Abstract
Lancelets are considered to take a key position in the evolution of lineages leading to vertebrates. Herein, a serpin from the lancelet Branchiostoma lanceolatum, Bl-Spn1, was identified that inhibits the PCs (proprotein convertases) PC1/3 and furin. The inhibitor forms SDS-stable complexes with either of its targets. Analysis of the inhibitor/furin reaction products by mass spectroscopy assigns the enzyme's cleavage position C-terminally to Met-Met-Lys-Arg↓ in the reactive site loop of Spn1, in concordance with the classical recognition/cleavage site of the principal vertebrate PCs. The inhibitor is equipped with a canonical ER (endoplasmic reticulum) retrieval signal, Lys-Asp-Glu-Leu (KDEL), marking the inhibitor as a guardian of the cellular secretory routes. Deletion of the ER retrieval signal results in the export of the inhibitor into the medium of transfected COS-7 cells, consistent with the assigned intracellular location. These results identify Bl-Spn1 as the first serpin that may inhibit PC1/3-like subtilases at their natural sites of action. Phylogenetic comparisons support a concept implying a general role for ER-residing serpins in the surveillance of subtilase-like enzymes along the constitutive and regulated secretory pathways of metazoans including a role in the defence of intruders that turn PCs to their propagation.
Keywords: Branchiostoma lanceolatum, furin, lancelet, proprotein convertase, secretory pathway, serpin
Abbreviations: dec-RVKR-cmk, decanoyl-Arg-Val-Lys-Arg-chloromethane; ER, endoplasmic reticulum; GST, glutathione S-transferase; MALDI–TOF, matrix-assisted laser-desorption ionization–time-of-flight; PC, proprotein convertase; hPC1/3, human PC1/3; mPC1/3, mouse PC1/3; pERTKR-AMC, L-pyroglutamyl-Arg-Thr-Lys-Arg-7-amido-4-methylcoumarin; poly(A)+ RNA, polyadenylated RNA; RSL, reactive site loop; SI, stoichiometry of inhibition; TEV, tobacco etch virus
INTRODUCTION
Proprotein convertases (PCs) control the maturation of a large variety of hormones, neuropeptides and other proteins transported to the extracellular space, the cell surface, or to subcellular sites via the secretory pathways [1–3]. The precisely regulated processing of these polypeptides is carried out by cleavage at specific consensus motifs of their precursors within the secretory routes of cells [4]. Moreover, many intracellular pathogens that propagate via intracellular transport pathways, including Ebola virus, HIV, measles virus, or Bacillus anthracis, exploit the activity of PCs for the processing of their envelope proteins or other components needed for infection. As a consequence, PCs are key players in many important (patho)-physiological processes including aberrant proliferation, embryo implantation and development, neurodegenerative processes, lipoprotein metabolism and infectious diseases [5]. In mammals, nine PCs with different substrate selectivity and tissue distribution have been identified. These enzymes are synthesized as inactive zymogens that are converted by autocleavage into the active form [6], in some cases assisted by other proteins [7], and then transferred to multiple subcellular sites associated with the secretory pathways, ranging from the ER (endoplasmic reticulum) via the Golgi apparatus and the trans-Golgi network to extracellular sites [2,8,9].
It is evident that the inadequate activity of PCs may result in severe defects. Thus cells must have evolved finely tuned mechanisms to regulate PCs. Surprisingly little is known about the cellular systems involved in regulating the activity of these enzymes. Intracellular pH and Ca2+ gradients have been found to modulate the (auto)-activation of PC proforms. Ectopic expression of the propeptide of furin results in a decrease of various furin-mediated processes in several transformed cell lines. Recently, a few natural proteinaceous PC inhibitors have been identified: 7B2 [10], proSAAS [11], CRES (cystatin-related epididymal spermatogenic protein) [12] and Spn4A [13–15]. Spn4A belongs to a superfamily of proteins – the serpins – that mostly consist of protease inhibitors that function as suicide substrates by presenting an RSL (reactive site loop) as bait to their targets. The serpins also include members that exert functions surprisingly distinct from protease inhibition [16,17].
Spn4A is a serpin that is derived by alternative splicing from the Drosophila melanogaster (fruitfly) gene spn4, which encodes multiple inhibitor isoforms due to the mutual exclusive use of four RSL coding exon cassettes [18]. The protein has been shown to inhibit human furin and amontillado, the putative Drosophila homologue of mammalian PC2. Spn4A is the first PC-targeted inhibitor shown to be equipped with all signals enabling access to the ER and the secretory cellular routes [13,18,19], strongly suggesting that the inhibitor and its target may encounter each other at their natural cellular locations. As yet, however, serpins with activity towards PCs and a subcellular location consistent with a guardian function in the secretory system have not been identified in other organisms, potentially arguing that some phylogenetic lineages may use alternative strategies for keeping PCs under control. In the present paper, we document the presence of a serpin with properties enabling the inhibition of PCs in the cellular vesicular transport and export system of the lancelet Branchiostoma lanceolatum, a model organism with great implications for developmental biology and the evolution of vertebrates [20]. The cephalochordate B. lanceolatum, also known as Amphioxus, is a marine animal that is closely related to vertebrates as indicated by their common classification in a phylum called chordates. These animals thus offer the possibility to elucidate some of the processes involved at the transition from invertebrates to vertebrates. Lancelets, for instance, have been used to study proILP, a proinsulin-like peptide that contains a C-peptide, and that is flanked by pairs of basic residues [21]. It has been demonstrated previously that Branchiostoma californiensis contains at least three PC genes that are presumably involved in processing of this and/or other hormones [22,23], one of which is believed to represent the orthologue of vertebrate PC1/3.
EXPERIMENTAL
Materials
Adult lancelets (B. lanceolatum) were obtained from the Alfred-Wegener-Institut für Polar- und Meeresforschung, Helgoland, Germany. The Oligotex Direct mRNA Mini kit (catalogue no. 72022), Omniscript reverse transcriptase (4 units/μl; catalogue no. 205110), and the DNeasy Tissue kit were obtained from Qiagen. Oligonucleotides were purchased from MWG Biotech. Sawady Taq DNA polymerase (5 units/μl; catalogue no. 01-1030) and Pfu DNA polymerase (2.5 units/μl; catalogue no. M7741) were obtained from PEQLAB and Promega respectively. Klentherm™ DNA polymerase was obtained from Genecraft (10 units/μl; catalogue no. GC-001). The GeneRacer™ kit was purchased from Invitrogen. The rabbit antiserum directed against purified GST (glutathione S-transferase)–Spn1 was from Pineda Antikörper Service (Berlin, Germany). Recombinant secreted mPC1/3 (mouse PC1/3; residues 84–753) and hPC1/3 (human PC1/3; residues 111–617) were obtained from Dr C. Lazure and N. Rabah (both from Neuropeptides Structure and Metabolism Laboratory, Clinical Research Institute of Montreal, University of Montreal, Montreal, ON, Canada) [24,25] and R&D Systems (catalogue no. 2810-SE) respectively. The sources of all other materials have been described in [13].
Spn1 cDNA synthesis, isolation of genomic DNA and sequencing
Single-stranded cDNA was synthesized (20 μl reactions) with Omniscript reverse transcriptase, using poly(A)+ RNA (polyadenylated RNA) isolated individually from snap-frozen adult lancelets. Serpin-specific cDNA fragments were PCR-amplified with degenerate ‘search’ primer pairs (Table 1) derived from conserved serpin signature motifs located within strand s3A (NAIY motif) and the RSL region (NEEGTEAAA motif) respectively. PCR reactions included an initial denaturation step (2 min at 94 °C), followed by an amplification programme encompassing 15 cycles, each consisting of denaturation (30 s at 94 °C), annealing [starting with a 30 s period at 65 °C in the first cycle, followed by a temperature gradient (1 °C decrease per cycle)], and extension (70 s at 68 °C). Then 30 cycles under constant cycling conditions (30 s at 94 °C, 30 s at 65 °C and 70 s at 70 °C) were performed, followed by a final 3 min period at 68 °C. PCR products were purified and re-amplified. A 509 bp fragment was subcloned and sequenced. The gene-specific primers Blr5′ and Blr3′ (Table 1) respectively were used in RACE (rapid amplification of cDNA ends) reactions [18] for full-length cDNA synthesis.
Table 1. Primers used for Spn1 cDNA amplification.
In the primers, S stands for a sense primer and A for an antisense primer. The position of primers refers to the Spn1 cDNA sequence. Bases not complementary to Spn1 cDNA (linker sequences etc.) are shown in italics.
| Primer | Position | Sequence (5′→3′) |
|---|---|---|
| NAIY | S, 679–707 | GTNCTNGTNAAYGCNATHTAYTTYAARGG |
| NEEGTEA | A, 1162–1187 | GCNGCNGCYTCNGTNCCYTCYTCATT |
| BLr5′ | A, 1113–1132 | GACTGACGTGAAGGTCACGC |
| BLr3′ | S, 807–828 | CCGGTTCAAACTCGCCTACGAC |
| Spn1_XhoI | S, 168–186 | CCATCTCGAGAATCTTTATTTTCAGGCAACTCTGGGAAGCTCC |
| Spn1_BamHI | A, 1319–1347 | CAGCCGGATCCTTACAGTTCATCCTTTGTAGTTAATCCTT |
Genomic DNA was prepared with the DNeasy Tissue kit as recommended by the supplier. Amplification of spn1 gene fragments was performed with Sawady Taq DNA polymerase and several sets of cDNA-derived primers. The overlapping PCR products were subcloned and sequenced. Sequences were aligned with CLUSTAL W [26], and intron positions were mapped to the serpin scaffold of mature human α1-antitrypsin [27].
Generation of plasmids, expression, purification and analysis of recombinant proteins
Spn1 (residues 16–407, Figure 1) was expressed in Escherichia coli, strain BL21 (DE3), as a GST fusion protein, using the pKM263 vector system. To this end, Spn1 cDNA was amplified using the primer pair Spn1_XhoI and Spn1_BamHI (Table 1), thereby creating a restriction fragment flanked by XhoI and BamHI sites. In the fusion construct, the serpin moiety is hooked C-terminally to GST adjacent to a TEV (tobacco etch virus) protease cleavage site. Protein induction and affinity purification on glutathione–Sepharose 4B beads followed a previously published procedure [13], except that the Hepes buffer used previously was replaced by 50 mM Tris/HCl (pH 8.0). After treatment with TEV protease, His-tagged contaminants (residual fusion protein, GST and TEV protease) were removed by chromatography on a Protino Ni 2000 column as described in [13]. Protein purity was assessed by Coomassie Brilliant Blue staining following SDS/PAGE (10% gels). The concentration of the purified protein was determined with a standard Bradford assay (Pierce). MALDI–TOF (matrix-assisted laser-desorption ionization–time-of-flight)-MS analyses were performed as outlined in [13].
Figure 1. Nucleotide sequence of the cDNA coding for Spn1 of B. lanceolatum and translated amino acid sequence.
Nucleotides preceding the poly(A) tail of two Spn1 mRNA variants with shortened 3′-untranslated region are indicated by white-on-black printing. The P1–P1′ bond (triangle) and the N-glycosylation signals (open boxes) are marked.
For COS-7 cell expression, Spn1 cDNAs coding for the complete protein (residues 1–407) or for the deletion mutant, Spn1-ΔKDEL (where KDEL is Lys-Asp-Glu-Leu), which lacks the four C-terminal amino acids, each flanked by NheI and EcoRI sites, were inserted into the NheI/EcoRI-cleaved pcDNA3.1(+) vector. COS-7 cells were transfected with Lipofectamine™ in 9 cm2 dishes [13]. At 36 h post-transfection, the supernatants were centrifuged (30 min, 13000 g) to remove debris. Cellular extracts were prepared by lysing cells (30 min) in 100 μl of ice-cold lysis buffer [50 mM Tris/HCl, pH 7.2, 1% Nonidet P40, 0.1% SDS, 150 mM NaCl, 2 mM EDTA and protease inhibitors (P8340, Sigma)]. The lysate was cleared by centrifugation (13000 g for 1 h at 4 °C). Equal volumes of the supernatants and the cleared lysates from each transfection sample were analysed by reducing SDS/PAGE and Western blotting.
Formation of complexes between Spn1 and target enzymes and enzyme inhibition kinetics
Purified Spn1 was incubated with titrated human furin at 30 °C in 100 mM Hepes (pH 7.5), 1 mM CaCl2 and 0.25% Triton X-100. Complex-formation assays with hPC1/3 were performed in 100 mM sodium acetate (pH 6.0), 5 mM CaCl2, 0.01% Triton X-100 at 30 °C. The reaction products were separated by reducing SDS/PAGE (10% gels) and analysed by Western blotting. The anti-Spn1 antiserum was used at a dilution of 1:20000.
Human furin activity was measured using pERTKR-AMC (L-pyroglutamyl-Arg-Thr-Lys-Arg-7-amino-4-methylcoumarin) as substrate. The concentration [E0] of catalytically active furin was determined by active site titration with dec-RVKR-cmk (decanoyl-Arg-Val-Lys-Arg-chloromethane), and the SI (stoichiometry of inhibition) was investigated with 3 units of the titrated enzyme and variable inhibitor concentrations (0–75 nM) as described in [13]. The activity of dec-RVKR-cmk titrated mPC1/3 was measured after a 6 h pre-activation period in 100 mM sodium acetate and 10 mM CaCl2 (pH 6.0), using the same substrate. The Km value for substrate cleavage in this reaction was 9 μM.
Progress curves for furin inhibition at different Spn1 concentrations [I] were analysed under pseudo-first-order conditions, using furin at a concentration of 2 nM and the substrate pERTKR-AMC [S] at 200 μM [28]. Inhibition curves for mPC1/3 were recorded with an enzyme concentration of 2.5 nM and a substrate concentration of 10 μM. The data were fitted to the following equation for slow tight-binding inhibition:
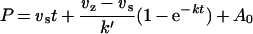 |
where P is the fluorescence, vz the initial velocity, vs the final steady-state velocity, k′ the apparent first-order rate constant and A0 the initial fluorescence. Non-linear regression provided k′, vs, vz and A0 for each [I]. By plotting k′ versus [I], kass (kass is the association rate constant) was obtained from linear regression, using a Km value of 20 μM for furin [13], and 9 μM for mPC1/3, according to the following equation [29]:
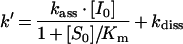 |
where kdiss is the dissociation rate constant.
RESULTS
Isolation of cDNAs coding for Spn1 from B. lanceolatum
Poly(A)+ RNA was isolated from an adult individual of B. lanceo-latum and full-length cDNAs were synthesized using the GeneRacer™ kit, following the manufacturer's instructions. In the initial screening for serpin-specific cDNAs, various PCR primer combinations derived from highly conserved regions of serpins (Table l) were used. Amplification reactions under high and moderate stringency conditions resulted in the isolation of several serpin-related core sequences (C. Bentele, O. Krüger and H. Ragg, unpublished work), including Spn1 cDNA fragments. Full-length cDNA synthesis finally resulted in the isolation of several Spn1 clones of varying size that differed in their 3′-untranslated regions (Figure 1).
The conceptual translation of the cDNA sequences reveals an open reading frame of 407 amino acids. Residues 1–22 constitute a signal peptide as proposed by the PSORT II program [30], suggesting that the protein may be transferred into the lumen of the ER. A second in-frame ATG, 14 residues further downstream, potentially encodes a signal peptide-depleted Spn1 variant. The sequence encompassing the putative positions P15 to P5′ within the RSL suggested an inhibitory role for Spn1 with Arg364-Ser365 as probable P1-P1′ positions. The C-terminal part of Spn1 extends beyond the serpin core sequence as indicated by sequence alignments (results not shown) and it is terminated by the sequence KDEL, suggesting the presence of an ER retention/retrieval signal. Asn72 and Asn384 could serve as carbohydrate attachment sites, since they are part of classical N-glycosylation signals.
Spn1 forms SDS-stable complexes with serine proteases and inhibits PCs
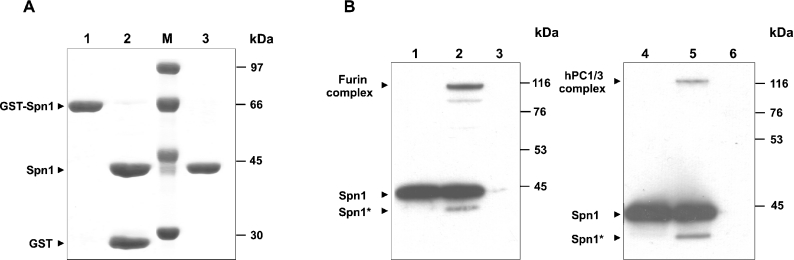
The arginine residue at the putative P1 position implicated that Spn1 might be an inhibitor of trypsin-like enzymes. However, two features of Spn1 strongly inferred PCs as possible interaction partners for the serpin. First, the N-terminal signal sequence in combination with the C-terminal KDEL sequence suggested its residence and physiological role in the secretory export system of the cell, one of the predominant territories of PCs. Secondly, the dipeptidyl sequence Lys-Arg at the suspected RSL positions P2 and P1 (residues 363 and 364) that typically precedes the recognition/cleavage site of classical vertebrate PCs, also implicated Spn1 as a potential suicide substrate inhibitor of these enzymes. To evaluate the target spectrum of Spn1, the inhibitor was recombinantly expressed in E. coli, purified (Figure 2A), and exposed to soluble recombinant human furin (molecular mass ∼53 kDa), hPC1/3 (molecular mass ∼63 kDa) or human α-thrombin respectively. Figure 2(B) shows that incubation of Spn1 with either of these PCs resulted in the formation of SDS-stable complexes with molecular masses of approx. 105 kDa (Spn1–furin) or approx. 120 kDa (Spn1–hPC1/3) respectively. Analogous results were obtained after exposure of Spn1 to mPC1/3 (results not shown). Part of the inhibitor molecules were cleaved by the enzymes, in accord with the branched-pathway mechanism of serpins [17]. Small amounts of Spn1–protease complexes were detected after several hours of incubation with thrombin, but most of the inhibitor molecules were cleaved (results not shown).
Figure 2. Isolation of recombinant Spn1 and analysis of complex formation with PCs.
(A) GST-fused Spn1 was purified from bacterial extracts via glutathione–Sepharose™ 4B (lane 1). After treatment with TEV protease (lane 2), His-tagged contaminants were removed by metal chelate affinity chromatography (lane 3). Proteins were stained with Coomassie Brilliant Blue after SDS/PAGE (10% gels). The positions of molecular mass markers (lane M) are given on the right. (B) Purified Spn1 was incubated with human furin or with hPC1/3 respectively. After SDS/PAGE (10% gels) under reducing conditions, the reaction products were blotted and analysed with antibodies directed against Spn1. Lanes 1 and 4, Spn1; lane 2, Spn1 incubated with furin; lane 3, furin; lane 5, Spn1 incubated with hPC1/3; lane 6, hPC1/3. Cleaved forms of Spn1 are marked by an asterisk. The signal at approx. 100 kDa in lane 2 is presumed to represent a partially degraded Spn1–furin complex. The positions and molecular masses of marker proteins are indicated on the right.
Identification of the P1-P1′ cleavage site
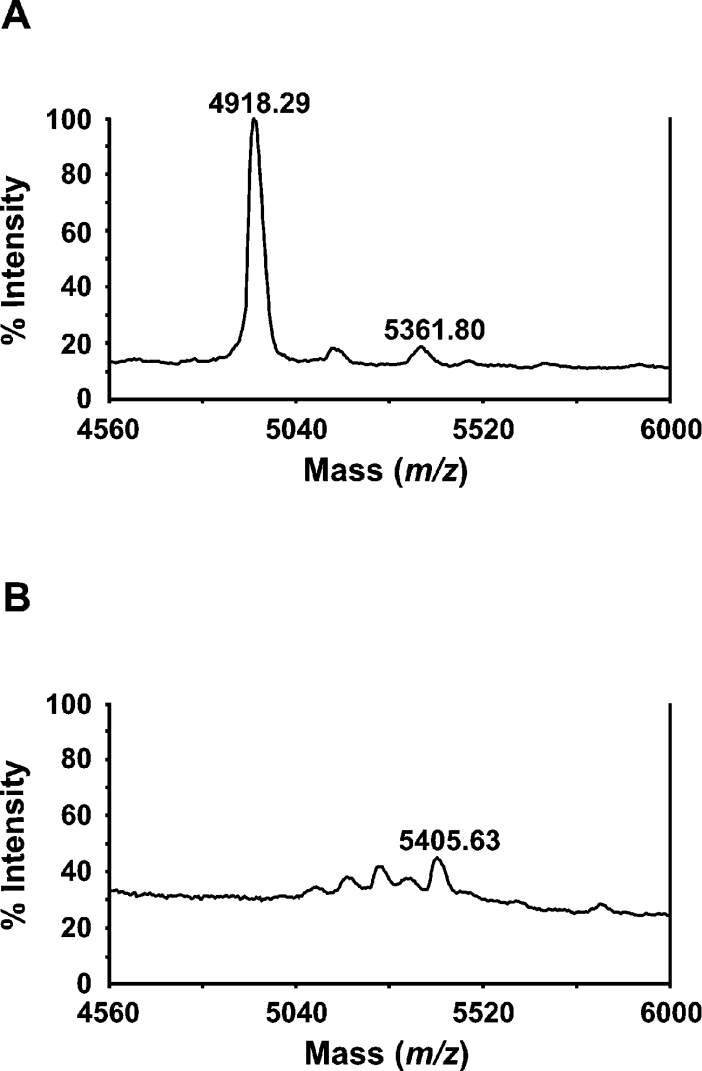
The cleavage sites for furin and hPC1/3 in the RSL region of Spn1 were analysed by MALDI–TOF-MS. To identify the scissile bond, purified recombinant Spn1 was exposed to furin or hPC1/3 respectively and the reaction products were subjected to fragment analysis. After furin treatment, a signal corresponding to a peptide with a molecular mass of 4918.29 Da was observed that locates the cleavage site between residues 364 and 365, identifying Arg and Ser as functional P1-P1′ residues of Spn1 (Figure 3). Analogous results were obtained for cleavage of Spn1 with hPC1/3 (results not shown).
Figure 3. Identification of the furin cleavage site.
Purified recombinant Spn1 (720 nM) was incubated (60 min at 30 °C) in the presence of furin (98 nM) followed by MALDI–TOF-MS analysis. The signal at m/z=4918.29 indicates cleavage between Arg364 and Ser365 in the RSL (A). The signal was not observed in the mass spectrum of the uncleaved inhibitor (B).
SI and rates of enzyme inhibition
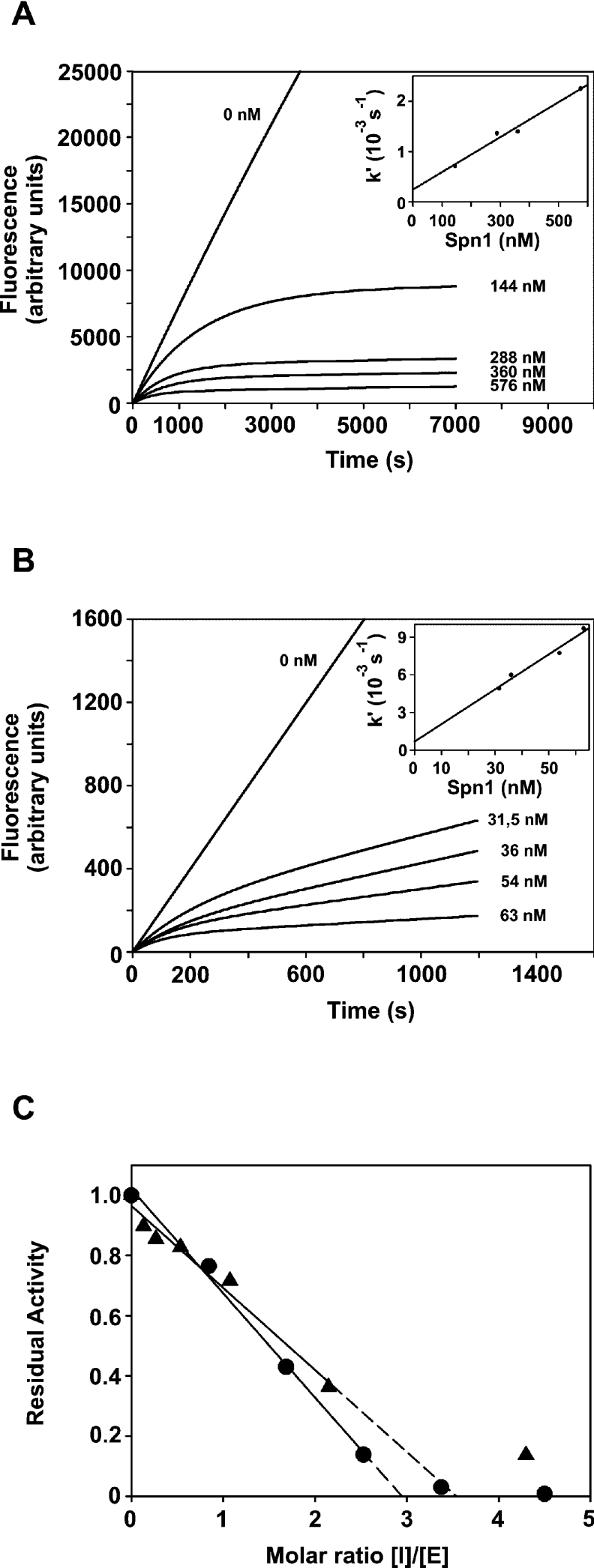
The branched-pathway mechanism of serpins can be characterized by two parameters, the SI value and the inhibition rate constants (Figure 4). To estimate the relative contributions of inhibitory and substrate pathways of Spn1–PC interaction, constant concentrations of active-site-titrated furin (25 nM) or mPC1/3 (206 nM) respectively were incubated with increasing amounts of Spn1 for 60 min at 30 or 25 °C respectively. Residual enzyme activities were detected by addition of the fluorogenic substrate pERTKR-AMC. The SI values for Spn1–furin and Spn1–mPC1/3 interaction were 2.9 and 3.5 respectively. The rates of inhibition of furin and mPC1/3 were determined with the progress curve method. To meet conditions for pseudo-first-order reaction kinetics, an at least 10-fold excess of inhibitor over enzymes was used. Non-linear regression analysis of furin inhibition by Spn1 gave an apparent kass of 3.8×104 M−1·s−1 (SI corrected value: 1.1×105 M−1·s−1). For mPC1/3, an apparent inhibition rate constant of 2.8×105 M−1·s−1 (SI corrected: 9.8×105 M−1·s−1) was determined.
Figure 4. Progress curves for inhibition of PCs by Spn1 and SI.
Human furin (2 nM) or mPC1/3 (2.5 nM) was added to a mixture consisting of the fluorogenic substrate pERTKR-AMC and different concentrations of Spn1. Residual enzyme activities were determined by measuring the change of fluorescence resulting from cleavage of the substrate at pH 7.5 by furin (A) or at pH 6.0 by mPC1/3 (B) respectively. The insets show that k′ depends linearly on the inhibitor concentration. The SI values (C) for interaction of Spn1 with dec-RVKR-cmk-titrated furin (●-●) or mPC1/3 (▲-▲) were determined by incubation of the enzymes with increasing amounts of the inhibitors. The residual enzyme activities were measured and the SI values were determined by linear regression analysis to extrapolate the Spn1/enzyme ratio that resulted in complete inhibition.
The KDEL sequence prevents export of the inhibitor from COS-7 cells
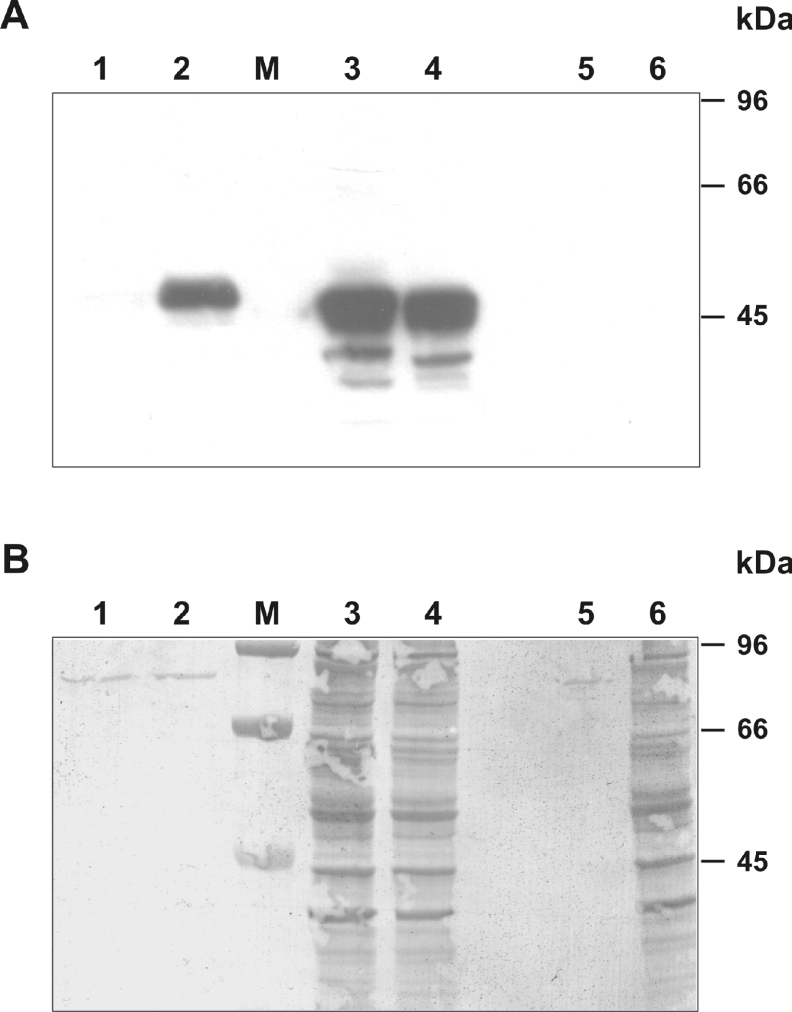
To demonstrate the significance of the C-terminal KDEL sequence as an element responsible for retention of Spn1 in the ER and retrieval from secretory organelles, a functional assay was used. Expression vectors coding for Spn1 containing or lacking the C-terminal tetrapeptide motif (Spn1-ΔKDEL) were introduced into COS-7 cells. Proteins from supernatants and extracts of transfected cells were collected 36 h post-lipofection, resolved by SDS/PAGE, and examined by Western blotting for the presence of Spn1 (Figure 5). In contrast with the medium of cells expressing the truncated serpin variant, there were no significant amounts of inhibitor molecules in the conditioned medium of cells transfected with a vector coding for the KDEL-containing Spn1 variant. Large amounts of Spn1 were detected in cellular extracts of transfected cells, irrespective of the presence or absence of the KDEL sequence in the transgene construct.
Figure 5. The C-terminal KDEL motif mediates intracellular retention of Spn1.
(A) Immunoanalysis of supernatants and cellular extracts of COS-7 cells expressing Spn1. COS-7 cells were transiently transfected with a plasmid coding for wild-type Spn1 or for a mutant (Spn1-ΔKDEL) lacking the C-terminal KDEL motif. Supernatants and cellular extracts were collected 36 h after DNA addition, resolved by SDS/PAGE (10% gels), and analysed by Western blotting, using anti-Spn1 antibodies. (B) After immunodetection, the blotting membrane was stained with Coomassie Brilliant Blue, to check for equal loading. Lanes 1 and 2, supernatants from cells transfected with wild-type Spn1 or Spn1-ΔKDEL respectively. Lanes 3 and 4, extracts from cells transfected with wild-type Spn1 and Spn1-ΔKDEL respectively. Lanes 5 and 6, supernatants and extracts of untransfected control cells. M, molecular mass markers.
Exon–intron structure of the spn1 gene and distribution of serpins with PC-inhibitory potential in the animal kingdom
Each of the lancelet serpin Spn1 and serpin Spn4A from D. melanogaster is equipped with signals mediating ER retrieval and act as an inhibitor of PCs, raising the possibility that they are orthologues. To examine this issue, the amino acid sequences of these serpins and the organizations of their genes were compared. On the protein level, lancelet Spn1 and fruitfly Spn4A share approx. 38% sequence identity. Pairwise comparison of Spn1 with the amino acid sequences of 27 conceptual serpins from D. melanogaster (see: ALIGN_000632.dat; ftp://ftp.ebi.ac.uk/pub/databases/embl/align/) indicated sequence identities between 19% (Spn1/ACP76A) and 36% (Spn1/CG9456) respectively.
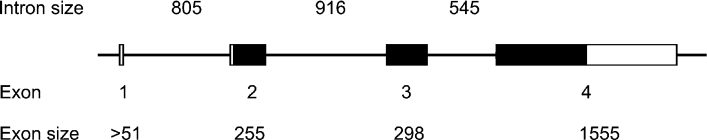
Oligonucleotide primers derived from various regions of the Spn1 cDNA sequence were used to amplify Spn1-related genomic sequences. Sequencing of the genomic fragments revealed the presence of a gene with a four exons/three introns organization (Figure 6). The first intron is located in the 5′-untranslated region, while intron 2 and intron 3 interrupt the coding region between residues 55 and 56, and after the first base coding for residue 155 respectively. Using mature α1-antitrypsin as a scaffold, intron 2 mapped to amino acid position 75c, while intron 3 (phase a) may be assigned to any of the amino acid positions between residues 174 and 176 of the human reference, due to sequence alignment ambiguities. However, it is clear that none of the B. lanceolatum spn1 gene introns can be superimposed with any of the introns mapping to the serpin core of fruitfly Spn4, which contains two introns located at positions 305c and 352a with respect to the human reference protein, and none of the two spn1 gene introns located in the serpin core maps to any of the positions identified for a total of 25 vertebrate serpins [27].
Figure 6. Exon–intron organization of the spn1 gene.
Exons are depicted as numbered bars. Protein coding parts are marked in black, and untranslated regions are denoted as open boxes. Introns are represented as lines. The genomic DNA co-ordinates indicated refer to the numbering used for the sequence deposited in DDBJ, EMBL, GenBank® and GSDB Nucleotide Sequence Databases (accession no. AJ889984).
We note that there are several nucleotide substitutions in the coding region of the Spn1 cDNA sequence and the spn1 gene sequence. Several of these differences are non-synonymous (gene versus cDNA changes: 7S>C, 43T>A, 66A>T, 94V>A, 213I>V, 256L>F, 276I>L, 330S>L). Since genomic DNA and cDNA were isolated from different individuals, these differences might represent allelic variations. Alternatively, two closely related genes may be present in the B. lanceolatum genome.
To evaluate the distribution of PC-inhibitory serpins in the animal kingdom, a database search was conducted. The survey revealed the presence of serpins containing an HDEL/RDEL signal or related sequences at their C-terminus and demonstrated or suspected anti-PC activity in a variety of species (Table 2), thus demonstrating their broad distribution in metazoans.
Table 2. Serpins with demonstrated or putative PC-inhibitory potential in metazoans and equipped with an ER retrieval signal.
GenBank® accession numbers are indicated in parentheses. Experimentally determined or presumed P1-P1′ positions are shown in boldface. For comparison, human α1-antitrypsin (A1AT) is included.
| Serpin (accession no.) | Species | RSL sequence | ER retrieval signal |
|---|---|---|---|
| Spn1 (AJ548509) | B. lanceolatum (lancelet) | GSEAAAATAVNMMKRSLDGETFFADHPF | KDEL |
| Spn4A (AJ428880) | D. melanogaster (fruitfly) | GTEAAAATGMAVRRKRAIMSPEEPIEFF | HDEL |
| XM_395991 | Apis mellifera (honeybee) | GTEAAATTAVSLRLRRCIYPEETEKFIV | RDEL |
| SRPN10 (AJ420785) | Anopheles gambiae (malaria mosquito) | GTEAAAATAAVVRVKRALINRLKVRLDH | HEEL |
| A1AT (P01009) | Homo sapiens (human) | GTEAAGAMFLEAIPMSIPPEVKFNKPFV | − |
DISCUSSION
A systematic search in B. lanceolatum has led to the identification of a serpin, Spn1, that inhibits PC1/3 and furin at physiologically relevant rates. The inhibitor is captured in SDS-stable complexes with its target enzymes, a hallmark feature of serine protease-inhibiting serpins. To our knowledge, Spn1 is the first serpin shown to exhibit activity towards PC1/3. The SI corrected values for the association rate constants were 9.8×105 and 1.1×105 M−1·s−1 for inhibition of mPC1/3 and furin respectively. Compared with Spn4A from D. melanogaster [13,15], the second-order rate constant for furin inhibition is considerably lower for the lancelet serpin. In this context, it may be of significance that the RSL loop of B. lanceolatum Spn1 is one residue shorter than that of Spn4A and other serpins implicated in inhibiting PCs (Table 2). It was demonstrated recently that deletion of one residue (Ala at P6) within the RSL of Spn4A significantly affects inhibition of amontillado, the fruitfly orthologue of human PC2, but not that of human furin [15]. However, for an adequate discussion of the properties of this inhibitor, several points must be considered. First, both inhibition rate constants and SI values may depend on the assay conditions [31–33]. Secondly, serpins – like all other proteins – have been selected to fulfil their role towards their natural targets, i.e. the species-specific PCs or target enzymes from host-specific pathogens. Lancelets contain at least three different PCs [22,23] and it will be interesting to analyse the inhibitory properties of Spn1 towards the suspected natural targets.
Serpins with PC-inhibitory potential and equipped with signals marking them as residents of the ER and compartments involved in cellular trafficking have now been identified in insects and cephalochordates. These inhibitors thus seem to be broadly distributed in the animal kingdom, suggesting a general role for these proteins as guardians of the secretory pathway that control PCs and possibly other serine proteases. A furin-inhibiting serpin [PI8 (protease inhibitor 8)] has also been identified in mammals, but it is not clear how it can enter the secretory pathways, since it is devoid of obvious signals mediating access to the cellular secretory pathways [34]. However, despite their similar functions and cellular locations, the evolutionary relationships between the PC-inhibiting serpins seem to be intricate, and the orthology of lancelet Spn1 and fruitfly Spn4 must be questioned for several reasons. First, the amino acid sequence identity between them does not significantly exceed the values for other fruitfly/Branchiostoma serpin pairs. Secondly, their genes have quite different exon–intron organizations and not a single intron position is shared. Thirdly, Spn4 and Spn1 depict different codons for Ser56 (α1-antitrypsin numbering). The TCN/AGY dichotomy at this position discriminates serpins of deuterostomes from those of protostomes [35] with only few exceptions. Accordingly, in Spn4 this position is coded by TCC (gene-specific residue: Ser71; see [18]), while in Spn1 it is represented by AGT (gene-specific position: Ser58; Figure 1). Another level of complexity is created by the now well-documented phenomenon of hooking multiple RSL coding exon cassettes to the serpin body, probably due to unequal crossing over events, which may obscure phylogenetic relationships further. Thus it seems possible that PC-inhibiting serpins in different taxonomic groups have evolved independently. Since furin and other PCs may be involved in the propagation of pathogenic viruses and bacteria, counteracting serpins may have a role in innate immunity, directed against pathogens that use the secretory cellular routes for their propagation. Apart from the role that PC-inhibiting serpins might fulfil in the control of subtilase-like enzymes and the possibilities they offer for unravelling the physiological functions of PCs, they also emerge as interesting blueprints and agents for pharmacological intervention in a variety of pathologies [36,37].
Acknowledgments
We thank Dr M. C. Letzel (University of Bielefeld) for advice and help in MS. We are grateful to Dr C. Lazure and N. Rabah for the gift of mPC1/3. This work was supported in part by Deutsche Forschungsgemeinschaft, Graduate Programme ‘Bioinformatics’ at the University of Bielefeld.
References
- 1.Nakayama K. Furin: a mammalian subtilisin/Kex2p-like endoprotease involved in processing of a wide variety of precursor proteins. Biochem. J. 1997;327:625–635. doi: 10.1042/bj3270625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomas G. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat. Rev. Mol. Cell Biol. 2002;3:753–766. doi: 10.1038/nrm934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rockwell N. C., Krysan D. J., Komiyama T., Fuller R. S. Precursor processing by kex2/furin proteases. Chem. Rev. 2002;102:4525–4548. doi: 10.1021/cr010168i. [DOI] [PubMed] [Google Scholar]
- 4.Steiner D. F. The proprotein convertases. Curr. Opin. Chem. Biol. 1998;2:31–39. doi: 10.1016/s1367-5931(98)80033-1. [DOI] [PubMed] [Google Scholar]
- 5.Taylor N. A., Van De Ven W. J., Creemers J. W. Curbing activation: proprotein convertases in homeostasis and pathology. FASEB J. 2003;17:1215–1227. doi: 10.1096/fj.02-0831rev. [DOI] [PubMed] [Google Scholar]
- 6.Anderson E. D., Molloy S. S., Jean F., Fei H., Shimamura S., Thomas G. The ordered and compartment-specific autoproteolytic removal of the furin intramolecular chaperone is required for enzyme activation. J. Biol. Chem. 2002;277:12879–12890. doi: 10.1074/jbc.M108740200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muller L., Cameron A., Fortenberry Y., Apletalina E. V., Lindberg I. Processing and sorting of the prohormone convertase 2 propeptide. J. Biol. Chem. 2000;275:39213–39222. doi: 10.1074/jbc.M003547200. [DOI] [PubMed] [Google Scholar]
- 8.Denault J., Bissonnette L., Longpre J., Charest G., Lavigne P., Leduc R. Ectodomain shedding of furin: kinetics and role of the cysteine-rich region. FEBS Lett. 2002;527:309–314. doi: 10.1016/s0014-5793(02)03249-0. [DOI] [PubMed] [Google Scholar]
- 9.Salvas A., Benjannet S., Reudelhuber T. L., Chrétien M., Seidah N. G. Evidence for proprotein convertase activity in the endoplasmic reticulum/early Golgi. FEBS Lett. 2005;579:5621–5625. doi: 10.1016/j.febslet.2005.09.029. [DOI] [PubMed] [Google Scholar]
- 10.Martens G. J., Braks J. A., Eib D. W., Zhou Y., Lindberg I. The neuroendocrine polypeptide 7B2 is an endogenous inhibitor of prohormone convertase PC2. Proc. Natl. Acad. Sci. U.S.A. 1994;91:5784–5787. doi: 10.1073/pnas.91.13.5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fricker L. D., McKinzie A. A., Sun J., Curran E., Qian Y., Yan L., Patterson S. D., Courchesne P. L., Richards B., Levin N., et al. Identification and characterization of proSAAS, a granin-like neuroendocrine peptide precursor that inhibits prohormone processing. J. Neurosci. 2000;20:639–648. doi: 10.1523/JNEUROSCI.20-02-00639.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cornwall G. A., Cameron A., Lindberg I., Hardy D. M., Cormier N., Hsia N. The cystatin-related epididymal spermatogenic protein inhibits the serine protease prohormone convertase 2. Endocrinology. 2003;144:901–908. doi: 10.1210/en.2002-220997. [DOI] [PubMed] [Google Scholar]
- 13.Oley M., Letzel M. C., Ragg H. Inhibition of furin by serpin Spn4A from Drosophila melanogaster. FEBS Lett. 2004;577:165–169. doi: 10.1016/j.febslet.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 14.Osterwalder T., Kuhnen A., Leiserson W. M., Kim Y. S., Keshishian H. Drosophila serpin 4 functions as a neuroserpin-like inhibitor of subtilisin-like proprotein convertases. J. Neurosci. 2004;24:5482–5491. doi: 10.1523/JNEUROSCI.5577-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richer M. J., Keays C. A., Waterhouse J., Minhas J., Hashimoto C., Jean F. The Spn4 gene of Drosophila encodes a potent furin-directed secretory pathway serpin. Proc. Natl. Acad. Sci. U.S.A. 2004;101:10560–10565. doi: 10.1073/pnas.0401406101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silverman G. A., Bird P. I., Carrell R. W., Church F. C., Coughlin P. B., Gettins P. G., Irving J. A., Lomas D. A., Luke C. J., Moyer R. W., et al. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J. Biol. Chem. 2001;276:33293–33296. doi: 10.1074/jbc.R100016200. [DOI] [PubMed] [Google Scholar]
- 17.Gettins P. G. W. Serpin structure, mechanism, and function. Chem. Rev. 2002;102:4751–4804. doi: 10.1021/cr010170+. [DOI] [PubMed] [Google Scholar]
- 18.Krüger O., Ladewig J., Köster K., Ragg H. Widespread occurrence of serpin genes with multiple reactive centre-containing exon cassettes in insects and nematodes. Gene. 2002;293:97–105. doi: 10.1016/s0378-1119(02)00697-2. [DOI] [PubMed] [Google Scholar]
- 19.Han J., Zhang H., Min G., Kemler D., Hashimoto C. A novel Drosophila serpin that inhibits serine proteases. FEBS Lett. 2000;468:194–198. doi: 10.1016/s0014-5793(00)01224-2. [DOI] [PubMed] [Google Scholar]
- 20.Holland L. Z., Laudet V., Schubert M. The chordate amphioxus: an emerging model organism for developmental biology. Cell. Mol. Life Sci. 2004;61:2290–2308. doi: 10.1007/s00018-004-4075-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan S. J., Cao Q. P., Steiner D. F. Evolution of the insulin superfamily: cloning of a hybrid insulin/insulin-like growth factor cDNA from amphioxus. Proc. Natl. Acad. Sci. U.S.A. 1990;87:9319–9323. doi: 10.1073/pnas.87.23.9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oliva A. A., Jr, Steiner D. F., Chan S. J. Proprotein convertases in amphioxus: predicted structure and expression of proteases SPC2 and SPC3. Proc. Natl. Acad. Sci. U.S.A. 1995;92:3591–3595. doi: 10.1073/pnas.92.8.3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oliva A. A., Jr, Chan S. J., Steiner D. F. Evolution of the prohormone convertases: identification of a homologue of PC6 in the protochordate amphioxus. Biochim. Biophys. Acta. 2000;1477:338–348. doi: 10.1016/s0167-4838(99)00283-6. [DOI] [PubMed] [Google Scholar]
- 24.Boudreault A., Gauthier D., Rondeau N., Savaria D., Seidah N. G., Chrétien M., Lazure C. Molecular characterization, enzymatic analysis, and purification of murine proprotein convertase-1/3 (PC1/PC3) secreted from recombinant baculovirus-infected insect cells. Protein Expr. Purif. 1998;14:353–366. doi: 10.1006/prep.1998.0964. [DOI] [PubMed] [Google Scholar]
- 25.Rabah N., Gauthier D. J., Gauthier D., Lazure C. Improved PC1/3 production through recombinant expression in insect cells and larvae. Protein Expr. Purif. 2004;37:377–384. doi: 10.1016/j.pep.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 26.Chenna R., Sugawara H., Koike T., Lopez R., Gibson T. J., Higgins D. G., Thompson J. D. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ragg H., Lokot T., Kamp P. B., Atchley W. R., Dress A. Vertebrate serpins: construction of a conflict-free phylogeny by combining exon–intron and diagnostic site analyses. Mol. Biol. Evol. 2001;18:577–584. doi: 10.1093/oxfordjournals.molbev.a003838. [DOI] [PubMed] [Google Scholar]
- 28.Morrison J. F., Walsh C. T. The behavior and significance of slow-binding enzyme-inhibitors. Adv. Enzymol. Relat. Areas Mol. Biol. 1988;61:201–301. doi: 10.1002/9780470123072.ch5. [DOI] [PubMed] [Google Scholar]
- 29.Bieth J. G. Theoretical and practical aspects of proteinase inhibition kinetics. Methods Enzymol. 1995;248:59–84. doi: 10.1016/0076-6879(95)48007-2. [DOI] [PubMed] [Google Scholar]
- 30.Nakai K., Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 1999;24:34–35. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- 31.Patston P. A., Gettins P., Beechem J., Schapira M. Mechanism of serpin action: evidence that C1 inhibitor functions as a suicide substrate. Biochemistry. 1991;30:8876–8882. doi: 10.1021/bi00100a022. [DOI] [PubMed] [Google Scholar]
- 32.Komiyama T., Gron H., Pemberton P. A., Salvesen G. S. Interaction of subtilisins with serpins. Protein Sci. 1996;5:874–882. doi: 10.1002/pro.5560050509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cole E. B., Miller D., Rometo D., Greenberg R. M., Brömme D., Cataltepe S., Pak S. C., Mills D. R., Silverman G. A., Luke C. J. Identification and activity of a lower eukaryotic serine proteinase inhibitor (serpin) from Cyanea capillata: analysis of a jellyfish serpin, jellypin. Biochemistry. 2004;43:11750–11759. doi: 10.1021/bi049020u. [DOI] [PubMed] [Google Scholar]
- 34.Dahlen J. R., Jean F., Thomas G., Foster D. C., Kisiel W. Inhibition of soluble recombinant furin by human proteinase inhibitor 8. J. Biol. Chem. 1998;273:1851–1854. doi: 10.1074/jbc.273.4.1851. [DOI] [PubMed] [Google Scholar]
- 35.Krem M. M., Di Cera E. Conserved Ser residues, the shutter region, and speciation in serpin evolution. J. Biol. Chem. 2003;278:37810–37814. doi: 10.1074/jbc.M305088200. [DOI] [PubMed] [Google Scholar]
- 36.Khatib A. M., Siegfried G., Chretien M., Metrakos P., Seidah N. G. Proprotein convertases in tumor progression and malignancy: novel targets in cancer therapy. Am. J. Pathol. 2002;160:1921–1935. doi: 10.1016/S0002-9440(10)61140-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fugère M., Day R. Cutting back on pro-protein convertases: the latest approaches to pharmacological inhibition. Trends Pharmacol. Sci. 2005;26:294–301. doi: 10.1016/j.tips.2005.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]