Abstract
The kinetics of proton transport through mammalian UCP1 (uncoupling protein 1) expressed in yeast mitochondria were measured. There was little or no UCP1 activity in the absence of added palmitate, but significant activity in its presence. The activator 4-HNE (4-hydroxy-2-nonenal) had little effect when added alone, but significantly enhanced proton conductance in the presence of added palmitate. Activation of the proton conductance of UCP1 was synergistic: proton conductance in the presence of both palmitate and 4-HNE was significantly greater than the sum of the individual effects. Mitochondria from control yeast transformed with empty vector showed no such synergy, showing that synergy is a property of UCP1. Activation by the 4-HNE analogue trans-cinnamate showed essentially the same characteristics as activation by 4-HNE. Mitochondria from brown adipose tissue also showed synergistic activation of GDP-sensitive proton conductance by palmitate and 4-HNE. These results show that reactive alkenals activate the proton conductance of UCP1 more strongly when fatty acids are also added, with implications for both mechanistic and physiological models of UCP1 activation.
Keywords: fatty acid, 4-hydroxy-2-nonenal (4-HNE), mitochondria, proton leak, Saccharomyces cerevisiae, uncoupling protein 1 (UCP1)
Abbreviations: ADIFAB, acrylodated intestinal fatty acid binding protein; FCCP, carbonyl cyanide p-trifluoromethoxyphenylhydrazone; 4-HNE, 4-hydroxy-2-nonenal; SOD, superoxide dismutase; TPMP+, triphenylmethylphosphonium cation; TTNPB, 4-[(E)-2-(5,6,7,8-tetrahydro-5,5,8,8-tetramethyl-2-naphthalenyl)-1-propenyl]benzoic acid; UCP, uncoupling protein
INTRODUCTION
Reactive oxygen species and their lipid peroxidation products (e.g. reactive alkenals) are historically considered deleterious, mainly due to their reactivity with nucleic acids and lipid bilayer components, leading to cellular damage [1]. It has been proposed, however, that these molecules can also act as secondary messengers in the cell, mediating important physiological processes [2]. Products of lipid peroxidation such as 4-HNE (4-hydroxy-2-nonenal) and related reactive alkenals have been identified as biological signals for the activation of mild mitochondrial uncoupling via UCPs (uncoupling proteins) [3]. The ancestral function of this activation may be to lower reactive oxygen species production by the respiratory chain complexes [4,5].
UCP1 is a well-characterized member of the UCP family. UCP1 catalyses adaptive thermogenesis in mammalian brown adipose tissue by greatly increasing the proton conductance of the mitochondrial inner membrane. When appropriately activated, the leak of protons through this proton conductance pathway uncouples substrate oxidation from phosphorylation of ADP to ATP, leading to fast oxygen consumption and ultimately to heat production [6]. The classic activators of UCP1 are fatty acids, although the proposed mechanisms by which they activate remain controversial.
The activation of UCPs by reactive alkenals was first discovered in connection with the observation that superoxide activates the proton conductance of UCPs [7]. More recently, it was proposed that superoxide, as well as the activators ubiquinone [8,9] and AAPH [2,2′-azobis-(2-methyl propionamidine) dihydrochloride; a carbon-centred radical generator] [10], work indirectly on UCPs by generating carbon-centred radicals on polyunsaturated fatty acyl chains of phospholipids in the mitochondrial inner membrane [10,11]. The lipid peroxides produced by oxidation of these radicals autocatalytically release 4-HNE, which is proposed to be the direct activator of the UCPs [10,12]. Other activators, such as trans-retinoate [13], trans-cinnamate, trans-retinal [3] and TTNPB {4-[(E)-2-(5,6,7,8-tetrahydro-5,5,8,8-tetramethyl-2-naphthalenyl)-1-propenyl]benzoic acid} [13,14], are proposed to act as 4-HNE analogues, although retinoids and TTNPB may have additional effects due to their similarity to fatty acids [12].
In the present paper, we analyse the interrelationships between fatty acid and reactive alkenal activation of UCP1 proton conductance in isolated mitochondria. For this purpose, we expressed the protein in wild-type yeast, and assayed proton transport activity in isolated mitochondria. We show that in the absence of fatty acids, 4-HNE has little or no activatory effect on the proton conductance of UCP1 in yeast mitochondria. However, when fatty acids are present to partially activate UCP1, addition of 4-HNE causes a significant additional increase in UCP1-dependent proton conductance. The same observation was made using mitochondria isolated from brown adipose tissue. The interpretation of these observations and other results in the literature leads to the proposal of a dual site model for UCP1 activation, in which fatty acids and reactive alkenals activate proton conductance synergistically.
EXPERIMENTAL
Expression of UCP1 in Saccharomyces cerevisiae
Cells of the S. cerevisiae wild-type strain CEN.PK2-1C [15] were transformed with either a mouse UCP1 expression construct or a plasmid containing an empty vector, using an Sc EasyComp transformation kit (Invitrogen). The UCP1 expression construct used in the present study (pBF307, modest UCP1 expression) and the empty vector plasmid (pBF254) were constructed and used previously [16,17].
Precultures of the transformed yeast cells were grown at 30 °C in an orbital shaker (220 rev./min; 18–24 h) in selective lactate medium {2% (v/v) DL-lactic acid [85% (w/w) syrup]/0.67% yeast nitrogen base without amino acids (Difco)/0.2% casamino acids (Difco)/0.12% (NH4)2SO4/0.1% KH2PO4/0.1% glucose/0.002% L-tryptophan/0.004% adenine}. Precultures were grown to high density [D600 (attenuance) 1–2] and then transferred to culture medium [22% DL-lactic acid (85% syrup)/1% yeast nitrogen base without amino acids/1% (w/v) D-galactose (Fischer Chemicals)/0.1% glucose (AnalaR; BDH)/0.05% CaCl2 (Fischer Chemicals)/0.05% NaCl/0.06% MgCl2/0.1% KH2PO4 (Fisons Chemicals, Loughborough, Leics., U.K.)/0.1% NH4Cl (AnalaR; BDH)/0.8% NaOH (AnalaR; BDH)/0.1% casamino acids/0.04% adenine/0.02% L-tryptophan, pH 5.5] for growth under adequate conditions for induction of UCP1 expression. Cultures were incubated at 30 °C in orbital shakers (230–250 rev./min).
Isolation of mitochondria
Yeast mitochondria were isolated according to a previously published method [11]. Yeast cells from cultures grown at 30 °C with a D600 of 0.8–1.1 were harvested by centrifugation at 2900 g (16 min) at 20–23 °C, resuspended in deionized water and re-centrifuged. Pellets were resuspended in a buffer containing 100 mM Tris/HCl and 20 mM dithiothreitol (pH 9.3), and incubated at 30 °C in an orbital shaker (220 rev./min; 10 min). Cells were re-centrifuged and washed twice in a buffer containing 100 mM Tris/HCl and 500 mM KCl (pH 7.0), and then resuspended in 1 ml of iso-osmotic spheroplasting buffer (40 mM citric acid/120 mM Na2HPO4/1.35 M sorbitol/1 mM EGTA, pH 5.8) per 1 g of cells. Lyticase was added (at 3 mg/g wet weight of cells), and the suspension was incubated at 30 °C (10–15 min) in an orbital shaker (220 rev./min). Subsequent steps were performed at 4 °C. Spheroplasts were pelleted, washed twice in approx. 30 ml of a buffer containing 10 mM Tris maleate, 0.75 M sorbitol, 0.4 M mannitol, 2 mM EGTA and 0.1% (w/v) defatted BSA (pH 6.8) and then resuspended in 30 ml of isolation buffer (10 mM Tris maleate/0.6 M mannitol/2 mM EGTA/1 mM EDTA/0.5 mM Na2HPO4/1% BSA, pH 6.8) with Complete™ protease inhibitor (one tablet per 40 ml of buffer; Boehringer Mannheim). The resuspended pellets were further homogenized by 12 passes with a Wesley Coe homogenizer (clearance 0.54 mm), and homogenates were centrifuged at 800 g (10 min). The supernatants were recovered by pipette, and the pellets were resuspended in a small volume of isolation buffer and re-centrifuged at 800 g (10 min), to collect mitochondria still retained in the pellet. Supernatants were combined and centrifuged three times at 800 g (10 min), and the pellets were discarded. The resultant supernatants were centrifuged at 11000 g (10 min). The mitochondrial pellets were washed in a buffer containing 10 mM Tris maleate and 0.65 M sorbitol, pH 6.8, and centrifuged at 11000 g. The final pellet was resuspended in 150–300 μl of the same buffer. The protein content of mitochondria was assayed using the Bio-Rad DC modified Lowry protein assay, with BSA as protein standard.
Brown adipose tissue mitochondria were isolated essentially as described in [3]. Briefly, interscapular, subscapular and aortic brown adipose tissues were taken from six 7–8-week-old female rats (housed at 25 °C). Subsequent steps were carried out at 4 °C. Tissue was placed in STE buffer (250 mM sucrose/5 mM Tris/HCl/2 mM EGTA, pH 7.4) supplemented with 1% defatted BSA, minced, homogenized in STE+BSA buffer with 10 strokes of a Dounce homogenizer and filtered through two layers of gauze. The filtrate was centrifuged at 8500 g (10 min) and the pellet was resuspended in STE+BSA and centrifuged at 700 g (10 min), and the supernatant was then centrifuged at 8500 g (10 min). The mitochondrial pellet was suspended in STE, re-centrifuged and finally resuspended in 500–600 μl of STE. Protein concentration was determined by the biuret method.
Proton leak kinetics of yeast mitochondria
The kinetic response of the proton conductance pathway to its driving force (i.e. the proton-motive force) is appropriately expressed as the relationship between the rate of oxygen consumption driving proton leak and the membrane potential (under conditions where the pH component of proton-motive force is converted into membrane potential). The mitochondrial membrane potential can be varied by titration with electron transport chain substrates (or alternatively by titration with respiratory chain inhibitors, in the presence of excess substrate), and proton leak kinetics under different situations can be studied.
Respiration rate and membrane potential were measured simultaneously at 30 °C, using electrodes sensitive to oxygen (Clark-type) and to the potential-dependent probe TPMP+ (triphenylmethylphosphonium cation) [18]. The oxygen electrode was calibrated with air-saturated electrode buffer, assumed to contain 424.8 nmol of O/ml at 30 °C [19]. Mitochondria (0.6 mg of protein/ml) were incubated in 2.5 ml of electrode buffer (20 mM Tris/HCl/450 mM sorbitol/100 mM KCl/0.5 mM EGTA/5 mM MgCl2/10 mM K2HPO4/0.1% defatted BSA, pH 6.8), after addition to the medium of 3 μM myxothiazol (inhibitor of the cytochrome bc1 complex), 10 μg/ml oligomycin (to prevent ATP synthesis, allowing all measured proton conductance to be due to proton leak processes) and 1 μg/ml nigericin (to collapse the pH difference across the mitochondrial inner membrane, leaving all proton-motive force in the form of membrane potential). The electrode chamber was closed, avoiding air bubbles, and subsequent additions were made by microsyringe through a small channel. The TPMP+ electrode was calibrated with sequential additions of 1 μM TPMP+, up to a final concentration of 4 μM. Mitochondria were incubated for a total of 4 min before the first addition of substrate. Respiration and membrane potential were progressively stimulated through successive steady states by addition of the respiratory substrate D-lactate (an electron donor to cytochrome c in yeast mitochondria), in steps up to 2.2 mM final concentration. FCCP (carbonyl cyanide p-trifluoromethoxyphenylhydrazone; 5 μM) was added at the end of each run to dissipate the membrane potential and release all TPMP+ to the medium, allowing linear baseline correction for any small electrode drift. A second addition of 5 μM FCCP allowed estimation of the small correction for direct effects of the uncoupler on the TPMP+ electrode signal.
For calculation of membrane potential, binding of TPMP+ to the mitochondrial membrane needs to be corrected for. The TPMP+ binding correction and matrix volume of haploid yeast mitochondria were measured by Dr E. J. Cornwall and Dr J. L. Pakay (MRC Dunn Human Nutrition Unit, Cambridge, U.K.) according to protocol 6 of [18], which corrects the accumulation ratio of radiolabelled TPMP+ to the accumulation ratio for 86Rb (which does not bind to the mitochondrial membrane) measured over a range of membrane potentials. The TPMP+ binding correction was 0.4, and the mitochondrial matrix volume was 1.8 μl/mg [20].
Proton leak kinetics of brown adipose tissue mitochondria
The proton leak kinetics of brown adipose tissue mitochondria were measured at 37 °C following the same principles described above. Electrode buffer was assumed to contain 406 nmol of O/ml at 37 °C [19]; the TPMP+ binding correction was from [18]. Brown adipose tissue mitochondria (0.35 mg of protein/ml) were incubated in 3.5 ml of assay medium (50 mM KCl/5 mM Hepes/1 mM EGTA/4 mM KH2PO4/1% defatted BSA, pH 7.2) and the TPMP+ electrode was inserted, sealing the chamber. The medium was supplemented with 80 ng/ml nigericin, 10 μg/ml oligomycin and the Complex I (NADH:ubiquinone oxido-reductase) inhibitor rotenone (5 μM). The TPMP+ electrode was calibrated as described above but using five 0.5 μM additions of TPMP+. Mitochondria were energized by addition of 10 mM glycerol 3-phosphate as substrate and, after 2.5 min, membrane potential was decreased in steps with sequential additions of cyanide (50 mM KCN in 0.5 M Hepes) (14.3–228.6 μM).
Free (unbound) palmitate levels in the medium
The free palmitate concentration in a medium containing BSA in the presence and absence of 4-HNE or trans-cinnamate was determined using the fluorescence probe ADIFAB (acrylodated intestinal fatty acid binding protein; Invitrogen), according to the supplier's instructions. Knowing the Kd of the conjugate for palmitate (320 nM at 37 °C [21]) and the fluorescence intensity of unbound indicator at 432 nm, the free (unbound) fatty acid concentration was calculated using the equation in [21].
Free fatty acids were measured using conditions approximating to those of measurements of proton leak kinetics. ADIFAB (0.2 μM) was added to 2 ml of electrode buffer with 0.01% defatted BSA in a quartz cuvette (10 mm×10 mm; Hellma) at 30 °C, and fluorescence was measured at 432 nm using an RF-5301PC spectrofluorophotometer (Shimadzu). Palmitate (0.5 μM) and 0.35 μM cinnamate or 32 μM 4-HNE were added to the cuvette, and changes from the initial fluorescence intensity were recorded 6 min after each addition.
Palmitate bound to mitochondria
Changes induced by 4-HNE and trans-cinnamate in palmitate bound to BSA in the medium were assessed indirectly by measuring the binding of palmitate to a convenient hydrophobic phase assumed to be in equilibrium with palmitate in the medium: fresh rat liver or kidney mitochondria (see [7,22] for isolation methods). Binding was determined according to modifications of a reported method [23] under conditions identical with those of proton leak measurements. Mitochondria (0.6 mg of protein) were added to microfuge tubes containing 1 ml of electrode buffer (containing 0.1% defatted BSA, 3 μM myxothiazol, 10 μg/ml oligomycin, 1 μg/ml nigericin and 4 μM TPMP+) at 30 °C, followed by 0.4 μCi/ml [U-14C]sucrose and 50 μM palmitate containing 1.5 μCi/ml [9,10(n)-3H]palmitate. Where appropriate, 150 μM 4-HNE or 150 μM cinnamate was also added. Tubes were vortex-mixed for 5 s and incubated at 30 °C for 7 min, mixing by inversion every 2 min. Mitochondria were centrifuged at 10000 g, and 50 μl of supernatant was recovered and placed in a scintillation vial. The remaining supernatant was discarded and any remaining liquid was carefully removed using tissue paper. The mitochondrial pellet was resuspended in 50 μl of 20% (v/v) Triton X-100, by vortex-mixing. The bottom of the microfuge tube containing the resuspended pellet was cut and placed into a scintillation vial. Scintillation cocktail (4 ml; Ultima Gold™; Packard Biosciences) was added to each vial (containing either supernatant or resuspended mitochondria), and mixed by inversion. Radioactivity was determined using a PerkinElmer Liquid Scintillation analyser, Tri-Carb 2800 TR, and from the data obtained, the concentration of palmitate bound to mitochondria (nmol per mg of mitochondrial protein) was determined.
Statistics
Points on proton leak curves represent average values (with S.E.M.) obtained for that point on different days. To construct proton leak rate bar graphs and allow statistical testing, the respiration rate under each condition was taken at the highest potential common to all conditions on a given day, i.e. the state 4 membrane potential in the presence of UCP1, alkenal and palmitate. Respiration values at this potential under other conditions were obtained by linear interpolation between flanking points. For empty vector, values were taken at the mean highest common potential for UCP1-containing yeast. Means for these values (with S.E.M.) are plotted in the bar graphs with the mean value for the highest common potential indicated, and differences between mean respiration values for different days were tested for significance by paired Student's t test, assuming equal variance.
Chemicals
Potassium D-lactate (stored at −20 °C) and sodium GDP (prepared fresh every day) were dissolved in deionized water; palmitic acid, trans-cinnamic acid, trans-hydrocinnamic acid, myxothiazol, nigericin, oligomycin and FCCP were dissolved in 100% ethanol and stored at −20 °C. Defatted BSA contained less than 0.005% (w/w) fatty acids. Chemicals were from Sigma unless stated otherwise. Radiochemicals were from Amersham International.
RESULTS
UCP1 activity in yeast mitochondria in the absence of added fatty acids
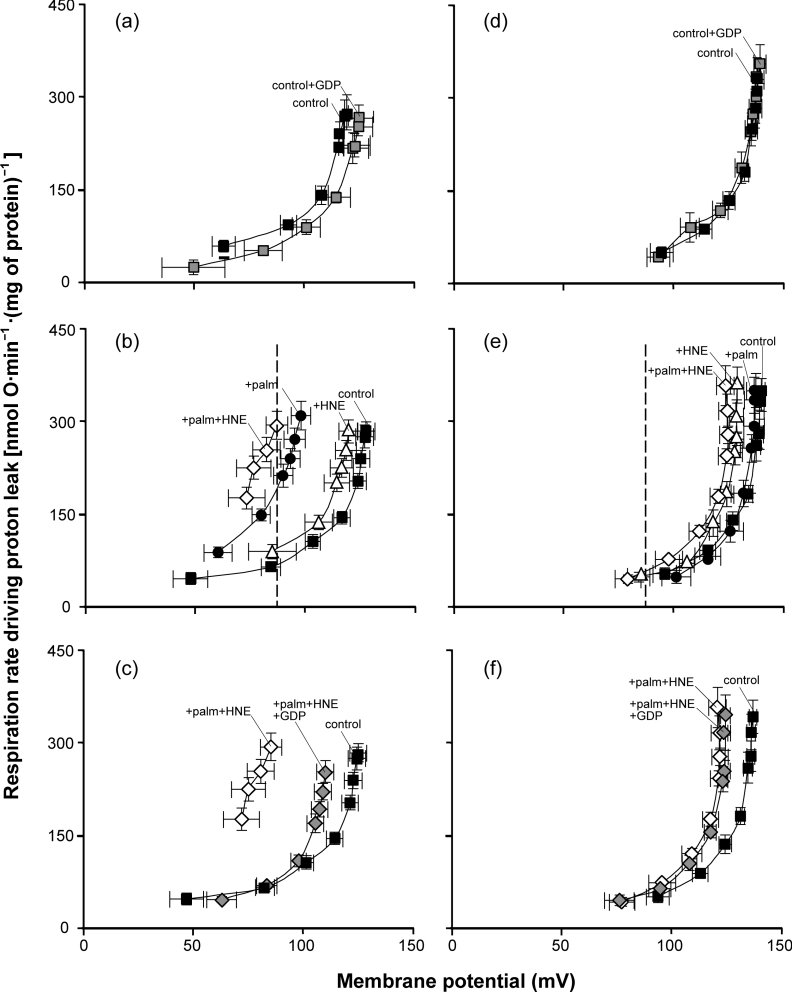
The proton leak kinetics of yeast mitochondria containing UCP1, measured as the oxygen consumption rate needed to drive proton leak as a function of the driving force, mitochondrial membrane potential, have been characterized previously [3,7,11,17]. Figure 1 shows the classic non-Ohmic proton leak kinetics of yeast mitochondria: proton leak rate increases steeply at higher membrane potentials. As previously reported [17], expression of modest amounts of mouse UCP1 from plasmid pBF307 did not greatly change the proton conductance. This is shown by the relatively small increase in proton leak rate at any given potential in mitochondria from yeast expressing UCP1 (‘UCP1-containing mitochondria’) (Figure 1a) compared with mitochondria from yeast transformed with empty vector (‘empty vector mitochondria’) (Figure 1b). UCP1 was almost inactive in yeast mitochondria under conditions of no added fatty acids or alkenals, since GDP, the classic UCP1 inhibitor, decreased the proton leak rate only marginally (P=0.10 at 87 mV; Figure 1a) compared with its large inhibitory effect on the proton leak rate through activated UCP1 (Figure 1c, see below).
Figure 1. Kinetics of proton leak in yeast mitochondria: effects of 4-HNE and palmitate (palm).
Mitochondria isolated from wild-type CEN.PK2-1C yeast containing (a–c) pBF307 (for UCP1 expression) or (d–f) an empty vector (pBF254), grown under the same conditions, were incubated in a medium containing 0.1% defatted BSA as described in the Experimental section. Where indicated, 1 mM GDP, 150 μM 4-HNE and/or 50 μM palmitate were present. Oxygen consumption and mitochondrial membrane potential were varied with D-lactate titration. Points represent means±S.E.M. for three to seven independent experiments. The dashed line marks 87 mV, the highest membrane potential common to all curves.
UCP1 activity in yeast mitochondria in the presence of added fatty acids
In the presence of inducers of proton transport through UCP1, such as palmitate, mitochondria from yeast expressing modest levels of UCP1 are known to have much greater proton conductance than empty vector controls under the same conditions [11,17,24]. Addition of GDP returns the higher proton conductance of UCP1-containing yeast mitochondria almost to the basal level seen with empty vector mitochondria. In contrast, addition of low concentrations of palmitate has no effect in empty vector controls, showing that the induced uncoupling effect is caused by UCP1 activity [3,7,11,17]. These effects of palmitate were replicated in the present study (Figures 1b and 1e), showing that mouse UCP1 in our yeast mitochondria was competent and required addition of fatty acid before large proton leak rates could be observed.
Figure 2 shows respiration rates (which are proportional to proton leak rates) at the highest membrane potential common to all proton leak curves in Figure 1, to allow easier visualization of differences in proton leak rate at the same driving force under different conditions. Figure 2 emphasizes the observation in Figure 1(b) that addition of palmitate activated the proton conductance of UCP1 in yeast mitochondria, and the observation in Figure 1(e) that palmitate, at the concentration used here, had no effect on the proton conductance of mitochondria lacking UCP1.
Figure 2. Proton leak rates of yeast mitochondria at fixed membrane potential: effects of 4-HNE and palmitate (palm).
Proton leak rates at fixed membrane potential were measured as the respiration rate driving proton leak at the highest membrane potential common to all curves on each day; mean values from different days ±S.E.M. are shown. The mean value for the highest common potential was 87 mV. Results shown are the same as those used to construct Figure 1. Values of P were calculated using paired Student's t test.
UCP1 activity in yeast mitochondria in the presence of 4-HNE
In the presence of fatty acids, 4-HNE activates GDP-sensitive uncoupling in yeast mitochondria containing UCP1, but not in empty vector mitochondria [3]. To investigate whether the increase in mitochondrial proton conductance in the presence of added 4-HNE depends on added fatty acids, the effect of 4-HNE on proton conductance was tested in the absence of added fatty acids. The results are shown in Figures 1(b) and 2. The addition of 4-HNE to mitochondria containing UCP1 induced only a marginal increase in proton conductance (Figures 1b and 2), which did not reach statistical significance (P=0.06). However, above approx. 120 mV, this increase was also seen in empty vector mitochondria (Figure 1e), so it mostly reflects UCP1-independent uncoupling in yeast mitochondria. Therefore, in the absence of palmitate, 4-HNE induces little or no proton conductance through UCP1.
UCP1 activity in yeast mitochondria in the presence of added fatty acids and 4-HNE
The kinetics of proton leak in the presence of palmitate and 4-HNE simultaneously were measured in UCP1-containing and empty vector mitochondria (Figures 1b, 1c, 1e, 1f and 2). Palmitate and 4-HNE added simultaneously to UCP1-containing mitochondria induced higher proton leak rates at any given membrane potential than palmitate alone (Figures 1b and 2). The increased proton conductance was fully sensitive to GDP up to approx. 120 mV (Figure 1c), above which non-specific effects of 4-HNE become obvious, and (except for these non-specific effects of 4-HNE) was absent from empty vector mitochondria (Figures 1e and 1f), indicating that it was mediated by UCP1.
Activation of UCP1 proton conductance by 4-HNE and palmitate is synergistic
Figure 2 analyses the increases in proton leak rate at 87 mV caused by palmitate and 4-HNE. The increase in proton leak rate induced in UCP1-containing mitochondria by simultaneous addition of 4-HNE and palmitate (+palm+HNE) was higher than that induced by palmitate alone (+palm) and greater than the 4-HNE-induced and palmitate-induced effects added together ([+palm]+[+HNE]) (P=0.03). There were no comparable effects in the empty vector mitochondria. This clearly shows that 4-HNE and palmitate activate proton conductance via UCP1 in a synergistic manner, and are not simply additive.
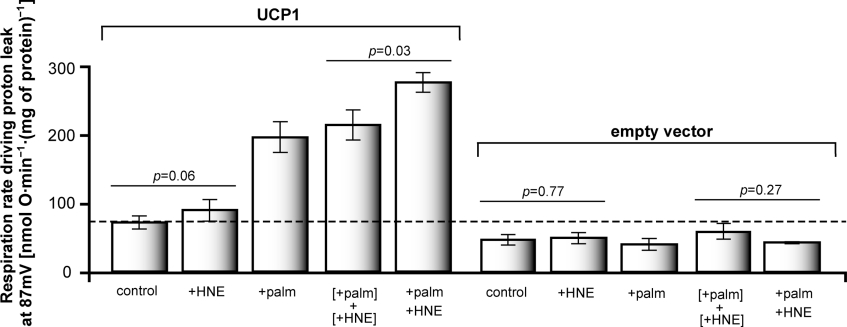
Fatty acids and cinnamate activate UCP1 synergistically
Compounds structurally related to 4-HNE that contain the reactive 2-alkenal group (e.g. trans-2-nonenoate, trans-nonenal, trans-retinoate, trans-retinal and trans-cinnamate) also induce an increase in GDP-sensitive proton conductance through UCPs [3]. However, other workers failed to observe activation of UCP1 in yeast mitochondria by cinnamate in the absence of added fatty acid [25]. To test whether the activation of UCP1 by trans-cinnamate, like activation by 4-HNE, is much more prominent in the presence of fatty acids, we investigated the interdependence of UCP1 activation by palmitate and cinnamate.
Figure 3 shows that activation of proton conductance through UCP1 by trans-cinnamate had essentially the same characteristics as activation by 4-HNE. Figures 3(a) and 3(c) show that in the absence of added palmitate, cinnamate caused a significant but rather small increase in proton conductance in mitochondria from yeast containing UCP1. Cinnamate plus palmitate induced a significantly greater increase in proton conductance than palmitate alone or the sum of the effects of palmitate and cinnamate added separately. The effect of cinnamate plus palmitate on proton conductance was sensitive to GDP (Figure 3b), indicating that it was catalysed by UCP1. Thus cinnamate and 4-HNE activate UCP1 with essentially the same characteristics, and the effect of each is strongly enhanced by the presence of fatty acid.
Figure 3. Kinetics of proton leak in yeast mitochondria: effects of trans-cinnamate (CA) and palmitate (palm).
Mitochondria isolated from wild-type CEN.PK2-1C yeast containing pBF307 (for UCP1 expression) were incubated in a medium containing 0.1% defatted BSA as described in the Experimental section. Where indicated, 1 mM GDP, 150 μM trans-cinnamate and/or 50 μM palmitate were present. (a, b) Oxygen consumption and mitochondrial membrane potential were varied with D-lactate titration. Points represent means±S.E.M. for five independent experiments. The dashed line marks 83 mV, the highest membrane potential common to all curves. (c) Proton leak rates were measured as the respiration rate driving proton leak at the highest membrane potential common to all curves on each day; mean values from different days ±S.E.M. are shown. The mean value for the highest common potential was 83 mV. Results shown are the same as those used to construct (a). Values of P were calculated using paired Student's t test.
4-HNE and cinnamate do not increase UCP1-dependent uncoupling by displacing palmitate from BSA
Since the palmitate concentrations used in the present study were subsaturating for UCP1 activation, there is a possibility that 4-HNE and cinnamate induced proton conductance through UCP1 in the presence of palmitate indirectly, by displacing palmitate bound to BSA. This would cause an increase in free palmitate in the medium, and a consequent increase in UCP1 proton conductance.
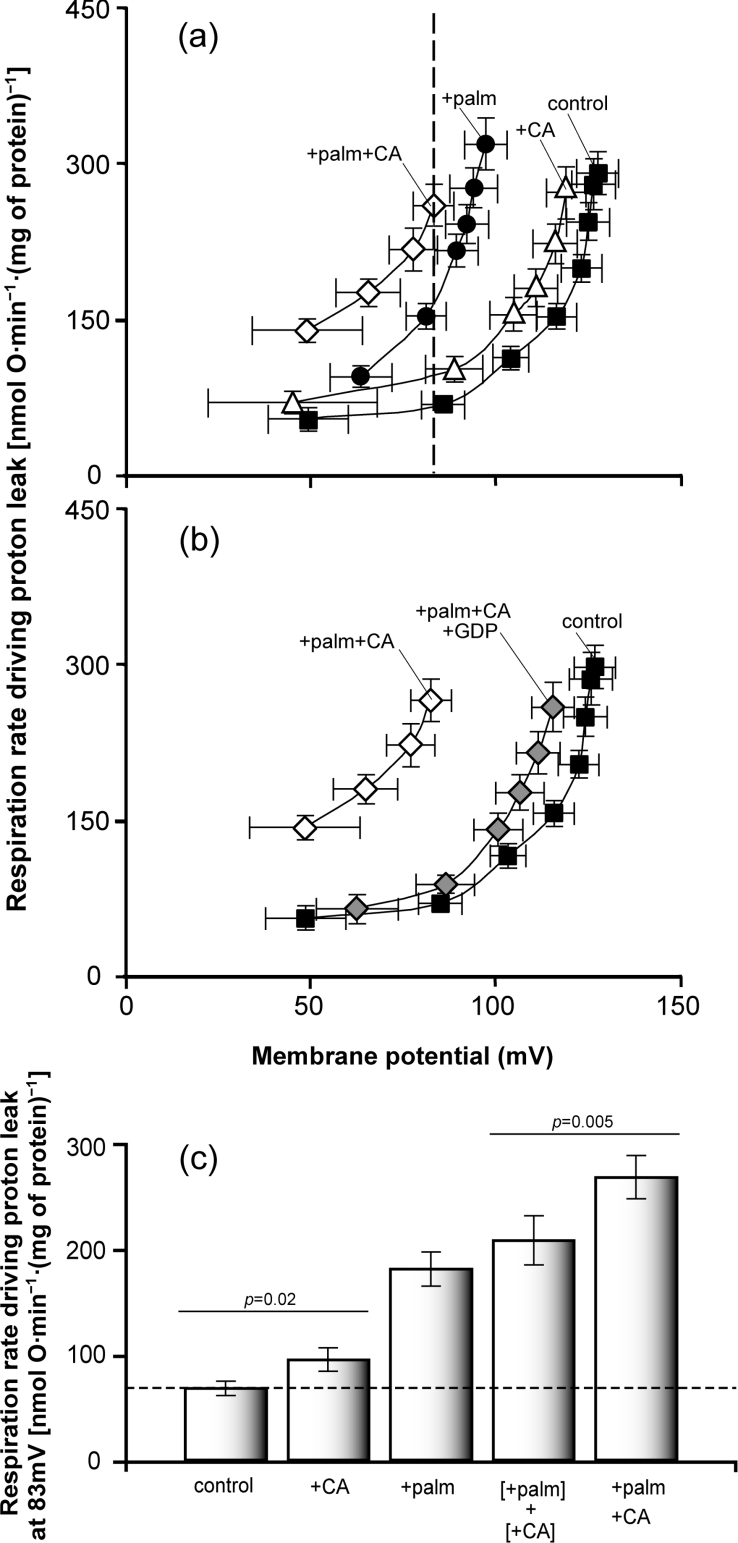
Hydrocinnamate is structurally similar to cinnamate but lacks the critical double bond at C-2, so it should not activate UCP1 directly [3]. However, the lack of the double bond should have little effect on any displacement of palmitate from BSA. We investigated whether trans-hydrocinnamate induced proton conductance in mitochondria with UCP1 in the presence of fatty acids. Figure 4(a) shows that palmitate plus hydrocinnamate did not induce greater proton conductance than palmitate alone, supporting the previous observations that the 2-alkenal group is a structural requirement for activation of UCPs by reactive alkenals, and suggesting that the activation by cinnamate was not caused by displacement of palmitate from BSA.
Figure 4. Tests of whether reactive alkenals activate UCP1 by displacing palmitate from BSA.
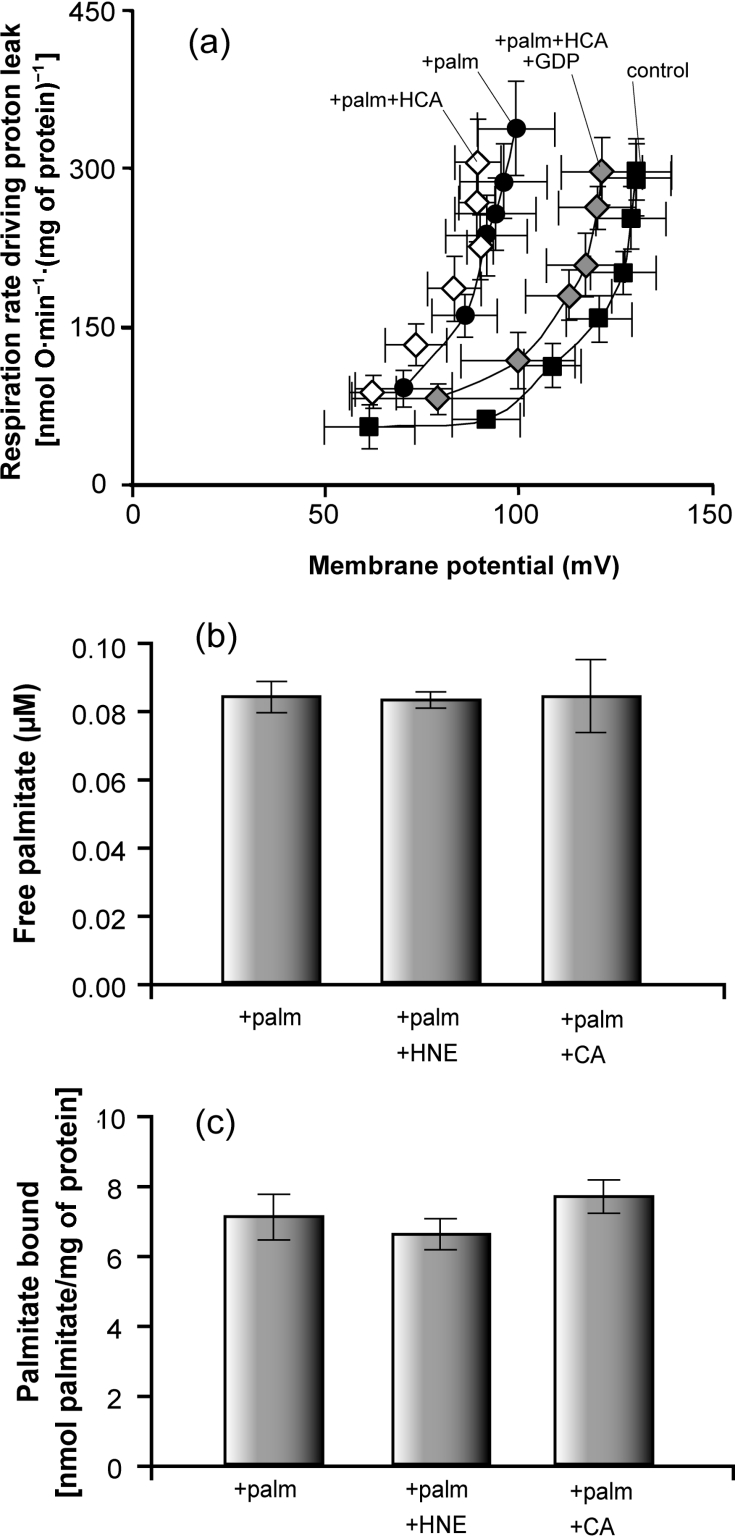
(a) Mitochondria isolated from wild-type CEN.PK2-1C yeast containing pBF307 (for UCP1 expression) were incubated in a medium containing 0.1% defatted BSA as described in the Experimental section. Where indicated, 1 mM GDP, 150 μM trans-hydrocinnamate (HCA) and/or 50 μM palmitate were present. Oxygen consumption and mitochondrial membrane potential were varied with D-lactate titration. Points represent means±S.E.M. for three independent experiments. (b) Free fatty acid (palmitate; palm) concentration was measured in electrode buffer (composition given in the Experimental section) containing 0.1% defatted BSA, using the fluorescent probe ADIFAB, in the presence of 0.5 μM palmitate, 32 μM 4-HNE and 0.35 μM trans-cinnamate where indicated. Bars represent means±S.E.M. for three to eight independent experiments. (c) Binding of radiolabelled palmitate to mitochondria. Mitochondria from rat liver and kidney were incubated at 0.6 mg of protein/ml in electrode buffer containing 0.1% defatted BSA in the presence of 50 μM palmitate (palm), 150 μM 4-HNE and/or 150 μM trans-cinnamate (CA) where indicated, and radiolabel associated with the mitochondrial pellet was measured. Bars represent means±S.E.M. for two (kidney) and three (liver) independent experiments, with three to six replicates per experiment (data from all experiments were pooled, as results showed no difference between liver and kidney). Methods used are described in the Experimental section.
A second way to test whether 4-HNE and cinnamate activate UCP1 indirectly by displacing palmitate from BSA is to assay for changes in free palmitate concentrations in BSA-containing media in the presence and absence of 4-HNE or cinnamate. Free palmitate was determined using the fluorescence probe ADIFAB. Figure 4(b) shows that neither 4-HNE (4-HNE/albumin molar ratio of 213, i.e. 21.3 times higher than this ratio during proton leak experiments) nor trans-cinnamate (cinnamate/albumin molar ratio of 2.3, the same as in proton leak experiments) induced an increase in free palmitate in the medium.
A third test is to assay for changes in palmitate bound to a hydrophobic phase in equilibrium with free palmitate. A convenient hydrophobic phase that can be separated out by centrifugation is provided by added freshly isolated mitochondria. Labelled palmitate picked up by added mitochondria will report any changes in free palmitate caused by displacement of labelled palmitate from BSA when 4-HNE or cinnamate is added. Figure 4(c) shows that palmitate binding to rat liver or kidney mitochondria in a medium containing 50 μM palmitate and 0.1% defatted BSA did not change significantly when 150 μM 4-HNE or 150 μM cinnamate was added. These results recap those obtained using yeast mitochondria in an independent study using a similar method [23].
These three independent methods show that 4-HNE and cinnamate do not displace fatty acids from BSA. Therefore these compounds probably increase the proton conductance of UCP1 in the presence of palmitate by directly interacting with the protein.
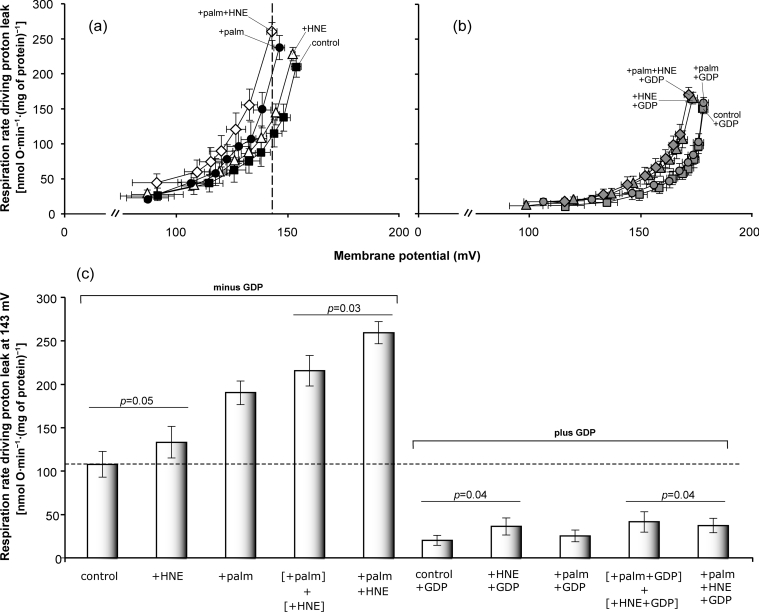
4-HNE and fatty acids activate UCP1 synergistically in mitochondria isolated from brown adipose tissue
To investigate whether palmitate is required for activation of UCP1 by alkenals in its native environment, the effects of palmitate and 4-HNE on proton conductance mediated by UCP1 were assessed in mitochondria isolated from rat brown adipose tissue. Figure 5(a) shows that the proton conductance of these mitochondria was highest when both palmitate and 4-HNE were present. Figure 5(b) shows that GDP prevented most of the activation, consistent with it being caused by UCP1. As with yeast mitochondria (Figures 1e and 1f), 4-HNE also had a small non-specific effect through GDP-insensitive pathways, particularly at higher membrane potentials (Figure 5b). Figure 5(c) shows that, at the highest common membrane potential, GDP-sensitive proton leak through UCP1 when both compounds were present (+palm+HNE) was significantly higher than the combined increase in GDP-sensitive UCP1 activity when the compounds were added separately ([+palm]+[+HNE]). Thus activation of proton conductance through UCP1 by 4-HNE and palmitate is synergistic in brown adipose tissue mitochondria just as it is in yeast mitochondria.
Figure 5. Kinetics of proton leak in rat brown adipose tissue mitochondria: effects of 4-HNE and palmitate (palm).
(a, b) Mitochondria from rat brown adipose tissue were incubated in a medium containing 1% (w/v) defatted BSA as described in the Experimental section. Where indicated, 0.5 mM GDP, 80 μM 4-HNE and/or 40 μM palmitate were present. (a) No GDP, (b) plus GDP. Oxygen consumption and membrane potential were varied by cyanide titration. (c) Proton leak rates at fixed membrane potential were measured as the respiration rate driving proton leak at the highest membrane potential common to all curves on each day; mean values from different days ±S.E.M. are shown. The mean value of the highest common potential was 143 mV. Data are the same as those used to construct (a, b). Values of P were calculated using paired Student's t test. Values represent means±S.E.M. for six independent experiments.
DISCUSSION
Our results (Figures 1–3) show that 4-HNE and its analogue trans-cinnamate are poor activators of proton conductance through UCP1 in the absence of palmitate. In the presence of palmitate, however, these compounds do activate UCP1. Activation is synergistic: the effect of 4-HNE and palmitate when they are both present is significantly greater than the sum of their effects when they are added separately. Synergistic activation depends on the presence of UCP1, since it is not seen in yeast mitochondria lacking mouse UCP1 and is prevented by the classic UCP1 inhibitor, GDP. Synergy is seen when UCP1 is present either in yeast mitochondria or in its native environment of brown adipose tissue mitochondria, suggesting that synergistic activation of the proton conductance of UCP1 by alkenals and fatty acids may be a normal attribute of UCP1 in its physiological context.
In trans-hydrocinnamate, the double bond at C-2 in trans-cinnamate is replaced by a single bond. Hydrocinnamate does not stimulate the proton conductance of UCP1 even in the presence of fatty acids (Figure 4), indicating that this double bond is important for activation of UCP1, in support of previous conclusions [3].
Tomás et al. [25] did not observe activation of UCP1 activity by added trans-cinnamate in yeast mitochondria. Our observations in Figure 3 replicate and explain this result, since the experiments in [25] were carried out in the absence of added fatty acids. We show in Figure 3 that palmitate greatly enhances the activation of UCP1 by cinnamate. In contrast with trans-cinnamate, all-trans-retinoate, which also contains the 2-alkenal group, is able to activate the proton conductance of UCP1 in both yeast and brown adipose tissue mitochondria, independently of added fatty acids [13,25]. Retinoate is a stronger activator than palmitate, and activation by retinoate and palmitate is neither additive nor synergistic [13]. We propose that this is because retinoate and other fatty acid-independent activators (such as the retinoid, TTNPB; see [13]) interact with UCP1 at two separate sites, acting as both fatty acids and reactive alkenals, explaining the lack of synergy with other activators that is seen with these bifunctional molecules (see also [12]). Cinnamate, despite its carboxy group, appears to be unable to interact with UCP1 as a fatty acid, and 4-HNE and other alkenals lack the required carboxy group, but both cinnamate and 4-HNE can react with UCP1 as reactive alkenals and activate UCP1 synergistically with competent fatty acids.
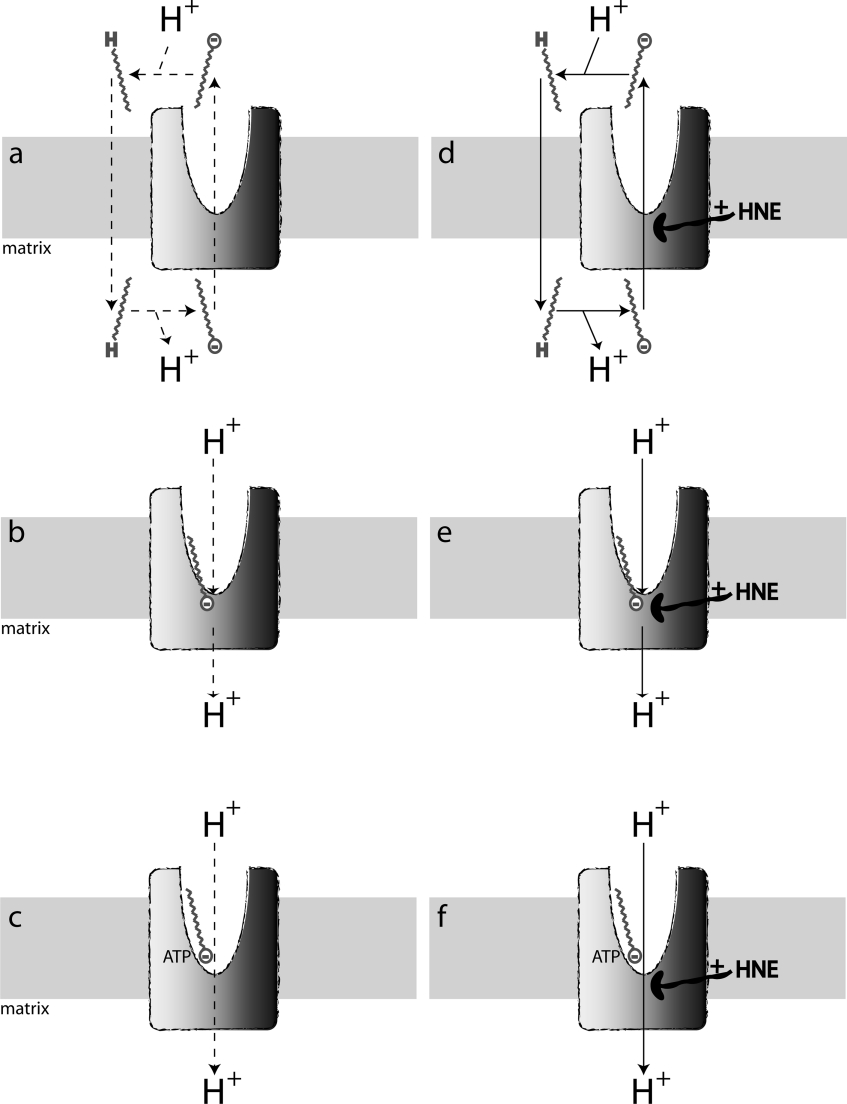
The proton conductance of UCP1 is greatly enhanced by fatty acids, but whether they are essential for proton transport by UCP1 is controversial. Fatty acids may be essential for the mechanism of proton conductance by UCP1 if this requires transmembrane fatty acid cycling [26,27] (Figure 6a) or if fatty acids complete a proton-binding pathway through UCP1 [28,29] (Figure 6b). Alternatively, fatty acids may not be required for the proton translocation mechanism, but may activate proton transport indirectly, by overcoming inhibition by purine nucleotides [30,31] (Figure 6c). To investigate proton transport by UCP1 while minimizing contaminating fatty acids, UCP1 was reconstituted into liposomes [8,28]. In this system, addition of fatty acids was required for protein-mediated proton transport activity, supporting the conclusion that fatty acids are obligatory for UCP1 function [8,28]. In parallel, other authors have tried to eliminate the influence of fatty acids on the proton conductance activity of UCP1 in isolated mitochondria, either by attempting to eradicate fatty acids from mitochondrial preparations [23,32] or by mathematically accounting for the influence of endogenous fatty acids on UCP1 activity [31]. Results from these studies suggest that UCP1 has native proton transport activity that does not require endogenous or added fatty acids.
Figure 6. Dual site models for activation of proton conductance through UCP1.
(a–c) Reactive alkenals absent; (d–f) reactive alkenals such as 4-HNE present. (a) UCP1 catalyses fatty acid anion export; then, fatty acids protonate, flip back to the matrix and deprotonate, causing net proton influx [26,27]. (b) UCP1 catalyses proton influx; fatty acids provide the carboxy group required to complete the proton translocation pathway [28,29]. (c) UCP1 catalyses proton influx; fatty acids alter binding of nucleotides so that they fail to inhibit proton transport [30,31]. (d–f) Same as (a–c) but 4-HNE alters the protein at a separate site from the fatty acids, perhaps by covalent reaction and/or conformational changes, to enhance the pathway of anion or proton translocation. The dashed lines in (a–c) indicate that it is unclear whether or not reactive alkenals are essential for the net proton translocation activity of UCP1 (see the text).
The results presented in this paper suggest a dual-site model of the regulation of UCP1 activity in which fatty acids and reactive alkenals interact synergistically but at separate sites (Figures 6d–6f). In this model, alkenals stimulate the proton conductance pathway, perhaps by altering the conformation of the protein. Synergy with fatty acids would arise either because fatty acids are needed for the UCP1 proton transport mechanism (Figures 6d and 6e) or because they are needed to overcome the inhibition by purine nucleotides (Figure 6f).
It is unclear whether activation by reactive alkenals is strictly required for the mechanism of UCP1 proton transport, or whether alkenals simply enhance activity. This is because it is very hard to assess the extent of endogenous UCP1 activation by these reactive alkenals in natural membranes. In such membranes, polyunsaturated fatty acyl chains are easily oxidized, releasing alkenals. Figure 1(b) confirms the extensive literature on brown adipose tissue mitochondria, showing that palmitate is able to activate UCP1 strongly in the absence of added alkenals. However, some lines of evidence suggest that both fatty acids and reactive alkenals are each strictly required for UCP1 activation in vitro. Echtay et al. [8] only observed proton uptake in liposomes containing UCP1 in the presence of both added fatty acids and ubiquinone. Since ubiquinone is not required in yeast containing UCP1, it may have been working indirectly in the liposomes by causing phospholipid oxidation and formation of (essential) reactive alkenals in this otherwise alkenal-free preparation [11].
In mitochondria, the activation of other UCPs by reactive alkenals and fatty acids appears to differ from the activation of UCP1 by these compounds. In brown adipose tissue and yeast mitochondria, UCP1 can be activated by fatty acids in the absence of added alkenals, which merely enhance the activation. In contrast, we have found that UCP2, UCP3, avian UCP and plant UCP in isolated mitochondria do not transport protons unless alkenals are added (or generated by addition of superoxide or other initiators of lipid peroxidation; see [12,33]), whereas fatty acids do not activate and have no effect on the activation by alkenals [3]. There are two possible explanations for these differences between UCP1 and the other UCPs. The affinity of UCP1 for fatty acids, nucleotides and alkenals may be different from the affinities of the other UCPs for these effectors. This might be because UCP1 has been selected for fatty-acid-mediated activation for adaptive thermogenesis, while the other UCPs have been selected for alkenal-mediated activation to protect against superoxide generation. Alternatively, the differences may only be apparent, and may reflect different levels of endogenous contaminating fatty acids, purine nucleotides and alkenals in different preparations. This second possibility is supported by observations in liposomes that UCP2 and UCP3 have the same absolute requirement for fatty acids and ubiquinone as UCP1 [8,9], and by differences between laboratories in the requirement for adding each of these effectors to achieve particular activation states of the different UCPs. For example, the requirement for ubiquinone in liposomes is not reported by all laboratories [26]; UCP1 in isolated mitochondria in the absence of effectors may be fully active [23,31] or significantly less than maximally activated (Figure 1), and alkenal activation of UCP1 sometimes requires added fatty acids (Figure 1) but sometimes does not [3]. This variability of response to effectors means that it is not currently possible to be sure whether or not UCP1 and the other UCPs are intrinsically different in their responses to fatty acids, nucleotides and alkenals.
What is the physiological relevance of alkenal activation of UCP1? We can speculate as follows, based on current models of UCP1 activation [6] and the results presented in this paper. In the resting brown adipocyte, endogenous concentrations of nucleotides (mainly ATP) keep UCP1 fully inhibited. Under acute adrenergic stimulation, the concentration of fatty acids rises and fatty acids overcome the inhibitory effect of nucleotides, either by providing the missing cofactor for UCP1 activity (Figures 6a and 6b) or by competing off the functional inhibition by ATP (Figure 6c), or both. In this situation, however, proton conductance is still lower than its full capacity. Fatty acid β-oxidation then provides the substrate for rapid electron transport, but this also generates superoxide [34] that oxidizes membrane phospholipids and produces a cascade of 4-HNE production, synergistically augmenting the original fatty acid activation of UCP1. Proton conductance through UCP1 only achieves its maximum when both fatty acids and reactive alkenals are present. When adrenergic stimulation is removed, fatty acid levels decrease quickly, inactivating UCP1. However, the alkenal activation might be more persistent, allowing a quicker and stronger response to subsequent adrenergic stimulation until the slower up-regulation of UCP1 gene expression leads to chronically increased UCP1 levels in cold adaptation. There is no information on the kinetics of the reversal of alkenal activation to allow evaluation of this hypothesis. A recent study [35] suggested that endogenously generated matrix superoxide does not regulate UCP1 activity and energy expenditure in vivo, since the activity of UCP1 was not altered in brown adipose tissue mitochondria from SOD2 (superoxide dismutase 2) transgenic mice, an animal model with increased SOD activity and lowered superoxide levels in the mitochondrial matrix. However, it may be that the endogenous initiator of UCP1 activation is hydroperoxide (protonated superoxide) within the inner membrane, where it will be unaffected by SOD in the matrix, so these experiments are not definitive.
In conclusion, our results show that alkenals and fatty acids activate the proton conductance of UCP1 synergistically in yeast and brown adipose tissue mitochondria, suggesting that activation of UCP1 is more subtle than previous models have suggested.
Acknowledgments
We thank Catherine F. Clarke (Department of Chemistry and Biochemistry, University of California, Los Angeles, CA, U.S.A.) for the yeast strain used in this study, and Julie Buckingham and Helen Boysen for technical assistance. This work was funded by the Medical Research Council (U.K.) (M.D.B. and N.P.), the Portuguese Government (Fundação para a Ciência ea Tecnologia) and the European Social Fund (T.C.E.).
References
- 1.Halliwell B., Gutteridge J. M. C. Oxford, U.K.: Oxford University Press; 1999. Free Radicals in Biology and Medicine. [Google Scholar]
- 2.Allen R. G., Tresini M. Oxidative stress and gene regulation. Free Radical Biol. Med. 2000;28:463–499. doi: 10.1016/s0891-5849(99)00242-7. [DOI] [PubMed] [Google Scholar]
- 3.Echtay K. S., Esteves T. C., Pakay J. L., Jekabsons M. B., Lambert A. J., Portero-Otin M., Pamplona R., Vidal-Puig A., Wang S., Roebuck S. J., et al. A signalling role for 4-hydroxy-2-nonenal in regulation of mitochondrial uncoupling. EMBO J. 2003;22:4103–4110. doi: 10.1093/emboj/cdg412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skulachev V. P. Uncoupling: new approaches to an old problem of bioenergetics. Biochim. Biophys. Acta. 1998;1363:100–124. doi: 10.1016/s0005-2728(97)00091-1. [DOI] [PubMed] [Google Scholar]
- 5.Brand M. D., Esteves T. C. Physiological functions of the mitochondrial uncoupling proteins UCP2 and UCP3. Cell Metab. 2005;2:85–93. doi: 10.1016/j.cmet.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 6.Cannon B., Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol. Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 7.Echtay K. S., Roussel D., St-Pierre J., Jekabsons M. B., Cadenas S., Stuart J. A., Harper J. A., Roebuck S. J., Morrison A., Pickering S., et al. Superoxide activates mitochondrial uncoupling proteins. Nature (London) 2002;415:96–99. doi: 10.1038/415096a. [DOI] [PubMed] [Google Scholar]
- 8.Echtay K. S., Winkler E., Klingenberg M. Coenzyme Q is an obligatory cofactor for uncoupling protein function. Nature (London) 2000;408:609–613. doi: 10.1038/35046114. [DOI] [PubMed] [Google Scholar]
- 9.Echtay K. S., Winkler E., Frischmuth K., Klingenberg M. Uncoupling proteins 2 and 3 are highly active H+ transporters and highly nucleotide sensitive when activated by coenzyme Q (ubiquinone) Proc. Natl. Acad. Sci. U.S.A. 2001;98:1416–1421. doi: 10.1073/pnas.98.4.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murphy M. P., Echtay K. S., Blaikie F. H., Asin-Cayuela J., Cocheme H. M., Green K., Buckingham J. A., Taylor E. R., Hurrell F., Hughes G., et al. Superoxide activates uncoupling proteins by generating carbon-centered radicals and initiating lipid peroxidation. J. Biol. Chem. 2003;278:48534–48545. doi: 10.1074/jbc.M308529200. [DOI] [PubMed] [Google Scholar]
- 11.Esteves T. C., Echtay K. S., Jonassen T., Clarke C. F., Brand M. D. Ubiquinone is not required for proton conductance by uncoupling protein 1 in yeast mitochondria. Biochem. J. 2004;379:309–315. doi: 10.1042/BJ20031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esteves T. C., Brand M. D. The reactions catalysed by the mitochondrial uncoupling proteins UCP2 and UCP3. Biochim. Biophys. Acta. 2005;1709:35–44. doi: 10.1016/j.bbabio.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 13.Rial E., González-Barroso M., Fleury C., Iturrizaga S., Sanchis D., Jimenez-Jimenez J., Ricquier D., Goubern M., Bouillaud F. Retinoids activate proton transport by the uncoupling proteins UCP1 and UCP2. EMBO J. 1999;18:5827–5833. doi: 10.1093/emboj/18.21.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krauss S., Zhang C. Y., Lowell B. B. A significant portion of mitochondrial proton leak in intact thymocytes depends on expression of UCP2. Proc. Natl. Acad. Sci. U.S.A. 2002;99:118–122. doi: 10.1073/pnas.012410699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Proft M., Kotter P., Hedges D., Bojunga N., Entian K. D. CAT5, a new gene necessary for derepression of gluconeogenic enzymes in Saccharomyces cerevisiae. EMBO J. 1995;14:6116–6126. doi: 10.1002/j.1460-2075.1995.tb00302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harper A. J. Cambridge, U.K.: University of Cambridge; 2002. Expression and characterization of the uncoupling proteins in yeast; p. 76. Ph.D. thesis. [Google Scholar]
- 17.Stuart J. A., Harper J. A., Brindle K. M., Jekabsons M. B., Brand M. D. A mitochondrial uncoupling artifact can be caused by expression of uncoupling protein 1 in yeast. Biochem. J. 2001;356:779–789. doi: 10.1042/0264-6021:3560779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brand M. D. Measurement of mitochondrial proton motive force. In: Brown G. C., Cooper C. E., editors. Bioenergetics – A Practical Approach. Oxford, U.K.: IRL Press; 1995. pp. 39–62. [Google Scholar]
- 19.Reynafarje B., Costa L. E., Lehninger A. L. O2 solubility in aqueous media determined by a kinetic method. Anal. Biochem. 1985;145:406–418. doi: 10.1016/0003-2697(85)90381-1. [DOI] [PubMed] [Google Scholar]
- 20.Cornwall E. J. Cambridge, U.K.: University of Cambridge; 2004. The role of the adenine nucleotide translocase in mitochondrial proton leak; pp. 100–104. Ph.D. thesis. [Google Scholar]
- 21.Richieri G. V., Ogata R. T., Kleinfeld A. M. A fluorescently labeled intestinal fatty acid binding protein. Interactions with fatty acids and its use in monitoring free fatty acids. J. Biol. Chem. 1992;267:23495–23501. [PubMed] [Google Scholar]
- 22.Rolfe D. F. S., Hulbert A. J., Brand M. D. Characteristics of mitochondrial proton leak and control of oxidative phosphorylation in the major oxygen-consuming tissues of the rat. Biochim. Biophys. Acta. 1994;1188:405–416. doi: 10.1016/0005-2728(94)90062-0. [DOI] [PubMed] [Google Scholar]
- 23.González-Barroso M. M., Fleury C., Bouillaud F., Nicholls D. G., Rial E. The uncoupling protein UCP1 does not increase the proton conductance of the inner mitochondrial membrane by functioning as a fatty acid anion transporter. J. Biol. Chem. 1998;273:15528–15532. doi: 10.1074/jbc.273.25.15528. [DOI] [PubMed] [Google Scholar]
- 24.Arechaga I., Raimbault S., Prieto S., Levi-Meyrueis C., Zaragoza P., Miroux B., Ricquier D., Bouillaud F., Rial E. Cysteine residues are not essential for uncoupling protein function. Biochem. J. 1993;296:693–700. doi: 10.1042/bj2960693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomás P., Jimenez-Jimenez J., Zaragoza P., Vuligonda V., Chandraratna R. A., Rial E. Activation by retinoids of the uncoupling protein UCP1. Biochim. Biophys. Acta. 2004;1658:157–164. doi: 10.1016/j.bbabio.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 26.Garlid K. D., Orosz D. E., Modriansky M., Vassanelli S., Jezek P. On the mechanism of fatty acid-induced proton transport by mitochondrial uncoupling protein. J. Biol. Chem. 1996;271:2615–2620. doi: 10.1074/jbc.271.5.2615. [DOI] [PubMed] [Google Scholar]
- 27.Skulachev V. P. Anion carriers in fatty acid-mediated physiological uncoupling. J. Bioenerg. Biomembr. 1999;31:431–445. doi: 10.1023/a:1005492205984. [DOI] [PubMed] [Google Scholar]
- 28.Winkler E., Klingenberg M. Effect of fatty acids on H+ transport activity of the reconstituted uncoupling protein. J. Biol. Chem. 1994;269:2508–2515. [PubMed] [Google Scholar]
- 29.Klingenberg M., Echtay K. S. Uncoupling proteins: the issues from a biochemist point of view. Biochim. Biophys. Acta. 2001;1504:128–143. doi: 10.1016/s0005-2728(00)00242-5. [DOI] [PubMed] [Google Scholar]
- 30.Huang S. G. Binding of fatty acids to the uncoupling protein from brown adipose tissue mitochondria. Arch. Biochem. Biophys. 2003;412:142–146. doi: 10.1016/s0003-9861(03)00031-6. [DOI] [PubMed] [Google Scholar]
- 31.Shabalina I. G., Jacobsson A., Cannon B., Nedergaard J. Native UCP1 displays simple competitive kinetics between the regulators purine nucleotides and fatty acids. J. Biol. Chem. 2004;279:38236–38248. doi: 10.1074/jbc.M402375200. [DOI] [PubMed] [Google Scholar]
- 32.Rial E., Aguirregoitia E., Jimenez-Jimenez J., Ledesma A. Alkylsulfonates activate the uncoupling protein UCP1: implications for the transport mechanism. Biochim. Biophys. Acta. 2004;1608:122–130. doi: 10.1016/j.bbabio.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 33.Brand M. D., Affourtit C., Esteves T. C., Green K., Lambert A. J., Miwa S., Pakay J. L., Parker N. Mitochondrial superoxide: production, biological effects, and activation of uncoupling proteins. Free Radical Biol. Med. 2004;37:755–767. doi: 10.1016/j.freeradbiomed.2004.05.034. [DOI] [PubMed] [Google Scholar]
- 34.St-Pierre J., Buckingham J. A., Roebuck S. J., Brand M. D. Topology of superoxide production from different sites in the mitochondrial electron transport chain. J. Biol. Chem. 2002;277:44784–44790. doi: 10.1074/jbc.M207217200. [DOI] [PubMed] [Google Scholar]
- 35.Silva J. P., Shabalina I. G., Dufour E., Petrovic N., Backlund E. C., Hultenby K., Wibom R., Nedergaard J., Cannon B., Larsson N. G. SOD2 overexpression: enhanced mitochondrial tolerance but absence of effect on UCP activity. EMBO J. 2005;24:4061–4070. doi: 10.1038/sj.emboj.7600866. [DOI] [PMC free article] [PubMed] [Google Scholar]