Abstract
The major antigen-adapted immune response protecting a vertebrate against virus infection is that mediated by CTLs (cytotoxic T-lymphocytes). CTLs destroy virus-infected cells, thereby containing the infection. They are activated by recognition of peptide antigens or epitopes, presented to them in the context of MHC I proteins. These epitopes are derived from proteolytic degradation of endogenously synthesized proteins, which is mediated by the proteasome. Augmentation of epitope presentation by MHC I is thought to be effected by the immunoproteasome, induced in response to IFN-γ (interferon-γ) in some cells, and constitutively expressed in others. In this issue of the Biochemical Journal, Remoli and colleagues describe the manipulation of the immunoproteasome by the Tat (transcriptional activation) protein of HIV. The authors show that Tat deregulates the balance of the three proteins, LMP2 (low-molecular-mass polypeptide 2), LMP7 and MECL1 (multicatalytic endopeptidase complex-like 1), which distinguish the immunoproteasome from the proteasome, and they provide a molecular explanation. Intracellular Tat sequesters IRF-1 (interferon-regulatory factor-1) from its cognate promoter element, where normally it associates with STAT1 (signal transducer and activator of transcription 1) to activate basal transcription of the LMP2 gene. LMP2 expression is inhibited as a consequence, skewing the stoichiometry of the immunoproteasome and changing its enzymatic activity. These findings provide a molecular account of an immunomodulatory activity of HIV: changing the peptide antigen profile of cells expressing or exposed to Tat. They may also provide an avenue for manipulating vaccine efficacy and specificity with Tat-based adjuvants.
Keywords: antigen processing, HIV, interferon, immunoproteasome, signal transducer and activator of transcription (STAT), Tat
Protein turnover in the cell is mediated by the proteasome. Proteins are targeted for degradation by the addition of a polyubiquitin chain and include those that are incompletely or incorrectly synthesized, the so-called DRiPs (defective ribosomal products), and those that are lethal, regulatory or produced in excess. Thus the ‘constitutive’ proteasome, expressed in most cell types, and the derivative immunoproteasome (see below) serve in the defence of the organism. They produce the antigenic peptides that associate in the endoplasmic reticulum with the MHC I proteins and are presented on the surface of the cell, where they are surveyed by patrolling CTLs (cytotoxic T-lymphocytes). In this way, cells that are infected, for example with virus, can be eliminated and the virus infection can be contained or eliminated.
One product of activated CTLs is the cytokine IFN-γ (interferon-γ), although it can also be produced by other cell types. This cytokine can function in a paracrine manner on bystander cells expressing its receptor. IFN-γ receptor ligation induces a signal transduction pathway: the JAK (Janus kinase)/STAT (signal transducer and activator of transcription) pathway. One consequence of activating this pathway is the up-regulation of the expression of three proteins: LMP2 (low-molecular-mass protein 2), LMP7 and MECL1 (multicatalytic endopeptidase complex-like 1). These three proteins replace their constitutively expressed homologues in emerging proteasomes to form the immunoproteasome. The immunoproteasome is thought to increase antigen presentation by MHC I, compared with the constitutive proteasome, since LMP2, LMP7 and MECL1 confer upon the immunoproteasome altered protein-cleavage specificity, facilitating MHC I antigen processing and presentation (reviewed in [1]).
Thus CTL recognition of foreign peptide, of viral origin in our example, presented by MHC I will activate cognate CTLs to eliminate the presenting cell. At the same time, paracrine IFN-γ will institute immunoproteasome assembly in bystander cells that may already have become infected by the virus, arming these cells with the ability to increase their antigenic peptide-presentation profile and concomitantly their chance of being detected by appropriate CTLs. Virus replication and spread might then be contained [2].
In some cell types, the immunoproteasome is constitutively expressed. In others, basal transcription of the genes encoding LMP2, LMP7 and MECL1 can occur, and can be induced by IFN-γ [3–6]. IFN-γ increases expression of the genes encoding LMP2, LMP7 and MECL1 via IRF-1 (interferon-regulatory factor-1), indicating the importance of this transcription factor in MHC I antigen processing and therefore presentation.
In the present issue of Biochemical Journal, Remoli et al. [7] begin to provide a molecular account of how the Tat (transcriptional activator) protein of HIV disrupts the protein content of the immunoproteasome, and therefore its enzymatic activity. These authors showed previously that Tat down-regulated LMP2 protein content in the immunoproteasome, but not LMP7 or MECL1 content [8].
In the context of HIV replication, Tat substantially increases transcription from the LTR (long terminal repeat) promoter of HIV. It does so by binding to a stem–loop structure, the TAR (transactivating-responsive region) that forms at the 3′-end of stalled LTR transcripts. Here it recruits cyclin T1 and CDK9 (cyclin-dependent kinase 9), which co-operate to hyperphosphorylate the CTD (C-terminal domain) of RNA polymerase II, increasing the processivity of the polymerase to transcribe the entire HIV genome. Tat may also facilitate transcription complex assembly at the pre-initiation step.
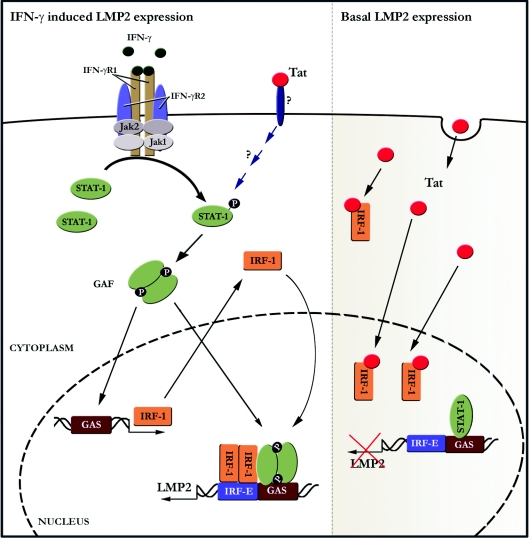
Basal transcription of the LMP2 gene is dependent upon both IRF-1 and unphosphorylated STAT1 binding to their cognate sequences in the promoter of the gene. Remoli et al. [7] show that intracellular Tat down-regulates the abundance of basal LMP2 protein at the transcriptional level. This effect is not due to inhibiting endogenous synthesis of IRF-1 or STAT1, but rather to inhibiting IRF-1 binding to the interferon-responsive elements in the LMP2 promoter (Figure 1, right-hand panel). The work by Remoli et al. [7] suggests that intracellular Tat interferes with the formation of the STAT1–IRF-1 complex, inhibiting IRF-1 binding to the LMP2 promoter. They postulate that sequestration of IRF-1 by Tat is likely to be due to direct protein–protein interaction between the two proteins, since this group has reported previously that the C-terminal domain of IRF-1 mediates direct contact with Tat [9]. Adenovirus E1A operates similarly, but instead of interacting with IRF-1 like Tat, it binds STAT1 [10].
Figure 1. HIV Tat mediates proteasome protein-cleavage specificity at the transcriptional level.
Left-hand panel: the binding of IFN-γ ligand to the IFN-γ receptor (IFN-γR1 and IFN-γR2) results in the activation of JAK1 and JAK2 followed by tyrosine phosphorylation of IFN-γR1. STAT1 is recruited and tyrosine-phosphorylated, enabling homodimerization, which forms the IFN-γ-activation factor (GAF) complex [2]. GAF binds to the IFN-γ-activation site (GAS) of the IFR-1 promoter to drive IRF-1 gene expression. In this way, IFN-γ induces LMP2 gene expression, since GAF also binds to GAS elements in the LMP2 promoter and the induced IRF-1 binds to its cognate IRF element (IRF-E) in this promoter. Remoli et al. [7] show that exogenous Tat protein induced IRF-1 expression within 5 h, yet their reporter gene assays indicate that intracellular Tat is not functional until 12 h after treating the cells. We speculate that IRF-1 is induced though the cell sensing extracellular Tat with a pattern-recognition receptor, which induces GAF formation, as part of the innate immune response. After 12 h, Tat has entered the cell and functions in the same way as de novo synthesized intracellular Tat, i.e. binding IRF-1 to destabilize the STAT1–IRF-1 complex and suppress LMP2 basal transcription (right-hand panel). Right-hand panel: the LMP2 gene requires both IRF-1 and non-phosphorylated STAT1 for basal expression. In this context, Tat protein interacts intracellularly with IRF-1, disrupting STAT1–IRF-1 complex formation and preventing basal LMP2 gene expression.
However, inhibition of basal transcription of the LMP2 gene could be overcome by treatment with IFN-γ, perhaps due to post-translational activation of STAT1, driving de novo synthesis of IRF-1 to levels that saturate Tat (Figure 1, left-hand panel). Nevertheless, expression of LMP7 and MECL1 is not down-regulated by Tat [8]. Why not, when both are also apparently transcriptionally regulated by IRF-1? Addressing this issue would be of interest, since perhaps other transcriptional control elements independent of, or interacting with, IRF-1 are involved and which dominate IRF-1 sequestration by Tat. In this regard, the extent of other activities of IRF-1, for example inducing expression of p21, could be investigated in the presence of intracellular Tat. However, although the levels of LMP7 and MECL1 proteins are not affected by Tat, their functions may be. Tat can interact with LMP7 and MECL (but not LMP2) [11]; it might modulate the enzymatic activities of these proteins at the post-translational level and therefore the output of the immunoproteasome.
Tat is produced from HIV-infected cells, and so Remoli et al. [7] treated cells with exogenous Tat protein. Surprisingly, IRF-1 expression was induced, with peak expression occurring 5 h post-treatment. In parallel, however, peak transcriptional activation of an LTR reporter plasmid by extracellular Tat did not peak for at least another 19 h. These studies suggest that cells treated with extracellular Tat might ‘sense’ this protein with an extracellular pattern-recognition receptor, such as a Toll-like receptor (Figure 1, left-hand panel), and that IRF-1 is induced as a consequence of a normal innate immune response. Once Tat enters the cell, it may then sequester IRF-1 to suppress LMP2 expression (Figure 1, right-hand panel).
In their study, Remoli et al. [7] provide another piece to the puzzle of the interaction between HIV and Tat. This group has demonstrated previously that IRF-1 is induced upon HIV infection and that it activates transcription from the HIV-1 LTR in the absence of Tat protein [9]. From these data, they hypothesized that IRF-1 plays a pivotal role in HIV transcription during the early stages of HIV infection of a cell and during virus reactivation [9]. Thus IRF-1 expression establishes a positive-feedback loop by providing the necessary transcriptional activation to drive production of Tat protein before virus transactivators are abundant [9]. Taken together, in the context of HIV replication, it appears that this positive-feedback loop is then negatively regulated by Tat sequestering IRF-1. However, whether sequestration of IRF-1 is either specific to the Tat protein of HIV-1 strain IIIB, or an activity common to all strains has still to be determined.
Importantly, the work of Remoli et al. [7] begins to provide us with a molecular understanding of this group's observation that modulation of the immunoproteasome by Tat alters the profile of the peptide antigens it produces. The consequence could be physiologically relevant, by shifting in vivo the profile of antigenic targets that are displayed to CTLs [8]. This shift is possible because the specific enzymatic activity of LMP2 is different to that of both LMP7 and MECL1 and changing the stoichiometry of the three proteins in the immunoproteasome will change the profile of peptide antigens it produces. As such, Tat might mediate an HIV immune evasion strategy, adjusting the antigenic profile of HIV-infected cells to one that is less likely to generate an effective anti-HIV CTL response, but this possibility needs to be determined empirically. Conversely, this group's observation that Tat promoted expression of subdominant antigenic peptides of another virus, EBV (Epstein–Barr virus) [8], led them to speculate that Tat might promote CTL clearance of virus-infected or tumour cells by exposing such peptides, which are less susceptible to mutation and therefore CTL escape.
Aside from the physiological relevance of Tat manipulation of immunoproteasome activity in the context of HIV infection, Remoli et al. [7] speculate that Tat might provide the molecular basis for fine-tuning the immune response as a vaccine adjuvant. Such forward-thinking is based on their own past successes with Tat-based anti-HIV vaccines [12], but potential exists to extrapolate the technology beyond HIV to vaccines against other viruses or tumour antigens. In this regard, thymic regulation of the CTL repertoire by the immunoproteasome is becoming increasingly accepted [13]. In conclusion, Remoli et al. [7] provide another example of how studying virus–host interactions furthers our understanding of the immune response.
References
- 1.Strehl B., Seifert U., Kruger E., Heink S., Kuckelkorn U., Kloetzel P. M. Interferon-γ, the functional plasticity of the ubiquitin–proteasome system, and MHC class I antigen processing. Immunol. Rev. 2005;207:19–30. doi: 10.1111/j.0105-2896.2005.00308.x. [DOI] [PubMed] [Google Scholar]
- 2.Goodbourn S., Didcock L., Randall R. E. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 2000;81:2341–2364. doi: 10.1099/0022-1317-81-10-2341. [DOI] [PubMed] [Google Scholar]
- 3.White L. C., Wright K. L., Felix N. J., Ruffner H., Reis L. F., Pine R., Ting J. P. Regulation of LMP2 and TAP1 genes by IRF-1 explains the paucity of CD8+ T cells in IRF-1−/− mice. Immunity. 1996;5:365–376. doi: 10.1016/s1074-7613(00)80262-9. [DOI] [PubMed] [Google Scholar]
- 4.Chatterjee-Kishore M., Kishore R., Hicklin D. J., Marincola F. M., Ferrone S. Different requirements for signal transducer and activator of transcription 1α and interferon regulatory factor 1 in the regulation of low molecular mass polypeptide 2 and transporter associated with antigen processing 1 gene expression. J. Biol. Chem. 1998;273:16177–16183. doi: 10.1074/jbc.273.26.16177. [DOI] [PubMed] [Google Scholar]
- 5.Namiki S., Nakamura T., Oshima S., Yamazaki M., Sekine Y., Tsuchiya K., Okamoto R., Kanai T., Watanabe M. IRF-1 mediates upregulation of LMP7 by IFN-γ and concerted expression of immunosubunits of the proteasome. FEBS Lett. 2005;579:2781–2787. doi: 10.1016/j.febslet.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 6.Foss G. S., Prydz H. Interferon regulatory factor 1 mediates the interferon-γ induction of the human immunoproteasome subunit multicatalytic endopeptidase complex-like 1. J. Biol. Chem. 1999;274:35196–35202. doi: 10.1074/jbc.274.49.35196. [DOI] [PubMed] [Google Scholar]
- 7.Remoli A. L., Marsili G., Perrotti E., Gallerani E., Ilari R., Nappi F., Cafaro A., Ensoli B., Gavioli R., Battistini A. Intracellular HIV-1 tat protein represses constitutive LMP2 transcription increasing proteasome activity by interfering with the binding of IRF-1 to STAT1. Biochem. J. 2006;396:371–380. doi: 10.1042/BJ20051570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gavioli R., Gallerani E., Fortini C., Fabris M., Bottoni A., Canella A., Bonaccorsi A., Marastoni M., Micheletti F., Cafaro A., et al. HIV-1 tat protein modulates the generation of cytotoxic T cell epitopes by modifying proteasome composition and enzymatic activity. J. Immunol. 2004;173:3838–3843. doi: 10.4049/jimmunol.173.6.3838. [DOI] [PubMed] [Google Scholar]
- 9.Sgarbanti M., Borsetti A., Moscufo N., Bellocchi M. C., Ridolfi B., Nappi F., Marsili G., Marziali G., Coccia E. M., Ensoli B., Battistini A. Modulation of human immunodeficiency virus 1 replication by interferon regulatory factors. J. Exp. Med. 2002;195:1359–1370. doi: 10.1084/jem.20010753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chatterjee-Kishore M., van Den Akker F., Stark G. R. Adenovirus E1A down-regulates LMP2 transcription by interfering with the binding of STAT1 to IRF1. J. Biol. Chem. 2000;275:20406–20411. doi: 10.1074/jbc.M001861200. [DOI] [PubMed] [Google Scholar]
- 11.Apcher G. S., Heink S., Zantopf D., Kloetzel P. M., Schmid H. P., Mayer R. J., Kruger E. Human immunodeficiency virus-1 Tat protein interacts with distinct proteasomal α and β subunits. FEBS Lett. 2003;553:200–204. doi: 10.1016/s0014-5793(03)01025-1. [DOI] [PubMed] [Google Scholar]
- 12.Cafaro A., Caputo A., Fracasso C., Maggiorella M. T., Goletti D., Baroncelli S., Pace M., Sernicola L., Koanga-Mogtomo M. L., Betti M., et al. Control of SHIV-89. 6P-infection of cynomolgus monkeys by HIV-1 Tat protein vaccine. Nat. Med. 1999;5:643–650. doi: 10.1038/9488. [DOI] [PubMed] [Google Scholar]
- 13.Osterloh P., Linkemann K., Tenzer S., Rammensee H.-G., Radsak M. P., Busch D. H., Schild H. Proteasomes shape the repertoire of T cells participating in antigen-specific immune responses. Proc. Natl. Acad. Sci. U.S.A. 2006;103:5042–5047. doi: 10.1073/pnas.0509256103. [DOI] [PMC free article] [PubMed] [Google Scholar]