Abstract
The breast cancer susceptibility gene, BRCA1, encodes a large nuclear phosphoprotein, the major isoform of which is 1863 amino acids in size. Structure–function studies have been largely restricted to the only two domains identified by homology searches: the RING (really interesting new gene) and BRCT (BRCA1 C-terminus) domains. However, we have recently reported the identification of a large central soluble region of BRCA1 (residues 230–534) that binds specifically to four-way junction DNA, a property that potentially facilitates its role in the repair of DNA lesions by homologous recombination. We have now used a combination of limited proteolysis and extension cloning to identify more accurately the DNA-binding region of BRCA1. Limited trypsinolysis of BRCA1-(230–534) resulted in the production of a soluble domain identified as residues 230–339. However, after cloning, expression and purification of this region, studies revealed that it was unable to bind to four-way junctions, suggesting that the DNA-binding activity, in part, resides within residues 340–534. A series of fragments extending from residue 340 were produced, and each was tested for its ability to bind to four-way junction DNA in gel retardation assays. In these experiments, residues 340–554 of BRCA1 were identified as the minimal DNA-binding region. We then went on to characterize the conformation of this region using CD spectroscopy and analytical centrifugation.
Keywords: breast cancer, breast cancer susceptibility protein 1 (BRCA1), circular dichroism, DNA-binding domain, four-way junction DNA, limited proteolysis
Abbreviations: BASC, BRCA1 genome surveillance complex; BRCA1, breast cancer susceptibility protein 1; BRCT, BRCA1 C-terminus; DTT, dithiothreitol; MALDI, matrix-assisted laser-desorption ionization; MSH, human MutS homologue; NBS, Nijmegen breakage syndrome protein; Ni-NTA, Ni2+-nitriloacetic acid; RAD, radiation-dependent; RING, really interesting new gene; TAE, Tris/acetate/EDTA
INTRODUCTION
The breast cancer susceptibility gene BRCA1 encodes a tumour suppressor, mutations in which are responsible for 45–76% of families with hereditary breast cancer [1–3]. Identified in 1993, its role as a tumour suppressor was discovered after loss of heterozygosity on chromosome locus 17q21 was observed in a number of families with a history of breast cancer [4,5].
The BRCA1 gene encodes a large protein of 1863 amino acids that functions as a scaffold co-ordinating protein–protein interactions and/or post-translational modifications that are required for effective signal transduction upon DNA damage. This is illustrated by the formation of the BASC (BRCA1 genome surveillance complex) [6] and the requirement of BRCA1 for the ATM (gene mutated in ataxia telangiectasia)- and ATR (Atm- and Rad3-related)-dependent phosphorylation of p53, c-Jun, NBS (Nijmegen breakage syndrome protein), CtIP [CtBP (C-terminus-binding protein)-interacting protein], Smc1 (sister maintenance chromosome protein 1) and Chk-2 [7], which are key regulators of the DNA-damage response. BASC is characterized by a high number of factors that sense and repair DNA lesions; for example, the MSH2–MSH6 (MSH is human MutS homologue) and RAD50 (radiation-dependent 50)–MRE11 (meiotic recombination protein 11)–NBS1 complexes. BRCA1 has also been identified as a component of other regulatory complexes, such as the RNA polymerase II holoenzyme [8] and a SWI/SNF chromatin remodelling complex [9], relating BRCA1 to transcriptional regulation and providing a link to ionizing-radiation-induced transcription-coupled repair.
Cells deficient in BRCA1 display defects in survival and proliferation [10,11], radiosensitivity [12], chromosomal abnormalities [13,14], G2/M checkpoint loss [15,16] and impaired homologous recombination repair [16], confirming its importance in maintaining the structural integrity of the genome.
Although the structure of BRCA1 is largely undefined, two domains have been identified [17]: these include an N-terminal zinc-binding RING (really interesting new gene) domain and two tandem copies of the BRCT (BRCA1 C-terminus) domain located at the C-terminus. These domains are now recognized motifs that are found in a number of proteins that function in DNA repair [18] and targeted protein degradation [19].
The central two-thirds of BRCA1 are thought to be largely unstructured [20] and thus possess the structural flexibility that is required for the numerous interactions that are associated with this region, many of which are involved in DNA repair, e.g. p53 [21,22], RAD50 [23], FANCA (Fanconi anaemia) [24] and RAD51 [25]. We and others have shown that this central region of BRCA1 also has interaction sites for distorted DNA structures that occur as a result of the repair of double-strand breaks by homologous recombination [26,27]. To determine the full significance of the DNA-binding activity of BRCA1, we report in the present paper the identification of the DNA-binding region more precisely, using a combination of limited proteolysis and extension cloning, and its characterization by CD and analytical centrifugation.
EXPERIMENTAL
Purification of BRCA1-(230–534)
Following published procedures [26], BRCA1-(230–534) was overexpressed in Escherichia coli BL21 (DE3) codon plus and purified to homogeneity from crude cell extracts by ion-exchange and Ni-NTA (Ni2+-nitrilotriacetate)-affinity chromatography.
Limited proteolysis of BRCA1-(230–534)
Samples for proteolytic digestion were prepared in 10 mM Tris/HCl, pH 7, 100 mM NaCl, 1 mM EDTA and 1 mM DTT (dithiothreitol) at a protein concentration of 0.2 mg/ml. Trypsin was added at a ratio of 1:500 or 1:1000 (w/w), and the reaction mixtures were incubated at 20 °C. Reactions were quenched at different time intervals by the addition of SDS loading dye and incubation at 95 °C for 5 min. The proteolytic products were analysed by SDS/15% PAGE and were visualized by Coomassie Blue staining.
Identification of BRCA1 domains
Proteolytic products to be N-terminally sequenced were separated on an SDS/15% polyacrylamide gel and electroblotted on to a PVDF membrane (the transfer buffer was 10 mM CAPS, pH 11, and 10% methanol). When the transfer was complete, the membrane was stained with Amido Black and destained with distilled water. Selected bands were excised and N-terminally sequenced by automated sequential Edman degradation using an ABI 476A gas/liquid-phase protein sequencer.
For analysis by MS, a 1 h 1:500 digest was scaled up and applied to a Zorbax 300SB-C3 reverse-phase column (4.6 mm×250 mm) equilibrated in 2% acetonitrile and 0.05% trifluoroacetic acid. The products were separated by application of an increasing gradient of acetonitrile (2–35% in 50 min, 35–60% in 20 min and 60–90% in 10 min). A sample of the peak fractions was vacuum-dried and analysed on an SDS/15% polyacrylamide gel, and selected fractions were electroblotted and N-terminally sequenced as described above. The remainder of the sample was vacuum-dried and analysed by MALDI (matrix-assisted laser-desorption ionization) MS. Spectra were recorded on a Bruker REFLEX III in linear mode.
Cloning, expression and purification of BRCA1 constructs
The four regions of BRCA1 encoding residues 230–347, 340–513, 340–554 and 340–595 were amplified by PCR from a plasmid template carrying the full-length BRCA1 sequence. The forward and reverse primers for each construct contained an NdeI site and a Xho1 site respectively (Table 1). The PCR products were phosphorylated and ligated directly into dephosphorylated SmaI-digested pUC18. After positive selection, recombinant pUC18 constructs were digested sequentially with XhoI and NdeI, and ligated into pET22b for eventual overexpression with a hexahistidine tag at the C-terminal end. The sequence and frame of each insert were confirmed by DNA sequencing.
Table 1. Primer sequences used to amplify BRCA1 protein fragments.
For each pair, the first sequence is the N-terminal primer and the second is the C-terminal primer.
| Construct | PCR primers |
|---|---|
| BRCA1-(230–347) | 5′-TGCCGACATATGGAGACGCATCTAACAAATAC-3′ |
| 5′-ACTCCGAAGCTTTCACTCGAGCAGGGGATCAGCATTCAGATCTACC-3′ | |
| BRCA1-(340–513) | 5′-TGCCGACATATGGTAGATCTGAATGCTGATCCC-3′ |
| 5′-ACTCCGAAGCTTTCACTCGAGAAGGCCTGATGTAGGTCTCC-3′ | |
| BRCA1-(340–554) | 5′-TGCCGACATATGGTAGATCTGAATGCTGATCCC-3′ |
| 5′-ACTCCGAAGCTTTCACTCGAGCTCTAGACCACTATTAGT-3′ | |
| BRCA1-(340–595) | 5′-TGCCGACATATGGTAGATCTGAATGCTGATCCC-3′ |
| 5′-ACTCCGAAGCTTTCACTCGAGTTCCATATTGCTTATACTGCTGC-3′ |
For expression, each of the pET22b constructs were transformed into E. coli BL21 (DE3) codon plus cells and grown in 500 ml of 2× YT (yeast tryptone) medium supplemented with ampicillin (50 μg/ml) at 37 °C. The cells were grown to a turbidity at 600 nm (D600) of 0.5–0.6, whereupon protein expression was induced by the addition of 1 mM isopropyl β-D-thiogalactoside. The cells were grown for an additional 3 h, harvested by centrifugation at 10000 g for 10 min and resuspended in 10 mM sodium phosphate, pH 6, 800 mM NaCl, 1 mM EDTA, 1 mM DTT and one Complete™ protease inhibitor tablet (Roche). The bacteria were lysed using a French press and the cell debris was removed from the cell lysate by centrifugation at 36000 rev./min for 1 h at 4 °C in a SW40 Ti rotor. The soluble cell lysate was absorbed on to an Ni-NTA column (Qiagen) and eluted with 300 mM imidazole. Bound fractions were dialysed in 10 mM phosphate, pH 6, 1 mM DTT and 1 mM EDTA, and were stored at −20 °C. BRCA1-(230–347) was purified further by ion-exchange chromatography using a Resource S column (Amersham Biosciences).
Identification of the DNA-binding region by gel retardation analysis
Four-way junction DNA was formed by incubation of equimolar concentrations of the oligonucleotides shown in Table 2 at 95 °C for 10 min and cooling slowly to 4 °C. The junction was incubated with increasing concentrations of protein (1, 2 and 6 μM) at room temperature (22 °C) in a total volume of 10 μl for 30 min. Binding buffer contained 10 mM Tris/HCl, pH 8.0, 1 mM DTT, 50 mM NaCl, 5% (v/v) glycerol and 1 mM EDTA. The samples were analysed in pre-electrophoresed 6% non-denaturing polyacrylamide TAE (40 mM Tris/acetate and 1 mM EDTA, pH 8.5) gels and visualized by ethidium bromide staining.
Table 2. Oligonucleotides used in the formation of four-way junction and duplex DNA.
| Substrate | Oligonucleotide | Sequence |
|---|---|---|
| Four-way junction | F1 | 5′-GAATTCAGCACGAGTCCTAACGCCAGATCT-3′ |
| F2 | 5′-AGATCTGGCGTTAGGTGATACCGATGCATC-3′ | |
| F3 | 5′-GATGCATCGGTATCAGGCTTACGACTAGTG-3′ | |
| F4 | 5′-CACTAGTCGTAAGCCACTCGTGCTGAATTC-3′ | |
| Duplex | D1 | 5′-GTCGACTTATGCCAAGTGGTACGTCTCCGT-3′ |
| D2 | 5′-ACGGAGACGTACCACTTGGCATAAGTCGAC-3′ |
Analysis of the DNA-binding specificity of BRCA1-(230–554)
Four-way junction and duplex DNA (formed by annealing equimolar concentrations of the oligonucleotides displayed in Table 2) (1 nmol) were end-labelled using [γ-32P]ATP and T4 polynucleotide kinase at 37 °C for 30 min. The reactions were inactivated by incubation on ice for 10 min, and the labelled DNA was purified from unincorporated ATP using Sephadex G50 spin columns (Amersham Biosciences). After ethanol precipitation, the labelled probes were resuspended in 10 μM unlabelled duplex or junction DNA, 10 mM Tris/HCl, pH 8.0, and 1 mM EDTA. Binding reactions were carried out as described above, where increasing amounts of protein (0, 0.5, 1, 2.5, 5, 10, 15, 25 and 50 μM) were incubated with a fixed concentration (1 μM) of labelled junction or duplex DNA. The free and bound components of the equilibria were resolved by electrophoresis in 6% non-denaturing polyacrylamide/TAE gels. The labelled components were visualized by autoradiography of the dried gels, and integrated band intensities were determined using Fluorchem 5500 imaging software (Alpha Innotech).
The binding data were fitted by non-linear regression, using ProFit (Quantum Soft), to the general quadratic expression, assuming a 1:1 stoichiometry:
 |
(1) |
Eqn 1 describes a heteromeric associative equilibrium [28] and relates θ (the fraction of bound DNA sites, [PD]/[Dt]) to the total protein concentration ([Pt]) at each point of a titration.
For competition gel-retardation analysis, the binding reaction contained 1 μM labelled four-way junction DNA, 5 μM protein, 50 mM NaCl, 10 mM Tris/HCl, pH 8.0, 0.5 mM EDTA, 1 mM DTT, 5% (v/v) glycerol and increasing concentrations of competitor duplex DNA in a final reaction volume of 10 μl. Competitor DNAs (0.5, 1, 5, 15, 25 and 50 μM) were added at the start of the incubation reaction. The reactions were analysed by electrophoresis in 6% non-denaturing polyacrylamide/TAE gels. Protein–DNA complexes were visualized by autoradiography.
CD spectroscopy
Far-UV (190–250 nm) CD spectra were recorded using a Jasco J-810 spectropolarimeter. Measurements were taken every 0.5 nm in a 1 mm pathlength cell. Buffer conditions were 10 mM sodium phosphate buffer, pH 7, and 50 mM NaCl at a protein concentration of 200 μg/ml. Spectra were corrected for buffer absorbance and represent an average of ten scans. Spectra were recorded in millidegrees and were converted into mean residue ellipticities.
Analytical ultracentrifugation
Sedimentation velocity experiments were performed using an Optima XL-A analytical ultracentrifuge (Beckman Instruments). Before centrifugation, BRCA1-(230–347) and BRCA1-(340–554) were dialysed exhaustively into 10 mM sodium phosphate buffer, pH 7, and 50 mM NaCl and made up to 1 mM tris-(2-carboxyethyl)-phosphine. Samples (400 μl, 1 mg/ml) were centrifuged in a 1.2 cm pathlength two-sector aluminium centrepiece cell, with sapphire windows, in a four-place An-60 Ti analytical rotor at 50000 rev./min for BRCA1-(230–347) and 47000 rev./min for BRCA1-(340–554) for 8 h at a temperature of 20 °C. Changes in solute concentration were detected by 280 nm absorbance scans. Whole-boundary analysis was performed using the program Sedfit version 8.7, and sedimentation coefficients were obtained using SVEDBERG (version 6.39). Using the freeware program Sednterp, the amino acid composition of the two proteins gave estimates of the partial specific volume (v) of 0.7076 ml/g for BRCA1-(230–347) and 0.7264 ml/g for BRCA1-(340–554), and hydration values of 0.4663 and 0.4636 g of water/g of protein respectively. Buffer densities and viscosities were also calculated using Sednterp.
RESULTS
Limited proteolysis of BRCA1-(230–534)
Previously, we reported the identification of a soluble fragment of BRCA1 (amino acids 230–534) that binds specifically to four-way junction DNA [26]. To determine the precise location of the DNA-binding domain, we have used a combination of limited proteolysis and MS. Limited proteolysis is an effective technique for probing the domain structure of large multifunctional proteins that are often refractory to structure determination by NMR and X-ray crystallography. Proteases preferentially cleave at exposed loops to produce a discrete pattern of fragments that represent isolated domains.
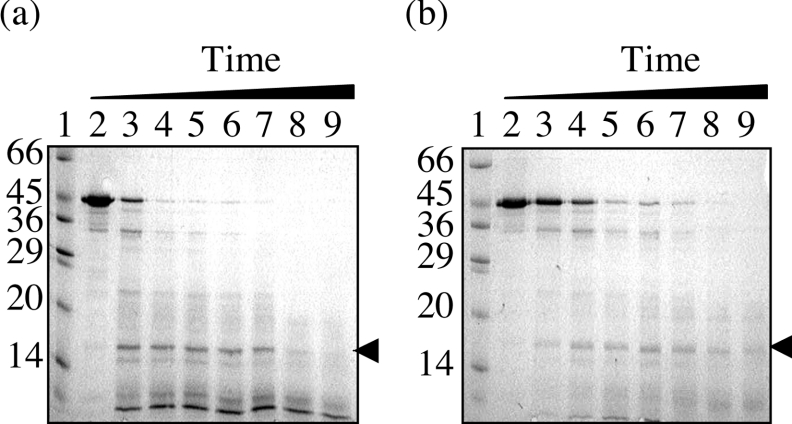
A time course of proteolysis was performed on BRCA1-(230–534) using low trypsin/substrate ratios of 1:500 and 1:1000. Analysis by SDS/PAGE (Figures 1a and 1b) revealed that the time-dependent disappearance of BRCA1-(230–534) was concomitant with the appearance of a single metastable fragment at approx. 15 kDa. The high protease resistance of this fragment suggests a solvent-inaccessible structure typical of a protein domain. To determine the location of this domain, it was transferred on to PVDF membrane and analysed by N-terminal sequencing. The first six residues were identified as ETDVN (a proportion of this sample retained the N-terminal methionine), identical with those of BRCA1-(230–534).
Figure 1. Limited proteolysis of purified BRCA1-(230–534).
Proteolysis was conducted at room temperature over a 60 min period, with trypsin/protein ratios of (a) 1:500 and (b) 1:1000. The digestion products were analysed by SDS/15% PAGE by comparison with molecular-mass standards (lanes 1; sizes are indicated in kDa). Lanes 2–9 are 0, 5, 10, 15, 20, 30, 40 and 60 min digests respectively. The fragments were visualized by Coomassie Blue staining, and the arrow indicates the stable domain.
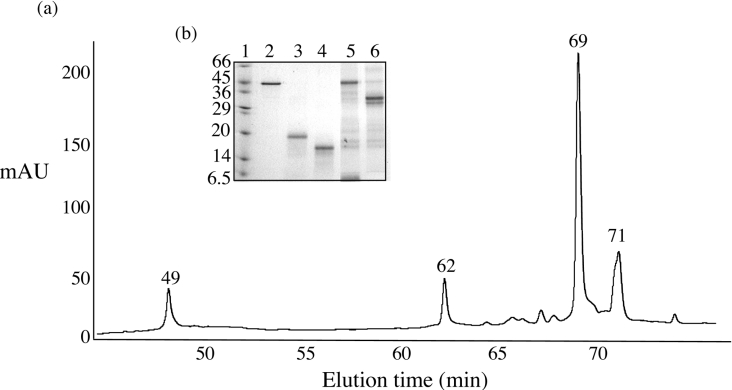
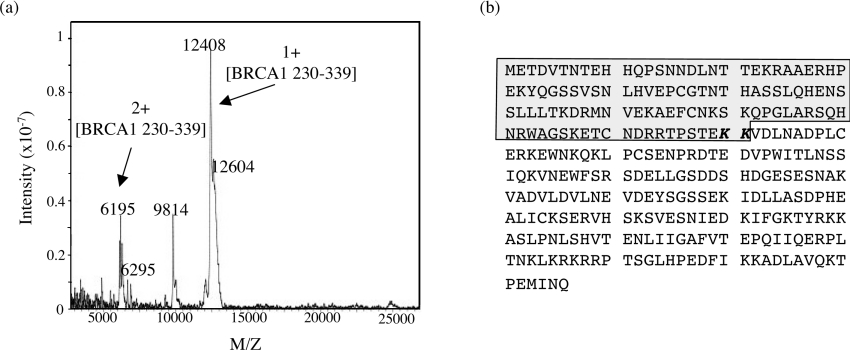
To assign the C-terminal cleavage site, the 1:500 digest was scaled up and was allowed to proceed for 1 h. The products were then adsorbed on to a C3 reverse-phase column and were separated with an increasing gradient of acetonitrile (Figure 2a). The fractions were analysed by SDS/PAGE (Figure 2b), and the first peak eluting at 49 min was identified by N-terminal sequencing as a self-cleavage product of trypsin. The BRCA1-(230–534)-derived domain eluted as a single peak after 62 min (Figure 2b). To determine the molecular mass and hence the C-terminal cleavage site, the purified domain was analysed by MALDI MS. The spectrum (Figure 3a) shows the presence of single and multiple charged ions for the domain, with m/z values that corresponded to heterogeneous cleavage at amino acids Lys338 and Lys339 respectively (Figure 3b). The broadness of the peaks was due to the heterogeneity of the masses produced as a result of multiple cleavages at KK (Lys-Lys) and a small proportion on the sample retaining the N-terminal methionine. However, the domain borders could be unambiguously assigned to amino acids 230–339 of BRCA1.
Figure 2. Purification of the domain derived from limited trypsinolysis by reverse-phase chromatography.
(a) Separation of the products of a scaled-up 1:500 digestion on a C3 reverse-phase column. Bound fractions were eluted with a gradient of acetonitrile (2–35% in 50 min, 35–60% in 20 min and 60–90% in 10 min) and monitored by their absorbance at 220 nm. mAU, milli-absorbance units. (b) Analysis of the peak fraction by SDS/15% PAGE. Lane 1 contains molecular-mass standards (sizes are indicated in kDa), lane 2 contains BRCA1-(230–534), and lanes 3–6 contain the contents of peaks eluting at 49, 62, 69 and 71 min respectively.
Figure 3. Mapping of the major proteolytic fragments of BRCA1-(230–534), resulting from limited trypsinolysis, by MALDI-TOF.
(a) Mass spectrum of peak 62. The heterogeneity observed in the peaks is due to cleavage at two adjacent lysine residues and a proportion of the sample retaining the N-terminal methionine. (b) The amino acid sequence of BRCA1-(230–534). The highlighted region was identified as a domain produced from limited trypsinolysis. This region has a calculated molecular mass of 12556 Da and thus domain boundaries are identified as residues 230–339.
Cloning expression and purification of BRCA1-(230–347)
The results presented above suggest that amino acids 230–339 constituted a soluble domain in BRCA1. To determine whether this domain was responsible for binding to four-way junction DNA, the region encoding amino acids 230–347 of BRCA1 was amplified by PCR and ligated into the expression vector pET22b. The construct contained codons for a few additional amino acids, extending the polypeptide at its C-terminus, which was assumed to aid correct folding of the domain in E. coli.
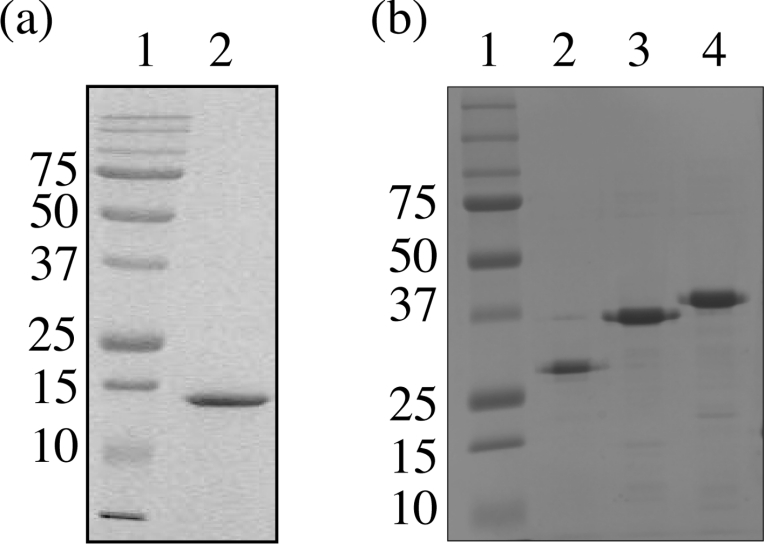
BRCA1-(230–347) was expressed with a C-terminal hexahistidine fusion in E. coli BL21 (DE3) codon plus cells, where expression levels approached 50% of the total cellular protein. BRCA1-(230–347) was found predominantly in the soluble supernatant fraction and was purified to apparent homogeneity by Ni-NTA-affinity chromatography and high-resolution cation-exchange chromatography (Figure 4a). N-terminal amino acid sequencing of the first eight amino acids confirmed that the polypeptide had the expected sequence METDVTN. Analysis by gel retardation assays revealed that BRCA1-(230–347) was unable to bind to four-way junction DNA (Figure 5a, lanes 11–13).
Figure 4. Purification of BRCA1 protein fragments.
All regions were expressed from the T7 promoter of pET22b in E. coli BL21 (DE3) codon plus cells. (a) Purified BRCA1-(230–347). After lysis, the soluble fraction was applied to an Ni-NTA-affinity column. Fractions containing BRCA1-(230–347) were pooled and purified further by cation-exchange chromatography. Purified BRCA1-(230–347) was analysed by SDS/15% PAGE by comparison with molecular-mass standards. (b) Electrophoretic analysis of purified BRCA1-(340–513), BRCA1-(340–554) and BRCA1-(340–595). All were purified directly from the soluble fraction of the cell lysate by Ni-NTA-affinity chromatography. Lane 1 contains molecular-mass standards (sizes are indicated in kDa), and lanes 2–4 contain residues 340–513, 340–354 and 340–395 of BRCA1 respectively. The mobility of each of the fragments is higher than predicted for their molecular masses, which are 20745, 25284 and 29787 for residues 340–513, 340–554 and 340–595 of BRCA1 respectively. This abnormal migration may be associated with the high proportion of acidic residues. All the fragments have pI values ranging from 5.4 to 5.9, and it has been reported that some acidic proteins bind less efficiently to SDS, resulting in higher than predicted mobilities [29].
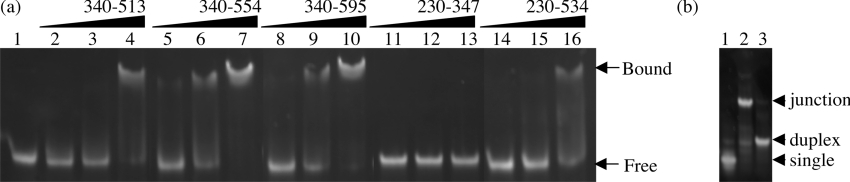
Figure 5. Gel retardation analysis of residues 340–513, 340–554, 340–595, 230–347 and 230–534 of BRCA1.
(a) Four-way junction DNA (2 μM) was incubated with an increasing concentration (1, 2 and 6 μM) of each region of BRCA1 and analysed by non-denaturing 6% PAGE. Lane 1, free DNA; lanes 2–4, BRCA1-(340–513); lanes 5–7, BRCA1-(340–554); lanes 8–10, BRCA1-(340–595); lanes 11–13, BRCA1-(230–347); lanes 14–16, BRCA1-(230–534). (b) Analysis of four-way junction formation by non-denaturing 8% PAGE. Lane 1, the F2 single strand of the junction; lane 2, four-way junction DNA; lane 3, duplex DNA. Bands were visualized by ethidium bromide staining.
Mapping the DNA-binding region of BRCA1
Since the N-terminal domain identified by trypsinolysis of BRCA1-(230–534) was unable to bind to four-way junction DNA, a series of C-terminal protein fragments were produced that extended from residue 340 to 595. Each region of DNA encoding these fragments was amplified by PCR and was cloned into the expression vector pET22b, such that all the resultant proteins were expressed with C-terminal hexahistidine fusions. All constructs produced soluble protein fragments when expressed in E. coli and were purified to approx. 98% homogeneity directly from the supernatant of clarified cell lysates using Ni-NTA-affinity chromatography (Figure 4b). The binding of each protein fragment to four-way junction DNA was analysed by comparison with BRCA1-(230–534) in gel retardation assays (Figure 5a). Formation of the four-way junction was confirmed by comparison of its mobility in non-denaturing polyacrylamide gels to the F2 single strand of the junction and to duplex DNA (Figure 5b). The shortest protein fragment, BRCA1-(340–513), bound to the junction with comparable affinity with that of BRCA1-(230–534) (compare lanes 4 and 16 in Figure 5a), confirming that the DNA-binding activity resides, at least in part, in the C-terminal portion of BRCA1-(230–534). Increasing the length of this fragment to residue 554 resulted in an increase in affinity as judged by the increase in fraction bound at 6 μM protein (compare lanes 4 and 7 in Figure 5a). Increasing the length to residue 595 resulted in no further increase in affinity (compare lanes 7 and 10 in Figure 5a), suggesting that the DNA-binding region of BRCA1 is contained within residues 340–554.
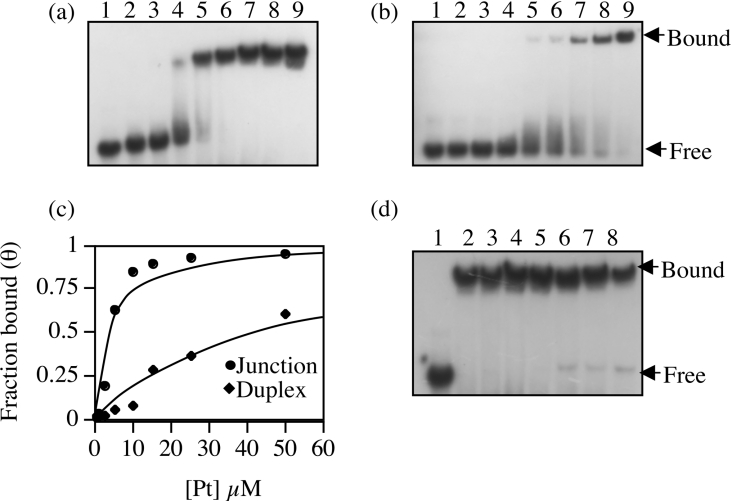
To determine whether BRCA1-(340–554) retained selectivity for four-way junction DNA, its binding constant for this substrate was determined and compared with that of duplex DNA. From the gel retardation assays shown in Figures 6(a) and 6(b), binding isotherms were generated by plotting fraction bound (θ) against total protein concentration, and the best fit to the quadratic expression described in the Experimental section was obtained with a Kd of 42±4.9 μM for duplex DNA and 3.03±0.263 μM for four-way junction DNA (Figure 6c). This demonstrates a greater than 10-fold increase in affinity of BRCA1-(340–554) for four-way junction DNA; thus the selectivity observed for BRCA1-(230–534) [26] is retained. This was confirmed further in a competition assay where a 50-fold excess of linear DNA was unable to compete for four-way junction binding (Figure 6d).
Figure 6. Analysis of the DNA-binding specificity of BRCA1-(340–554).
Binding to (a) four-way junction DNA and (b) duplex DNA was compared by gel retardation analysis. In both assays, 0, 0.5, 1, 2.5, 5, 10, 15, 25 and 50 μM protein (lanes 1–9 respectively) was incubated with 1 μM DNA. The free and bound components of the equilibria were resolved by non-denaturing 6% PAGE and were visualized by autoradiography. (c) The fraction bound (θ) for each binding reaction was plotted against the total protein concentration. The solid line is the curve of best fit used to estimate the dissociation constants of binding to both substrates. (d) Competition gel retardation analysis of BRCA1-(340–554) bound to four-way junction. Lane 1, free DNA; lane 2, no competitor; lanes 3–8, 0.5, 1, 5, 15, 25 and 50 μM unlabelled duplex competitor DNA. The concentration of labelled four-way junction DNA and BRCA1-(350–534) were 1 μM and 5 μM respectively. The bound and free components were visualized by autoradiography.
Conformational analysis of BRCA1-(230–347) and BRCA1-(340–554)
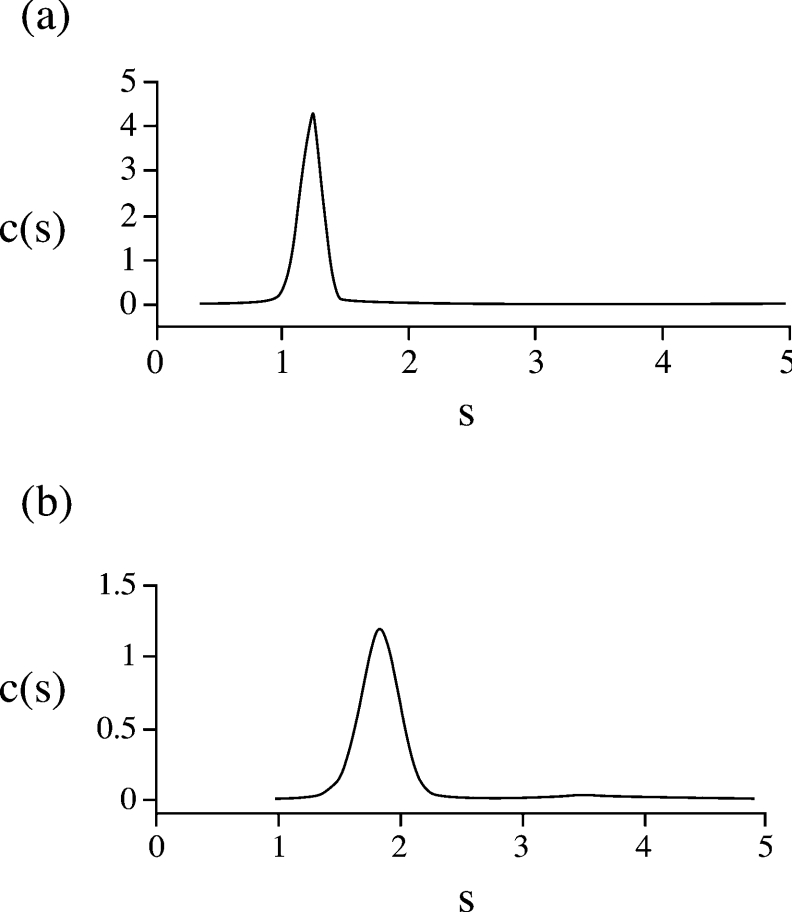
The solution conformation of both BRCA1-(230–347) and BRCA1-(340–554) was analysed by sedimentation velocity and CD spectroscopy. The continuous size distribution functions c(s) [30] derived from the sedimentation velocity data revealed one discrete species for both BRCA1-(230–347) (Figure 7a) and BRCA1-(340–554) (Figure 7b). Apparent sedimentation coefficients (s0) were obtained using SVEDBERG [31]. These were corrected for concentration and standard conditions (s020,W) to give values of 1.22 S and 1.88 S for BRCA1-(230–347) and BRCA1-(340–554) respectively.
Figure 7. Continuous size distribution analysis of (a) BRCA1-(230–347) and (b) BRCA1-(340–554).
The calculated concentration distributions c(s) are plotted against the sedimentation coefficients (s).
To estimate the molecular masses for the two proteins, the program Sedfit was used to model the sedimentation concentration profiles using c(M) boundary analysis, and from this molecular masses of 15939 Da for BRCA1-(230–347) and 26500 Da for BRCA1-(340–554) were calculated. These are in good approximation to the molecular masses calculated from their amino acid sequences, which are 14458 Da and 25284 Da respectively. This indicates that, at a concentration of 1 mg/ml, both molecules are monomeric.
The frictional coefficients (f) were calculated from the sedimentation coefficients using eqn (2):
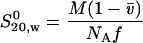 |
(2) |
where M is the mass calculated from the sequence, v is the partial specific volume and ρ is the solvent density. This was compared with the theoretical frictional coefficients of an unhydrated sphere of equal mass to give frictional ratios (f/f0) of 1.91 for BRCA1-(230–347) and 1.68 for BRCA1-(340–554). The measured frictional ratio is the product of the frictional ratios due to hydration (f/f0)hydration and shape (f/f0)shape:
 |
(3) |
where δ is the degree of hydration (estimated from Sednterp; see the Experimental section). Thus frictional ratios due to shape alone can be calculated and were found to be 1.6 for BRCA1-(230–347) and 1.4 for BRCA1-(340–554). These indicate that BRCA1-(230–347) is much more elongated than BRCA1-(340–554), which has a more compact structure.
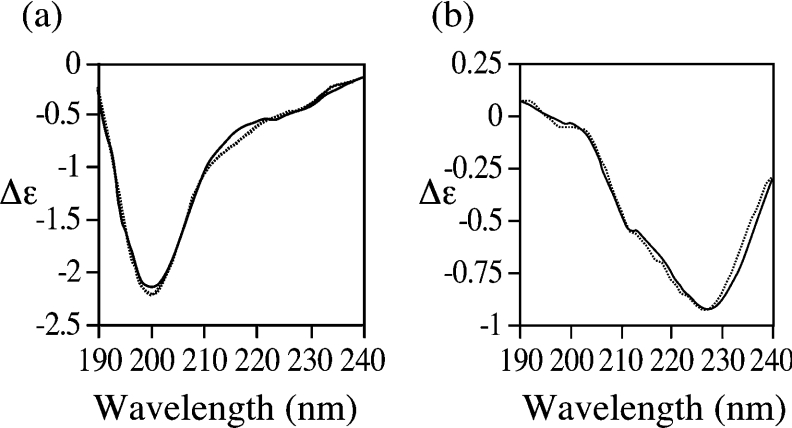
These data are reinforced by CD spectroscopy. The far-UV CD spectrum of BRCA1-(230–347) (Figure 8a) is identical with that of BRCA1-(230–534) [26] with a prominent negative band at 195 nm and a shoulder at 227 nm. These features are characteristic of a predominantly disordered conformation with the presence of short and/or distorted β-strands [32,33]. The CD spectrum of residues 340–534 reveals a much less disordered structure and is dominated by a minimum at 227 nm (Figure 8b), indicating the presence of more ordered β-sheet structures [33]. This is confirmed by analysis of the data using the CD fitting program CDSSTR [34], which predicts 6% α-helix, 37% β-sheet, 25% turn and 32% disordered structures.
Figure 8. Far-UV CD spectra of (a) BRCA1-(230–347) and (b) BRCA1-(340–554).
The CD signals are expressed in molar units (Δϵ). The observed and fitted CD spectra are represented by the continuous and broken line respectively.
DISCUSSION
BRCA1 is a large nuclear phosphoprotein that plays a multifaceted role in maintaining genome integrity and transcriptional regulation. Like many large multifunctional proteins, the different functions of BRCA1 are likely to be dispersed throughout the protein in discrete folded domains. However, to date, only two domains have been identified and characterized, the RING and BRCT domains [17], which account for only 16% of the total protein. Studies to define the domain structure have been restricted largely by its significant lack of similarity to any other protein and its immense size; by way of comparison, it is almost three times the size of haemoglobin. Therefore, having identified and characterized a soluble fragment of BRCA1 (residues 230–534) that binds specifically to four-way junction DNA [26], we have now used a combination of limited proteolysis, MS and extension cloning to dissect the domain structure of BRCA1-(230–534) and define more accurately the DNA-binding region.
The results presented touch upon several aspects of the structure–function relationship of BRCA1. Limited proteolysis demonstrates the existence of a stable domain, residues 230–339. Analysis of this region by CD suggests that it is largely unfolded and we therefore would expect it to be susceptible to proteolysis. However, localized areas of β-sheet folding are also predicted, suggesting that this region may be partly folded, explaining the observed resistance to trypsin digestion. The existence of such partly folded domains in BRCA1 is not surprising, as they possess the inherent flexibility required to support the regulation and stabilization of large multiprotein complexes such as BASC. An increasing number of proteins/domains, over a hundred to date, also display such highly flexible structures under physiological conditions and, like BRCA1, many of these proteins serve as scaffolds to support the regulated assembly of large multiprotein complexes [35].
Gel retardation analysis of BRCA1-(230–347) revealed that it was not essential for DNA binding. This suggested that the DNA-binding domain was, in part, located in residues 347–534. To locate the binding region more precisely, a number of protein fragments were generated, and residues 340–554 were shown to contain the DNA-binding domain. A comparison of the dissociation constants for binding to duplex and four-way junction DNA revealed that BRCA1-(340–554) also retained the specificity observed for BRCA1-(230–534), which has been used to suggest a more direct role for BRCA1 in the repair of double-strand breaks by homologous recombination [26]. The dissociation constant for four-way junction binding was estimated to be 3.3 μM. While this may be considered to be weak affinity binding, it is possible that one or more of the numerous interactions that occur within this region of BRCA1 enhance its affinity and may be a mechanism by which the function of BRCA1 in DNA repair is regulated.
Sedimentation studies revealed that residues 340–534 of BRCA1 form a more compact structure than BRCA1-(230–347), with a (f/f0)shape of 1.4 compared with 1.6, suggesting a more ordered conformation. This was confirmed by CD analysis in which the β-sheet component is much more apparent. In the limited proteolysis study, residues 340–534 were not stable to trypsin digestion, suggesting a largely unfolded structure: this was confirmed by comparison of the CD spectra of BRCA1-(230–347) with that of BRCA1-(230–534) (reported previously in [26]). As the degree of disorder is the same in both it suggests that residues 340–534 are also largely disordered. Therefore the addition of 20 C-terminal residues aids the correct folding of this region into a functional β-sheet DNA-binding domain. While numerous helical DNA-binding motifs have been observed, there is a precedent for β-sheet DNA interactions; for example, the eukaryotic proteins, TBP (TATA-binding protein) and transcription factor T domain, [36,37] and the prokaryotic proteins, IHF (integration host factor) and Met J repressor [38,39].
Recently, Mark et al. [20] showed that 16 fragments across the central region of BRCA1 were largely disordered. Owing to the lack of any defined homology for this region, the cloning of these fragments was based on secondary-structure predictions, which do not define exact domain boundaries. Therefore, while their conclusion that the central region of BRCA1 is largely unfolded is certainly true for residues 340–534, accurate domain mapping for the DNA-binding region shows that there are structured regions within the centre of BRCA1.
The identification of the DNA-binding domain of BRCA1 simplifies structural studies to provide a framework in which to evaluate the mechanistic consequences of hereditary mutations. Ultimately, it may also lead to a functional screen for such mutations. In addition, we anticipate that the results described in this paper will lead to a more complete understanding of the relationships between the biochemical and in vivo behaviour of BRCA1.
Acknowledgments
This work was supported by the Biotechnology and Biological Sciences Research Council (BBSRC), Breast Cancer Campaign and the Engineering and Physical Sciences Research council (EPSRC).
References
- 1.Easton D. F., Ford D., Bishop D. T. Breast and ovarian cancer incidence in BRCA1-mutation carriers. Breast Cancer Linkage Consortium. Am. J. Hum. Genet. 1995;56:265–271. [PMC free article] [PubMed] [Google Scholar]
- 2.Ford D., Easton D. F., Stratton M., Narod S., Goldgar D., Devilee P., Bishop D. T., Weber B., Lenoir G., Chang-Claude J., et al. Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The Breast Cancer Linkage Consortium. Am. J. Hum. Genet. 1998;62:676–689. doi: 10.1086/301749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rowell S., Newman B., Boyd J., King M. C. Inherited predisposition to breast cancer and ovarian cancer. Am. J. Hum. Genet. 1994;55:861–865. [PMC free article] [PubMed] [Google Scholar]
- 4.Neuhausen S. L., Marshall C. J. Loss of heterozygosity in familial tumors from three BRCA1-linked kindreds. Cancer Res. 1994;54:6069–6072. [PubMed] [Google Scholar]
- 5.Miki Y., Swensen J., Shattuck-Eidens D., Futreal P. A., Harshman K., Tavtigian S., Liu Q., Cichran C., Bennett L., Ding W., et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y., Cortez D., Yazdi P., Neff N., Elledge S. J., Qin J. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- 7.Foray N., Marot D., Gabriel A., Randrianarison V., Carr A. M., Perricaudet M., Ashworth A., Jeggo P. A subset of ATM- and ATR-dependent phosphorylation events requires the BRCA1 protein. EMBO J. 2003;22:2860–2871. doi: 10.1093/emboj/cdg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson S. F., Schlegel B. P., Nakajima T., Wolpin E. S., Parvin J. D. BRCA1 protein is linked to the RNA polymerase II holoenzyme complex via RNA helicase A. Nat. Genet. 1998;19:254–256. doi: 10.1038/930. [DOI] [PubMed] [Google Scholar]
- 9.Bochar D. A., Wang L., Beniya H., Kinev A., Xue Y., Lane W. S., Wang W., Kashanchi F., Shiekhattar R. BRCA1 is associated with a human SWI/SNF-related complex: linking chromatin remodeling to breast cancer. Cell. 2000;102:257–265. doi: 10.1016/s0092-8674(00)00030-1. [DOI] [PubMed] [Google Scholar]
- 10.Gowen L. C., Johnson B. L., Latour A. M., Sulik K. K., Koller B. H. Brca1 deficiency results in early embryonic lethality characterized by neuroepithelial abnormalities. Nat. Genet. 1996;12:191–194. doi: 10.1038/ng0296-191. [DOI] [PubMed] [Google Scholar]
- 11.Hakem R., de la Pompa J. L., Sirard C., Mo R., Woo M., Hakem A., Wakeham A., Potter J., Reitmair A., Billia F., et al. The tumor suppressor gene Brca1 is required for embryonic cellular proliferation in the mouse. Cell. 1996;85:1009–1023. doi: 10.1016/s0092-8674(00)81302-1. [DOI] [PubMed] [Google Scholar]
- 12.Shen S. X., Weaver Z., Xu X., Li C., Weinstein M., Chen L., Guan X. Y., Ried T., Deng C. X. A targeted disruption of the murine Brca1 gene causes gamma-irradiation hypersensitivity and genetic instability. Oncogene. 1998;17:3115–3124. doi: 10.1038/sj.onc.1202243. [DOI] [PubMed] [Google Scholar]
- 13.Xu X., Weaver Z., Linke S. P., Li C., Gotay J., Wang X. W., Harris C. C., Ried T., Deng C. X. Centrosome amplification and a defective G2–M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol. Cell. 1999;3:389–395. doi: 10.1016/s1097-2765(00)80466-9. [DOI] [PubMed] [Google Scholar]
- 14.Welcsh P. L., Owens K. N., King M. C. Insights into the functions of BRCA1 and BRCA2. Trends Genet. 2000;16:69–74. doi: 10.1016/s0168-9525(99)01930-7. [DOI] [PubMed] [Google Scholar]
- 15.Larson J. S., Tonkinson J. L., Lai M. T. A BRCA1 mutant alters G2–M cell cycle control in human mammary epithelial cells. Cancer Res. 1997;57:3351–3355. [PubMed] [Google Scholar]
- 16.Moynahan M. E., Chiu J. W., Koller B. H., Jasin M. Brca1 controls homology-directed DNA repair. Mol. Cell. 1999;4:511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 17.Koonin V. F., Altschul S. F., Bork P. BRCA1 protein products: functional motifs. Nat. Genet. 1996;13:266–267. doi: 10.1038/ng0796-266. [DOI] [PubMed] [Google Scholar]
- 18.Bork P., Hofmann K., Bucher P., Neuwald A. F., Altschul S. F., Koonin E. V. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 1997;11:68–76. [PubMed] [Google Scholar]
- 19.Pickart C. M. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 20.Mark W. Y., Liao J. C., Lu Y., Ayed A., Laister R., Szymczyna B., Chakrabartty A., Arrowsmith C. H. Characterization of segments from the central region of BRCA1: an intrinsically disordered scaffold for multiple protein–protein and protein–DNA interactions? J. Mol. Biol. 2005;345:275–287. doi: 10.1016/j.jmb.2004.10.045. [DOI] [PubMed] [Google Scholar]
- 21.Zhang H., Somasundaram K., Peng Y., Tian H., Zhang H., Bi D., Weber B. L., El-Deiry W. S. BRCA1 physically associates with p53 and stimulates its transcriptional activity. Oncogene. 1998;16:1713–1721. doi: 10.1038/sj.onc.1201932. [DOI] [PubMed] [Google Scholar]
- 22.Chai Y. L., Cui J., Shao N., Shyam E., Reddy P., Rao V. N. The second BRCT domain of the BRCA1 protein interacts with p53 and stimulates transcription from the p21WAF1/CIP1 promoter. Oncogene. 1999;18:263–268. doi: 10.1038/sj.onc.1202323. [DOI] [PubMed] [Google Scholar]
- 23.Zhong Q., Chen C. F., Li S., Chen Y., Wang C. C., Xiao J., Chen P. L., Sharp Z. D., Lee W. H. Association of BRCA1 with the hRad50–hMre11–p95 complex and the DNA damage response. Science. 1999;285:747–750. doi: 10.1126/science.285.5428.747. [DOI] [PubMed] [Google Scholar]
- 24.Folias A., Matkovic M., Bruun D., Reid S., Hejna J., Grompe M., D'Andrea A., Moses R. BRCA1 interacts directly with the Fanconi anemia protein FANCA. Hum. Mol. Genet. 2002;11:2591–2597. doi: 10.1093/hmg/11.21.2591. [DOI] [PubMed] [Google Scholar]
- 25.Scully R., Chen J., Plug A., Xiao Y., Weaver D., Feunteun J., Ashley T., Livingston D. M. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell. 1997;88:265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- 26.Sturdy A., Naseem R., Webb M. Purification and characterisation of a soluble N-terminal fragment of the breast cancer susceptibility protein BRCA1 J. Mol. Biol. 2004;340:469–475. doi: 10.1016/j.jmb.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 27.Paull T. T., Cortez D., Bowers B., Elledge S. J., Gellert M. Direct DNA binding by Brca1. Proc. Natl. Acad. Sci. U.S.A. 2001;98:6086–6091. doi: 10.1073/pnas.111125998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor I. A., McIntosh P. B., Pala P., Treiber M. K., Howell S., Lane A. N., Smerdon S. J. Characterization of the DNA-binding domains from the yeast cell-cycle transcription factors Mbp1 and Swi4. Biochemistry. 2000;39:3943–3954. doi: 10.1021/bi992212i. [DOI] [PubMed] [Google Scholar]
- 29.Takano E., Maki M., Nori H., Hatanaka N., Marti T., Titani K., Kannagi R., Ooi T., Murachi T. Pig heart calpastatin: identification of repetitive domain structures and anomalous behavior in polyacrylamide gel electrophoresis. Biochemistry. 1988;27:1964–1972. doi: 10.1021/bi00406a024. [DOI] [PubMed] [Google Scholar]
- 30.Stafford W. F., III Boundary analysis in sedimentation transport experiments: a procedure for obtaining sedimentation coefficient distributions using the time derivative of the concentration profile. Anal. Biochem. 1992;203:295–301. doi: 10.1016/0003-2697(92)90316-y. [DOI] [PubMed] [Google Scholar]
- 31.Philo J. S. A method for directly fitting the time derivative of sedimentation velocity data and an alternative algorithm for calculating sedimentation coefficient distribution functions. Anal. Biochem. 2000;279:151–163. doi: 10.1006/abio.2000.4480. [DOI] [PubMed] [Google Scholar]
- 32.Gursky O., Aleshkov S. Temperature dependent β-sheet formation β amyloid Aβ1–40 peptide in water: uncoupling β-structure folding from aggregation. Biochim. Biophys. Acta. 2000;1476:93–102. doi: 10.1016/s0167-4838(99)00228-9. [DOI] [PubMed] [Google Scholar]
- 33.Venyaminov S. Y., Yang J. T. Determination of protein secondary structure. In: Fasman G. D., editor. Circular Dichroism and the Conformational Analysis of Biomolecules. New York: Plenum Press; 1996. pp. 69–108. [Google Scholar]
- 34.Sreerama N., Woody R. W. Estimation of protein secondary structure from CD spectra: comparison of CONTIN, SELCON and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 35.Tompa P. Intrinsically unstructured proteins. Trends Biochem. Sci. 2002;27:527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 36.Kosa P. F., Ghosh G., Dedecker B. S., Sigler P. B. The 2.1Å crystal structure of an archaeal preinitiation complex: TATA box-binding protein/transcription factor (II)B core/TATA box. Proc. Nat. Acad. Sci. U.S.A. 1997;94:6042–6047. doi: 10.1073/pnas.94.12.6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muller C. W., Herrmann B. G. Crystallographic structure of the T domain–DNA complex of the Brachyury transcription factor. Nature (London) 1997;389:884–888. doi: 10.1038/39929. [DOI] [PubMed] [Google Scholar]
- 38.Somers W. S., Phillips S. E. V. Crystal structure of the met repressor-operator complex at 2.8 Å resolution reveals DNA recognition by β-strands. Nature (London) 1992;359:387–393. doi: 10.1038/359387a0. [DOI] [PubMed] [Google Scholar]
- 39.Rice P. A., Yang S. W., Mizuuchi K., Nash H. A. Crystal structure of an IHF–DNA complex: a protein-induced DNA U-turn. Cell. 1996;87:1295–1306. doi: 10.1016/s0092-8674(00)81824-3. [DOI] [PubMed] [Google Scholar]