Abstract
Thermotoga maritima invertase (β-fructosidase), a member of the glycoside hydrolase family GH-32, readily releases β-D-fructose from sucrose, raffinose and fructan polymers such as inulin. These carbohydrates represent major carbon and energy sources for prokaryotes and eukaryotes. The invertase cleaves β-fructopyranosidic linkages by a double-displacement mechanism, which involves a nucleophilic aspartate and a catalytic glutamic acid acting as a general acid/base. The three-dimensional structure of invertase shows a bimodular enzyme with a five bladed β-propeller catalytic domain linked to a β-sandwich of unknown function. In the present study we report the crystal structure of the inactivated invertase in interaction with the natural substrate molecule α-D-galactopyranosyl-(1,6)-α-D-glucopyranosyl-β-D-fructofuranoside (raffinose) at 1.87 Å (1 Å=0.1 nm) resolution. The structural analysis of the complex reveals the presence of three binding-subsites, which explains why T. maritima invertase exhibits a higher affinity for raffinose than sucrose, but a lower catalytic efficiency with raffinose as substrate than with sucrose.
Keywords: crystal structure, exo-glycosidase, β-fructosidase, glycosidase GH32 family, invertase, retaining mechanism
Abbreviations: Aa, Aspergillus awamori; NCS, non-crystallographic symmetry; R.M.S.D., root mean square deviation; TBAC, Terrific Broth with ampicillin and chloramphenicol; Tm, Thermotoga maritima
INTRODUCTION
Many bacteria and fungi, and approx. 15% of flowering plants synthesize fructans, which are fructose oligosaccharides and polysaccharides synthesized from sucrose, the end product of photosynthesis [1]. The breakdown products of these storage oligosaccharides and polysaccharides, fructose and glucose, are important components of plant signalling pathways and function as a key sensors of the nutritional status of plants. Thus the enzymes that cleave sucrose and fructans play a pivotal role in controlling plant cell differentiation and development [2,3]. These enzymes, called invertases or β-fructosidases, are found in family 32 (GH32) of the classified glycoside hydrolases (http://afmb.cnrs-mrs.fr/CAZY) [4].
Over 50 years ago Koshland and Stein [5] showed that yeast invertase-mediated hydrolysis of sucrose proceeds with overall retention of the β configuration of fructose. Subsequently, the catalytic residues of yeast invertase have been identified as aspartic acid, D23, acting as the nucleophile [6], and glutamic acid, E204, which serves as the general catalytic acid/base [7] in the double-displacement mechanism. Recently, the crystal structures of three GH32 enzymes, Thermotoga maritima (Tm) invertase [8], exo-inulinase from Aspergillus awamori [9] and a plant fructan 1-exohydrolase from Cichorium intybus [10] have all been reported. The three-dimensional structures of these three enzymes revealed a similar bimodular arrangement with a catalytic-domain folding in a rather unusual five-bladed β-propeller linked to a C-terminal β-sandwich domain of unknown function. So far, the fivebladed β-propeller fold has only been reported for tachylectin [11] and for two other glycoside hydrolase families, GH43 [12] and GH68 [13]. This structural resemblance has reinforced the establishment of an evolutionary relationship between retaining (GH32 and GH68) and inverting (GH43) enzymes [8,8a].
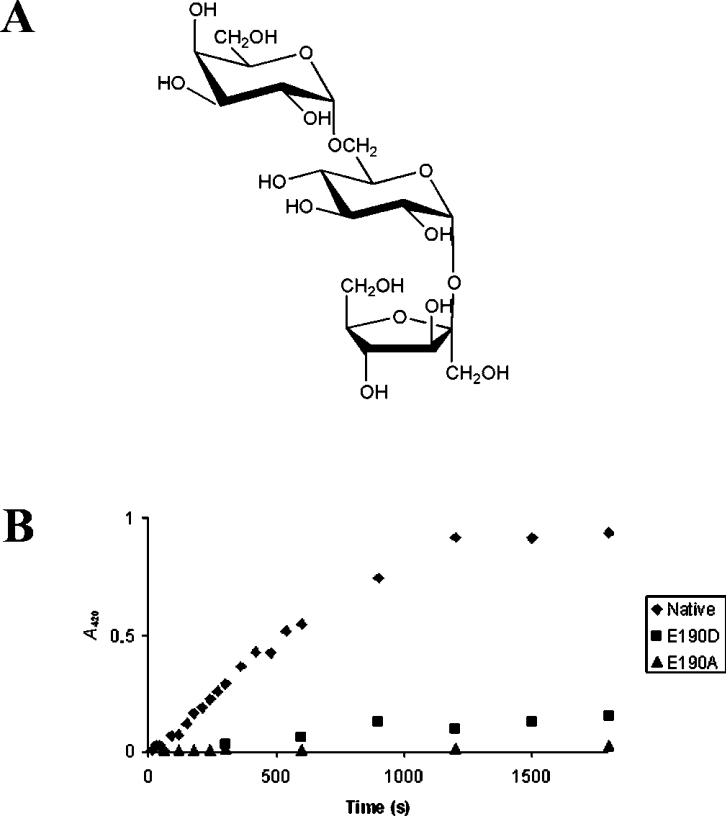
The exo-inulinase from A. awamori (Aa exo-inulase) was crystallized in the presence of fructose and thus permited the identification of residues that constitute the fructose-binding pocket of the active site [9]. To complete the structural analysis of the active sites of GH32 family enzymes, we needed to obtain and characterize an enzyme–substrate complex that incorporated and extended beyond the cleavage site. For this purpose, the enzyme was inactivated by site directed mutagenesis[14] and was co-crystallized or saturated with the substrate. Here we describe the crystal structure of a Tm invertase mutant E190D (inv-E190D) in complex with the trisaccharide raffinose [α-D-galactopyranosyl-(1,6)-α-D-glucopyranosyl-β-D-fructofuranoside; Figure 1A]. This substrate molecule is readily cleaved by the Tm invertase that can also use sucrose and fructofuranose polymers such as inulin as substrates [15]. In addition we discuss the structure of the enzyme–substrate complex in view of the enzyme's specificity and we describe the detailed structure of the three binding-subsites that comprise the active-site pocket of Tm invertase.
Figure 1. (A) Schematic representation of the substrate molecule α-D-galactopyranosyl-(1,6)-α-D-glucopyranosyl-β-D-fructofuranoside (raffinose). (B) Comparison of the activity between native invertase (◆) with mutants’ inv-E190D (■) and inv-E190A (▲).
Glucose release after sucrose hydrolysis was assayed by the colorimetric reaction of the reducing ends with ferricyanide. The activity was monitored by the decrease in A420 as a function of time (scale in seconds).
MATERIAL AND METHODS
Mutagenesis, protein expression and purification of the inactivated mutant inv-E190A
The plasmid pINV that was obtained by cloning the native invertase gene (bfrA) into the N-terminal His-tag encoding expression plasmid pDET17 [8] (Invitrogen) was subsequently used as a template for mutagenesis using the QuickChange Mutagenesis Kit (Strategene). Amplifications were carried out using the primers E190AF (5′ CCACAAAAGAAATAGCGTGTCCCGATCTTGTGAG 3′) and E190AR (5′ CTCACAAGATCGGGACACGCTATTTCTTTTGTGG 3′) to mutate Glu190 into alanine (substitutions are underlined), and the primers E190DF (5′ CCACAAAAGAAATAGATTGTCCCGATCTTGTGAG 3′) and E190DR (5′ CTCACAAGATCGGGACAATCTATTTCTTTTGTGG 3′) to mutate Glu190 into aspartate. Protein expression and purification were carried out essentially as described previously for the native invertase [8]. Briefly, a single colony of Escherichia coli, BL21pLysS, containing the pINV-E190A plasmid was used to inoculate TBAC (Terrific Broth supplemented with 100 μg/ml ampicillin and 34 μg/ml chloramphenicol). The culture was incubated at 37 °C and was then used to inoculate 3 litres of TBAC. Induction was performed by adding 0.5 mM isopropyl-1-thio-β-D-galactopyranose when D600 reached 0.8 nm. After incubation at 37 °C for another 4 h the cultures were pelleted and then submitted to the lysis procedure. After lysis the supernatant containing the soluble proteins was separated from the pellet by centrifugation (20000 g for 30 min at 4 °C) and then filtered before two-step purification by nickel affinity chromatography and gel filtration on a Sephadex column (Amersham Biosciences). The fractions containing the protein were pooled and concentrated to 11 mg/ml on ultrafiltration styrene acrylonitrile membranes (Millipore).
Activity assays
Invertase activity was determined following the method of Kidby and Davidson [16] by measuring the release of reducing sugars by the ferricyanide method. Invertase was incubated at 75 °C in 100 mM sodium acetate buffer (pH 5.5) and 120 mM sucrose [15]. Aliquots (100 μl) of the sample were taken after different incubation times. The enzymatic reaction was revealed by mixing samples with 1 ml of reaction buffer [1 mM K3Fe(CN)6, 130 mM sodium acetate and 5 mM NaOH] and by heating the samples for 7 min at 95 °C. The activity was monitored by the decrease in A420 as a function of time.
Protein crystallization and X-ray diffraction data collection
The crystallization conditions for inv-E190D and inv-E190A were similar to those of the native protein [8]. Crystals of suitable size for X-ray diffraction experiments were grown in 15–17% PEG [poly(ethylene glycol) 1000], 150 mM Li2SO4 and 100 mM sodium citrate at pH 4.2, by the vapour-diffusion method using hanging drops. Co-crystallization experiments were set up under the same crystallization conditions except that the protein samples (100 μl of 11 mg/ml of protein) were mixed with 5 mM substrate solutions. All crystals that were tested failed to display interpretable electron density. Crystals of inv-E190D and inv-E190A were then soaked for 2 min in mother liquor saturated with D-(+)-raffinose (Sigma–Aldrich) and supplemented with 15% glycerol and then immediately flash frozen in a cryogenic nitrogen stream at 100 K. Trials with other substrate molecules, such as sucrose, kestose or nystose led to diffracting crystals but at lower resolution and none displayed interpretable electron density in the active-site pocket (results not shown).
Data collection was carried out at the ESRF [(European Synchrotron Radiation Facility), Grenoble, France] on beamline ID14-EH2. The only successful data set that gave a substrate–enzyme complex structure was collected from an inv-E190D crystal that had been soaked in raffinose as indicated above. The data collection statistics are reported in Table 1. The crystals of inv-E190D bound with raffinose belonged to space group P21 with unit cell parameters a=94.5 Å (1 Å=0.1 nm), b=114.7 Å, c=130.0 Å and β=98.90°. The asymmetric unit contains six monomers giving a VM value of 2.2 Å·Da−1 and a 43% solvent content [17].
Table 1. Summary of data collection and refinement statistics.
| Data collection | invE190–raffinose complex |
|---|---|
| X-ray source | ESRF (ID14-EH2) |
| Wavelength (Å) | 0.934 |
| Resolution range (Å) | 32.1–1.87 (1.94–1.87) |
| Space group | P 21 |
| Rsym | 0.040 (0.18) |
| Completeness (%) | 97.7 (91.7) |
| I/σI | 16.2 (5.5) |
| Number of unique reflections | 220454 (20270) |
| Refinement statistics | |
| Resolution range (Å) | 32.1–1.87 (1.94–1.87) |
| Rcryst (%) | 19.8 (21.7) |
| Rfree (%) | 22.9 (26.1) |
| Overall B-factor (Å2) | 25.5 |
| R.M.S.D. in bond lengths (Å) | 0.006 |
| R.M.S.D. in bond angles (°) | 1.103 |
| Ramachandran plot | |
| Most favoured regions (%) | 88.3 |
| Additionally allowed regions (%) | 10.7 |
Structural refinement
The structural model of inv-E190D was refined with REFMAC5 [18], using NCS (non-crystallographic symmetry) restraints and starting with the co-ordinates of the native invertase structure. The refinement cycles were alternated with intermittent manual rebuilding with TURBO-FRODO [19]. Individual B-factors were refined after the application of a TLS (translation, libration and screw-rotation) correction [20]. Water molecules were added by ARP/wARP (automated refinement procedure/weighted ARP) [21]. The final model is composed of six copies of the protein residues 2–432 (2592 amino acids), four SO42− ions, one buffer molecule (sodium citrate), six raffinose molecules and a total of 1890 water molecules, which led to R and Rfree [22] values of 19.8 and 22.9% respectively. Ramachandran statistics (PROCHECK [23]) indicated that for the overall structure of the six molecules present in the asymmetric unit, 88.3% of the atoms are in the most favoured region and 10.7% are in additionally allowed regions. Details of refinement statistics are summarized in Table 1.
PDB accession numbers
The co-ordinates and structural factors have been deposited in the PDB under accession numbers 1W2T and r1W2Tsf for the inv-E190D mutant protein in complex with raffinose.
RESULTS AND DISCUSSION
Quality of the model and overall structure of the inactivated invertase
The structural analysis of glycoside hydrolases in complex with substrate molecules requires their inactivation before crystallization. For this purpose, two active site mutants of Tm invertase, inv-E190D and inv-E190A, were produced and tested for their activity towards sucrose (Figure 1B). Inv-E190D retains 14% of native invertase activity, whereas no activity was detectable for inv-E190A. Both Tm mutants crystallized with the same space group, P21, as the native protein, and in each case the asymmetric unit contained six independent invertase molecules. Both inv-E190D and inv-E190A mutants were used for soaking and co-crystallization experiments but only inv-E190D, in complex with raffinose, gave suitable high-resolution crystallographic data and is described below. Apparently, the glutamic acid to alanine mutation affects the binding properties of the enzyme, whereas in the inv-E190D mutant replacement by an aspartate residue helps to maintain the electronegative microenvironment, and binding to the substrate molecules is less affected.
The inv-E190D–raffinose structure was refined to a final R factor of 19.8%, and an Rfree factor of 22.9%, applying medium NCS restraints to the six independent molecules of the asymmetric unit. The refined model for inv-E190D in complex with raffinose (PDB entry 1W2T) is composed of 431 amino acid residues for each independent molecule, a total of 1891 water molecules, four SO42−, a buffer (sodium citrate) molecule and one molecule of raffinose in each active site. The raffinose molecules are clearly defined by the electron density in all active sites. Overall the crystal structure of the mutant enzyme–substrate complex is essentially identical to that of the wild-type structure [8] and to the uncomplexed mutant (results not shown).
No significant conformational change is observed in the presence of the substrate molecules. The R.M.S.D. (root mean square deviation) between the co-ordinates of the wild-type structure and the complex is 0.195 Å for 431 Cα atoms. All other data quality and refinement parameters are listed in Table 1.
Structural comparison of Tm invertase with Ci fructan exohydrolase and Aa exo-inulinase
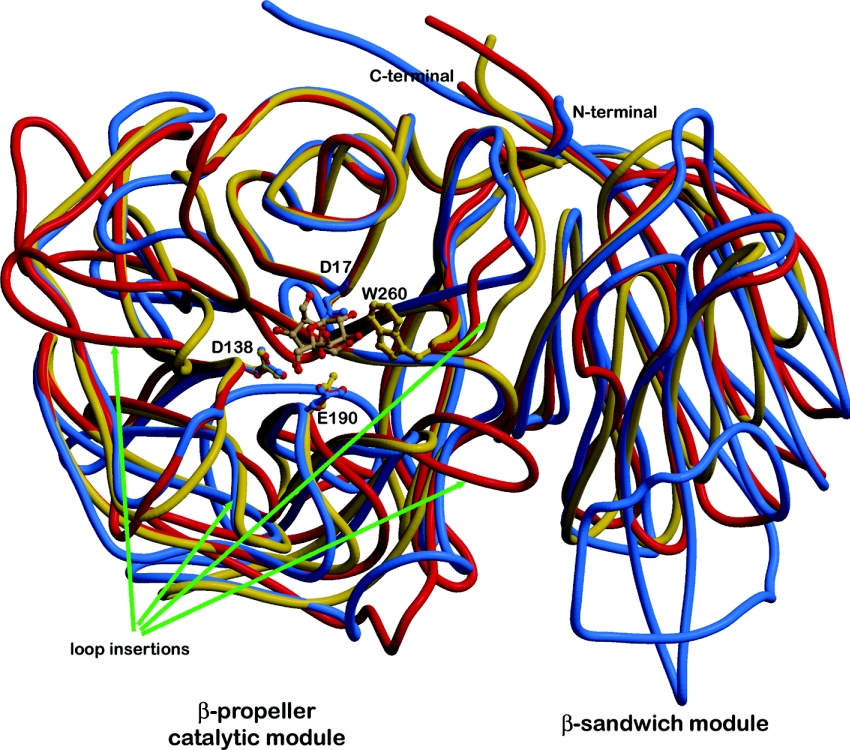
The superposition of the two complexed structures (inv-E190D with raffinose; PDB ID, 1W2T and Aa exo-inulinase with fructose; PDB ID, 1Y9G) leads to a R.M.S.D. of 1.46 Å for 403 Cα atoms. This is in the same order of magnitude as that reported for the two native structures, which is 1.6 Å for 392 Cα atoms [9]. As expected, the most divergent regions contain substantial insertions and deletions in the loops surrounding the active-site cleft (Figure 2). In particular, Tm invertase lacks a loop of 11 residues, situated after Glu101 (Asp147 in Aa exo-inulase). Together with a second loop insertion from Asn265 to Gly274 in Aa exo-inulinase that corresponds to the loop from Thr208 to Asn 211 in Tm invertase (for sequence alignment see Nagem et al. [9] and Alberto et al. [8]), located on the opposite side of the active-site cleft, these loops are most likely to define substrate specificity. In Tm invertase both of these loops contain residues involved in substrate binding (as described below). Tm invertase contains only one loop-insertion at the surface close to the active-site cleft, namely from residues Gly155 to Gly161 (Ala208 to His211 in Aa exo-inulinase), but none of these residues bind to the substrate molecule trapped in the active site of Tm invertase.
Figure 2. Ribbon representation of the superimposition of the three three-dimensional structures of GH32 family enzymes.
Tm invertase is coloured in red, Ci fructan 1-exohydrolase in blue and Aa inolinase in yellow. The arrows highlight specific loop insertions that most probably are responsible for modifying the substrate specificities of the three enzymes. Figures 2, 3a and 4 were produced with Molscript [30,31] and rendered with Raster3D.
Three binding subsites in the active-site pocket of Tm invertase
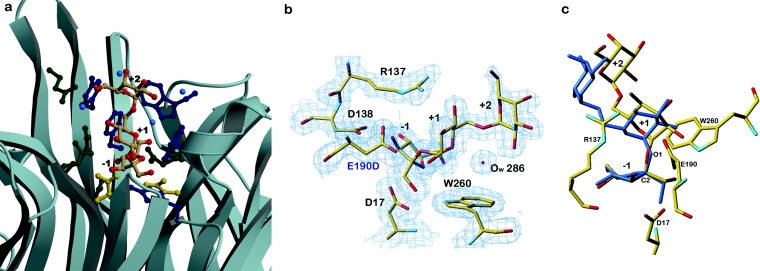
Tm invertase is a typical exo-enzyme [24] and displays a 30 Å deep and 18 Å wide active-site pocket. The identity of the two residues that form the catalytic machinery, Asp17 and Glu190 in the case of Tm invertase, is now well established [6,7,9] and the putative roles for the different conserved stretches [25] in the sequences of GH32 family enzymes have recently been described [8,9]. The present structural analysis of the inactivated invertase–raffinose complex allows a much improved view of the substrate-binding site, in particular across the cleavage site. A single raffinose molecule is bound between the blades of each β-propeller module, defining three substrate binding subsites −1, +1 and +2 (Figure 3; subsite nomenclature adapted from Davies et al. [26] with cleavage taking place between −1 and +1, and the β-fructofuranosyl unit undergoing catalysis placed at subsite −1). For all six independent molecules within the asymmetric unit, the active sites and binding patterns are essentially the same and only one is described below.
Figure 3. Localization and structural arrangement of raffinose bound in the active-site pocket in Tm invertase.
(a) Ribbon representation of the active-site pocket in Tm invertase. The raffinose molecule trapped in the active site is shown in coloured atoms as a stick-representation. (Oxygen atoms are coloured in red). Catalytic residues are shown in gold, aromatic residues in dark blue and other residues in green. Blue balls are water molecules. The substrate-binding subsites are annotated from −1 to +2. (b) Electron density calculated after the final refinement step for the Tm invertase inv-E190D in complex. The Fourier map (2Fo–Fc) is contoured at 1σ showing the trapped substrate molecule present in the active site of inv-E190D. (c) Stick representation of raffinose in complex with Tm inv-E190D (standard atom type colours) superimposed on to the crystal structure of raffinose pentahydrate (in blue). Figures 3(b) and 3(c) were prepared with TURBO-FRODO [19].
The overall structure of the bound raffinose molecule is highly similar to that of raffinose in the crystalline state [27,28]. The only difference (Figure 3c) is a change of the torsion angle around the glycosidic linkage that undergoes catalysis (C2′–O1 in Figure 3c) by approx. 6°. This change is certainly induced by the replacement of the hydrogen-bonding pattern in the raffinose pentahydrate crystal structure by hydrogen bonds formed with the enzyme. At this stage the importance of this torsion for the catalytic mechanism is unclear. The superimposition of each of the three monosaccharide units of the raffinose crystal structure on the enzyme-bound raffinose molecule did not reveal any notable ring distortion. This is not surprising, since the fructofuranosyl moiety does not require extensive distortion to place the glycosidic linkage in axial orientation for direct in-line nucleophilic attack by the enzyme [29].
The present complex of Tm inv-E190D with raffinose also supports the model that places sucrose in the active site of Tm invertase [8] that we had inferred from the presence of a glycerol molecule and from superimposition of the levansucrase–sucrose complex [13]. The disaccharide substrate modelled for Tm invertase perfectly superimposes on to the β-D-fructofuranosyl-α-D-galactopyranosyl-(1→6) moiety of raffinose, demonstrating that active-site residues such as Arg137, Glu188 or Trp260 (as described below) are responsible for the torsion angle geometry of the trapped substrate molecules.
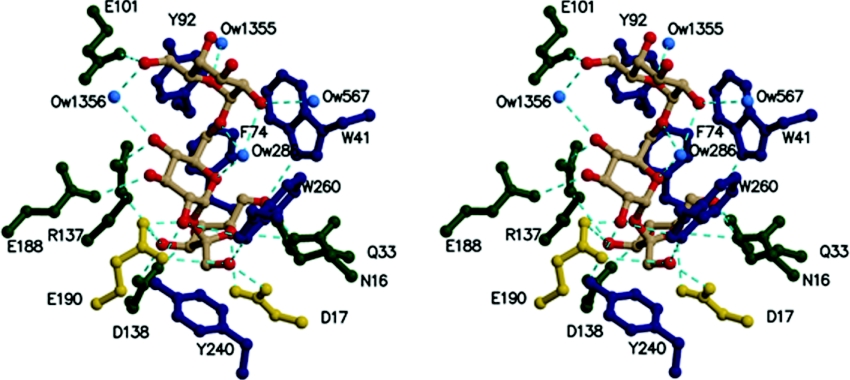
The fructose moiety is located in subsite −1 at the deepest end of the active-site pocket, and is held in place by numerous amino acid residues, which form hydrogen bonds to its hydroxyl groups (Table 2). Of these, Trp260, Trp41, Gln33, Asn16 and Asp138 are clearly responsible for substrate specificity and recognition of the fructose moiety that is to be cleaved: Trp260 forms a hydrogen bond to O1′ of the fructose moiety, Trp41, Gln33 and Asn16 are within hydrogen bond distance from O5′, and Asp138 forms two bridging hydrogen bonds with O3′ and O4′ respectively (Figure 4). The same hydrogen bonding pattern has also recently been described for fructose bound with the Aa exo-inulinase [9] and most of these residues are conserved throughout GH32 family enzymes.
Table 2. Protein-substrate distances, strong hydrogen bonds are indicated in bold.
| Substrate atom | Protein atom | Distance (Å) | |
|---|---|---|---|
| Fructose | O'1 | Asp17 Oδ1 | 2.77 |
| Trp260 Nϵ1 | 3.07 | ||
| Glu190 Oϵ1 | 3.53 | ||
| O'2 | Asp17 Oδ2 | 3.11 | |
| Asn16 Nδ2 | 3.26 | ||
| O'3 | Arg137 Nϵ | 3.55 | |
| Asp138 Oδ2 | 2.65 | ||
| Glu190 Oϵ2 | 3.05* | ||
| O'4 | Asp138 Oδ1 | 2.78 | |
| O'5 | Gln33 Oϵ1 | 2.74 | |
| Trp41 Nϵ1 | 3.07 | ||
| Asn16 Nδ2 | 3.01 | ||
| Glucose | O1 | Glu190 Oϵ2 | 2.67* |
| Glu190 Oϵ1 | 2.74* | ||
| O2 | Thr208 Oγ1 | 3.39 | |
| Trp260 Nϵ1 | 3.44 | ||
| O3 | Glu188 Oϵ1 | 3.23 | |
| O4 | Arg137 Nη2 | 2.86 | |
| Wat1356 O | 3.38 | ||
| O5 | Wat286 O | 2.89 | |
| O6 | Wat286 O | 2.99 | |
| Galactose | O2 | Wat567 O | 3.28 |
| Wat286 O | 2.87 | ||
| O5 | Wat1355 O | 2.73 | |
| O6 | Wat1356 O | 2.68 | |
| Glu101 Oϵ1 | 3.67 |
*, The distance to Glu190 was inferred from the superposition of the complexed mutant structure on the native structure.
Figure 4. Stereographic view of the catalytic site of invertase in complex with raffinose.
Catalytic residues are shown in gold, aromatic residues in dark blue and other residues in green. Hydrogen bonds are in cyan. Ow designates water molecules.
The central α-linked glucose unit of raffinose in subsite +1 is surrounded by three conserved aromatic residues present throughout the GH32 family, these are Trp65, Phe74 and Trp260. Nevertheless, none of these aromatic residues are involved in typical hydrophobic stacking interactions with the glucose unit. This may be important to allow adaptation to binding of other sugar units, such as fructose units of poly-fructofuranose and inulin for example, which are also substrates cleaved by Tm invertase. Interestingly, the structure of the plant fructan 1-exohydrolase [10] revealed that Trp260 has no equivalent. Furthermore, this residue is part of a two-residue loop-insertion (Figure 2) that is not present in most plant invertases and that obstructs the access to a cleft formed at the interface of the β-propeller catalytic module and the C-terminal β-sandwich module. Verhaest et al. [10] suggested that this cleft may play an important role in polymer substrate-binding. At the level of binding subsite +1, three hydrogen bonds are also formed between the glucose sugar unit and the enzyme residues. Namely, Arg137 is hydrogen bonded to O4, Glu188 to O3 and Thr208 to O2 (distances are listed in Table 2). Interestingly, Arg137 is part of the highly conserved sequence motif RDP and together with the neighbouring Asp138 that binds to the fructose hydroxyl groups certainly plays a crucial role in substrate binding and recognition. It appears to have a different conformational position to the equivalent residue (Arg188) in Aa exoinulinase, however, no conformational change is observed compared with the native uncomplexed Tm invertase structure. Glu188 and Thr208 are not conserved throughout invertase or β-fructosidase enzymes and therefore seem to be important in defining substrate specificity for this particular enzyme.
The terminal α-linked galactose unit of raffinose is located in subsite +2 (Figures 3 and 4), which is situated in the active-site pocket close to the protein's surface. This suggests that for longer-chain substrates, any additional sugar unit would not be bound to the protein, but would dangle out into the solvent. Nevertheless, the enzyme is also capable of cleaving shorter substrates such as sucrose, or longer polysaccharide chains of fructose, such as nystose, kestose or derivative polymers of inulin [15]. Unfortunately, several attempts to crystallize Tm invertase with nystose, kestose or longer fructofuranose chains were unsuccessful. The crystallographic data for inactivated Tm invertase in complex with sucrose (results not shown) did reveal electron density in the active site, but this was too diffuse to reliably construct a bound substrate molecule. This being the case, the diffuse density might be due to either high disorder of the bound molecules or to the fact that the affinity of the crystal structure for sucrose molecules is too low for them to become trapped. Indeed, the Km value reported for Tm invertase is best when raffinose is the substrate (Km=15 mM), followed by inulin (Km=19 mM) whereas the Km with sucrose as substrate is 4-fold greater than that for raffinose with a value of 64 mM [15]. However, when it comes to catalytic efficiency, the situation is reversed with kcat/Km values of 4.1×104, 3.1×104 and 9.3×103 M−1·s−1 for sucrose, inulin and raffinose as substrates respectively. Thus filling the +2 binding subsite with the α-galactose moiety of raffinose perhaps increases the affinity of Tm invertase for the substrate, but probably also reduces the catalytic efficiency of Tm invertase by making the disaccharide product more ‘sticky’.
The hydrogen-bonding pattern around subsite +2 involves only one direct protein–substrate contact between Glu101 Oϵ1 and the galactose O6 hydroxyl group. However, several strong hydrogen bonds are mediated by water molecules: Tyr92 binds via a water molecule (Tyr Oη–Ow 1355; 3.20 Å) to the O5 ring oxygen of the galactose unit and Glu101 Oϵ2 via Ow 1356 (a distance of 2.64 Å) again to the O6 hydroxyl group. Neither Tyr92 or Glu101 are conserved in GH32 family enzymes and are most probably responsible for substrate specificity in this particular enzyme.
Determination of the structures of more enzyme–substrate complexes covering the large diversity of enzymes is still required to depict in detail the fine-tuning of substrate specificities that occur in the GH32 family. The present structural analysis of the inactivated Tm invertase in complex with raffinose certainly represents a first achievement that has helped to identify several loops and specific residues that appear to be important for determining substrate specificity. Subsequent investigation by mutagenesis experiments should shed further light on the structural determinants responsible for substrate specificity within the GH32 enzyme family.
Acknowledgments
The authors thank Dr Wolfgang Liebl (Georg-August-Universität, Göttingen, Germany) for generously providing T. maritima genomic DNA. We also thank the staff of the European Synchrotron Radiation source for provision of beam time and for technical assistance at the beamline ID14-EH2. We thank Dr G. Sulzenbacher, Dr C. Bignon and Dr A. Gruez for helpful discussions. This work was partly funded by grant QLK5-CT-2001-00443 (EDEN) from the European Commission.
References
- 1.Ritsema T., Smeekens S. Fructans: beneficial for plants and humans. Curr. Opin. Plant Biol. 2003;6:223–230. doi: 10.1016/s1369-5266(03)00034-7. [DOI] [PubMed] [Google Scholar]
- 2.Sturm A., Tang G. Q. The sucrose-cleaving enzymes of plants are crucial for development, growth and carbon partitioning. Trends Plant Sci. 1999;4:401–407. doi: 10.1016/s1360-1385(99)01470-3. [DOI] [PubMed] [Google Scholar]
- 3.Sturm A. Invertases. Primary structures, functions, and roles in plant development and sucrose partitioning. Plant Physiol. 1999;121:1–8. doi: 10.1104/pp.121.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 1991;280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koshland D. E., Jr, Stein S. S. Correlation of bond breaking with enzyme specificity; cleavage point of invertase. J. Biol. Chem. 1954;208:139–148. [PubMed] [Google Scholar]
- 6.Reddy V. A., Maley F. Identification of an active-site residue in yeast invertase by affinity labeling and site-directed mutagenesis. J. Biol. Chem. 1990;265:10817–10820. [PubMed] [Google Scholar]
- 7.Reddy A., Maley F. Studies on identifying the catalytic role of Glu-204 in the active site of yeast invertase. J. Biol. Chem. 1996;271:13953–13957. doi: 10.1074/jbc.271.24.13953. [DOI] [PubMed] [Google Scholar]
- 8.Alberto F., Bignon C., Sulzenbacher G., Henrissat B., Czjzek M. The three-dimensional crystal structure of invertase (β-fructosidase) from Thermotoga maritima reveals a bimodular arrangement and an evolutionary relationship between retaining and inverting glycosidases. J. Biol. Chem. 2004;279:18903–18910. doi: 10.1074/jbc.M313911200. [DOI] [PubMed] [Google Scholar]
- 8a.Naumoff D. G. β-Fructosidase superfamily: homology with some α-L-arabinases and β-D-xylosidase. Proteins. 2001;42:66–76. doi: 10.1002/1097-0134(20010101)42:1<66::aid-prot70>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 9.Nagem R. A. P., Rojas A. L., Golubev A. M., Korneeva O. S., Eneyskaya E. V., Kulminskaya A. A., Neustroev K. N., Polikarpov I. Crystal structure of exo-inulinase from Aspergillus awamori: the enzyme fold and structural determinants of substrate recognition. J. Mol. Biol. 2004;344:471–480. doi: 10.1016/j.jmb.2004.09.024. [DOI] [PubMed] [Google Scholar]
- 10.Verhaest M., Ende W. V. d., Roy K. L., Ranter C. J. D., Laere A. V., Rabijns A. X-ray diffraction structure of a plant glycosyl hydrolase family 32 protein: fructan 1-exohydrolase IIa of Cichorium intybus. The Plant Journal. 2005;41:400–411. doi: 10.1111/j.1365-313X.2004.02304.x. [DOI] [PubMed] [Google Scholar]
- 11.Beisel H. G., Kawabata S., Iwanaga S., Huber R., Bode W. Tachylectin-2: crystal structure of a specific GlcNAc/GalNAc-binding lectin involved in the innate immunity host defense of the Japanese horseshoe crab Tachypleus tridentatus. EMBO J. 1999;18:2313–2322. doi: 10.1093/emboj/18.9.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nurizzo D., Turkenburg J. P., Charnock S. J., Roberts S. M., Dodson E. J., McKie V. A., Taylor E. J., Gilbert H. J., Davies G. J. Cellvibrio japonicus alpha-L-arabinanase 43A has a novel five-blade β-propeller fold. Nat. Struct. Biol. 2002;9:665–668. doi: 10.1038/nsb835. [DOI] [PubMed] [Google Scholar]
- 13.Meng G., Futterer K. Structural framework of fructosyl transfer in Bacillus subtilis levansucrase. Nat. Struct. Biol. 2003;10:935–941. doi: 10.1038/nsb974. [DOI] [PubMed] [Google Scholar]
- 14.Davies G. J., Mackenzie L., Varrot A., Dauter M., Brzozowski A. M., Schülein M., Withers S. G. Snapshots along an enzymatic reaction coordinate: analysis of a retaining beta-glycoside hydrolase. Biochemistry. 1998;37:15280–15287. doi: 10.1021/bi981315i. [DOI] [PubMed] [Google Scholar]
- 15.Liebl W., Brem D., Gotschlich A. Analysis of the gene for beta-fructosidase (invertase, inulinase) of the hyperthermophilic bacterium Thermotoga maritima, and characterisation of the enzyme expressed in Escherichia coli. Appl. Microbiol. Biotechnol. 1998;50:55–64. doi: 10.1007/s002530051256. [DOI] [PubMed] [Google Scholar]
- 16.Kidby D. K., Davidson D. J. A convenient ferricyanide estimation of reducing sugars in the nanomole range. Anal. Biochem. 1973;55:312–325. doi: 10.1016/0003-2697(73)90323-0. [DOI] [PubMed] [Google Scholar]
- 17.Matthews B. W. Solvent content in protein crystals. J. Mol. Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 18.CCP4. The CCP4 (Collaborative Computational Project Number 4) suite: programs for protein crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 19.Roussel A., Cambillau C. Silicon Graphics Geometry Partners Directory 88. CA, U.S.A.: Mountain View; 1991. TURBO-FRODO program. [Google Scholar]
- 20.Winn M. D., Isupov M. N., Murshudov G. N. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. Sect. D Biol. Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 21.Perrakis A., Sixma T. K., Wilson K. S., Lamzin V. S. wARP: improvement and extension of crystallographic phases by weighted averaging of multiple-refined dummy atomic models. Acta Crystallogr. Sect. D Biol. Crystallogr. 1997;53:448–455. doi: 10.1107/S0907444997005696. [DOI] [PubMed] [Google Scholar]
- 22.Brünger A. T. Free R-value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature (London) 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 23.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 24.Davies G., Henrissat B. Structures and mechanisms of glycosyl hydrolases. Structure. 1995;3:853–859. doi: 10.1016/S0969-2126(01)00220-9. [DOI] [PubMed] [Google Scholar]
- 25.Pons T., Olmea O., Chinea G., Beldarrain A., Marquez G., Acosta N., Rodriguez L., Valencia A. Structural model for family 32 of glycosyl-hydrolase enzymes. Proteins. 1998;33:383–395. doi: 10.1002/(sici)1097-0134(19981115)33:3<383::aid-prot7>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 26.Davies G. J., Wilson K. S., Henrissat B. Nomenclature for sugar-binding subsites in glycosyl hydrolases. Biochem. J. 1997;321:557–559. doi: 10.1042/bj3210557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berman H. M. The crystal structure of a trisaccharide, raffinose pentahydrate. Acta Crystallogr. Sect. B Struct. Sci. 1970;26:290–299. doi: 10.1107/s0567740870002273. [DOI] [PubMed] [Google Scholar]
- 28.Jeffrey G. A., Huang D. B. The hydrogen bonding in the crystal structure of raffinose pentahydrate. Carbohydr. Res. 1990;206:173–182. doi: 10.1016/0008-6215(90)80058-b. [DOI] [PubMed] [Google Scholar]
- 29.Davies G., Ducros V. M.-A., Varrot A., Zechel D. L. Mapping the conformational itinerary of β-glycosidases by X-ray crystallography. Biochem. Soc. Trans. 2003;31:523–527. doi: 10.1042/bst0310523. [DOI] [PubMed] [Google Scholar]
- 30.Merritt E. A., Murphy M. E. P. Raster3D Version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 31.Kraulis P. J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]