total cellular RNA was isolated using TRIzol® reagent according to the manufacturer's instructions. Routinely, 20 μg of total cellular RNA per lane was size-fractionated through a 1% agarose gel containing 6% (v/v) formaldehyde. The RNA was then transferred to a Hybond-N nylon membrane and immobilized by short UV light illumination. Blots were prehybridized in prehybridization solution [10% (w/v) dextran sulphate, 5× Denhardt's (1× Denhardt's is 0.02% Ficoll 400, 0.02% polyvinylpyrrolidone and 0.02% BSA), 50% (v/v) deionized formamide, 75 μg/ml salmon sperm DNA and 0.1% SDS] for 4–24 h at 45 °C, and hybridized with the radiolabelled specific probe directly added to the prehybridization solution for 48–72 h at 45 °C. Then the blot was washed twice in 2× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) at 65 °C for 30 min each, and exposed to a Kodak X-OMAT film.
Probes
The cDNA probes for human tyrosinase and PKC-β were purchased from A.T.C.C. cDNAs were radiolabelled with [α-32P]dCTP (3000 Ci/mmol; New England Nuclear), using a commercially available oligo-labelling kit (Pharmacia LKB Biotechnology).
RT (reverse transcriptase)–PCR
Total RNA was isolated from subconfluent cultures of human melanocytes and 2 μg of RNA was reverse-transcribed into cDNA. cDNA (0.1 μg) was then amplified with 15 pmol of each forward and reverse primer. All PCR reactions were terminated at the 29 cycles that fell within the exponential phase of amplification. Denaturation was performed at 95 °C for 30 s, primer annealing at 65 °C for 1 min and DNA polymerization at 72 °C for 1 min in a thermal cycle (MJ Research, Waltham, MA, U.S.A.). Specific primers for MITF-M, MITF-A, MITF-B, MITF-H and MITF-E (Table 1) were used to determine the expression of MITF isoforms. Each PCR product was then sequenced and confirmed against published reports [13,22].
Table 1. Primer sequences for MITF isoforms.
| Isoform | Primer sequences |
|---|---|
| M | F1: 5′-ATGCTGGAAATGCTAGAATACAGT-3′ |
| R1: 5′-ATCATCCATCTGCATGCAC-3′ | |
| A | F1: 5′-ATGCAGTCCGAATCGGGGATCGTG-3′ |
| R1: 5′-ATCATCCATCTGCATGCAC-3′ | |
| H | F1: 5′-ATGGAGGCGCTTAGATTTGAGATG-3′ |
| R1: 5′-ATCATCCATCTGCATGCAC-3′ | |
| E | F1: 5′-CCAGATACACAGACAGTCAC-3′ |
| R1: 5′-ATCATCCATCTGCATGCAC-3′ | |
| B | F1: 5′-GATACCCCCAAAGTGCAAACGAAG-3′ |
| R1: 5′-ACACAAATTATCAGAAGAGGCCCACC-3′ |
Vectors
Expression vectors for MITF-M and MITF-A were provided by Dr Shigeki Shibahara at Tohoku University in Japan [13], and dn-MITF was provided by Dr David E. Fisher at the Dana Farber Institute (Harvard Medical School, Boston, MA, U.S.A.) [38]. PKC-β promoter–CAT (chloramphenicol acetyltransferase) constructs (PKC-β/CAT) were obtained from Dr Dennis Cooper at the University of Florida at Miami [35]. Tyrosinase promoter/CAT construct was provided by Dr Gunther Schutz at Institute of Cell and Tumor Biology, German Cancer Research Center, Heidelberg, Germany [39,40].
Transient transfection
In order to introduce dn-MITF, FuGene6 was used according to the instructions of the manufacturer. Routinely, melanocytes were passaged on the day before the transfection at 60–80% confluence and 2–5 μg of DNA was introduced into a 100 mm dish. Cells were harvested 48–72 h after transfection. To transfect Cos-7 cells, cells were plated at 5×105 cells/60 mm plate 24 h prior to transfection, and 0.4 μg of promoterless Renilla luciferase reporter construct was co-transfected to assess and normalize differences in the transfection efficiencies among the plates. Cells were harvested at 48 h after the transfection, and CAT and luciferase assays were performed according to the manufacturer's instructions. Results were expressed as the ratio of CAT activity to luciferase activity.
cAMP determination
The intracellular level of cAMP was measured using Cyclic AMP Enzyme Immunoassay kit from BIOMOL.
RESULTS
IBMX induces the protein level of PKC-β
The cAMP-dependent pathway has been shown to be a major positive regulator of melanogenesis [1,6,41–46]. cAMP induces these melanogenic proteins at least in part through MITF [15,24]. Because PKC-β plays a key role during melanogenesis [32,34], we asked whether increasing the intracellular level of cAMP in cultured melanocytes also increases the expression of PKC-β, as seen with other key melanogenic proteins.
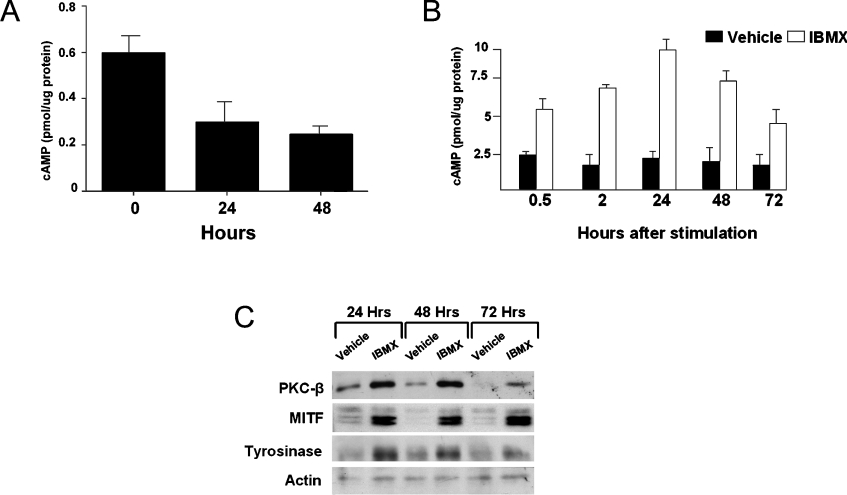
Because human melanocytes are customarily grown in M199 medium supplemented with growth factors, BPE and dbcAMP [47], or with choleragen [34], the basal intracellular level of cAMP in melanocytes is customarily elevated. To avoid masking effects of cAMP on the expression of PKC-β by an already elevated basal level of cAMP, paired cultures of subconfluent human melanocytes were re-fed with serum-free DMEM for 24 and 48 h, and the intracellular level of cAMP was measured. The intracellular level of cAMP was maximally reduced within 24 h of re-feeding with serum-free DMEM (Figure 1A). Therefore cells were routinely re-fed with serum-free DMEM prior to treatment with IBMX, a cAMP-elevating agent. Interestingly, in the absence of strong stimulators of cAMP, a wide range of basal levels of cAMP were found in melanocytes cultured from different foreskin donors, as also observed by others [48]. In our cultures, the basal concentration of cAMP ranged from 0.2 to 3.0 pmol/μg of protein.
Figure 1. IBMX increases the protein levels of PKC-β, MITF and tyrosinase.
(A) Paired cultures of melanocytes were re-fed with serum-free DMEM. Cells were harvested and intracellular level of cAMP was measured, 24 and 48 h after introducing serum-free DMEM. In three independent experiments, the intracellular level of cAMP was decreased significantly at both 24 h (P<0.03) and 48 h (P<0.01). One representative experiment of the three separate experiments is presented. (B) Paired cultures of melanocytes were depleted of cAMP and treated with vehicle or 100 μM IBMX or vehicle alone. The levels of intracellular cAMP were measured at the indicated time points. In two independent experiments, IBMX increased the level of intracellular cAMP within 30 min, with maximal increase at 24 h, and over 72 h. (C) Paired cultures were immunoblotted for PKC-β, MITF and tyrosinase. For the loading control, the membrane was immunoblotted for actin. A representative result from three independent experiments is presented. Densitometric analysis normalized against the loading control showed that the IBMX increased the level of all proteins at all three time points: PKC-β by 198±42% (P<0.05, 24 h), 246±70% (P<0.05, 48 h) and 535±150% (P<0.05, 72 h); tyrosinase by 285±1.0% (P<0.001, 24 h), 231±92% (P<0.05, 48 h), 254±70% (P<0.05, 72 h); and MITF by 160±50% (P<0.04, 24 h), 367±36% (P<0.01, 48 h) and 290±40% (P<0.01, 72 h).
To determine whether IBMX induces the expression of PKC-β, basal levels of cAMP were first reduced and cells were then treated with 100 μM IBMX or vehicle alone (DMSO) with fresh provision of medium. Paired plates were harvested at 0.5, 2, 24, 48 and 72 h after treatment to determine the intracellular level of cAMP. IBMX increased intracellular levels of cAMP as expected (Figure 1B). Maximal induction of >3-fold was observed after 24 h, and the cAMP level remained elevated even 72 h after IBMX treatment. In paired dishes, the protein levels of MITF, PKC-β and tyrosinase were determined by immunoblot analysis using monoclonal antibodies specific for each protein. In the vehicle-treated group, the levels of tyrosinase and PKC-β proteins were higher at 24 h than at 48 and 72 h, as frequently observed in this melanocyte culture system (H.-Y. Park and B. A. Gilchrest, unpublished work). Nevertheless, within 24 h, IBMX induced the level of PKC-β as well as MITF, and also induced tyrosinase within 48 h (Figure 1C). The induction of all three proteins persisted through 72 h after the IBMX treatment. These results suggest that cAMP co-ordinately regulates the expression of PKC-β, tyrosinase and MITF.
IBMX induces the level of PKC-β mRNA
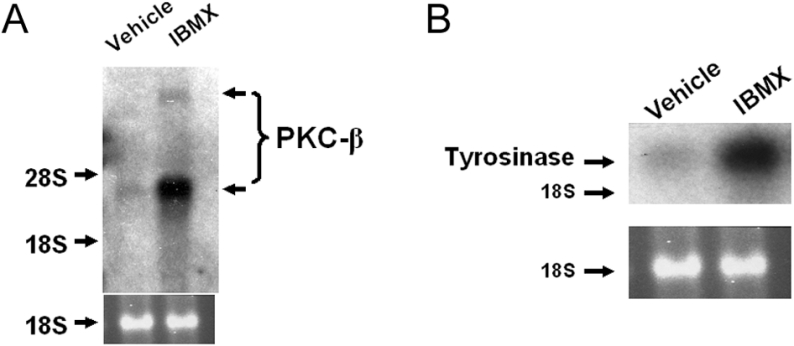
If MITF transcriptionally increases expression of PKC-β when the intracellular level of cAMP is elevated, then the mRNA level of PKC-β should also be induced by cAMP. To determine whether the cAMP-dependent pathway increases the level of PKC-β mRNA, basal levels of cAMP were first lowered as described above and the paired cultures of melanocytes were then treated with 100 μM IBMX or vehicle alone for 48 h. Cells were harvested and Northern-blot analysis was performed using cDNA specific for PKC-β. IBMX induced the two known transcripts of PKC-β mRNA (Figure 2A), consistent with the cAMP-dependent pathway regulating transcription of PKC-β through MITF. In parallel, the level of tyrosinase was also examined. The IBMX-induced increases in the intracellular level of cAMP also increased the level of tyrosinase mRNA (Figure 2B).
Figure 2. IBMX increases the levels of PKC-β and tyrosinase mRNAs.
Paired cultures of melanocytes were depleted of cAMP and treated with 100 μM IBMX or vehicle alone. After 48 h, cells were harvested and Northern-blot analysis was performed using specific cDNA against PKC-β (A) and tyrosinase (B). The 18 S RNA band serves as a loading control. In two independent experiments, IBMX induced PKC-β mRNA levels (768±132%), and in three independent experiments, IBMX induced tyrosinase mRNA (377±100%).
MITF is induced by IBMX prior to tyrosinase and PKC-β
Published DNA sequences of the promoter region of PKC-β reveal at least two E-boxes, at −26 and −110 nt positions [35], suggesting that MITF may directly modulate the transcription of PKC-β. Moreover, if the cAMP-dependent pathway up-regulates the expression of PKC-β through MITF, then cAMP should induce the level of MITF prior to inducing PKC-β. To test this hypothesis, paired cultures of human melanocytes were first depleted of intracellular level of cAMP as before and treated with either 100 μM IBMX or vehicle alone for 4 and 6 h. MITF was strongly induced within 4 h of IBMX treatment, whereas PKC-β and tyrosinase remained unaffected by IBMX even up to 6 h of treatment (Figure 3). Induction of MITF thus preceded the induction of PKC-β and tyrosinase, a result consistent with the possibility that cAMP induces the level of PKC-β through MITF.
Figure 3. IBMX increases the protein level of MITF.
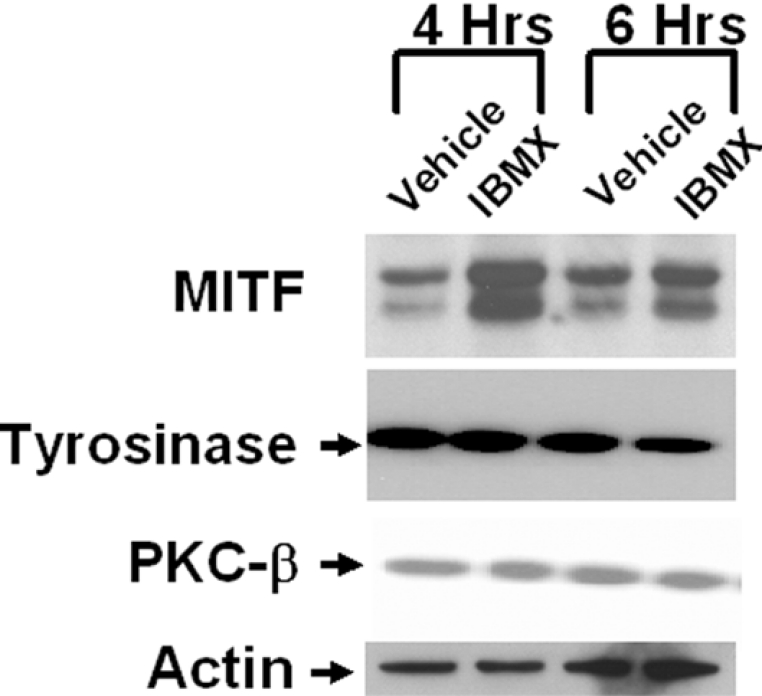
Paired cultures of melanocytes were depleted of cAMP and treated with vehicle or 100 μM IBMX for 4 and 6 h. Cells were harvested and immunoblot analysis was performed using monoclonal antibodies specific for MITF, tyrosinase and PKC-β. For the loading control, the membrane was immunoblotted for actin. A representative immunoblot from one of two independent experiments is shown. Densitometric analysis showed that IBMX caused an increase in the level of MITF by approx. 560±60% and approx. 400±35% over 4 and 6 h respectively, but no significant changes were observed for PKC-β and tyrosinase.
dn-MITF reduces the protein levels of tyrosinase and PKC-β
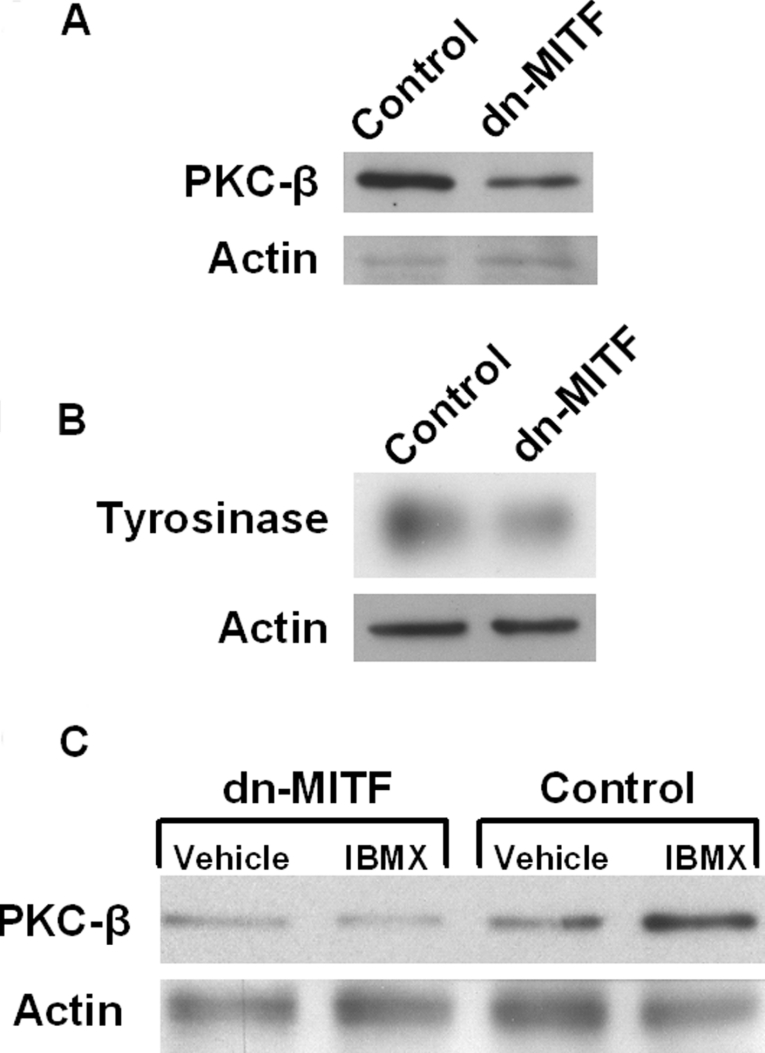
To explore whether MITF is involved in the expression of PKC-β, a set of subconfluent melanocyte cultures was transfected with dn-MITF or a control vector. Cells were harvested 48 h after the transfection, and the levels of PKC-β and tyrosinase proteins were examined by immunoblot analysis using specific monoclonal antibodies against each protein. dn-MITF reduced the level of PKC-β and tyrosinase (Figures 4A and 4B). As determined by densitometric analysis of three separate experiments, PKC-β level was reduced by 63±3.7% (P<0.001) and tyrosinase level was reduced by 39.7±16.9% (P<0.027). These results confirm that MITF up-regulates PKC-β and are consistent with direct transcriptional regulation via MITF binding to the known E-boxes in the PKC-β promoter.
Figure 4. dn-MITF reduced the basal protein levels of PKC-β and tyrosinase.
Paired cultures of melanocytes were transfected with dn-MITF or control vector. Cells were harvested after 48 h and immunoblotted for PKC-β (A) and tyrosinase (B), with actin as a loading control. Densitometric analysis of three separate experiments showed that the level of PKC-β was decreased by 63±3.7% (P<0.001) and the level of tyrosinase was decreased by 39.7±16.9% (P<0.027) in dn-MITF-transfected cells when compared with control vector-transfected cells. (C) Paired cultures of melanocytes were transfected with dn-MITF or control vector. Then, cells were treated with 100 μM IBMX or vehicle alone to determine whether dn-MITF inhibited IBMX-induced increases in PKC-β. Densitometric analysis showed that IBMX induced the level of PKC-β in control vector-transfected cells by 240%, whereas IBMX did not induce PKC-β in dn-MITF-transfected cells.
In order to confirm that MITF plays a critical role in the cAMP-induced increases in PKC-β expression, a paired culture of subconfluent melanocytes was transfected with dn-MITF or control vector, after which cultures in each group were treated with 100 μM IBMX or vehicle alone. As expected, in control vectortransfected cells, IBMX induced the PKC-β protein (Figure 4C). However, transfection of dn-MITF blocked IBMX-induced increase in PKC-β (Figure 4C), further demonstrating that MITF plays a critical role in IBMX-induced increase in PKC-β.
MITF-M up-regulates the transcriptional activity of the PKC-β promoter
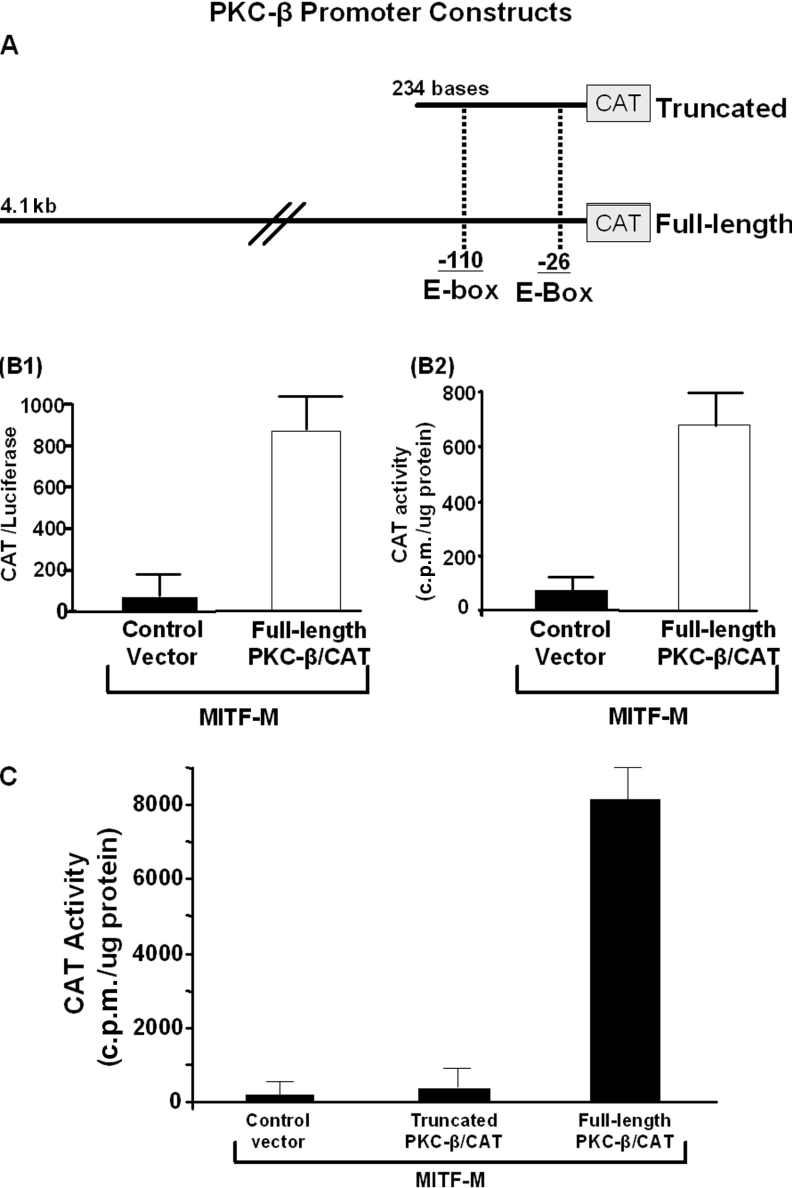
To determine whether MITF-M transcriptionally up-regulates PKC-β, a CAT-reporter construct containing the full-length 4.1 kb PKC-β upstream promoter or, in separate experiments, a truncated promoter (Figure 5A) and an expression vector for MITF-M, were co-transfected into Cos-7 cells. Renilla luciferase was also introduced to assess the transfection efficiencies among samples. Untransfected Cos-7 cells did not express detectable levels of MITF (results not shown). Cells were harvested 48 h after the transfection. MITF-M up-regulated full-length PKC-β promoter activity approx. 60-fold (Figure 5B1), suggesting that MITF-M is an important transcription factor for endogenous PKC-β. When Renilla luciferase was not co-transfected to allow for normalization of transfection rates and the result was expressed as CAT activity (c.p.m./μg of protein), a similar induction was observed (Figure 5B2), suggesting a minimal difference in transfection efficiencies between the samples. Therefore, in subsequent transfection experiments, results were expressed as CAT activity (c.p.m./μg of protein).
Figure 5. MITF up-regulates the transcription of PKC-β.
(A) Diagram of the PKC-β promoter constructs. (B) Cos-7 cells were co-transfected with MITF-M and full-length PKC-β/CAT. (B1) In one set of Cos-7 cells, promoterless Renilla luciferase was co-transfected to assess the transfection efficiencies; 48 h after the transfection, cells were harvested, CAT and luciferase activities were determined and results were expressed as CAT/luciferase. (B2) In another set of Cos-7 cells, Cos-7 cells were co-transfected with MITF-M and PKC-β/CAT and results were expressed as CAT activity (c.p.m./μg of protein). Both approaches showed similar results when transfections were performed with (two separate experiments) and without (two separate experiments) Renilla luciferase. (C) Cos-7 cells were co-transfected with MITF-M and full-length PKC-β/CAT or truncated PKC-β/CAT; 48 h after the transfection, cells were harvested and CAT assay was performed. In three separate transfections, MITF-M failed to transactivate truncated PKC-β/CAT.
To determine whether the presence of E-boxes alone is sufficient to up-regulate PKC-β promoter activity by MITF, the severely truncated PKC-β promoter that nevertheless contains both known E-boxes was co-transfected with MITF-M. The full-length PKC-β promoter was co-transfected in paired cultures as a control. As expected, the full-length construct was fully activated in the presence of MITF-M, but despite the presence of E-boxes, the truncated construct was inactive (Figure 5C). These results suggest that, as for tyrosinase [49], MITF-M alone is not sufficient to upregulate PKC-β transcription.
Only the MITF-M isoform up-regulates transcription of PKC-β
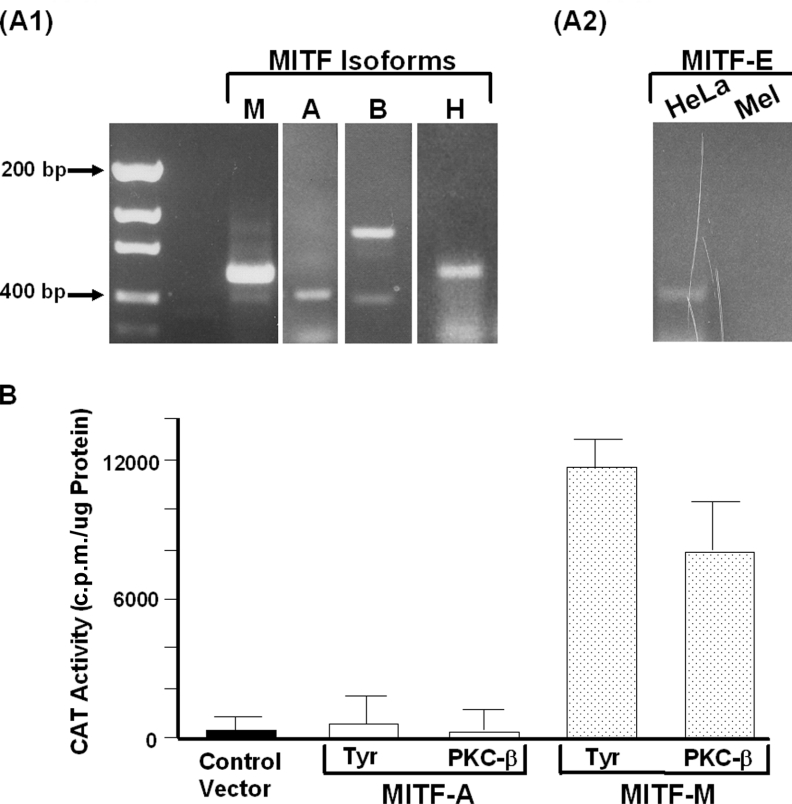
Because MITF has multiple isoforms, we asked if other forms are expressed in melanocytes. RT–PCR revealed that MITF-M, MITF-B, MITF-H and MITF-A, but not MITF-E, are expressed in human melanocytes (Figures 6A1 and 6A2). Because MITF-A and PKC-β are expressed together in many cell types [50–52], we examined the ability of MITF-A to up-regulate transcription of PKC-β as well as tyrosinase, a classic melanogenic protein. Paired cultures of Cos-7 cells were transfected with MITF-M or MITF-A isoforms with the full-length CAT construct of PKC-β or a full-length CAT construct of tyrosinase. Only MITF-M was capable of up-regulating the transcription of PKC-β and tyrosinase (Figure 6B).
Figure 6. MITF-A does not up-regulate transcription of PKC-β.
(A) To determine the expression of MITF isoforms, RT–PCR was performed using RNA isolated from melanocytes and specific primers against MITF-M, MITF-A, MITF-B, MITF-H and MITF-E. Each PCR product was sequenced to confirm its identity. MITF-M, MITF-A, MITF-B and MITF-H were readily detected in melanocytes (A1). RT–PCR using primers specific for the MITF-E failed to detect an expression in melanocytes (Mel), whereas MITF-E expression was easily detected in HeLa cells (A2), a cell type known to express MITF-E [60]. (B) Cos-7 cells were co-transfected with MITF-M or MITF-A with full-length PKC-β/CAT or tyrosinase/CAT. In three separate experiments, MITF-A failed to transactivate tyrosinase/CAT and PKC-β/CAT, whereas MITF-M transactivated both PKC-β/CAT and tyrosinase/CAT.
DISCUSSION
The cAMP-dependent pathway has been shown to be a key modulator of melanogenesis. Recent studies show that cAMP-induced increases in the expression of many melanogenic proteins are mediated through MITF [13,15]. We demonstrate that the cAMP-dependent pathway also up-regulates the expression of PKC-β via MITF. Similar to tyrosinase and TRP-1, the PKC-β promoter also lacks the DNA consensus sequences for CRE [35], but the PKC-β promoter contains at least two E-boxes [35], a DNA consensus sequence known to interact with MITF [12,13]. The co-transfection of MITF-M and a full-length PKC-β promoter into Cos-7 cells resulted in transactivation of the PKC-β promoter. Consistent with the hypothesis that MITF-M is a key transcription factor for PKC-β, transfection of dn-MITF into cultured melanocytes reduced the level of PKC-β. As expected, the level of tyrosinase was also reduced. However, dn-MITF did not completely abolish the expression of PKC-β and tyrosinase. This may be due to the inability of dn-MITF to completely block the activity of endogenous MITF or, alternatively, MITF may not be the only transcription factor regulating the expression of PKC-β.
Our results also suggest that loss of MITF transcriptional activity decreases tyrosinase activity proportionally to its effect on PKC-β protein levels, rather than to its effect on tyrosinase protein level. This would be consistent with the lack of correlation between tyrosinase activity and the level of tyrosinase protein in melanocytes [32,36,53,54] and the observation that depletion of PKC-β decreases or abolishes melanogenesis in human melanocytes [32,34]. Moreover, many amelanotic melanoma cell lines express ample tyrosinase [32,54,55] but lack PKC-β expression [55]. While some of the discrepancies between the expression of tyrosinase and the lack of melanin in melanoma cells may be attributed to mis-regulation in post-translational modification of tyrosinase, such as incomplete glycosylation of tyrosinase [56], in at least some amelanotic melanoma cells, re-introduction of PKC-β rescues melanin production [32]. These results suggest that activation of tyrosinase by PKC-β may be the rate-limiting event during melanogenesis.
In one experiment in which melanocytes were transfected with dn-MITF or control vector, tyrosinase activity was measured in parallel. Interestingly, the level of decrease in tyrosinase activity in the cells transfected with dn-MITF paralleled the level of decrease in PKC-β, but not the level of decrease in tyrosinase by dn-MITF. In this experiment, in dn-MITF-transfected cells, the level of PKC-β was decreased by 53% and the level of tyrosinase was decreased by 24%, whereas tyrosinase activity measured in parallel was decreased by 55% (9826 c.p.m./μg of protein in control vector-transfected cells versus 5563 c.p.m./μg of protein in dn-MITF-transfected cells). This result further supports the idea that the level of PKC-β may be rate-limiting for tyrosinase activity.
MITF-M transactivated the full-length promoter PKC-β promoter, but not a truncated PKC-β promoter, despite the fact that the latter construct contained both E-boxes. This result suggests that MITF-M is an important but not sufficient factor for transactivating transcription of the PKC-β gene. In this context, it is interesting that MITF-M is a key transcription factor for tyrosinase, but is not alone sufficient to transactivate transcription of the tyrosinase gene [49]. Transcription factors that work with MITF-M to up-regulate PKC-β transcription are yet to be elucidated, although binding sites for AP-2 (activating protein-2) and Smad (Sma and Mad protein) are present in PKC-β promoter [35].
It is also possible that MITF does not directly bind to the PKC-β promoter. Because cells are harvested 48 h after transfection of melanocytes with dn-MITF, dn-MITF may have affected the level of other transcription factors required for PKC-β expression. However, the fact that dn-MITF blocks the IBMX-induced increase in PKC-β demonstrates that MITF plays a key role, direct or indirect, in mediating the cAMP-induced increase in PKC-β.
The MITF-M isoform has been associated with melanocytic cells [18,23], and has been thought to be the key transcription factor for melanogenic proteins. However, all MITF isoforms have identical DNA-binding sites [13] and potentially their activities overlap. Moreover, determination of the expression of MITF isoforms in a particular cell type has been difficult due to the lack of MITF antibodies specific for each isoform. The currently available antibody against MITF has been shown to cross-react with at least M, A and H isoforms of MITF [18,52,57]. Our present results using RT–PCR demonstrate that, in addition to MITF-M, human melanocytes express MITF-A, MITF-B and MITF-H but not MITF-E. However, when full-length PKC-β promoter construct was co-transfected with MITF-A or MITF-M isoforms, only MITF-M was capable of up-regulating PKC-β transcription. These results suggest that, in spite of sharing a high percentage of homology, each MITF isoform exerts specific biological actions. Whether this is because they recruit different co-activators, have differential affinities for binding to E-boxes, or other unique properties, is yet to be elucidated. Regardless, the biological function of MITF-A, MITF-B and MITF-H in melanocytes needs to be further explored.
MITF has also been shown to be involved in melanocyte survival [38]. It specifically up-regulates the anti-apoptotic protein Bcl-2 [38] and is often overexpressed in melanomas, as determined by immunohistochemistry [57–59], suggesting it may be a reliable diagnostic marker for melanoma [57–59]. Unfortunately, the only antibody available for MITF-M cross-reacts with other MITF isoforms [18,52,57]. When we transfected MITF-M and MITF-A into Cos-7 cells, identical bands were detected by immunoblot analysis (results not shown). Therefore, at present, one cannot determine which of the MITF isoforms are up-regulated in melanomas.
In summary, our results demonstrate that the expression of PKC-β, the kinase responsible for activating tyrosinase, is up-regulated by cAMP via the key transcription factor MITF-M. These results strongly support the co-ordinate regulation of multiple melanogenic proteins in melanocytes and establish that the PKC and cAMP pathways interact in the regulation of mammalian pigmentation.
Acknowledgments
We thank Dr David E. Fisher for providing dn-MITF construct and the monoclonal antibody against MITF. We also thank Dr Shigeki Shibahara and Dr Dennis Cooper for providing MITF expression vectors and PKC-β/CAT constructs respectively. We thank Mr Shaun Peterson for the excellent technical assistance.
References
- 1.Abdel-Malek Z., Swope V. B., Suzuki I., Akcali C., Harriger M. D., Boyce S. T., Urabe K., Hearing V. J. Mitogenic and melanogenic stimulation of normal human melanocytes by melanotropic peptides. Proc. Natl. Acad. Sci. U.S.A. 1995;92:1789–1793. doi: 10.1073/pnas.92.5.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fuller B. B., Lunsford J. B., Iman D. S. Alpha-melanocyte-stimulating hormone regulation of tyrosinase in Cloudman S-91 mouse melanoma cell cultures. J. Biol. Chem. 1987;262:4024–4033. [PubMed] [Google Scholar]
- 3.Fuller B. B., Viskochil D. H. The role of RNA and protein synthesis in mediating the action of MSH on mouse melanoma cells. Life Sci. 1979;24:2405–2415. doi: 10.1016/0024-3205(79)90448-x. [DOI] [PubMed] [Google Scholar]
- 4.Halaban R., Pomerantz S. H., Marshall S., Lambert D. T., Lerner A. B. Regulation of tyrosinase in human melanocytes grown in culture. J. Cell Biol. 1983;97:480–488. doi: 10.1083/jcb.97.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halaban R., Pomerantz S. H., Marshall S., Lerner A. B. Tyrosinase activity and abundance in Cloudman melanoma cells. Arch. Biochem. Biophys. 1984;230:383–387. doi: 10.1016/0003-9861(84)90121-8. [DOI] [PubMed] [Google Scholar]
- 6.Korner A., Pawelek J. Activation of melanoma tyrosinase by a cyclic AMP-dependent protein kinase in a cell-free system. Nature (London) 1977;267:444–447. doi: 10.1038/267444a0. [DOI] [PubMed] [Google Scholar]
- 7.Pawelek J. M. Studies on the Cloudman melanoma cell line as a model for the action of MSH. Yale J. Biol. Med. 1985;58:571–578. [PMC free article] [PubMed] [Google Scholar]
- 8.Cone R. D., Lu D., Koppula S., Vage D. I., Klungland H., Boston B., Chen W., Orth D. N., Pouton C., Kesterson R. A. The melanocortin receptors: agonists, antagonists, and the hormonal control of pigmentation. Recent Prog. Horm. Res. 1996;51:287–317. discussion 318. [PubMed] [Google Scholar]
- 9.Cone R. D., Mountjoy K. G., Robbins L. S., Nadeau J. H., Johnson K. R., Roselli-Rehfuss L., Mortrud M. T. Cloning and functional characterization of a family of receptors for the melanotropic peptides. Ann. N.Y. Acad. Sci. 1993;680:342–363. doi: 10.1111/j.1749-6632.1993.tb19694.x. [DOI] [PubMed] [Google Scholar]
- 10.Connor M., Christie M. D. Opioid receptor signalling mechanisms. Clin. Exp. Pharmacol. Physiol. 1999;26:493–499. doi: 10.1046/j.1440-1681.1999.03049.x. [DOI] [PubMed] [Google Scholar]
- 11.Edelman A. M., Blumenthal D. K., Krebs E. G. Protein serine/threonine kinases. Annu. Rev. Biochem. 1987;56:567–613. doi: 10.1146/annurev.bi.56.070187.003031. [DOI] [PubMed] [Google Scholar]
- 12.Saito H., Yasumoto K., Takeda K., Takahashi K., Yamamoto H., Shibahara S. Microphthalmia-associated transcription factor in the Wnt signaling pathway. Pigment Cell Res. 2003;16:261–265. doi: 10.1034/j.1600-0749.2003.00039.x. [DOI] [PubMed] [Google Scholar]
- 13.Shibahara S., Takeda K., Yasumoto K., Udono T., Watanabe K., Saito H., Takahashi K. Microphthalmia-associated transcription factor (MITF): multiplicity in structure, function, and regulation. J. Investig. Dermatol. Symp. Proc. 2001;6:99–104. doi: 10.1046/j.0022-202x.2001.00010.x. [DOI] [PubMed] [Google Scholar]
- 14.Widlund H. R., Fisher D. E. Microphthalamia-associated transcription factor: a critical regulator of pigment cell development and survival. Oncogene. 2003;22:3035–3041. doi: 10.1038/sj.onc.1206443. [DOI] [PubMed] [Google Scholar]
- 15.Bertolotto C., Abbe P., Hemesath T. J., Bille K., Fisher D. E., Ortonne J. P., Ballotti R. Microphthalmia gene product as a signal transducer in cAMP-induced differentiation of melanocytes. J. Cell Biol. 1998;142:827–835. doi: 10.1083/jcb.142.3.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bertolotto C., Bille K., Ortonne J. P., Ballotti R. In B16 melanoma cells, the inhibition of melanogenesis by TPA results from PKC activation and diminution of microphthalmia binding to the M-box of the tyrosinase promoter. Oncogene. 1998;16:1665–1670. doi: 10.1038/sj.onc.1201685. [DOI] [PubMed] [Google Scholar]
- 17.Lin C. B., Babiarz L., Liebel F., Roydon Price E., Kizoulis M., Gendimenico G. J., Fisher D. E., Seiberg M. Modulation of microphthalmia-associated transcription factor gene expression alters skin pigmentation. J. Invest. Dermatol. 2002;119:1330–1340. doi: 10.1046/j.1523-1747.2002.19615.x. [DOI] [PubMed] [Google Scholar]
- 18.Yasumoto K., Yokoyama K., Takahashi K., Tomita Y., Shibahara S. Functional analysis of microphthalmia-associated transcription factor in pigment cell-specific transcription of the human tyrosinase family genes. J. Biol. Chem. 1997;272:503–509. doi: 10.1074/jbc.272.1.503. [DOI] [PubMed] [Google Scholar]
- 19.Goding C. R. Mitf from neural crest to melanoma: signal transduction and transcription in the melanocyte lineage. Genes Dev. 2000;14:1712–1728. [PubMed] [Google Scholar]
- 20.Tachibana M., Takeda K., Nobukuni Y., Urabe K., Long J. E., Meyers K. A., Aaronson S. A., Miki T. Ectopic expression of MITF, a gene for Waardenburg syndrome type 2, converts fibroblasts to cells with melanocyte characteristics. Nat. Genet. 1996;14:50–54. doi: 10.1038/ng0996-50. [DOI] [PubMed] [Google Scholar]
- 21.Tassabehji M., Newton V. E., Read A. P. Waardenburg syndrome type 2 caused by mutations in the human microphthalmia (MITF) gene. Nat. Genet. 1994;8:251–255. doi: 10.1038/ng1194-251. [DOI] [PubMed] [Google Scholar]
- 22.Hershey C. L., Fisher D. E. Genomic analysis of the Microphthalmia locus and identification of the MITF-J/Mitf-J isoform. Gene. 2005;347:73–82. doi: 10.1016/j.gene.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 23.Fuse N., Yasumoto K., Suzuki H., Takahashi K., Shibahara S. Identification of a melanocyte-type promoter of the microphthalmia-associated transcription factor gene. Biochem. Biophys. Res. Commun. 1996;219:702–707. doi: 10.1006/bbrc.1996.0298. [DOI] [PubMed] [Google Scholar]
- 24.Price E. R., Horstmann M. A., Wells A. G., Weilbaecher K. N., Takemoto C. M., Landis M. W., Fisher D. E. Alpha-melanocyte-stimulating hormone signaling regulates expression of microphthalmia, a gene deficient in Waardenburg syndrome. J. Biol. Chem. 1998;273:33042–33047. doi: 10.1074/jbc.273.49.33042. [DOI] [PubMed] [Google Scholar]
- 25.Fang D., Tsuji Y., Setaluri V. Selective down-regulation of tyrosinase family gene TYRP1 by inhibition of the activity of melanocyte transcription factor, MITF. Nucleic Acids Res. 2002;30:3096–3106. doi: 10.1093/nar/gkf424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gordon P. R., Gilchrest B. A. Human melanogenesis is stimulated by diacylglycerol. J. Invest. Dermatol. 1989;93:700–702. doi: 10.1111/1523-1747.ep12319900. [DOI] [PubMed] [Google Scholar]
- 27.Allan A. E., Archambault M., Messana E., Gilchrest B. A. Topically applied diacylglycerols increase pigmentation in guinea pig skin. J. Invest. Dermatol. 1995;105:687–692. doi: 10.1111/1523-1747.ep12324466. [DOI] [PubMed] [Google Scholar]
- 28.Park H. Y., Gonzalez S., Lee J., Middelkamp-Hup M. A., Kapasi S., Peterson S., Gilchrest B. A. Topical application of a PKC inhibitor reduces skin and hair pigmentation. J. Invest. Dermatol. 2004;122:159–166. doi: 10.1046/j.0022-202X.2003.22134.x. [DOI] [PubMed] [Google Scholar]
- 29.Niles R. M., Loewy B. P. Induction of protein kinase C in mouse melanoma cells by retinoic acid. Cancer Res. 1989;49:4483–4487. [PubMed] [Google Scholar]
- 30.Park H. Y., Gilchrest B. A. Reduction of melanogenic activity and responsiveness to alpha-melanocyte stimulating hormone during serial passage of melanoma cells. J. Cutan. Med. Surg. 1996;1:4–9. [Google Scholar]
- 31.Park H. Y., Russakovsky V., Ao Y., Fernandez E., Gilchrest B. A. Alpha-melanocyte stimulating hormone-induced pigmentation is blocked by depletion of protein kinase C. Exp. Cell Res. 1996;227:70–79. doi: 10.1006/excr.1996.0251. [DOI] [PubMed] [Google Scholar]
- 32.Park H. Y., Russakovsky V., Ohno S., Gilchrest B. A. The beta isoform of protein kinase C stimulates human melanogenesis by activating tyrosinase in pigment cells. J. Biol. Chem. 1993;268:11742–11749. [PubMed] [Google Scholar]
- 33.King R. A., Townsend D., Oetting W. S. Inherited hypopigmentary disorders. In: Levine N., editor. Pigmentation and Pigmentary Disorders. Boca Raton, FL: CRC Press; 1993. pp. 297–336. [Google Scholar]
- 34.Park H. Y., Perez J. M., Laursen R., Hara M., Gilchrest B. A. Protein kinase C-beta activates tyrosinase by phosphorylating serine residues in its cytoplasmic domain. J. Biol. Chem. 1999;274:16470–16478. doi: 10.1074/jbc.274.23.16470. [DOI] [PubMed] [Google Scholar]
- 35.Patel N. A., Chalfant C. E., Yamamoto M., Watson J. E., Eichler D. C., Cooper D. R. Acute hyperglycemia regulates transcription and posttranscriptional stability of PKCbetaII mRNA in vascular smooth muscle cells. FASEB J. 1999;13:103–113. doi: 10.1096/fasebj.13.1.103. [DOI] [PubMed] [Google Scholar]
- 36.Naeyaert J. M., Eller M., Gordon P. R., Park H. Y., Gilchrest B. A. Pigment content of cultured human melanocytes does not correlate with tyrosinase message level. Br. J. Dermatol. 1991;125:297–303. doi: 10.1111/j.1365-2133.1991.tb14161.x. [DOI] [PubMed] [Google Scholar]
- 37.Pomerantz S. H. Tyrosine hydroxylation catalyzed by mammalian tyrosinase: an improved method of assay. Biochem. Biophys. Res. Commun. 1964;16:188–194. doi: 10.1016/0006-291x(64)90359-6. [DOI] [PubMed] [Google Scholar]
- 38.McGill G. G., Horstmann M., Widlund H. R., Du J., Motyckova G., Nishimura E. K., Lin Y. L., Ramaswamy S., Avery W., Ding H. F., et al. Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell (Cambridge, Mass.) 2002;109:707–718. doi: 10.1016/s0092-8674(02)00762-6. [DOI] [PubMed] [Google Scholar]
- 39.Beermann F., Ruppert S., Hummler E., Schutz G. Tyrosinase as a marker for transgenic mice. Nucleic Acids Res. 1991;19:958. doi: 10.1093/nar/19.4.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kluppel M., Beermann F., Ruppert S., Schmid E., Hummler E., Schutz G. The mouse tyrosinase promoter is sufficient for expression in melanocytes and in the pigmented epithelium of the retina. Proc. Natl. Acad. Sci. U.S.A. 1991;88:3777–3781. doi: 10.1073/pnas.88.9.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lerner A. B., Moellmann G., Varga V. L., Halaban R., Pawelek J. Action of melanocyte-stimulating hormone on pigment cells. Cold Spring Harbor Conf. Cell Prolif. 1979;6:187–197. [Google Scholar]
- 42.Lerner A. B., McGuire J. S. Effect of alpha- and betamelanocyte stimulating hormones on the skin colour of man. Nature (London) 1961;189:176–179. doi: 10.1038/189176a0. [DOI] [PubMed] [Google Scholar]
- 43.Pawelek J., Wong G., Sansone M., Morowitz J. Molecular biology of pigment cells. Molecular controls in mammalian pigmentation. Yale J. Biol. Med. 1973;46:430–443. [PMC free article] [PubMed] [Google Scholar]
- 44.Pawelek J. M. Factors regulating growth and pigmentation of melanoma cells. J. Invest. Dermatol. 1976;66:201–209. doi: 10.1111/1523-1747.ep12482134. [DOI] [PubMed] [Google Scholar]
- 45.Wong G., Pawelek J. Control of phenotypic expression of cultured melanoma cells by melanocyte stimulating hormones. Nat. New Biol. 1973;241:213–215. doi: 10.1038/newbio241213a0. [DOI] [PubMed] [Google Scholar]
- 46.Wong G., Pawelek J. Melanocyte-stimulating hormone promotes activation of pre-existing tyrosinase molecules in Cloudman S91 melanoma cells. Nature (London) 1975;255:644–646. doi: 10.1038/255644a0. [DOI] [PubMed] [Google Scholar]
- 47.Park H. Y., Gilchrest B. A. Signaling pathways mediating melanogenesis. Cell. Mol. Biol. (Noisy-le-Grand, France) 1999;45:919–930. [PubMed] [Google Scholar]
- 48.Abdel-Malek Z., Swope V. B., Pallas J., Krug K., Nordlund J. J. Mitogenic, melanogenic, and cAMP responses of cultured neonatal human melanocytes to commonly used mitogens. J. Cell. Physiol. 1992;150:416–425. doi: 10.1002/jcp.1041500226. [DOI] [PubMed] [Google Scholar]
- 49.Gaggioli C., Busca R., Abbe P., Ortonne J. P., Ballotti R. Microphthalmia-associated transcription factor (MITF) is required but is not sufficient to induce the expression of melanogenic genes. Pigment Cell Res. 2003;16:374–382. doi: 10.1034/j.1600-0749.2003.00057.x. [DOI] [PubMed] [Google Scholar]
- 50.Amae S., Fuse N., Yasumoto K., Sato S., Yajima I., Yamamoto H., Udono T., Durlu Y. K., Tamai M., Takahashi K., et al. Identification of a novel isoform of microphthalmia-associated transcription factor that is enriched in retinal pigment epithelium. Biochem. Biophys. Res. Commun. 1998;247:710–715. doi: 10.1006/bbrc.1998.8838. [DOI] [PubMed] [Google Scholar]
- 51.Fuse N., Yasumoto K., Takeda K., Amae S., Yoshizawa M., Udono T., Takahashi K., Tamai M., Tomita Y., Tachibana M., et al. Molecular cloning of cDNA encoding a novel microphthalmia-associated transcription factor isoform with a distinct amino-terminus. J. Biochem. (Tokyo) 1999;126:1043–1051. doi: 10.1093/oxfordjournals.jbchem.a022548. [DOI] [PubMed] [Google Scholar]
- 52.Oboki K., Morii E., Kataoka T. R., Jippo T., Kitamura Y. Isoforms of mi transcription factor preferentially expressed in cultured mast cells of mice. Biochem. Biophys. Res. Commun. 2002;290:1250–1254. doi: 10.1006/bbrc.2002.6332. [DOI] [PubMed] [Google Scholar]
- 53.Jimenez M., Kameyama K., Maloy W. L., Tomita Y., Hearing V. J. Mammalian tyrosinase: biosynthesis, processing, and modulation by melanocyte-stimulating hormone. Proc. Natl. Acad. Sci. U.S.A. 1988;85:3830–3834. doi: 10.1073/pnas.85.11.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salopek T. G., Jimbow K. Induction of melanogenesis during the various melanoma growth phases and the role of tyrosinase, lysosome-associated membrane proteins, and p90 calnexin in the melanogenesis cascade. J. Investig. Dermatol. Symp. Proc. 1996;1:195–202. [PubMed] [Google Scholar]
- 55.Gilhooly E. M., Morse-Gaudio M., Bianchi L., Reinhart L., Rose D. P., Connolly J. M., Reed J. A., Albino A. P. Loss of expression of protein kinase C beta is a common phenomenon in human malignant melanoma: a result of transformation or differentiation? Melanoma Res. 2001;11:355–369. doi: 10.1097/00008390-200108000-00006. [DOI] [PubMed] [Google Scholar]
- 56.Svedine S., Wang T., Halaban R., Hebert D. N. Carbohydrates act as sorting determinants in ER-associated degradation of tyrosinase. J. Cell Sci. 2004;117:2937–2949. doi: 10.1242/jcs.01154. [DOI] [PubMed] [Google Scholar]
- 57.Chang K. L., Folpe A. L. Diagnostic utility of microphthalmia transcription factor in malignant melanoma and other tumors. Adv. Anat. Pathol. 2001;8:273–275. doi: 10.1097/00125480-200109000-00004. [DOI] [PubMed] [Google Scholar]
- 58.Dorvault C. C., Weilbaecher K. N., Yee H., Fisher D. E., Chiriboga L. A., Xu Y., Chhieng D. C. Microphthalmia transcription factor: a sensitive and specific marker for malignant melanoma in cytologic specimens. Cancer. 2001;93:337–343. doi: 10.1002/cncr.9049. [DOI] [PubMed] [Google Scholar]
- 59.Samija I., Lukac J., Maric-Brozic J., Kusic Z. Microphthalmia-associated transcription factor and tyrosinase as markers of melanoma cells in blood of patients with melanoma. Croat. Med. J. 2004;45:142–148. [PubMed] [Google Scholar]
- 60.Vachtenheim J., Novatna H. Expression of genes for microphthalmia isoforms, Pax3 and MSG1, in human melanomas. Cell. Mol. Biol. (Noisy-le-Grand, France) 1999;45:1075–1082. [PubMed] [Google Scholar]