Abstract
The Tat protein is the transcriptional activator of HIV-1 gene expression, which is not only essential for viral replication, but also important in the complex HIV-induced pathogenesis of AIDS, as both an intracellular and an extracellular released protein. Accordingly, Tat is able to profoundly affect cellular gene expression, regulating several cellular functions, also in non-infected cells. We showed recently that Tat induces modification of immunoproteasomes in that it up-regulates LMP7 (low-molecular-mass polypeptide 7) and MECL1 (multicatalytic endopeptidase complex-like 1) subunits and down-modulates the LMP2 subunit, resulting in a change in the generation and presentation of epitopes in the context of MHC class I. In particular, Tat increases presentation of subdominant and cryptic epitopes. In the present study, we investigated the molecular mechanism responsible for the Tat-induced LMP2 down-regulation and show that intracellular Tat represses transcription of the LMP2 gene by competing with STAT1 (signal transducer and activator of transcription 1) for binding to IRF-1 (interferon-regulatory factor-1) on the overlapping ICS-2 (interferon consensus sequence-2)–GAS (γ-interferon-activated sequence) present in the LMP2 promoter. This element is constitutively occupied in vivo by the unphosphorylated STAT1–IRF-1 complex, which is responsible for the basal transcription of the gene. Sequestration of IRF-1 by intracellular Tat impairs the formation of the complex resulting in lower LMP2 gene transcription and LMP2 protein expression, which is associated with increased proteolytic activity. On the other hand, extracellular Tat induces the expression of LMP2. These effects of Tat provide another effective mechanism by which HIV-1 affects antigen presentation in the context of the MHC class I complex and may have important implications in the use of Tat for vaccination strategies.
Keywords: gene regulation, HIV-1 infection, interferon-regulatory factor-1 (IRF-1), signal transducer and activator of transcription 1 (STAT1), Tat, transcription factor
Abbreviations: ChIP, chromatin immunoprecipitation; CTL, cytotoxic T-lymphocyte; DC, dendritic cell; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GAS, γ-interferon-activated sequence; ICS, interferon consensus sequence; IFN, interferon; IRF-1, IFN-regulatory factor-1; IRF-E, IFN-responsive element; LMP, low-molecular-mass polypeptide; LTR, long terminal repeat; MECL1, multicatalytic endopeptidase complex-like 1; STAT1, signal transducer and activator of transcription 1; SBE, STAT-binding element; TAP1, transporter associated with antigen processing 1; TK, thymidine kinase; WCE, whole-cell extract
INTRODUCTION
The complex pathogenesis of AIDS and related syndromes involves factors that are able to also act on non-infected cells, and one such a factor is Tat. The HIV-1 transactivator protein Tat is a ∼14/15 kDa protein produced early after infection and before virus integration [1] and it is required for productive virus replication [2,3]. During acute infection, Tat is also released from acutely infected cells into the extracellular environment and is taken up by neighbouring cells, where it can increase virus infectivity and modulate cellular functions [3–5].
It has been shown that native Tat protein also displays immunomodulatory features. In particular, Tat targets and is very efficiently taken-up by monocyte-derived DCs (dendritic cells) and, after internalization, induces DC maturation, resulting in an increase in allogeneic and antigen-specific presentation by DCs and high activation of Th1 responses against recall antigens [6]. Furthermore, Tat modulates CTL (cytotoxic T-lymphocyte) epitope hierarchy by modifying the catalytic subunit composition of immunoproteasomes [7].
The proteasome is the main provider of peptide ligands for MHC class I molecules, degradation of cell proteins and generation of MHC class I-presented peptides [8]. During an immune response to pathogens, pro-inflammatory cytokines are released and induce the replacement of the constitutively expressed activesite subunits of the proteasome (delta, MB1 and Z) with LMP2 (low-molecular-mass polypeptide 2), LMP7 and MECL1 (multicatalytic endopeptidase complex-like 1). These proteasomes are called immunoproteasomes and display a marked change in the cleavage preference and in the production of T-cell epitopes [9–11]. LMP2, LMP7 and MECL1 are also constitutively expressed in specific cell types, such as DCs and B-cells [10,11].
Immunoproteasomes are very efficient for the generation of specific CTL epitopes, and it has been shown that substitution of standard β-subunits with LMP2, LMP7 and MECL1 subunits improves the production of peptide antigens with the correct C-termini for binding to MHC class I [9–12]. On the other hand, there is evidence, both in humans and mice, that the presence of LMP2 may inhibit the presentation of specific peptide antigens [10,12,13], therefore the control of its expression is particularly important for the specificity of the antigen presented.
Transcription of the LMP2 gene is regulated by a bidirectional promoter that also controls transcription of the TAP1 (transporter associated with antigen processing 1) gene [14]. The requirement of transcription factors for the expression of the two genes is, however, different [15]. In particular, basal, as well as cytokine-induced LMP2 expression, involves both IRF-1 [IFN (interferon)-regulatory factor-1] and STAT1 (signal transducer and activator of transcription 1) binding as a complex to a composite, partially overlapping, sequence which contains the IRF-E (IFN-responsive element), also referred to as ICS-2 (IFN consensus sequence-2), and the GAS (γ-IFN-activated sequence) site [15,16]. STAT1 is a major transcription factor in the IFN-α/β and IFN-γ signal transduction pathways which lead to the activation of antiviral, antiproliferative and immunomodulatory functions (for a review, see [17]). Stimulation by IFN-γ results in STAT1 activation through phosphorylation, followed by homodimerization and binding to sequences originally termed GAS and more recently referred to as SBE (for STAT-binding element) [18]. IFN-γ also strongly induces transcription of the IRF-1 gene through binding of the SBE in the IRF-1 promoter by activated STAT1 [19].
IRF-1 was originally identified as a protein that binds the cis-acting DNA elements in the IFN-β promoter and the ISRE (IFN-stimulated response element) also referred as IRF-E in the promoters of IFN-α/β-stimulated genes [20]. Thus far, the intensive functional analyses carried out on it have revealed a remarkable functional diversity of this transcription factor in the regulation of cellular responses, through the modulation of different sets of genes, depending on the cell type, state of the cell and/or the nature of the stimuli [21]. Studies in knockout mice implicated IRF-1 in the regulation of various immune processes, such as T-cell selection and maturation, leukaemogenic development and autoimmunity. Impairment in CD8+ cell maturation, defective Th1 responses, exclusive Th2 differentiation, impaired macrophage production of interleukin 12 and maturation of natural killer cells have all been observed in immune cells from IRF-1−/− mice [21–24]. Specifically, the impaired CD8+ response observed in the IRF-1−/− mice has been ascribed to the greatly reduced expression of TAP1 and LMP2 [25]. Indeed, it has been demonstrated that IRF-1 is essential for both basal and IFN-γ-induced expression of LMP2 [16,25].
In the context of HIV-1 infection, we reported previously that IRF-1 is produced early upon virus entry and co-operates with Tat in amplifying virus gene transcription and replication [26,27]. Owing to the ability of Tat to induce modification of immunoproteasomes by up-regulating LMP7 and MECL1 subunits and down-regulating the LMP2 subunit [7], we sought to investigate whether the effect of Tat on LMP2 expression may involve the STAT1–IRF-1-mediated regulation of LMP2 gene transcription. Discerning LMP2 regulation in Tat-exposed cells will provide essential information both for the control of antigen presentation in virally infected cells and for the use of Tat in vaccination strategies.
MATERIALS AND METHODS
Cells and treatments
Jurkat T-cells were grown in RPMI 1640 medium (Bio-Whittaker) containing 10% (v/v) foetal calf serum and antibiotics. Jurkat T-cells expressing Tat have been described previously [28] and were cultured in medium supplemented with 800 μg/ml neomycin (Sigma–Aldrich). Where indicated, cells were treated with 100–500 units/ml rIFN-γ (recombinant IFN-γ) (PeproTech) for the indicated time.
HIV-1 Tat protein
HIV-1 Tat from the human T-lymphotropic virus type IIIB isolate (BH10 clone) was expressed in Escherichia coli and purified by heparin-affinity chromatography and HPLC, as described previously [6]. Different lots of Tat were used with reproducible results, and, in all cases, the protein was active and endotoxin concentration was undetectable (detection threshold: 0.05 endotoxin unit/g).
Purification of proteasomes
Proteasomes were prepared by affinity chromatography as described previously [29]. Fractions containing proteasomes were combined and dialysed against 25 mM Tris/HCl, pH 7.5, and protein concentration was determined using the bicinchoninic acid method (Pierce).
Northern blot
RNA (20 μg) prepared using an RNeasy kit (Qiagen) was run on a denaturing agarose gel containing formaldehyde, transferred on to Hybond-N membranes (Amersham Biosciences) and hybridized to random primed 32P-labelled LMP2 and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) probes. The LMP2 probe was obtained by PCR amplification using the primers 5′-TGTGCACTCTCTGGTTCAGC-3′ and 5′-TGCTGCATCCACATAACCAT-3′.
Western blot assay
Equal amounts of purified proteasomes were separated by SDS/12% PAGE and electroblotted on to Protran nitrocellulose membranes (Schleicher & Schuell Microscience). Blots were probed with specific antibodies for α2 and LMP2 subunits (Affiniti Research Products) and developed using the ECL® (enhanced chemiluminescence) system (Amersham Biosciences). Cell extracts (20–50 μg), prepared as described in [26], were separated by SDS/10% PAGE. Blots were incubated with polyclonal antibodies against IRF-1 (sc-497; Santa Cruz Biotechnology; 1:200 dilution) or against STAT1 (sc-346; Santa Cruz Biotechnology; 1:200 dilution) and then with horseradish-peroxidase-coupled anti-rabbit secondary antibody (Amersham Biosciences; 1:2000 dilution) using the ECL® system.
DNA-affinity purification assay
DNA-affinity purification assays were performed as described previously [26]. Biotinylated oligonucleotides corresponding to the LMP2 IRF-E (−209 nt to −175 nt) and (−197 nt to −178 nt) [25] or a synthetic IRF-1-binding site (C13) (5′-GATCAAGTGAAAGTGAAAGTG-3′) or the GAS present on the IRF-1 promoter (5′-GATCGATTTCCCCGAAAT-3′) were mixed with 200 μg of WCEs (whole-cell extracts) prepared as described in [26], and the complexes formed were pulled-down with magnetic beads (Streptavidin MagneSphere® Paramagnetic Particles; Promega). Bound material eluted by boiling in SDS sample buffer (62.5 mM Tris/HCl, pH 6.8, 5% glycerol, 1% SDS, 0.05% 2-mercaptoethanol and 0.5% Bromophenol Blue) was analysed by Western Blot with antibodies against IRF-1 and STAT1 as above.
Immunoprecipitation and immunoblot analysis
WCEs (300 μg), prepared as described in [26], were incubated overnight with 1 μg of polyclonal anti-IRF-1 antibody. Cell extracts were then incubated with Ultralink immobilized Protein A/G–Sepharose (Pierce) for 2 h at room temperature (25 °C). After extensive washing, immunoprecipitates were eluted by boiling the beads for 5 min in 1×SDS sample buffer. Eluted proteins were separated by SDS/10% PAGE and subjected to Western blot analysis with a goat polyclonal anti-STAT1 antibody.
Transfection experiments and enzymatic assays
Transient transfection experiments were performed using the FuGENE 6 Transfection Reagent (Roche Diagnostics) according to the manufacturer's protocol. The constructs encoding a portion of the wild-type LMP2 promoter and a mutated version (mt LMP2) cloned upstream of the luciferase reporter gene, were a gift from Dr Kenneth L. Wright (H. Lee Moffitt Cancer Center and Research Institute, Department of Chemistry and Molecular Biology, University of South Florida, Tampa, FL, U.S.A.) and are described in [30]. The HIV-1 LTR (long-terminal repeat) and the CMV (cytomegalovirus)–Tat constructs were described previously [31]. The TK (thymidine kinase)–Renilla luciferase plasmid was co-transfected and used as a control for transfection efficiency. Reagents from Promega were used to assay extracts for luciferase activity in a Lumat LB9501 luminometer (E&G Berthold).
ChIP (chromatin immunoprecipitation) assay
The protocol used for ChIP was as described in [32] with some modifications. In brief, 3×107 cells were cross-linked by incubation with 1% formaldehyde for 10 min and, after fixation, cells were resuspended in lysis buffer (5 mM Pipes, pH 8, 85 mM KCl and 0.5% Nonidet P40 with protease inhibitors) and were Dounce-homogenized. Nuclei were resuspended in sonication buffer (0.1% SDS, 10 mM EDTA, 50 mM Tris/HCl, pH 8, and 0.5% deoxycholic acid with protease inhibitors) and were sonicated to an average length of 1000 bp. Pre-cleaned chromatin from 1×107 cells was incubated in immunoprecipitation buffer (sonication buffer plus 150 mM LiCl) overnight at 4 °C with 3 μg of anti-IRF-1 and anti-STAT1 polyclonal antibodies. DNA–protein complexes were collected with Ultralink immobilized Protein A/G–Sepharose followed by sequential washes with immunoprecipitation buffer, low-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris/HCl, pH 8.1, and 150 mM NaCl), high-salt buffer (0.25 M LiCl, 1% Nonidet P40, 1% deoxycholate, 1 mM EDTA and 10 mM Tris/HCl, pH 8.1) and TE (10 mM Tris/HCl and 1 mM EDTA, pH 8.0). Samples were then eluted and, after reverse cross-linking, proteins were digested with protease K and RNA was removed by RNase A. DNA was purified with QIAquick PCR purification kit (Qiagen) and resuspended in 30 μl of water, and 4 μl was used for PCR analysis performed with the primers given in [16a]. PCR products were analysed on a 1.0% agarose gel.
RESULTS
Endogenously expressed Tat inhibits the expression of the LMP2 subunit of immunoproteasomes by repressing the LMP2 promoter basal activity
The expression of the LMP2 subunit in immunoproteasomes purified from two different clones (JSL34 and JSL3) of Jurkat T-cells stably transfected with a vector expressing Tat, or with the empty vector [28] was markedly inhibited in both clones expressing Tat [7] (see Supplementary Figure S1 at http://www.BiochemJ.org/bj/396/bj3960371add.htm).
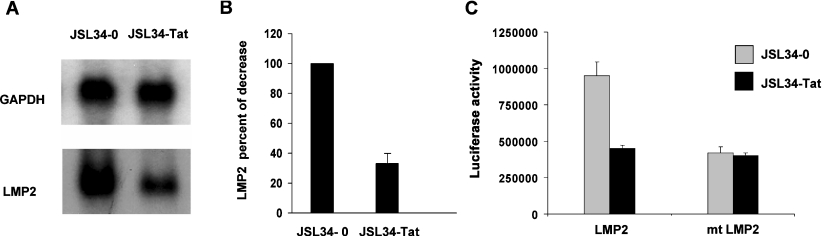
In order to evaluate at which level Tat affects LMP2 expression, Northern blot experiments were performed using the JSL34 clone (Figure 1A). A clear reduction (of approx. 70%; Figure 1B) in LMP2 mRNA expression was detected in Tat-expressing cells compared with control cells, indicating that Tat may exert its inhibitory effect at the transcriptional level. To determine whether Tat is able to inhibit LMP2 promoter activity, cells transduced with the empty vector or Tat-expressing JSL34 cells were transiently transfected with a LMP2–luciferase construct, wild-type LMP2 or mutated in the IRF-E consensus sequence (mt LMP2) [30]. As shown in Figure 1(C), the basal LMP2 promoter activity was substantially reduced (by approx. 50%) in cells constitutively expressing Tat, as compared with cells expressing the empty vector. Conversely, the basal expression of the LMP2 promoter construct mutated in the IRF-E element was substantially decreased as compared with the wild-type construct, but its transcription was not affected by the expression of Tat. These results indicate that Tat represses the transcriptional activity of the LMP2 promoter, and this repression is mediated by the IRF-E consensus sequence.
Figure 1. Endogenously expressed Tat transcriptionally represses LMP2 expression.
(A) Northern blot analysis of total RNA extracted form JSL34-0 and JSL34-Tat cells. One representative experiment out of three performed is shown. (B) Intensity of specific bands from three independent experiments was quantified by densitometry, and the results are expressed as means±S.D. after normalization with GAPDH. The normalized control was assigned a numerical value of 100. (C) The wild-type or mutated LMP2 promoter linked to the luciferase reporter gene was transiently transfected in Jurkat T-cells expressing the empty vector (JSL34-0) or the Tat-expressing vector (JSL34-Tat). After 24 h, WCEs were processed for luciferase activity. Means±S.D. from three separate experiments were calculated after normalization with the TK–Renilla activity.
Tat interferes with the binding of transcription factors to the LMP2 promoter
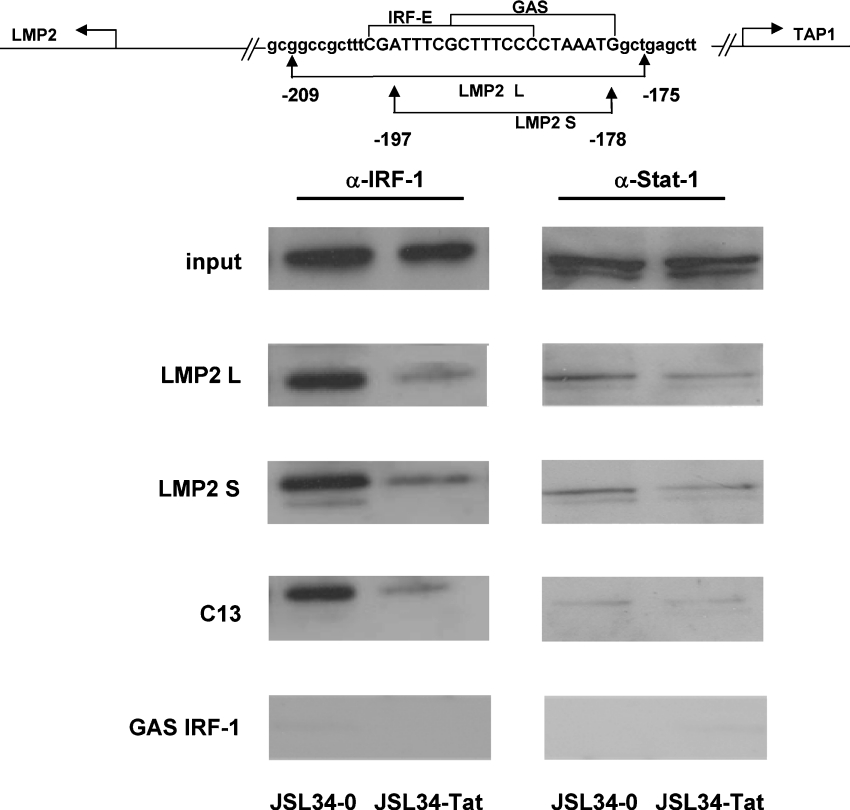
It has been reported previously [15,16] that constitutive expression of the LMP2 gene requires that unphosphorylated STAT1 and IRF-1 bind to the overlapping IRF-E (ICS2)–GAS element (Figure 2, top panel). As shown in Figure 1(C), this element is also the target of the Tat effect on LMP2 expression. To assess whether the inhibitory effect of Tat on LMP2 transcription is due to interference with the binding of transcription factors to this element, DNA-affinity purification assays were performed. WCEs from both control and Tat-expressing cells were incubated with biotinylated LMP2 IRF-E–GAS probes spanning nucleotides −178 nt to −197 nt (LMP2 S) and −175 nt to −209 nt (LMP2 L) respectively. The isolated complexes were then examined by immunoblotting against STAT1 and IRF-1. As shown in Figure 2, a decreased binding of IRF-1 to the cognate DNA sequences was evident in cell extracts from Tat-expressing cells, as compared with control cells transfected with the empty control vector. Binding of unphosphorylated STAT1 was also detected with the two DNA probes, but only a slight decrease in protein binding was detected in extracts from Tat-expressing cells.
Figure 2. Tat interferes with the binding of IRF-1 to the LMP2 promoter.
A schematic representation of the 200 bp fragment of the proximal LMP2 promoter and the sequence of the two oligonucleotides (LMP2 L and LMP2 S) used in the DNA pull-down assays is shown in the top panel. Lower panels: biotinylated oligodeoxynucleotides containing the LMP2 IRF-E (ICS-2)–GAS, the C13 and the GAS sequence on the IRF-1 promoter (see the Materials and methods section), coupled to Streptavidin MagneSphere®, were incubated with extracts from JSL34-0 and JSL34-Tat cells. Bound proteins were eluted from the beads by boiling in sample buffer and analysed by Western blotting with specific antibodies against IRF-1 and unphosphorylated STAT1. input indicates the level of endogenous IRF-1 and STAT1 in the cell extracts (50 μg). One representative experiment out of three performed is shown.
Interestingly, the inhibitory effect of Tat on IRF-1 binding was also observed when the specific synthetic IRF-1 consensus binding site (C13) was incubated with the same cell extracts. Moreover, as expected, unphosphorylated STAT1 bound neither to an oligonucleotide corresponding to the GAS sequence present on the IRF-1 promoter nor to the C13 oligonucleotide. Western blot analysis of WCEs (Figure 2, input) indicated that comparable amounts of STAT1 and IRF-1 were present in Jurkat T-cells irrespective of the presence of Tat.
This result confirms the specificity of the binding observed on the LMP2 promoter sequences and indicates that Tat specifically decreases the binding of IRF-1 to its consensus binding site on the LMP2 promoter.
Tat interferes with the formation of the STAT1–IRF-1 complex
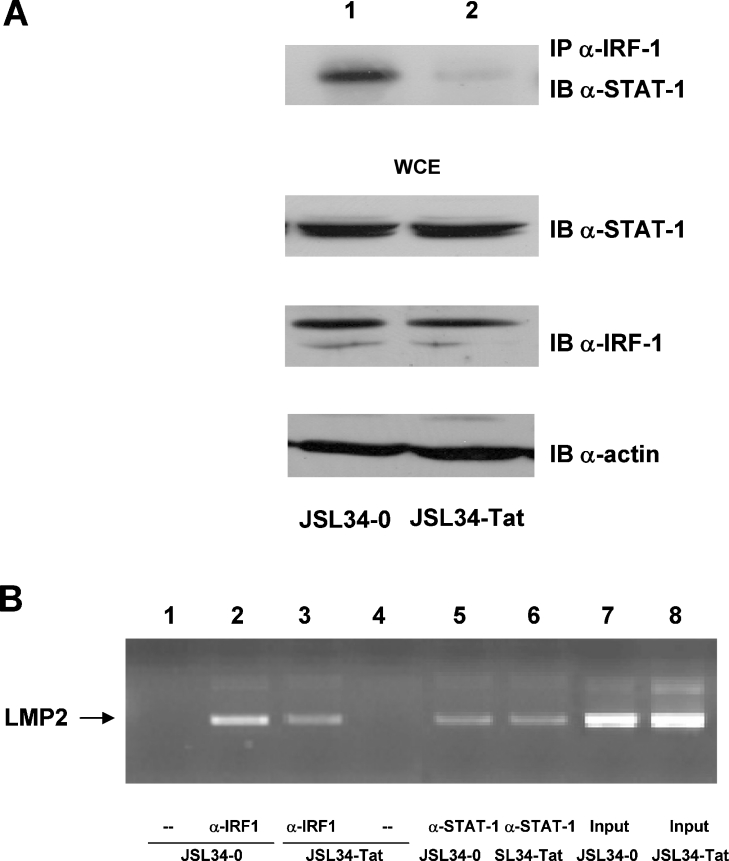
It has been demonstrated previously that IRF-1 and STAT1 form a complex while binding to the LMP2 promoter and also associate in vivo [33]. Similarly, by using in vitro and in vivo systems, we reported previously that IRF-1 and Tat physically interact intracellularly, and form an active transcriptional complex on the HIV-1 LTR promoter [26]. To demonstrate that the expression of Tat in Jurkat T-cells interferes with the formation of the STAT1–IRF-1 complex, co-immunoprecipitation experiments were performed with cell extracts from Tat-expressing and control cells. Anti-IRF-1 antibodies were used for immunoprecipitation followed by immunoblotting with anti-STAT1 antibodies. As shown in Figure 3(A), in cells transfected with the empty vector (lane 1), the STAT1 protein was readily detected in the anti-IRF-1 immunocomplexes, whereas almost undetectable STAT1 levels were present in immunoprecipitates from Tat-expressing cells (lane 2). Equal amounts of the two proteins were present in the cell extracts from both cell types before the immunoprecipitation assay, as demonstrated by immunoblot of the WCEs. These results clearly indicate that Tat does not affect the expression of STAT1 or IRF-1, but interferes with the formation of the STAT1–IRF-1 complex.
Figure 3. Tat impairs the formation of the STAT1–IRF-1 complex.
(A) WCEs (300 μg) from control or Tat-expressing Jurkat T-cells (lanes 1 and 2), were immunoprecipitated (IP) with anti-IRF-1 antibody (α-IRF-1). Immunoprecipitated complexes were separated by SDS/10% PAGE and were subsequently immunoblotted (IB) with anti-STAT1 antibody (α-STAT-1) as indicated. WCEs (30 μg) were separated on SDS/10% PAGE and probed with anti-STAT1 or anti-IRF-1 antibodies. One representative experiment out of three performed is shown. (B) Proteins were cross-linked to DNA in control and Tat-expressing Jurkat T-cells, and DNA-bound IRF-1 and STAT1 were immunoprecipitated. The chromatin fragments were amplified by PCR using primers for LMP2 IRF-E (ISC-2)–GAS (see the Materials and methods section).
Tat inhibits the STAT1–IRF-1 interaction on the LMP2 promoter
To demonstrate the functional relevance of Tat on the formation of the STAT1–IRF-1 complex in vivo, assembly of endogenous factors on the regulatory region of LMP2 was investigated by ChIP. Proteins were cross-linked to DNA, and sonicated chromatin was immunoprecipitated with specific anti-STAT1 and anti-IRF-1 antibodies. Fragments were amplified by PCR using specific oligonucleotide primers flanking the IRF-E (ICS-2)–GAS of the LMP2 promoter and were visualized on agarose gels. As shown in Figure 3(B), the LMP2 IRF-E (ICS-2)–GAS was amplified from DNA co-immunoprecipitated with both anti-IRF-1 and anti-STAT1 antibodies (lanes 2 and 3, and 5 and 6 respectively). However, a reproducible lower level of endogenous IRF-1 binding to the 5′ regulatory region of LMP2 was observed in Tat-expressing cells as compared with control cells (Figure 3B, lane 3 compared with lane 2). Conversely, immunoprecipitation with anti-STAT1 antibodies did not reveal significant differences between Tat-expressing and control cells (Figure 3B, lanes 5 and 6).
Taken together, the results obtained with pull-down, co-immunoprecipitation and ChIP assays demonstrate that the intracellularly expressed Tat effectively impairs the formation of the complex between IRF-1 and STAT1 on the LMP2 promoter, leading to a decreased expression of LMP2 in Tat-expressing cells.
Biphasic regulation of LMP2 gene expression by Tat
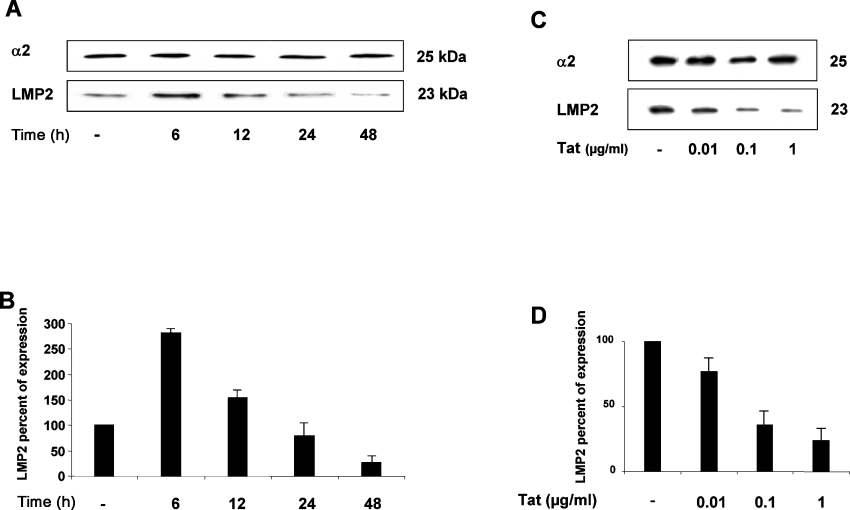
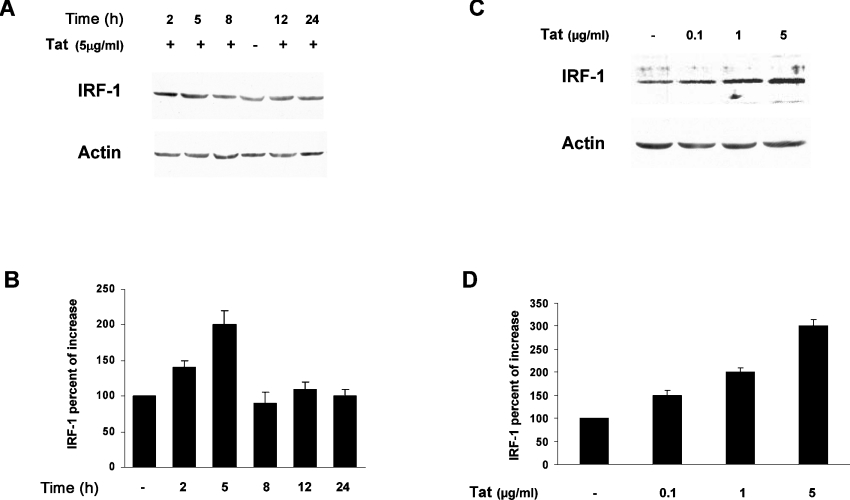
As the HIV-1 Tat protein produced in infected cells is also released extracellularly and is then taken up by the cell [4–6,34], we tested the effect of exogenously added Tat on LMP2 gene expression in kinetic experiments. Jurkat T-cells were cultured in the presence or in the absence of 5 μg/ml of the native Tat protein, and, at different time points, WCEs were collected. LMP2 expression was then determined by immunoblotting with anti-LMP2 antibody (Figure 4A). Surprisingly, at 6 h after Tat addition, an increase in the LMP2 expression of approx. 3-fold was clearly evident. Conversely, starting at 12 h onwards, a progressive decrease in LMP2 expression occurred. Almost undetectable levels of LMP2 were present after 48 h of the addition of Tat (Figure 4B). The inhibitory effect of Tat was dose-dependent as shown in Figures 4(C) and 4(D) at 24 h. These data indicate that Tat affects LMP2 expression with biphasic kinetics.
Figure 4. Kinetics of LMP2 expression in cells treated with Tat protein.
(A) Jurkat T-cells were treated for the indicated times with 5 μg/ml native Tat protein. Equal amounts of proteasomes (1 μg) were fractionated by SDS/10% PAGE, transferred on to nitrocellulose filters and probed with specific antibodies for the α2 and LMP2 subunits. One representative experiment out of three performed is shown. (B) The intensity of specific bands was measured by densitometry and the means±S.D. for three independent experiments is reported as a percentage of LMP2 expression in Tat-treated cells relative to control cells after normalization with α2. (C) Jurkat T-cells were treated for 24 h with increasing doses of Tat. Equal amounts of proteasomes were analysed as in (A). (D) Results are expressed as in (B).
To investigate further this dual effect, we performed a series of experiments aimed to evaluate whether the mechanisms responsible for LMP2 down-regulation in cells constitutively expressing Tat (Figures 1–3) were also responsible for the stimulation of LMP2 observed at early time points after Tat was exogenously added. We first determined, at different time points, expression of IRF-1 in Tat-treated cultures. It has, in fact, been demonstrated that an increase in IRF-1 is responsible for the IFN-γ-induced up-regulation of LMP2 during the cytokine-induced replacements of proteasome subunits [25]. Accordingly, we have found that Jurkat T-cells constitutively expressing IRF-1 show a clear increase in LMP2 protein expression (A. Battistini and R. Gavioli, unpublished work). We therefore asked whether the LMP2 increase observed between 0 and 6 h after Tat addition could be ascribed to a stimulation of IRF-1 expression, which, in turn, could lead to an increased STAT1–IRF-1 complex and LMP2 transcription stimulation. Indeed, as shown in Figure 5(A) starting at 2 h after Tat addition an increase in IRF-1 expression was evident, which peaked at 5 h and thereafter returned to basal levels (Figure 5B). Specifically, the Tat-induced IRF-1 expression was dose-dependent and also occurred at very low doses at 5 h as shown in Figures 5(C) and 5(D).
Figure 5. Kinetics of IRF-1 expression in cells treated with Tat protein.
(A) Jurkat T-cells were treated for the indicated times with 5 μg/ml native Tat protein. WCEs (30 μg) were analysed by immunoblotting with specific anti-IRF-1 antibodies. One representative experiment out of three performed is shown. (B) The intensity of the specific band from three independent experiments was measured by densitometry as the percentage of increases relative to control cells after normalization with actin. Results are means±S.D. (C) Jurkat T-cells were treated for 5 h with increasing doses of Tat, and equal amounts of WCEs were analysed as in (A). (D) Results were quantified and expressed as in (B).
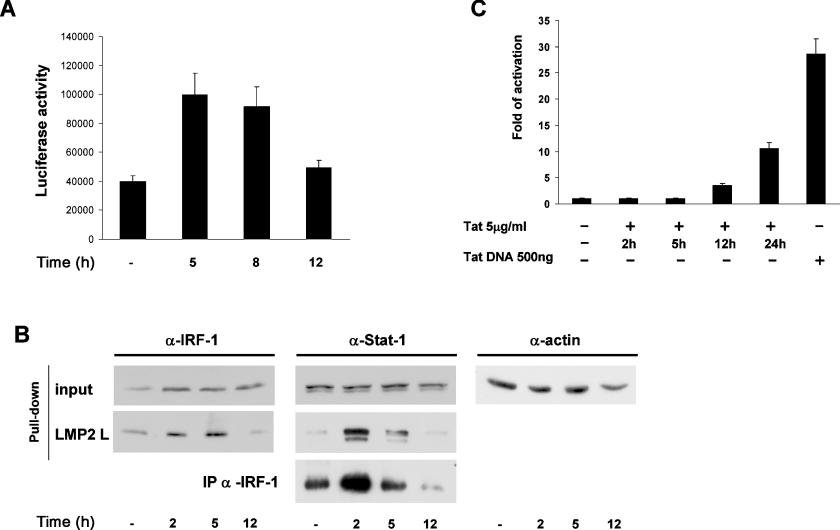
We also measured LMP2 promoter activity after Tat addition on a kinetic base. As shown in Figure 6(A), an increase of approx. 2.5-fold was observed at early time points, but, starting at 8 h onwards, a progressive decrease occurred. Pull-down assays and co-immunoprecipitation experiments with cell extracts purified at different time points after Tat addition (Figure 6B) demonstrated that the stimulatory effect of Tat at early time points was determined by the formation of an increased complex between IRF-1 and STAT1, as assessed by immunoprecipitation, bound to the overlapping IRF-E–GAS element on the LMP2 promoter (pull-down assays).
Figure 6. Biphasic regulation of the LMP2 gene expression by Tat.
(A) The LMP2 promoter linked to the luciferase reporter gene was transiently transfected in Jurkat T-cells. After 24 h, cells were treated with 5 μg/ml native Tat protein, and, at the indicated times, WCEs were processed for luciferase activity. Means±S.D. for three separate experiments were calculated after normalization with the TK–Renilla activity. (B) Biotinylated oligodeoxynucleotides containing the LMP2 IRF-E (ICS-2)–GAS (see the Materials and methods section), coupled to Streptavidin MagneSphere®, were incubated with nuclear extracts from Jurkat T-cells treated for the indicated times with 5 μg/ml native Tat protein. Bound proteins were eluted from the beads by boiling in sample buffer and analysed by Western blotting with specific antibodies against IRF-1 and unphosphorylated STAT1. input indicates the level of endogenous IRF-1 and STAT1 in the WCEs (20 μg). Immunoprecipitation assay: WCEs (300 μg) from control or Tat-treated cells were immunoprecipitated with anti-IRF-1 antibody (α-IRF-1). Immunoprecipitated complexes were separated by SDS/10% PAGE, and subsequently probed with anti-STAT1 antibody (α-Stat-1). One representative experiment of three is shown. (C) HIV-1 LTR construct (1 μg) (see the Materials and methods section) linked to the luciferase reporter gene was transiently transfected in Jurkat T-cells. After 12 h, cells were treated with 5 μg/ml native Tat protein or transfected with 500 ng of a Tat expression vector. At the indicated time points, WCEs were processed for the luciferase activity. Means±S.D. for three separate experiments were calculated after normalization with the TK–Renilla activity.
Considering that internalization of exogenously added Tat is different with respect to cell type, being of a few minutes in DCs [7], but requiring several hours in other cell types [34], we sought to determine the kinetics of cellular internalization of Tat in our experimental model. Since detection of Tat protein by immunological methods is very difficult, we indirectly monitored the internalized Tat by measuring its transactivation potential. For this purpose, Jurkat T-cells were transfected with an HIV-1 LTR–luciferase construct treated with 5 μg/ml Tat, and, at different time points, luciferase activity was measured. As shown in Figure 6(C), until 12 h after Tat addition, no LTR transcription was evident. Starting at 12 h, a progressive increase in LTR transactivation was observed instead, indicating that, in Jurkat T-cells, a discrete and detectable internalization of Tat requires at least 12 h.
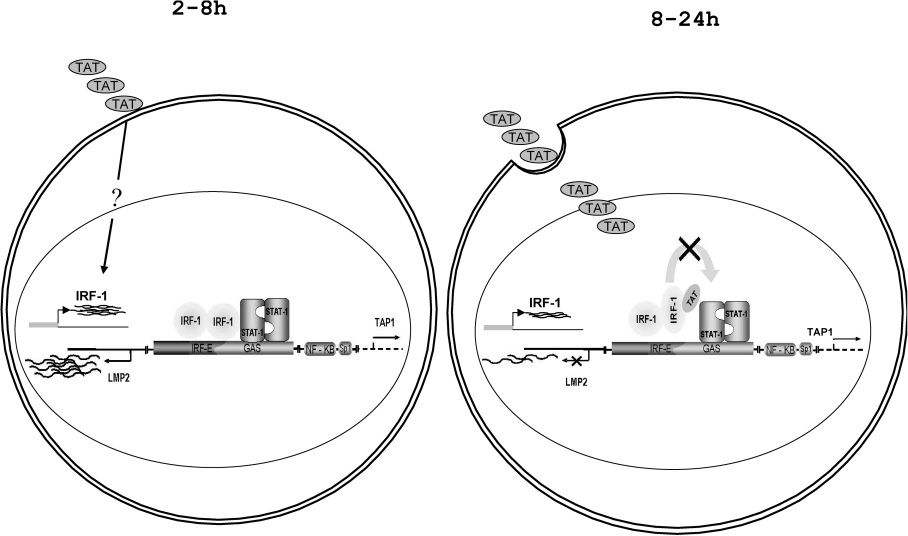
Taken together, these results lead us to conclude that the biphasic effect of Tat on LMP2 expression, i.e. stimulatory at early time points and inhibitory later on, mirrors that of IRF-1 and is dependent on the extracellular compared with intracellular localization of Tat (see also Figure 8).
Figure 8. Schematic representation of the effect of Tat on LMP2 transcription.
The promoter region between the LMP2 and TAP1 genes that regulates the transcription of both genes and the transcription factor binding is shown. A complex between IRF-1 and STAT1 binding a partially overlapping consensus binding site is responsible for both basal and induced LMP2 transcription. Tat exerts a biphasic effect on LMP2 expression: before internalization, a stimulation of IRF-1 expression by an unknown mechanism(s) results in LMP2 stimulation; in a second phase after Tat internalization, Tat competes with STAT1 for binding to IRF-1, impairing the formation of the complex and leading to LMP2 repression.
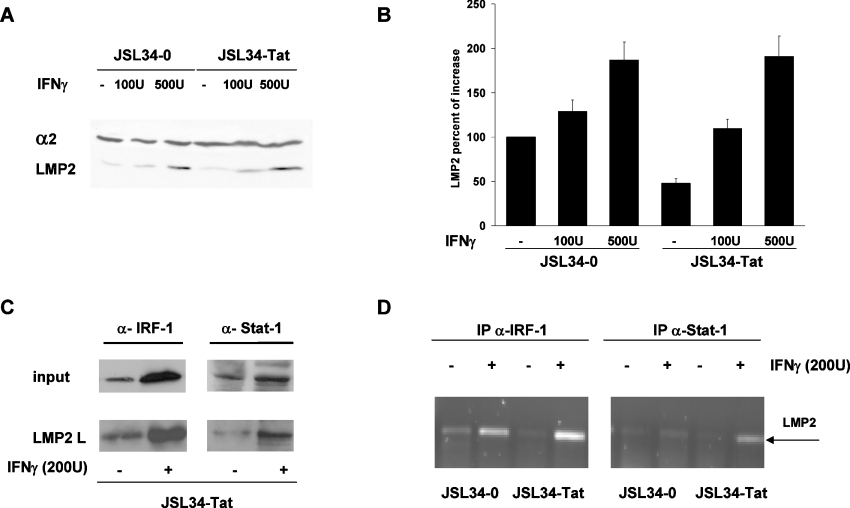
Intracellular Tat does not overcome the IFN-γ-induced stimulation of LMP2 expression
It is known that IFN-γ substantially induces the expression of LMP2 through IRF-1 transcription stimulation [25]. We therefore asked whether intracellular Tat was also able to inhibit the IFN-γ-induced expression of LMP2. Control or Tat-expressing cells were treated with two doses of IFN-γ for 24 h and Western blot analysis of cell extracts was performed. As shown in Figures 7(A) and 7(B), the inhibitory effect of Tat on LMP2 expression was reversed by high doses of IFN-γ, suggesting that Tat was unable to counteract the stimulatory effect of IFN-γ on LMP2 expression. Accordingly, pull-down assays (Figure 7C) showed a substantial increase in the binding of both IRF-1 and STAT1 to the IRF-E (ICS-2)–GAS element on the LMP2 promoter irrespective of Tat expression. This increase was comparable with that observed in control cells (results not shown). The inability of Tat to counteract the strong stimulatory effect of IFN-γ on LMP2 expression was also demonstrated in vivo using ChIP. An increased binding of IRF-1 and STAT1 to the LMP2 promoter in IFN-γ-treated cells was, in fact, observed irrespective of Tat expression (Figure 7D).
Figure 7. IFN-γ overcomes the inhibitory effect of Tat on LMP2 expression.
(A) Equal amounts of proteasomes (1 μg) were purified from JSL34-0 and JSL34-Tat cells untreated or treated with the indicated amount of IFN-γ for 24 h, fractionated by SDS/10% PAGE, transferred on to nitrocellulose filters, and probed with specific antibodies for the α2 and LMP2 subunits. (B) The intensity of the specific band from three independent experiments was measured by densitometry as the percentage increase relative to control cells after normalization with α2. Results are means±S.D. (C) JSL34-Tat cells were treated with 200 units of IFN-γ for 4 h and WCEs were subjected to DNA pull-down assays with the biotinylated oligodeoxynucleotide containing the IRF-E (ICS-2)–GAS sequence on the LMP2 promoter. Bound proteins were analysed as in Figure 2. (D) JSL34-0 and JSL34-Tat cells where treated with IFN-γ as indicated and subjected to ChIP as described in the Materials and methods section. Proteins were cross-linked to DNA, and DNA-bound IRF-1 and STAT1 were immunoprecipitated. Samples were amplified by PCR using primers for the LMP2 IRF-E (ICS-2)–GAS. U, units.
DISCUSSION
Perturbation of the proteasome system by viral infection, cell transformation or pharmacological treatments is often a key event in the modulation of the immune response [29,33,35–37]. The proteasome, in fact, plays a critical role in the degradation of intracellular proteins and in the generation of the majority of antigenic peptides presented by the MHC class I complex [8]. It is therefore not surprising that many intracellular pathogens, including viruses, have developed strategies to alter its functions [38]. We showed previously that the HIV-1 Tat protein modifies proteasome composition by up-regulating the LMP7 and MECL1 subunits and down-regulating the LMP2 subunit, leading to increased presentation of cryptic and subdominants CTL epitopes [7]. Manipulation of the 26 S proteasome is one of the intracellular activities of Tat, and direct interaction of Tat with components of the proteasome has been described previously [39]. Specifically, Tat interacts with both LMP7 and MECL-1 subunits of the proteasome [36] and inhibits the proteolytic activity of the 20 S proteasome by competing with PA28 regulator for binding to 20 S [40]. In the present study, we have identified another mechanism that is exploited by intracellular Tat to interfere with the proteasome activity, demonstrating that Tat decreases the LMP2 gene expression at the transcriptional level by impairing the formation of the STAT1–IRF-1 complex, which is crucial to LMP2 gene transcription. Transcription of the LMP2 gene is regulated by a bidirectional promoter, which also controls transcription of the TAP1 gene [14]. Despite the co-ordinated regulation of human TAP1 and LMP2 in several cell lines [14], the requirement of transcription factors for the transcription of the two genes is, however, different. It has, in fact, been reported that the binding of IRF-1 or STAT1 to the composite element IRF-E–GAS in the LMP2 promoter is sufficient for the regulation of TAP1; the binding of both factors is instead required for LMP2 basal transcription [15]. In the present study, we show that Tat-mediated repression of LMP2 gene expression is dependent upon the specific DNA sequence element containing the overlapping IRF-E–GAS element, which binds the STAT1–IRF-1 complex. Mutations in the IRF-1-binding site, in fact, not only reduced constitutive activity of the LMP2 promoter, but also completely abolished the effect of Tat (Figure 1C). The absolute requirement of IRF-1 binding to its consensus sequence for both basal and cytokine-induced transcription has been defined previously and is consistent with the extremely low LMP2 gene expression in IRF-1−/− mice [25]. Of note, the effect of Tat on LMP2 expression resembles that of IRF-1 deficiency; however, the effect of Tat does not decrease IRF-1 expression, since WCEs from control or Tat-expressing cells contain the same amount of IRF-1 (Figure 2). Instead, Tat interferes with the formation of the STAT1–IRF-1 complex as demonstrated using immunoprecipitation experiments (Figure 3) where anti-IRF-1 antibodies are no longer effective in immunoprecipitating STAT1 in Tat-expressing cells. The impairment in the formation of the STAT1–IRF-1 complex is due to the capacity of Tat and IRF-1 to interact physically as we demonstrated previously and depends upon the C-terminal region of IRF-1 [26]. Of note, the same region of IRF-1 is also involved in the binding with STAT1 [34], therefore Tat can interfere directly with the STAT1–IRF-1 interaction by occupying the C-terminus of IRF-1. The impairment in the formation of the STAT1–IRF-1 complex, in Tat-expressing cells, then results in a substantial reduction in the amount of IRF-1 bound to the LMP2 promoter, as demonstrated both in DNA pull-down assays (Figure 2) and in vivo in ChIP assays (Figure 3), and leads to reduced transcription of the gene. Intriguingly, the Tat-induced LMP2 down-regulation is not effective in IFN-γ-treated cells (Figure 7). IFN-γ induces LMP2 through a substantial induction of IRF-1 expression and STAT1 phosphorylation, which, in turn, also contributes to increase further IRF-1 amounts [20]. So, from a mechanistic point of view, we can speculate that phosphorylated STAT1 and/or higher amounts of IRF-1 form a more stable/abundant complex, thereby impairing the Tat–IRF-1 binding, or hindering the Tat effect.
During a productive infection, Tat is released by infected cells and is efficiently taken up by neighbouring cells. In this way, Tat can affect both infected and non-infected cells. Interestingly, in the present paper, we report that, when Tat is added exogenously, early on and before internalization (Figure 4), induction of LMP2 occurs instead of repression, and this effect is mediated directly by a stimulation of IRF-1 expression (Figure 5). Results in Figure 6, indeed, demonstrate that the Tat-stimulated IRF-1 is fully functional in that it increasingly binds to its consensus sequence and stimulates the LMP2 promoter. Even if standard ChIP analysis is not suitable for the detection of small variations in transcription factor binding, pull-down and luciferase assays clearly indicate that the increase in IRF-1 observed early after Tat addition is sufficient to stimulate LMP2 transcription before Tat internalization. Although the biological significance of IRF-1 stimulation by exogenous Tat has not yet been defined, we suggest that this early effect of Tat released extracellularly can be regarded as a mechanism of Tat-increased HIV-1 infectivity. Accordingly, we reported previously that, during the early phases of HIV-1 infection, IRF-1 is able to induce LTR transcription before expression of Tat [26]. IRF-1 stimulation in non-infected cells by extracellular released Tat can, therefore, contribute to the T-cell activation necessary to allow HIV-1 replication both in de novo infected cells and during reactivation from latency. In this respect, even if, as suggested in [41], the full biological implications of circulating Tat protein cannot be entirely determined until more reliable methods to measure amounts of viral protein in vivo are developed, in our experiments, the lowest effective dose of the protein (Figures 4C and 5C) is close to those measured in the plasma of some infected patients [41].
In the context of HIV-1 infection, we demonstrated previously that the Tat–IRF-1 interaction resulted in co-operation between the two transcription factors in inducing HIV-1 gene transcription [26]. In the present study, we identified a new output of this interaction, which targets a cellular gene, leading to its transcriptional repression. Interestingly, a mechanism of LMP2 down-regulation, similar to that described in the present paper, is exerted by the adenovirus early protein E1A, which interferes with the formation of the STAT1–IRF-1 complex by occupying domains of STAT1 that bind to IRF-1 [33]. In a specular fashion, we report that Tat interferes with the formation of the STAT1–IRF-1 complex by occupying the domain of IRF-1 that is involved in STAT1 binding. Sequestration of transcription factors on a cellular promoter can therefore be regarded as another mechanism exploited by Tat to affect the host immune response (Figure 8).
In this respect, we have shown previously that the Tat protein can be successfully used in vaccination strategies in that it protects monkeys against challenge with pathogenic SHIV (simian immunodeficiency virus–HIV chimaera), and this protection correlates with the Th1 response and CTL activity [42]. Therefore, when considering the use of Tat in vaccine strategies, since IRF-1 is a potent inducer of a Th1 response [24], it could be regarded as an effector of Tat activities. In addition, and more intriguingly, IRF-1 could itself act as a genetic adjuvant, as reported for some model antigens [43]. In a second phase, when Tat enters the cells, the sequestration of IRF-1 and the repression of LMP2 expression would result in a variation in the proteolytic activity of the proteasome with a variation in the presentation of immunodominant compared with subdominant epitopes, as described previously [7]. Changes in immunodominance may be particularly relevant since an increase in the presentation of subdominant epitopes may be beneficial for the elimination of virally infected cells. Indeed, it is well established that immunodominant epitopes are very prone to mutation and to viral escape, while subdominant epitopes are more stable and they may induce protection [44].
In conclusion, our results provide a logical base to explain some of the events through which Tat leads to a modulation of the cell transcriptome. Tat as a multifaceted protein is an important factor in the complex pathogenesis of AIDS; however, the biological implications of the present study, together with the reported efficacy of the use of Tat both as antigen and as adjuvant in vaccination strategies, can be regarded as a bright way of hijacking a key virus regulator to turn its activity to the host's advantage.
Acknowledgments
We thank Sabrina Tocchio for secretarial and editorial assistance and Roberto Gilardi for preparing graphs. This work was supported by Institutional grants, Italian grants from the ISS (Istituto Superiore di Sanità)/NIH (National Institutes of Health) and AIDS Project and from The Italian Ministry of Health to A.B. and B.E., and AIDS Project and Associazione Italiana per la Ricerca sul Cancro to R.G.E.G. is supported by a Ph.D. fellowship awarded by Ferrara University.
References
- 1.Wu Y., Marsh J. W. Selective transcription and modulation of resting T cell activity by preintegrated HIV DNA. Science. 2001;293:1503–1506. doi: 10.1126/science.1061548. [DOI] [PubMed] [Google Scholar]
- 2.Jeang K.-T., Gatignol A. Comparison of regulatory features among primate lentiviruses. Curr. Top. Microbiol. Immunol. 1994;188:123–144. doi: 10.1007/978-3-642-78536-8_7. [DOI] [PubMed] [Google Scholar]
- 3.Chang H.-K., Gallo R. C., Ensoli B. Regulation of cellular gene expression and function by the human immunodeficiency virus type 1 Tat protein. J. Biomed. Sci. 1995;2:189–202. doi: 10.1007/BF02253380. [DOI] [PubMed] [Google Scholar]
- 4.Huigen M. C., Kamp W., Nottet H. S. Multiple effects of HIV-1 trans-activator protein on the pathogenesis of HIV-1 infection. Eur. J. Clin. Invest. 2004;34:57–66. doi: 10.1111/j.1365-2362.2004.01282.x. [DOI] [PubMed] [Google Scholar]
- 5.Ensoli B., Buonaguro L., Barillari G., Fiorelli V., Gendelman R., Morgan R. A., Wingfield P., Gallo R. C. Release, uptake, and effects of extracellular human immunodeficiency virus type 1 Tat protein on cell growth and viral transactivation. J. Virol. 1993;67:277–287. doi: 10.1128/jvi.67.1.277-287.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fanales-Belasio E., Moretti S., Nappi F., Bacillari G., Micheletti F., Cafaro A., Ensoli B. Native HIV-1 Tat protein targets monocyte-derived dendritic cells and enhances their maturation, function, and antigen-specific T cell responses. J. Immunol. 2002;168:197–206. doi: 10.4049/jimmunol.168.1.197. [DOI] [PubMed] [Google Scholar]
- 7.Gavioli R., Gallerani E., Fortini C., Fabris M., Bottoni A., Canella A., Bonaccorsi A., Marastoni M., Micheletti F., Cafaro A., et al. HIV-1 tat protein modulates the generation of cytotoxic T cell epitopes by modifying proteasome composition and enzymatic activity. J. Immunol. 2004;173:3838–3843. doi: 10.4049/jimmunol.173.6.3838. [DOI] [PubMed] [Google Scholar]
- 8.Rock K. L., Goldberg A. L. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu. Rev. Immunol. 1999;17:739–779. doi: 10.1146/annurev.immunol.17.1.739. [DOI] [PubMed] [Google Scholar]
- 9.Sijts A. J., Standera S., Toes R. E., Ruppert T., Beekman N. J., van Veelen P. A., Ossendorp F. A., Melief C. J., Kloetzel P. M. MHC class I antigen processing of an adenovirus CTL epitope is linked to the levels of immunoproteasomes in infected cells. J. Immunol. 2000;164:4500–4506. doi: 10.4049/jimmunol.164.9.4500. [DOI] [PubMed] [Google Scholar]
- 10.Morel S., Levy F., Burlet-Schiltz O., Brasseur F., Probst-Kepper M., Peitrequin A. L., Monsarrat B., van Velthoven R., Cerottini J. C., Boon T., et al. Processing of some antigens by the standard proteasome but not by the immunoproteasome results in poor presentation by dendritic cells. Immunity. 2000;12:107–117. doi: 10.1016/s1074-7613(00)80163-6. [DOI] [PubMed] [Google Scholar]
- 11.Van den Eynde B. J., Morel S. Differential processing of class-I-restricted epitopes by the standard proteasome and the immunoproteasome. Curr. Opin. Immunol. 2001;13:147–153. doi: 10.1016/s0952-7915(00)00197-7. [DOI] [PubMed] [Google Scholar]
- 12.Schmidtke G., Eggers M., Ruppert T., Groettrup M., Koszinowski U. H., Kloetzel P. M. Inactivation of a defined active site in the mouse 20S proteasome complex enhances major histocompatibility complex class I antigen presentation of a murine cytomegalovirus protein. J. Exp. Med. 1998;187:1641–1646. doi: 10.1084/jem.187.10.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen W., Norbury C. C., Cho Y., Yewdell J. W., Bennink J. R. Immunoproteasomes shape immunodominance hierarchies of antiviral CD8+ T cells at the levels of T cell repertoire and presentation of viral antigens. J. Exp. Med. 2001;193:1319–1326. doi: 10.1084/jem.193.11.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wright K. L., White L. C., Kelly A., Beck S., Trowsdale J., Ting J. P. Coordinate regulation of the human TAP1 and LMP2 genes from a shared bidirectional promoter. J. Exp. Med. 1995;181:1459–1471. doi: 10.1084/jem.181.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chatterjee-Kishore M., Kishore R., Hicklin D. J., Marincola F. M., Ferrone S. Different requirements for signal transducer and activator of transcription 1α and interferon regulatory factor 1 in the regulation of low molecular mass polypeptide 2 and transporter associated with antigen processing 1 gene expression. J. Biol. Chem. 1998;273:16177–16183. doi: 10.1074/jbc.273.26.16177. [DOI] [PubMed] [Google Scholar]
- 16.Chatterjee-Kishore M., Wright K. L., Ting J. P., Stark G. R. How Stat1 mediates constitutive gene expression: a complex of unphosphorylated Stat1 and IRF1 supports transcription of the LMP2 gene. EMBO J. 2000;19:4111–4122. doi: 10.1093/emboj/19.15.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calo V., Migliavacca M., Bazan V., Macaluso M., Buscami M., Gebbia N., Russo A. STAT proteins: from normal control of cellular events to tumorigenesis. J. Cell. Physiol. 2003;197:157–168. doi: 10.1002/jcp.10364. [DOI] [PubMed] [Google Scholar]
- 18.Bach E. A., Aguet M., Schreiber R. D. The IFNγ receptor: a paradigm for cytokine receptor signalling. Annu. Rev. Immunol. 1997;15:563–591. doi: 10.1146/annurev.immunol.15.1.563. [DOI] [PubMed] [Google Scholar]
- 19.Pine R., Canova A., Schindler C. Tyrosine phosphorylated p91 binds to a single element in the ISGF2/IRF-1 promoter to mediate induction by IFNα and IFNγ, and is likely to autoregulate the p91 gene. EMBO J. 1994;13:158–167. doi: 10.1002/j.1460-2075.1994.tb06245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujita T., Sakakibara J., Sudo Y., Miyamoto M., Kimura Y., Taniguchi T. Evidence for a nuclear factor(s), IRF-1, mediating induction and silencing properties to human IFN-β gene regulatory elements. EMBO J. 1988;7:3397–3405. doi: 10.1002/j.1460-2075.1988.tb03213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taniguchi T., Ogasawara K., Takaoka A., Tanaka N. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 2001;19:623–655. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- 22.Matsuyama T., Kimura T., Kitagawa M., Pfeffer K., Kawakami T., Watanabe N., Kundig T. M., Amakawa R., Kishihara K., Wakeham A., et al. Targeted disruption of IRF-1 or IRF-2 results in abnormal type I IFN gene induction and aberrant lymphocyte development. Cell. 1993;75:83–97. [PubMed] [Google Scholar]
- 23.Ogasawara K., Hida S., Azimi N., Tagaya Y., Sato T., Yokochi-Fukuda T., Waldmann T. A., Taniguchi T., Taki S. Requirement for IRF-1 in the microenvironment supporting development of natural killer cells. Nature (London) 1998;391:700–703. doi: 10.1038/35636. [DOI] [PubMed] [Google Scholar]
- 24.Lohoff M., Ferrick D., Mittrucker H. W., Duncan G. S., Bischof S., Rollinghoff M., Mak T. W. Interferon regulatory factor-1 is required for a T helper 1 immune response in vivo. Immunity. 1997;6:681–689. doi: 10.1016/s1074-7613(00)80444-6. [DOI] [PubMed] [Google Scholar]
- 25.White L. C., Wright K. L., Felix N. J., Ruffner H., Reis L. F., Pine R., Ting J. P. Regulation of LMP2 and TAP1 genes by IRF-1 explains the paucity of CD8+ T cells in IRF-1−/− mice. Immunity. 1996;5:365–376. doi: 10.1016/s1074-7613(00)80262-9. [DOI] [PubMed] [Google Scholar]
- 26.Sgarbanti M., Borsetti A., Moscufo N., Bellocci M. C., Ridolfi B., Nappi F., Marsili G., Marziali G., Coccia E. M., Ensoli B., Battistini A. Modulation of human immunodeficiency virus 1 replication by interferon regulatory factors. J. Exp. Med. 2002;195:1359–1370. doi: 10.1084/jem.20010753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Battistini A., Marsili G., Sgarbanti M., Ensoli B., Hiscott J. IRF regulation of HIV-1 long terminal repeat activity. J. Interferon Cytokine Res. 2002;22:27–37. doi: 10.1089/107999002753452638. [DOI] [PubMed] [Google Scholar]
- 28.Caputo A., Sodroski J. G., Haseltine W. A. Constitutive expression of HIV-1 tat protein in human Jurkat T cells using a BK virus vector. J. Acquired Immune Defic. Syndr. 1990;3:372–379. [PubMed] [Google Scholar]
- 29.Gavioli R., Frisan T., Vertuani S., Bornkamm G. W., Masucci M. G. c-myc overexpression activates alternative pathways for intracellular proteolysis in lymphoma cells. Nat. Cell Biol. 2001;3:283–288. doi: 10.1038/35060076. [DOI] [PubMed] [Google Scholar]
- 30.Dovhey S. E., Ghosh N. S., Wright K. L. Loss of interferon-γ inducibility of TAP1 and LMP2 in a renal cell carcinoma cell line. Cancer Res. 2000;60:5789–5796. [PubMed] [Google Scholar]
- 31.Marsili G., Remoli A. L., Sgarbanti M., Battistini A. Role of acetylases and deacetylase inhibitors in IRF-1-mediated HIV-1 long terminal repeat transcription. Ann. N. Y. Acad. Sci. 2004;1030:636–643. doi: 10.1196/annals.1329.074. [DOI] [PubMed] [Google Scholar]
- 32.Weinmann A. S., Bartley S. M., Zhang T., Zhang M. Q., Farnham P. J. Use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol. Cell. Biol. 2001;21:6820–6832. doi: 10.1128/MCB.21.20.6820-6832.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chatterjee-Kishore M., van Den Akker F., Stark G. R. Adenovirus E1A down-regulates LMP2 transcription by interfering with the binding of stat1 to IRF1. J. Biol. Chem. 2000;275:20406–20411. doi: 10.1074/jbc.M001861200. [DOI] [PubMed] [Google Scholar]
- 34.Fittipaldi A., Ferrari A., Zoppe M., Arcangeli C., Pellegrini V., Beltram F., Giacca M. Cell membrane lipid rafts mediate caveolar endocytosis of HIV-1 Tat fusion proteins. J. Biol. Chem. 2003;278:34141–34149. doi: 10.1074/jbc.M303045200. [DOI] [PubMed] [Google Scholar]
- 35.Turnell A. S., Grand R. J., Gorbea C., Zhang X., Wang W., Mymryk J. S., Gallimore P. H. Regulation of the 26S proteasome by adenovirus E1A. EMBO J. 2000;19:4759–4773. doi: 10.1093/emboj/19.17.4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andre P., Groettrup M., Klenerman P., de Giuli R., Booth B. L., Jr, Cerundolo V., Bonneville M., Jotereau F., Zinkernagel R. M., Lotteau V. An inhibitor of HIV-1 protease modulates proteasome activity, antigen presentation, and T cell responses. Proc. Natl. Acad. Sci. U.S.A. 1998;95:13120–13124. doi: 10.1073/pnas.95.22.13120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu Z., Zhang Z., Doo E., Coux O., Goldberg A. L., Liang T. J. Hepatitis B virus X protein is both substrate and a potential inhibitor of the proteasome complex. J. Virol. 1999;73:7231–7240. doi: 10.1128/jvi.73.9.7231-7240.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ploegh H. L. Viral strategies of immune evasion. Science. 1998;280:248–253. doi: 10.1126/science.280.5361.248. [DOI] [PubMed] [Google Scholar]
- 39.Huang X., Seifert U., Salzmann U., Henklein P., Preissner R., Henke W., Sijts A. J., Kloetzel P. M., Dubiel W. The RTP site shared by the HIV-1 Tat protein and the 11S regulator subunit alpha is crucial for their effects on proteasome function including antigen processing. J. Mol. Biol. 2002;323:771–782. doi: 10.1016/s0022-2836(02)00998-1. [DOI] [PubMed] [Google Scholar]
- 40.Apcher G. S., Heink S., Zantopf D., Kloetzel P. M., Schmid H. P., Mayer R. J., Kruger E. Human immunodeficiency virus-1 Tat protein interacts with distinct proteasomal α and β subunits. FEBS Lett. 2003;553:200–204. doi: 10.1016/s0014-5793(03)01025-1. [DOI] [PubMed] [Google Scholar]
- 41.Xiao H., Neuveut C., Tiffany H. L., Benkirane M., Rich E. A., Murphy P. M., Jeang K. T. Selective CXCR4 antagonism by Tat: implications for in vivo expansion of coreceptor use by HIV-1. Proc. Natl. Acad. Sci. U.S.A. 2000;97:11466–1171. doi: 10.1073/pnas.97.21.11466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cafaro A., Titti F., Fracasso C., Maggiorella M. T., Baroncelli S., Caputo A., Goletti D., Borsetti A., Pace M., Fanales-Belasio E., et al. Vaccination with DNA containing tat coding sequences and unmethylated CpG motifs protects cynomolgus monkeys upon infection with simian/human immunodeficiency virus (SHIV89.6P) Vaccine. 2001;19:2862–2877. doi: 10.1016/s0264-410x(01)00002-0. [DOI] [PubMed] [Google Scholar]
- 43.Sasaki S., Amara R. R., Yeow W. S., Pitha P. M., Robinson H. L. Regulation of DNA-raised immune responses by cotransfected interferon regulatory factors. J. Virol. 2002;76:6652–6659. doi: 10.1128/JVI.76.13.6652-6659.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feltkamp M. C., Vreugdenhil G. R., Vierboom M. P., Ras E., van der Burg S. H., Ter S. J., Melief C. J., Kast W. M. Cytotoxic T lymphocytes raised against a subdominant epitope offered as a synthetic peptide eradicate human papillomavirus type 16-induced tumors. Eur. J. Immunol. 1995;25:2638–2642. doi: 10.1002/eji.1830250935. [DOI] [PubMed] [Google Scholar]