Abstract
CpG islands are important in the protection of adjacent housekeeping genes from de novo DNA methylation and for keeping them in a transcriptionally active state. However, little is known about their capacity to protect heterologous genes and assure position-independent transcription of adjacent transgenes or retroviral vectors. To tackle this question, we have used the mouse aprt CpG island to flank a Rous sarcoma virus (RSV)-derived reporter vector and followed the transcriptional activity of integrated vectors. RSV is an avian retrovirus which does not replicate in mammalian cells because of several blocks at all levels of the replication cycle. Here we show that our RSV-derived reporter proviruses linked to the mouse aprt gene CpG island remain undermethylated and keep their transcriptional activity after stable transfection into both avian and nonpermissive mammalian cells. This effect is most likely caused by the protection from de novo methylation provided by the CpG island and not by enhancement of the promoter strength. Our results are consistent with previous finding of CpG islands in proximity to active but not inactive proviruses and support further investigation of the protection of the gene transfer vectors from DNA methylation.
Cytosine methylation in CpG dinucleotides is an important mechanism of transcriptional regulation in vertebrates. Especially in the genome of mammalian somatic cells, the distribution of CpG dinucleotides, and the pattern of methylation are bimodal. In the major part of the genome, CpGs are underrepresented, dispersed, and predominantly methylated. Approximately 1–2% of the genome consists of nonmethylated CpG-rich stretches (CpG islands) that are typically 0.5–2 kb in length and usually associated with housekeeping genes (1, 2). This bimodal somatic methylation pattern is probably established by general de novo methylation, which skips CpG islands and adjacent gene promoters and leaves them unmethylated, and/or by active demethylation of these sequences. Sp1 sites within the core sequence of CpG islands were shown to be required to prevent methylation (3, 4).
Rous sarcoma virus (RSV) is an avian retrovirus that does not replicate in mammalian host cells. The nonpermissiveness of heterologous hosts is caused by several blocks of the replication cycle. Mammalian cells lack specific receptors for virus entry and do not promote correct splicing of retroviral mRNA, cleavage of viral polyproteins, or assembly of infectious virus particles on the inner surface of the cell membrane (reviewed in ref. 5). In addition to these obstacles, RSV proviral DNA is usually transcriptionally suppressed after integration into the mammalian genome. Fewer than 0.1% of RSV provirus-containing mammalian cells displays morphological transformation by virtue of the v-src oncogene (6, 7). The vast majority of RSV-infected cells harbors transcriptionally silent proviruses with undetectable amount of viral RNA (7, 8). Even in transformed mammalian cells, the proviral expression often tends to spontaneous silencing and segregation of revertant cell clones with nontransformed phenotypes (9–11). This proviral silencing is clearly an epigenetic event because proviral DNA is transiently expressed after transfection into mammalian cells (12, 13), and silenced proviruses can be rescued by molecular cloning and transfer into permissive cells (11). Transcriptionally silenced proviruses were found to be methylated (10, 11) and CpG methylation of RSV proviral constructs has an inhibitory effect on provirus expression (13, 14). Moreover, this inhibitory effect is directed to the proviral long terminal repeats (LTR), containing the promoter/enhancer sequences, because the Moloney murine leukemia virus (MoMLV) LTR inserted instead of the RSV LTR is not suppressed in this way (13).
The transcriptional activity of the provirus depends strongly on the cellular DNA sequences adjacent immediately to the provirus. We have described the presence of nonspecific gene silencer region 5′ to the simplified provirus composed of LTR-v-src-LTR in the tumor-derived cell line H-19 with a high rate of spontaneous morphological reversion (15). Comparison of cellular sequences flanking active and inactive RSV proviruses in rat cells showed the presence of CpG island-specific restriction sites close to the transcriptionally active but not inactive proviruses (16). In fact, the existence of blocks preventing viral replication in nonpermissive cells, as well as provirus silencing, were recognized first on the model of RSV transformed and revertant cells (reviewed in ref. 17) and provided a paradigm for other retrovirus–cell interaction, viral persistence included.
Several strategies were applied with the aim of overcoming the inhibitory effects of DNA methylation and the site of integration against proviral vectors or transgenes. Insertion of a chromatin insulator fragment from the chicken β-globin locus into both LTRs of a MoMLV-derived vector increased the probability of vector expression at random chromosomal integration sites (18). This was accompanied by a decreased level of proviral DNA methylation. The same chromatin insulator was used to protect murine stem cell-derived vectors in primary myelopoietic progenitor cells both in vitro and in vivo in transplanted mice and demonstrated its capacity to protect gene therapy vectors from chromosomal position effects (19). Similarly, a MoMLV-derived vector modified by insertion of the human interferon γ gene scaffold attachment region was shown to be long-term transcriptionally active and nonmethylated in T cells in vitro (20, 21). In the present study, we report a protective effect of the mouse adenin phosphoribosyl transferase (aprt) gene CpG island on the adjacent RSV provirus integrated randomly in nonpermissive mammalian cells. We suggest that the effect of the CpG island is bidirectional and consists in preventing de novo DNA methylation and not in enhancement of the promoter strength.
Materials and Methods
Animals and DNA Inoculation.
Syrian hamsters (Mesocricetus auratus) were bred at the Institute of Molecular Genetics (Prague). They have been inbred for >40 generations by brother × sister matings and characterized by the acceptance of skin grafts. Plasmid DNAs were prepared on Qiagen columns, linearized by SspI endonuclease outside of the proviral and CpG island sequences, and diluted in PBS-A. Newborn hamsters were inoculated s.c. with 1–25 μg of plasmid DNA in 0.1 ml, and incidence and latency of sarcoma induction were scored (13). Chicken embryo fibroblasts (CEFs) were prepared from 10-day Brown Leghorn embryos, phenotype C/E, by standard procedures.
Construction of Proviral Reporters.
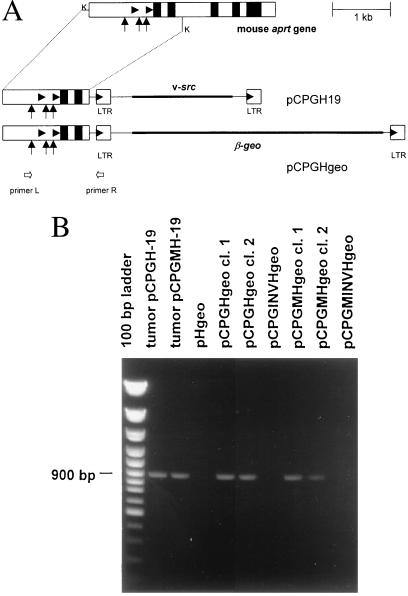
Our original proviral structure H-19 containing RSV LTR, v-src, LTR (22) was used for construction of proviral reporters. A 5.8-kb HindIII fragment of provirus together with flanking sequences was cloned in pUC19 vector, and flanking sequences were eliminated with the exception of 34 bp upstream (KpnI site) and 99 bp downstream (Eco47III site) of the provirus (construct pH-19KE). Plasmid pCPGH-19 was created by sticky-end ligation of 1.7-kb KpnI fragment of the wild-type mouse aprt gene CpG island cloned in pABS (3). Plasmid pCPGMH-19 was created by analogical insertion of a mutated mouse aprt gene CpG island cloned in pAZM2. In parallel, the CpG island of the mouse aprt gene was cloned upstream of the RSV LTR-driven reporter gene β-geo (plasmid pHgeo). Briefly, the v-src sequence in pH-19KE was replaced by β-geo coding sequence. The 3.9-kb HindIII–XbaI fragment from the pSAβgeo plasmid (23) was blunt-end ligated into NcoI and SauI sites (positions 760 and 2517, respectively) of pH-19KE. The 4-kb NcoI–SauI fragment of pH-19KE was treated with mung-bean nuclease to destroy the v-src AUG codon within the NcoI site. KpnI fragments (1.7 kb in length) of the wild-type and mutated aprt CpG islands were ligated in the KpnI site upstream of the LTR. Both orientations of the CpG islands were found and verified by DNA sequencing. Plasmids were denoted pCPGHgeo (wild-type CpG island), pCPGMHgeo (mutated CpG island), pCPGINVHgeo (wild-type CpG island in inverse orientation), and pCPGMINVHgeo (mutated CpG island in inverse orientation). Schematic representation of final proviral constructs is shown in Fig. 1A.
Figure 1.
Cloning of the aprt gene CpG island and RSV-based reporter proviruses. (A) Map of the mouse aprt gene and construction of reporter constructs pCPGH-19 and pCPGHgeo. Filled boxes represent exons of the aprt gene. Vertical arrows denote the position of three Sp1 sites in the CpG island. Filled arrowheads denote transcription starts of the aprt gene and 5′ LTR. Block arrows denote position of primers used for PCR. The 1.7-kb fragment was cloned upstream of the v-src- or β-geo-bearing proviral structures. K, KpnI. (B) PCR-based control of the junction between CpG island and 5′ proviral sequences. The diagnostic 896-bp fragment is amplified from L and R primers in DNA samples from tumors or cell clones containing the provirus with direct, but not inverse orientation of the CpG island.
Colony-Forming Assay.
Dishes with subconfluent NIL-2 cells were transfected with β-geo reporter plasmids by using the DOTAP liposomal transfection reagent {N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methylsulfate; Roche Molecular Biochemicals} according to the manufacturer's instruction. Selection with 400 μg/ml neomycin (G418; Sigma) was introduced 24 h after transfection and carried out for 10 days. Colonies of neo-resistant cell clones were grown for a further 10 days in 100 μg/ml neomycin, stained in situ with 1 mg/ml X-gal, counted, and photographed.
PCR.
Colonies of NIL-2 selected with neomycin were picked up, grown separately in 100 μg/ml neomycin (G418; Sigma), and DNA from cells was extracted. DNAs were also prepared from sarcomas induced by plasmids pCPGH-19 and pCPGMH-19. For detection of unaltered and properly oriented CpG island and of provirus we used primer L complementary to nucleotides 640–659 of the mouse aprt and primer R complementary to nucleotides 221–240 of the U3 region of LTR according to the sequence of the H-19 provirus (18). Sequences are 5′-TTGAGGTAGGGATGCTTGTG-3′ and 5′-CTTACTACCACCAATCGGCA-3′, respectively. Conditions for PCR were as follows: 95°C for 1 min, 55°C for 1 min, 72°C for 1 min (30 cycles).
β-Gal Assay.
Subconfluent cultures of NIL-2 cells were transfected with 2 μg of β-geo reporter plasmid DNA by using the DOTAP lipofection reagent (Roche Molecular Biochemicals). Cell lysates were prepared by three consecutive freeze–thawing cycles 48 h after transfection and β-galactosidase activities in 30 μl of the lysate were assayed in 0.1 M sodium phosphate (pH 7.5), 0.1 M MgCl2, and 1 mg/ml o-nitrophenyl-β-d-galactopyranoside. Reaction mixtures of 300 μl were incubated for 1 h at 37°C, stopped by adding 500 μl 1 M Na2CO3, and the optical density of yellow color was measured at a wavelength of 420 nm. Enzyme activities were finally normalized to protein concentration measured by Bradford dye assay (Bio-Rad protein assay).
Bisulfite Cytosine Methylation Analysis.
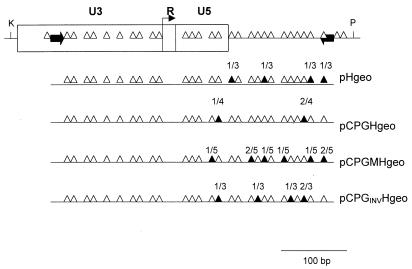
DNA samples for bisulfite analysis were digested by restriction endonuclease HindIII. Bisulfite treatment of DNA was done according to Olek et al. (24). PCR of the upper strand was performed with primers complementary to the U3 region of RSV LTR and the leader region (Fig. 2). Sequences of the primers are as follows: 5′-CAGTGAGCTCGTTTTATAAGGAAAGAAAAG-3′ (upper) and 5′-CAGTGTCGACCAACTTCTACCCTCCTAAAC-3′ (lower), respectively. Upper primer contains T and lower primer A instead of C in positions complementary to nonmethylable C, i.e., C out of CpG dinucleotides. Sequences complementary to the proviral U3 and leader are in italics; cloning sites SacI and SalI are in bold. PCRs were carried out with ca. 300 ng of DNA in one agarose bead at 95°C for 45 s, 58°C for 30 s, and 72°C for 30 s (35 cycles). PCR products (418 bp) were cloned into pUC-19 and sequenced by using universal pUC/M13 forward primer.
Figure 2.
CpG methylation pattern of the LTR region in actively integrated proviral reporters. Positions of CpG dinucleotides within RSV LTR and the adjacent leader region are depicted at the top. Summary results of CpG methylation analysis of individual β-geo proviral reporters is shown at the bottom. Open triangles, nonmethylated CpG residues; filled triangles, methylated CpG residues with the number of clones with methylated CpG/total number of clones analyzed; black block arrows, primers used for PCR and cloning. K, KpnI; P, PstI.
GenBank Accession Numbers.
Results
CpG Island Linked to the Provirus Prevents Transcriptional Silencing.
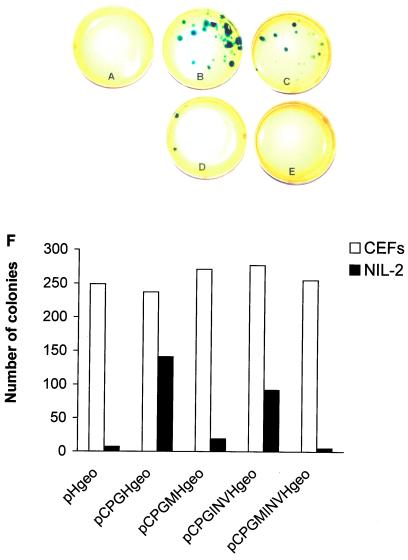
We have chosen the aprt gene CpG island for provirus DNA protection because its antimethylation effect was defined previously (3, 4, 25) and an inactive CpG island with mutated Sp1 sites is available as a negative control. We have fused a 1.7-kb fragment of the mouse aprt gene comprising the CpG island, the promoter region, the transcription start, and two exons of the coding sequence with the proviral structure RSV LTR, β-geo, LTR (Fig. 1A). Both wild-type and mutated CpG island with all three Sp1 sites destroyed (3) were used for transcription in both orientations (pHgeo, pCPGHgeo, pCPGMHgeo, pCPGINVHgeo, pCPGMINVHgeo; see Materials and Methods). These constructs were transfected into Syrian hamster cell line NIL-2 and CEFs. Cell clones bearing stably integrated and transcriptionally active proviruses were selected for neo resistance and visualized by an in situ β-gal colony assay. In NIL-2 cells, transfection of the provirus without CpG island produced active proviruses with low or zero incidence as illustrated (Fig. 3A) and summarized from several independent experiments (Fig. 3F), whereas proviruses with the adjacent CpG island in the 5′–3′ orientation remained active at least 30-times more frequently (Fig. 3 B and F). The inverted 3′–5′ orientation of the CpG island decreased the numbers of colonies with active proviruses to a slight, but reproducible extent (Fig. 3 C and F). The mutant CpG island protects integrated proviruses with low efficiency in the 5′–3′ orientation and is completely inefficient in the inverted orientation (Fig 3 D, E, and F). In contrast to NIL-2 cells, high numbers of β-gal positive neo-resistant colonies were detected in CEFs regardless of the wild-type or mutant form of the CpG island and its orientation (Fig. 3F). The presence of an unaltered provirus with the CpG island in proper orientation was checked by PCR using primers complementary to the CpG island (L primer) and to the U3 region of LTR (R primer). As expected, only DNAs from cell clones arising from the transfection of pCPGHgeo and pCPGMHgeo constructs give the specific 896-bp fragment (Fig. 1B). The results in NIL-2 cells suggest that the proviruses integrated in mammalian genomes are usually inactivated with the exception of those located in rare chromosomal sites permissive for the RSV LTR-driven expression. The presence of the CpG island close to the 5′ end of the provirus prevents its inactivation, probably by an antimethylation effect. The presence of intact Sp1 sites within the CpG island is important and proper orientation of the CpG island also plays some role in this protection.
Figure 3.
Colony-forming assay in NIL-2 cells transfected with aprt CpG island containing proviral constructs. (A) pHgeo, (B) pCPGHgeo, (C) pCPGINVHgeo, (D) pCPGMHgeo, (E) pCPGMINVHgeo. (F) The total number of neo-resistant β-gal positive colonies summarized from three independent colony-forming assays in NIL-2 cells and in CEFs. All three assays were done in triplicate.
Insertion of the CpG Island Does Not Increase the LTR Promoter Strength.
By inserting the CpG island of the mouse aprt gene 5′ to the proviral LTR, we have added a promoter/enhancer region and part of the coding sequence in proximity to the proviral regulatory sequences. To test the effect of such an insertion on the promoter strength of the RSV LTR we performed β-galactosidase assays on cell cultures transfected transiently with β-geo proviral reporter constructs. As shown in Fig. 4A, we have not found any significant differences in the levels of β-gal expression between cultures transfected with the β-geo provirus alone or adjacent to the wild-type or mutant CpG island in both orientations. These data indicate clearly that the CpG island does not improve transcriptional initiation, but acts in some protective way as an antimethylation tag. Furthermore, we have assayed the β-gal activities in several independent cell clones expanded from single neo-resistant colonies after stable transfection of NIL-2 cells by β-geo reporters. We have found high, intermediate, and low β-gal activities in several clones bearing the provirus protected by the wild-type CpG island. Among the rare clones bearing the provirus alone or provirus adjacent to the mutant or inverted CpG island, we have also found a broad spectrum of β-gal expression (Fig. 4B). We conclude that RSV-based proviral reporters integrated in hamster cells remain under the strong influence of flanking cellular sequences, despite the antisilencing effects of functional CpG islands.
Figure 4.
Transcriptional activities of proviral β-geo reporters after transient transfection and after integration in the host genome. (A) Relative β-gal activities in CEFs and NIL-2 cells 48 h after the transfection of β-geo proviral reporters. β-gal activity of pHgeo-transfected CEFs is calculated as 100%. Data represent the mean ± SEM of three parallels. (B) Relative β-gal activities in NIL-2 cell clones with stably integrated β-geo proviral reporters. β-gal activity of transiently pHgeo-transfected NIL-2 cells is calculated as 100%. Data represent the means of two parallels.
Undermethylation of Transcribed Proviruses.
To test whether the transcriptional activity of proviruses correlates with undermethylation in their regulatory sequences, we analyzed the upstream LTRs for methylation at CpG residues in several neo-resistant and β-gal positive cell clones from the previous experiment by using the bisulfite technique (24). Irrespective of the presence or absence of the CpG island, its orientation or mutation in Sp1 sites, the LTRs were almost completely demethylated in cells expressing the LTR-driven β-geo reporter, with rare methylated CpGs scattered over the R and U5 regions of LTR and leader sequence (Fig. 2). We suggest that the unmethylated status of at least some CpGs within the 5′ LTR is a prerequisite of RSV transcriptional activity in mammalian cells irrespective of the presence of a CpG island. However, the proximity of the CpG island and its proper orientation exert an important protective antimethylation effect and increase the proportion of active/inactive proviruses.
Protection by the CpG Island in Vivo.
To test whether the aprt CpG island protects the adjacent provirus in vivo we used the oncogenic proviral structure LTR-v-src-LTR as a reporter of proviral activity. We have shown previously that this proviral structure efficiently induces sarcomas when injected s.c. into chickens in the form of naked plasmid DNA (26). In Syrian hamsters, however, injection of this DNA does not induce any tumors even with high doses (13). We have cloned both wild-type and mutant forms of aprt CpG island upstream of the LTR-v-src-LTR provirus (plasmids pCPGH-19 and pCPGMH-19, respectively) and injected these constructs into newborn Syrian hamsters. The oncogenic provirus protected with wild-type CpG island induced sarcomas in 48% of DNA-inoculated animals with a mean latency of 36 weeks (Table 1). The provirus with a mutant form of the CpG island also produced sarcomas with similar latency, but with the incidence of only 4%. Cleavage of the plasmid DNA within the v-src coding sequence disrupts the sarcoma induction activity, showing clearly that cell transformation is mediated by v-src and not by position effects of the introduced CpG island. Again, the provirus alone did not induce any sarcomas even in doses five times higher. These in vivo experiments suggest again that the CpG island is able to preserve the transcriptional activity of the adjacent provirus.
Table 1.
Tumor induction in hamsters by H19 proviral DNA with adjacent CpG island
| Inoculated plasmid* | Frequency of hamsters with sarcomas† | Latency, weeks‡ |
|---|---|---|
| pCPGH-19 (1–5 μg) | 31/64 (48%) | 8–65 (36) |
| pCPGMH-19 (2.5–5 μg) | 2/51 (4%) | 10–50 |
| pCPGH-19 (2.5 μg) dig. PvuII | 0/9 (<11%) | — |
| pH-19 (25 μg) | 0/10 (<10%) | — |
Plasmid DNA was linearized by SspI digestion at the unique site in the plasmid vector. Digestion by PvuII which cleaves three times within the v-src coding sequence was used as a control.
Frequency of hamsters with sarcomas at the site of inoculation is expressed as the number of hamsters developing sarcomas/number of hamsters inoculated.
The mean latencies (in weeks) are given in parentheses. Inoculated hamsters were followed for 80 weeks. The ten hamsters inoculated with the high dose of pH-19 plasmid DNA were followed for two years.
Discussion
The well documented concept that DNA methylation in vertebrates evolved as an additive mechanism of developmental gene-expression control (2) has been challenged by an opinion that the main role of DNA methylation is to inactivate transcription of foreign sequences, transposons, and retroviruses (27). Despite the fact that our results cannot resolve this dispute they support a notion that intragenomic parasites invading the host have to overcome the generally suppressive influences of the surrounding DNA. Evasion from such suppressive influences might represent one of several important factors which determine the reshaping of retroviral genomes required for their adaptation to and coevolution with a new host species. An example of such coevolution between the parasite and its host DNA might be the gradual loss of methylable CpGs from genomes of HIV-1 and HIV-2 (28). RSV, because of its avian origin and high number of CpGs within LTR and leader sequences, can be regarded as not adjusted to efficient expression in the mammalian host and, therefore, it is usually silenced.
CpG islands of housekeeping genes can serve as refugia of proviral expression when active antimethylation strategies are absent. The range of CpGs protective influence is probably limited; active proviruses were previously found in the vicinity (up to 2 kb) of CpG islands (16) and, in our study, we have observed incomplete protection in the U5 part of the LTR (Fig. 2), i.e., approximately 1 kb downstream of the Sp1 region of the CpG island. Protection from de novo methylation and gene silencing by the aprt CpG island is far from being absolute and suppressive influences of the surrounding genome persist. First, the levels of proviral expression vary in different clones bearing the same proviral reporter integrated independently (Fig. 4B). Second, the number of β-geo-expressing NIL-2 colonies induced by reporter provirus with the wild-type CpG island does not reach the number of colonies induced in CEFs and, similarly, the incidence of in vivo sarcomas in hamsters induced by injection of pCPGH-19 is lower than in the case of MoMLV LTR-driven v-src (13). Third, during the long-term cultivation of β-geo-expressing colonies we have observed low incidence of β-gal negative progenitors (variegation), which indicates position effects at the clonal level. Similar position-effect variegation persisted in experiments with retroviral vectors equipped with chicken β-globin chromatin insulator (18, 19) even after ex vivo preselection of retrovirally transduced stem cells (29).
Several issues should be considered to establish the observed protection from methylation as a general approach. For instance, we have been using only one CpG island from the mouse genome, but testing additional CpG islands of mouse and Syrian hamster origin will be required to generalize our recent results. In addition, we should also discuss the possibility of preferential integration in GC-rich genomic regions, which might be influenced by the presence of a GC-rich insert. In our experiments, we have tightly controlled this possibility by using the mutant inactivated version of the CpG island for comparison with the wild-type CpG island. Because point mutations in three critical Sp1 sites almost abolished the protective effect of aprt CpG island, it seems highly unlikely that the used CpG island might have facilitated vector insertion in the CpG island-corresponding genomic region. However, analysis of loci in which CpG island-equipped reporter vs. CpG island-free reporter integrate should provide the more conclusive answer.
Transcriptional suppression of RSV proviruses strongly correlates with proviral DNA methylation (10, 11) and, therefore, RSV is a very suitable tool for these studies. In summary, we have shown that mouse aprt gene CpG island is able to protect adjacent nonhomologous genes such as the RSV-based proviral reporters. Provided that the CpG island sequences necessary for the prevention of de novo methylation reside in a short manageable element, as shown recently (30), our approach might enable the construction of RSV-based retroviral vector for mammalian cells.
Acknowledgments
We thank D. Macleod and A. P. Bird for providing us with the clone of mouse aprt CpG island and for encouraging comments on our results; J. Walter for kind help with the bisulfite analysis and for communicating results before publication; P. Soriano for providing us with the construct pSAβgeo; and A. Razin for valuable discussion. This study was supported by Grants 312/97P082 (to J.H.) and 312/98/0825 (to J.S.) from the Grant Agency of the Czech Republic.
Abbreviations
- RSV
Rous sarcoma virus
- LTR
long terminal repeat
- MoMLV
Moloney murine leukemia virus
- aprt
adenin phosphoribosyltransferase
- CEF
chicken embryo fibroblasts
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Bird A P. Nature (London) 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 2.Bird A P. Trends Genet. 1995;11:94–100. doi: 10.1016/S0168-9525(00)89009-5. [DOI] [PubMed] [Google Scholar]
- 3.Macleod D, Charlton J, Mullins J, Bird A P. Genes Dev. 1994;8:2282–2292. doi: 10.1101/gad.8.19.2282. [DOI] [PubMed] [Google Scholar]
- 4.Brandeis M, Frank D, Keshet I, Siegfried Z, Mendelsohn M, Nemes A, Temper V, Razin A, Cedar H. Nature (London) 1994;371:435–438. doi: 10.1038/371435a0. [DOI] [PubMed] [Google Scholar]
- 5.Svoboda J. Gene. 1998;206:153–163. doi: 10.1016/s0378-1119(97)00571-4. [DOI] [PubMed] [Google Scholar]
- 6.Boettiger D. Cell. 1974;3:71–76. doi: 10.1016/0092-8674(74)90042-7. [DOI] [PubMed] [Google Scholar]
- 7.Varmus H E, Guntaka R V, Deng C-T, Bishop J M. Cold Spring Harbor Symp Quant Biol. 1975;39:987–996. doi: 10.1101/sqb.1974.039.01.113. [DOI] [PubMed] [Google Scholar]
- 8.Green A R, Searle S, Gillespie D A F, Bissell M, Wyke J A. EMBO J. 1986;5:707–711. doi: 10.1002/j.1460-2075.1986.tb04271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiswell D J, Enrietto P J, Evans S, Quade K, Wyke J A. Virology. 1982;116:428–440. doi: 10.1016/0042-6822(82)90137-4. [DOI] [PubMed] [Google Scholar]
- 10.Searle S, Gillespie D A F, Chiswell D J, Wyke J A. Nucleic Acids Res. 1984;12:5193–5210. doi: 10.1093/nar/12.13.5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hejnar J, Svoboda J, Geryk J, Fincham V J, Hák R. Cell Growth Differ. 1994;5:277–285. [PubMed] [Google Scholar]
- 12.Overbeek P A, Lai S-P, VanQuill K R, Westphal H. Science. 1986;231:1574–1577. doi: 10.1126/science.3006249. [DOI] [PubMed] [Google Scholar]
- 13.Hejnar J, Plachý J, Geryk J, Machon̆ O, Trejbalová K, Guntaka R V, Svoboda J. Virology. 1999;255:171–181. doi: 10.1006/viro.1998.9597. [DOI] [PubMed] [Google Scholar]
- 14.Guntaka R V, Gowda S, Wagner H, Simon D. FEBS Lett. 1987;221:332–336. doi: 10.1016/0014-5793(87)80951-1. [DOI] [PubMed] [Google Scholar]
- 15.Machon̆ O, Hejnar J, Hájková P, Geryk J, Svoboda J. Gene. 1996;174:9–17. doi: 10.1016/0378-1119(96)00200-4. [DOI] [PubMed] [Google Scholar]
- 16.Fincham V J, Wyke J A. J Virol. 1991;65:461–463. doi: 10.1128/jvi.65.1.461-463.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Svoboda J. Intervirology. 1986;26:1–60. doi: 10.1159/000149682. [DOI] [PubMed] [Google Scholar]
- 18.Rivella S, Callegari J A, May C, Tan C, Sadelain M. J Virol. 2000;74:4679–4687. doi: 10.1128/jvi.74.10.4679-4687.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emery D W, Yannaki E, Tubb J, Stamatoyannopoulos G. Proc Natl Acad Sci USA. 2000;97:9150–9155. doi: 10.1073/pnas.160159597. . (First published July 25, 2000; 10.1073/pnas.160159597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agarval M, Austin T W, Morel F, Chen J, Bohnlein E, Plavec I. J Virol. 2000;74:3720–3728. doi: 10.1128/jvi.72.5.3720-3728.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dang Q, Auten J, Plavec I. J Virol. 2000;74:2671–2678. doi: 10.1128/jvi.74.6.2671-2678.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bodor J, Poliak E, Pichrtová J, Geryk J, Svoboda J. Nucleic Acids Res. 1989;17:8869. doi: 10.1093/nar/17.21.8869. (lett.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friedrich G, Soriano P. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- 24.Olek A, Oswald J, Walter J. Nucleic Acids Res. 1996;24:5064–5066. doi: 10.1093/nar/24.24.5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mummaneni P, Bishop P L, Turker M S. J Biol Chem. 1993;268:552–558. [PubMed] [Google Scholar]
- 26.Svoboda J, Plachý J, Hejnar J, Karakoz I, Guntaka R V, Geryk J. Immunogenetics. 1992;5:309–315. doi: 10.1007/BF00189893. [DOI] [PubMed] [Google Scholar]
- 27.Yoder J A, Walsh C P, Bestor T H. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 28.Nyce J W. In: Epigenetic Mechanisms of Gene Regulation. Russo V E, Martienssen R A, Riggs A D, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 561–574. [Google Scholar]
- 29.Kalberer C P, Pawliuk R, Imren S, Bachelot T, Takekoshi K J, Fabry M, Eaves C J, London I M, Humphries R K, Leblouch P. Proc Natl Acad Sci USA. 2000;97:5411–5415. doi: 10.1073/pnas.100082597. . (First published May 2, 2000; 10.1073/pnas.100082597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siegfried Z, Eden S, Mendelsohn M, Feng X, Tsuberi B-Z, Cedar H. Nat Genet. 1999;22:203–206. doi: 10.1038/9727. [DOI] [PubMed] [Google Scholar]