Abstract
NHE3 (Na+/H+ exchanger 3) is essential for Na+ absorption in the ileum and is expressed in a cell-specific manner in the apical membrane of the intestinal epithelial cells. In the present study, we report the stimulatory effect of PMA on the hNHE3 (human NHE3) transcription. Pretreatment with actinomycin D or cycloheximide blocked the up-regulation of the NHE3 mRNA by PMA, indicating that the increased level of NHE3 mRNA expression is regulated by transcriptional activation and is dependent on de novo protein synthesis. 5′-Deletion of the promoter region and transfection analysis in C2BBe1 cells revealed that the PMA effect is mediated through a GC-rich DNA region between nt −88 and −69. Gel mobility-shift assays demonstrated that in nuclear extracts from C2BBe1 cells grown under the basal growth conditions, Sp1 (stimulating protein-1) and Sp3 interact with this GC-rich DNA region, while, in PMA-treated nuclear extracts, PMA-induced EGR-1 (early growth response gene product 1) transcription factor binds to the same site. Binding of EGR-1 diminished the Sp1 and Sp3 interactions with this promoter region significantly. Co-transfection of Sp1 or Sp3 into SL2 cells activated the NHE3-reporter constructs, suggesting that Sp1 and Sp3 act as positive regulators of the NHE3 expression. In addition, overexpression of EGR-1 was sufficient to transactivate the NHE3-reporter gene activity, and knockdown of EGR-1 with gene-specific small interfering RNA resulted in inhibition of the PMA-induced up-regulation of the endogenous NHE3 mRNA expression. Furthermore, the PKC (protein kinase C) inhibitor chelerythrine chloride did not affect PMA-induced NHE3 promoter activity, suggesting that PMA stimulation of the hNHE3 gene expression may be PKC-independent.
Keywords: C2BBe1 cells, early growth response gene product 1 (EGR-1), Na+/H+ exchanger, PMA, small interfering RNA, stimulating protein-1 (Sp1)
Abbreviations: ADA, adenosine deaminase; EGR-1, early growth response gene product 1; FBS, foetal bovine serum; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GMSA, gel mobility-shift assay; NHE, Na+/H+ exchanger; hNHE, human NHE; PDGF, platelet-derived growth factor; PKC, protein kinase C; RT, reverse transcriptase; siRNA, small interfering RNA; Sp1, stimulating protein-1
INTRODUCTION
A total of nine isoforms of NHEs (Na+/H+ exchangers) (NHE1–NHE9) have been identified to date. Functionally, these proteins have been shown to be involved in pH homoeostasis, trans-epithelial ion and water absorption, cell proliferation and differentiation, as well as cell volume regulation [1–3]. The NHE1 is ubiquitously expressed and its protein product localized to the basolateral membrane of polarized epithelial cells. The NHE2 and NHE3 isoforms are localized to the apical surface of intestinal and kidney epithelial cells and are involved in vectorial Na+ absorption (for reviews, see [4–6]). NHE1, NHE2 and NHE3 have been the most characterized isoforms and shown to be expressed in the mammalian intestine. The role for the two apical exchangers seemingly involved in Na+ absorption is intriguing, but has not yet been fully defined. Studies in the rat and rabbit have suggested that the NHE2 isoform may be involved in basal Na+ transport, while the NHE3 isoform may be the predominant sodium-absorbing isoform responsive to regulatory signals. Although considerable information on regulation of NHEs by protein kinases, accessory factors and cytoskeleton has been published [7–9], until recently very little information was available on transcriptional regulation of the NHE isoforms, especially the NHE2 and NHE3 isoforms. Now, evidence is slowly emerging to indicate that transcriptional regulation of these isoforms may play an important role in intestinal physiology and pathophysiology. Earlier studies extensively focused on transcriptional regulation of NHE1. For example, chronic acidosis, hyperplastic agonists such as serum, PDGF (platelet-derived growth factor) and fibroblast growth factors have been shown to be involved in transcriptional regulation of NHE1 [10–12]. Previous studies have shown a stimulatory effect for sodium butyrate in the NHE3 mRNA and protein expression [13], and promoter activity [14]. Glucocorticoid administration has been shown to increase the levels of mRNA for NHE3 without changing the levels for NHE1 and NHE2 [15].
Our studies have focused on defining the factors that are involved in regulating the expression of the hNHE2 (human NHE2) and hNHE3 genes [16,17]. We recently cloned the hNHE3 promoter and analysed the promoter activity by utilizing the luciferase reporter gene [17]. To gain further understanding of the mechanisms involved in the transcriptional regulation of the hNHE3 gene, we report in the present study the regulation of the NHE3 promoter activity in control and PMA-stimulated C2BBe1 cells.
EXPERIMENTAL
Materials
All chemicals including PMA, chelerythrine chloride, 4α-PMA, actinomycin D and cycloheximide were purchased from Sigma (St. Louis, MO, U.S.A.) or Fisher Scientific (Pittsburgh, PA, U.S.A.); restriction endonucleases and other modifying enzymes were obtained from either New England Biolabs (Beverly, MA, U.S.A.), Gibco BRL (Gaithersburg, MD, U.S.A.) or Promega (Madison, WI, U.S.A.); polyclonal anti-human Sp1 (stimulating protein-1), Sp2, Sp3 and EGR-1 (early growth response gene product 1) antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA, U.S.A.); JM109 competent cells and the luciferase assay system were from Promega. [γ-32P]ATP (3000 Ci/mmol) was from Amersham (Arlington Heights, IL, U.S.A.).
Molecular techniques
DNA manipulations, including restriction-enzyme digestion, ligation, plasmid isolation and transformation, were carried out by standard methods [18].
RNA extraction and RT (reverse transcriptase)–PCR analysis
Total RNA was isolated by the RNazol (Tel-Test, Friendswood, TX, U.S.A.) according to the manufacturer's instructions. Semi-quantitative RT–PCR was performed using 5 μg of total RNA, which was reverse-transcribed in a 20 μl reaction mixture containing 100 ng of oligo(dT) using a SuperScript II RT cDNA synthesis kit (Invitrogen, Carlsbad, CA, U.S.A.) as recommended by the supplier. After incubation for 60 min at 42 °C and 10 min at 75 °C, reverse-transcription products were stored at −20 °C until needed. PCR amplifications were performed with 2 μl of the reverse-transcription products as templates and gene-specific primers (forward primer, 5′-CAGACCTGGCTTCTGAACCGC-3′, and reverse primer, 5′-CTCAGCCACGTAGCTGATGGCATCC-3′) for hNHE3 gene [19]. As a control, GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA was co-amplified to determine the accuracy of the amount of cDNA in each sample. The amplification cycles were 17 cycles with only hNHE3 primers, at which time GAPDH primers were added to the reaction mixtures and amplification continued for an additional 16 cycles. These numbers of cycles were determined to be within the exponential range. PCR was performed in a PerkinElmer/Cetus DNA cycler, with thermostable DNA polymerase rTth (Boehringer Mannheim, Indianapolis, IN, U.S.A.) with an initial denaturation period of 90 s at 95 °C, followed by amplification cycles at 94 °C 30 s, 56 °C 30 s and 68 °C 45 s with a final elongation period for 4 min at 68 °C. One-tenth volume of the PCR reactions was resolved on a 1.5% (w/v) agarose gel with ethidium bromide staining and photographed.
Reporter plasmid construction
Plasmids used for functional analysis of the NHE3 promoter activity were generated using pGL2-Basic (Promega) that contains a promoter-less luciferase reporter gene and have been described previously [17]. Three 5′-deletion constructs of p−95/+5, plasmids p−88/+5, p−76/+5 and p−69/+5 were generated by PCR amplifications using p−319/+131 as a template and the forward primers 5′-GAACCTCGAGCGGCGGGGGCGGGCAGGC-3, 5′-GAACCTCGAGGCAGGCTCCGCCCCGG-3′ and 5′-GAACTCGAGTCCGCCCCGGGGCGGGAG-3′ for deletions to positions −88, −76 and −69 respectively and a common reverse primer 5′-GAACAAGCTTGTACCGGCTACAGTCCG-3′. For subcloning purposes, the forward primers contained nucleotide recognition site for restriction enzyme XhoI and the reverse primer harboured a HindIII restriction site (shown in boldface). After PCR amplifications, the amplicons were digested with restriction enzymes XhoI and HindIII, gel-purified and cloned in pGL2-Basic vector digested with the same enzymes. The new clones were sequenced to rule out the presence of PCR-introduced artefacts.
Cell culture and transfections
C2BBe1 cell line, a subclone of the Caco-2 cells, was cultured and maintained as described in [16]. For transfection studies, cells (1.5×105) were seeded into 12-well plates and co-transfected the next day (80–90% confluent) with NHE3-reporter constructs and pSV-βgal using Lipofectamine™-2000 reagent (Invitrogen). The latter plasmid served as an internal control for transfection efficiency. A total of 2.0 μg of DNA/well, at a ratio of 4:1 for experimental versus pSV-βgal, was used for each transfection. After cells were incubated for 4 h with the DNA/transfection mixture, the media were replaced with complete media, and 48 h post-transfection, cell lysates were prepared and assayed for luciferase and β-galactosidase activity using a kit from Promega. Luciferase activity was assayed using TD 20/20 luminometer (Promega) and normalized to β-galactosidase activity. For EGR-1 co-transfection experiments, C2BBe1 cells were transfected with 1 μg of p−95/+5 or p−319/+131 NHE3 promoter-reporter constructs, 10 ng of pTK-RL (Promega) as an internal control and 0.25–1.0 μg of pAC-hEGR-1 expression vector. The total transfected DNA concentration was maintained constant with an empty vector. The firefly luciferase activity was assayed with a Dual Luciferase Assay system (Promega) in a TD 20/20 luminometer and normalized to Renilla luciferase activity. For PMA treatments, after transfection cells were placed in serum-reduced media [0.5% FBS (foetal bovine serum)] for 24 h prior to addition of PMA (100 nM) for 16 h, and 48 h post-transfection, cells were processed for enzymatic assays as described above. Control cells were kept in the serum-reduced media for the duration of the experiment. Addition of the vehicle (DMSO) at concentrations carried over by the treatments (1:100000 dilution) did not have an effect on untreated cells. EGR-1 expression vector (pAC-hEGR-1) containing the human EGR-1 cDNA was provided by Dr John Monroe (University of Pennsylvania, Philadelphia, PA, U.S.A.). To investigate whether PKC (protein kinase C) is involved in NHE3 activation in response to the PMA, transfected cells were incubated in serum-reduced media for 24 h prior to treatments and then pretreated with the PKC inhibitor chelerythrine chloride (2 μM) for 60 min. After this period, the cells were incubated in the presence or absence of PMA (100 nM) along with the inhibitor for 16 h. As a control for PMA effect, the transfected cells were also treated with an inactive PMA analogue, 4-α PMA (100 nM), for the same time as with PMA. At 48 h after transfections, cells were processed as described above with a single Luciferase Assay system (Promega). All transfection experiments were performed in triplicate and repeated at least three times.
siRNA (small interfering RNA) transfection
Gene silencing was performed using human EGR-1 sequence-specific duplex siRNA (Santa Cruz Biotechnology) and a control non-silencing siRNA (sense strand UUCUCCGAACGUGUCACGUdTdT and antisense strand ACGUGACACGUUCGGAGAAdTdT) from Qiagen. C2BBe1 cells were transfected in suspension using a published method [20] with some modifications. Briefly, for each transfection reaction in two separate tubes, 28 nM siRNA or 2 μl of siLentFect reagent (Bio-Rad Laboratories, Hercules, CA, U.S.A.) was mixed with 50 μl of the serum-free medium Optimem (Invitrogen) and incubated for 5 min at room temperature (∼25 °C). After this time, the contents of the two tubes were combined and allowed to form siRNA–siLentFect complexes for 20 min at room temperature. Just before transfection, C2BBe1 cells (80–90% confluence) were trypsinized, collected by slow centrifugation (3000 g) for 5 min, washed with serum-free media twice, collected and resuspended in Optimem. A 900 μl aliquot of the resuspended cells was combined with the siRNA–siLentFect mix, plated in a 12-well tissue culture dish at a density of 1.5×105 cells/cm2 and placed in a 37 °C, 5% CO2 incubator for 4–5 h. After this time, the media were replaced with normal growth media. At least 24 h prior to PMA treatments, transfected cells were incubated in serum-reduced media (0.5% FBS) and supplemented with PMA (100 nM) for 2, 4 and 6 h prior to harvesting the cells. Total proteins and RNA were extracted 48 h post-transfection. The protein concentrations were determined by Bradford assay (Bio-Rad), and silencing efficiency of the EGR-1 siRNA was examined by Western-blot analysis as described previously by us [21], using anti-EGR-1 antibody (Santa Cruz Biotechnology). RT–PCR of the total RNA was performed as described above.
Transfection of Drosophila SL2 cells
For these experiments, 0.5 or 1.0 μg of Sp1 or Sp3 expression vectors (pPacSp1 and pPacSp3) was co-transfected along with 1.0 μg of the test plasmid, p−95/+5 or p−319/+131, into SL2 cells. The total amount of DNA was maintained at 2.5 μg in each experiment by adjusting the empty vector, pPac0, concentration. DNA was mixed with 5 μl of Lipofectamine™-2000 in serum-free media and incubated at room temperature for 20–30 min. The DNA/Lipofectamine™ mixture was then added to 6-well dishes containing 2×106 cells/well. After 4 h, the DNA/Lipofectamine™ mixture was removed and replaced by SL2 media [Schneider's Drosophila medium supplemented with 10% (v/v) heat-inactivated FBS (Invitrogen)]. The cells were lysed in Reporter Lysis Buffer (Promega) 48 h post-transfection and protein concentration was determined by Bradford assay (Bio-Rad). The luciferase activity was measured and normalized to total cellular proteins. These transfections were carried out in duplicate and repeated at least three times. Expression vector pPacSp1 and the empty vector pPac0 were provided by Dr R. Tjian (University of California at Berkeley); pPacSp3 and pPacSp4 were provided by Dr G. Suske (Institute of Molecular Biology and Tumor Research, Marburg, Germany).
GMSA (gel mobility-shift assay)
All oligonucleotides for GMSA were synthesized by Invitrogen, Life Technologies. Complementary oligonucleotides were made double-stranded by heating to 95 °C for 5 min and slow cooling to 25 °C in TE buffer (10 mM Tris/HCl, pH 7.5, and 1 mM EDTA, pH 8.0). The sequences of the top strand of the probes containing EGR-1/Sp1 or Sp1 motif (shown in boldface) were 5′-TGCGCGGCGGGGGCGGGCAGGCTC-3′, 5′-GGCGTG-CGCGGGCGGGGCGGGCGTGCC-3′ and 5′-GCGCTGTGCTCCCCACGCCCCAGGAA-3′, which spanned from nt −91 to −68, −148 to −122 and −270 to −245 respectively. The probe was end-labelled with T4-polynucleotide kinase and [γ-32P]ATP (Amersham) and purified using G-25 quick spin mini columns (Boehringer Mannheim). Nuclear proteins were prepared as previously described [16]. DNA–protein binding reactions were performed in binding buffer [50 mM Tris/HCl, pH 7.5, 1 mM MgCl2, 0.5 mM EDTA, 0.5 mM dithiothreitol, 50 mM NaCl, 1 μg/sample poly(dI-dC)·(dI-dC) and 5% (v/v) glycerol] and 30000 c.p.m. of the probe. Reactions were initiated by addition of the nuclear proteins (5 μg) to the probe and incubation for 20 min at room temperature prior to electrophoresis on a native 5% (w/v) polyacrylamide gel in 0.5×TBE running buffer (1× TBE is 45 mM Tris/borate/1 mM EDTA, pH 8.3). Gels were dried and visualized by autoradiography. In competition assays, the unlabelled competitor oligonucleotides were added to the reaction 10 min before the addition of the labelled probe. Supershift assays were performed by addition of 1 μl (2 μg/μl) of the appropriate antibody (Santa Cruz Biotechnology) after the initial 20 min incubation with the labelled probe, and then further incubation for 30 min at room temperature.
Statistical analyses
Results are expressed as means±S.D. Differences between group means were analysed by Student's t test. Differences were considered significant at P<0.05.
RESULTS
PMA treatment stimulates hNHE3 gene expression at the transcriptional level
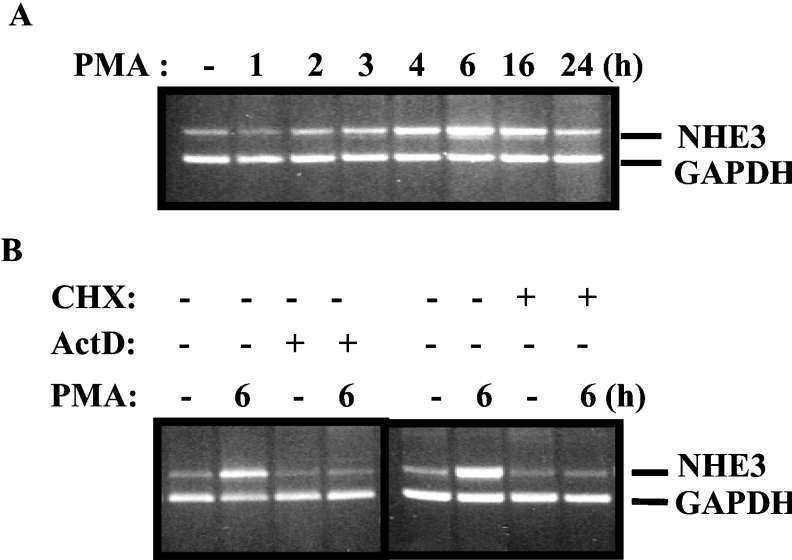
NHE3 is predominantly expressed at the apical membrane of the epithelial intestinal cells. Acute PMA treatment has been reported to decrease NHE activity of the NHE3 isoform [22,23]. To examine whether PMA affects the NHE3 mRNA expression, we performed semi-quantitative RT–PCR experiments. C2BBe1 cells were serum-starved for 24 h and then treated with 100 nM PMA for various time periods as indicated on the top of Figure 1(A). Total cell RNA was obtained from untreated and PMA-treated cells and subjected to reverse transcription and subsequent PCR amplification using hNHE3-specific primers. The results from this experiment showed that NHE3 mRNA expression was relatively low in untreated cells and mRNA levels increased gradually after PMA treatment, reaching a maximum level after 6 h and gradually decreased by 24 h.
Figure 1. hNHE3 mRNA expression is induced in response to PMA.
Total RNA was prepared from C2BBe1 cells grown to confluence and serum-starved for 24 h prior to treatment with PMA (100 nM) for 1, 2, 3, 4, 6, 16 and 24 h. For reverse-transcription reactions, 5 μg of total RNA from all time points was reverse-transcribed using an oligo(dT) as primer and SuperScript II. One-tenth of the reverse-transcription reaction products was used for PCR amplifications utilizing the NHE3 and an internal control, GAPDH, gene-specific primers. (A) RT–PCR of NHE3 induction by PMA. Duration of the PMA treatments is shown on the top of the gel. (B) RT–PCR products of cells treated with PMA for 6 h or treated with the transcription inhibitor actinomycin D (5 μM/ml) and the protein synthesis inhibitor cycloheximide (CHX; 15 μg/ml) in the absence or presence of PMA as indicated. Cells were pretreated with actinomycin D and cycloheximide for 30 and 60 min respectively prior to PMA addition.
To examine the mechanism by which PMA treatment leads to increased NHE3 mRNA expression, C2BBe1 cells were pretreated with the transcription inhibitor actinomycin D (5 μM/ml) or the protein synthesis inhibitor cycloheximide (15 μg/ml), and the cells were then incubated in the presence or absence of PMA. Total RNA was prepared and subsequently RT–PCR analysis was performed. In the continuous presence of actino-mycin D, PMA treatment (6 h) did not result in enhanced NHE3 mRNA levels, suggesting that the responsiveness to PMA is at the transcriptional level and PMA does not increase the stability of the NHE3 mRNA (Figure 1B). In addition, the lack of increased NHE3 mRNA expression in the presence of cycloheximide (Figure 1B) suggests that stimulation of NHE3 expression by PMA is dependent on new protein synthesis. Therefore these observations indicate that the up-regulation of the hNHE3 mRNA expression by PMA is mediated at the transcriptional level and requires de novo protein synthesis.
PMA induces the hNHE3 promoter activity
To examine the effect of PMA treatment on hNHE3 promoter activity, we initially focused on characterization of the −1507/+131 promoter region. C2BBe1 cells were transiently transfected with p−1507/+131 in which the NHE3 promoter region is fused to the promoter-less luciferase gene in pGL2-Basic. After 24 h serum starvation, the transfected cells were exposed to PMA (100 nM) for different time periods. Exposure to PMA for 6 and 8 h showed a minor stimulatory effect on reporter gene activity; however, after 16 and 24 h, luciferase activity increased dramatically over that exhibited by pGL2-Basic. Therefore, in all subsequent experiments, cells were treated with PMA for 16 h. These results revealed that chronic exposure to PMA results in increased NHE3 promoter activity and suggested that potential cis-element(s) responsible for the PMA effect are present in this DNA region.
Identification of the PMA-responsive region in the hNHE3 promoter
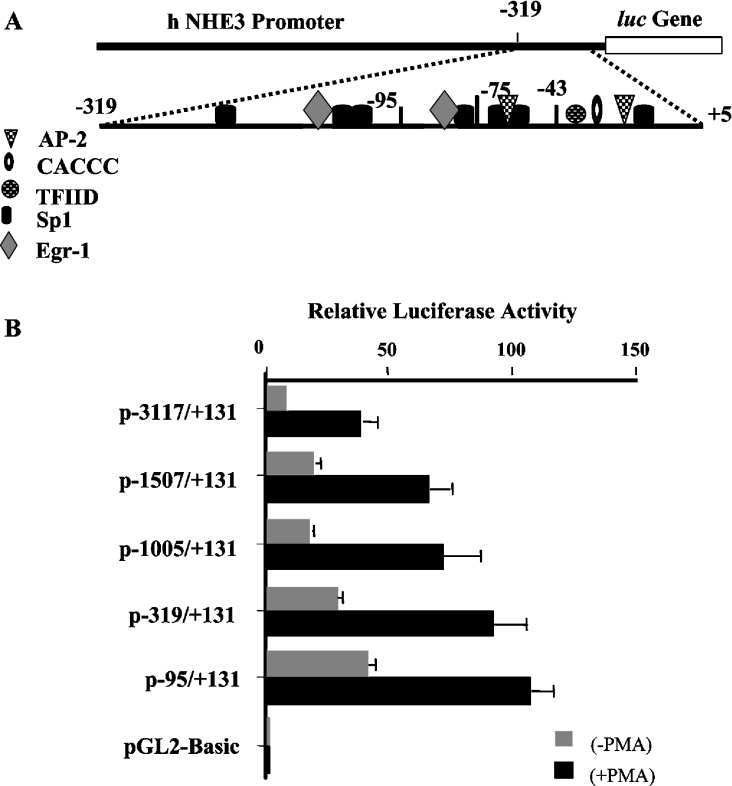
To identify specific DNA regions of the promoter that were responsible for PMA-induced stimulation of the NHE3 promoter activity, a series of 5′-truncated plasmids containing progressive deletion in the full-length promoter construct, p−3117/+131, [17] were transiently transfected into C2BBe1 cells. Analysis of the luciferase activity of the deletion constructs in control and PMA-treated cells showed a 2–3-fold increase in promoter activity after PMA treatment (Figure 2). Deletion to position −1507 resulted in a 2-fold increase in basal reporter gene activity, suggesting the presence of potential inhibitory elements in these regions; however, removal of this region showed no effect on PMA-induced promoter activity. Further deletion to position −1005 did not show any changes on the reporter gene expression. Additional removal of the promoter region to positions −319 and −95 led to a 20 and 30% increase in promoter activity respectively (Figure 2). These results indicated that PMA-responsive element(s) is located within the shortest construct, −95/+131.
Figure 2. Functional analysis of various 5′-deletion constructs in C2BBe1 cells in the presence and absence of PMA.
(A) A schematic representation of the NHE3 proximal promoter region and the location of the potential transcription-factor-binding sites are shown. (B) 5′-Deletion constructs were transiently transfected into C2BBe1 cells. Cells were serum-starved in 0.5% serum for 24 h prior to addition of PMA (100 nM) for 16 h (PMA-induced NHE3 promoter activity) or kept in serum-starvation media for the same duration (basal NHE3 promoter activity). Transfected cells were harvested 48 h post-transfection and cell lysates were prepared as described in the Experimental section. Luciferase activity was measured and normalized to β-galactosidase activity as an internal control for variations in transfection efficiency and presented relative to the normalized activity of the promoter-less pGL2-Basic. Error bars represent the S.E.M. (n=3). AP-2, activator protein-2; TFIID, transcription factor IID complex.
The EGR-1/Sp1 motif is essential for basal and PMA-induced NHE3 promoter expression
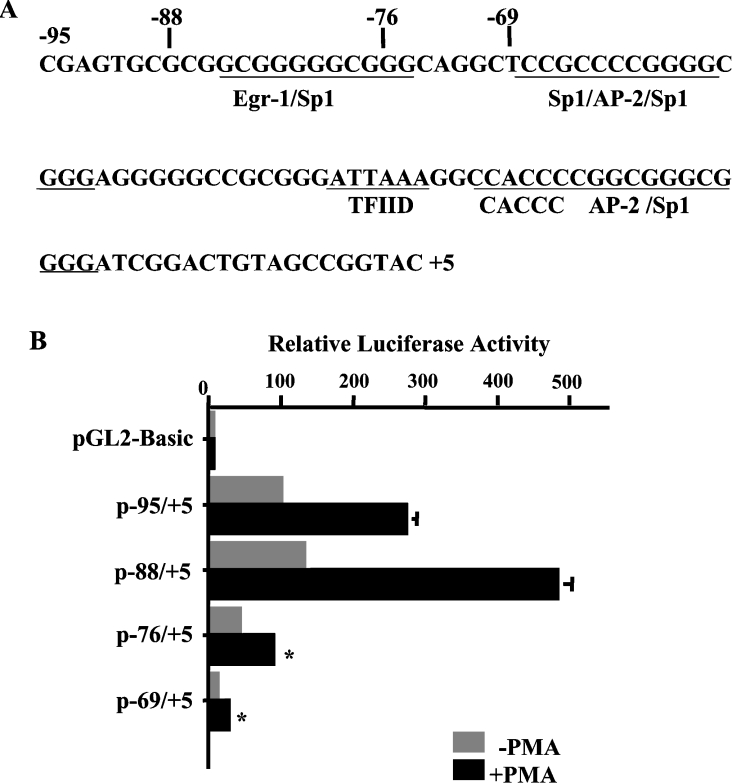
To map the location of the PMA-response element on the proximal promoter region, we next analysed p−95/+5 [17] and three additional 5′-deletions of this construct, p−88/+5, p−76/+5 and p−69/+5. We have shown previously that 5′-untranslated region sequences from position +5 to +131 do not play a role in NHE3 promoter activity, as deletion of this region in p−95/+131 did not affect the reporter gene expression [17]. The nucleotide sequence of the hNHE3 promoter region from bp −95 to +5 is shown in Figure 3(A), and the location of the potential cis-elements is indicated. As shown in Figure 3(B), both p–95/+5 and p−88/+5 displayed high levels of the basal and PMA-stimulated reporter activity, while p−76/+5 and p−69/+5 exhibited a 60 and 90% loss of basal promoter activity respectively and almost total elimination of the PMA-induced stimulation. Thus up-regulation of the hNHE3 promoter activity in response to PMA appears to be mediated, at least in part, through the GC-box at −88 to −69 promoter region. This region harbours an overlapping binding site for EGR-1 and Sp1 (Figure 3A). Another sequence similar to Sp1-binding site also overlaps with EGR-1 site at the 5′-end. Although this Sp1 site is truncated in p−88/+5, the plasmid shows high promoter activity. Thus we speculate that this sequence is not involved in interaction with Sp1/Sp3. A cis-element composed of 11 nt, 5′-GCGGGGGCGGG-3, which contains the overlapping EGR-1/Sp1 binding sites, resides at bp −85 to −75 (Figure 3A). This motif has been shown to be the target of PMA-response factors in a number of genes and various cell types. Furthermore, we have recently shown [21] that an overlapping EGR-1/Sp1 motif in the promoter of the hNHE2 gene mediates the stimulatory effect of PMA on the NHE2 gene expression. The 11 nt EGR-1/Sp1 motif in the −85 to −75 position of the NHE3 promoter is 100% identical with the core sequence of the hNHE2 PMA-response element and displays similar DNA–protein interactions [21].
Figure 3. cis-element EGR-1/Sp1 constitutes a PMA-response motif.
(A) The nucleotide sequence of the hNHE3 promoter region from bp −95 to +5 is shown and the location of the potential cis-elements is indicated. (B) 5′-Deletion constructs harbouring further deletion in the NHE3 proximal promoter region were transiently transfected into C2BBe1 cells, as indicated in the legend to Figure 2, and were similarly either treated with PMA or not treated. After 48 h, the cell lysates were prepared and luciferase activities were determined. Luciferase activity was corrected for transfection efficiency using β-galactosidase activity and presented relative to the normalized activity of the pGL2-Basic vector. Results are presented as the means±S.E.M. (n=3). *, Significantly different from the p−88/+5 treated with PMA, P<0.05.
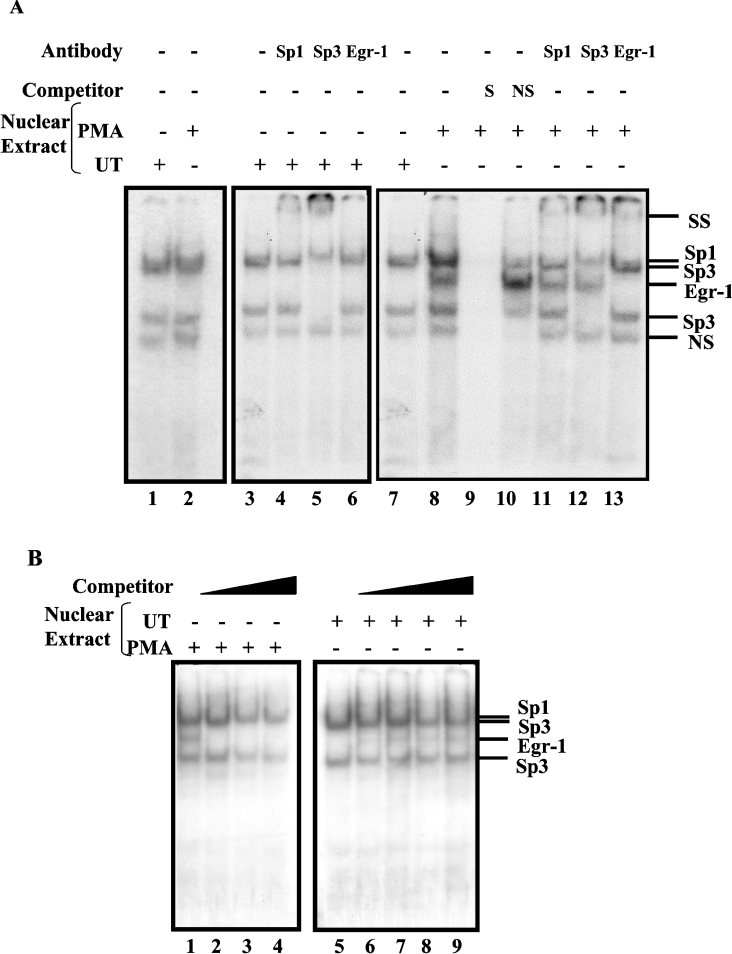
Basal and PMA-inducible nuclear proteins bind to the hNHE3 promoter
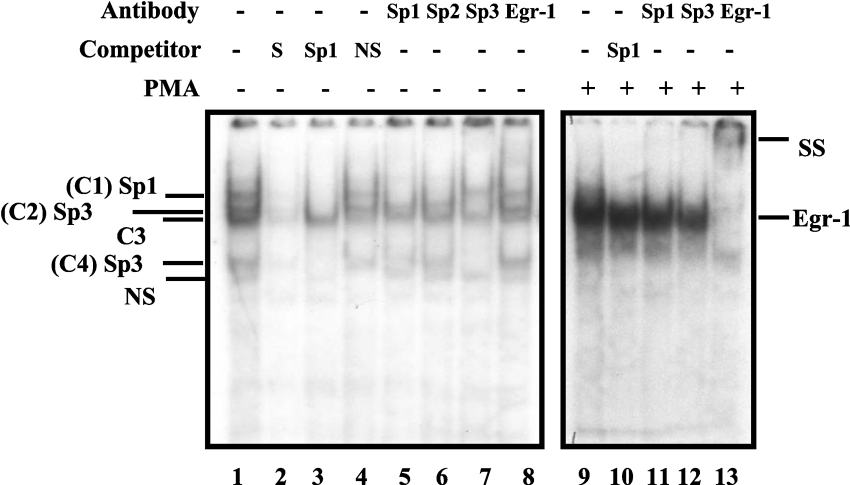
We have recently reported that EGR-1 mRNA and protein expression are induced by PMA in C2BBe1 cells [21]. In addition, we have shown that PMA-induced EGR-1 is involved in transcriptional up-regulation of the hNHE2 promoter. Thus, to determine whether the PMA-dependent activation of the NHE3 gene is also mediated by similar mechanisms, we first carried out a series of GMSAs. In these experiments, a double-stranded oligonucleotide (−91 to −68) spanning the EGR-1/Sp1 motif was utilized as an end-labelled probe and was incubated with nuclear extracts from the control or PMA-treated cells. Four specific DNA–protein complexes (C1–C4) were detected when nuclear extracts from the control cells were incubated with the probe (Figure 4, lane 1), whereas, in nuclear extracts from PMA-treated cells, a single novel DNA–protein complex was present (Figure 4, lane 9). The binding specificity of these complexes was examined by competition experiments where excess unlabelled specific probe or an unlabelled oligonucleotide carrying the Sp1 consensus-binding sequence was used in these assays (Figure 4, lanes 2 and 3 respectively). In these experiments, Sp1 oligonucleotide competed out complexes C1, C2 and C4, suggesting that the protein components of these complexes were Sp1-related. The identities of the proteins present in these complexes were established by supershift assays (Figure 4, lanes 5–8). An anti-Sp1 antibody removed complex C1, identifying the protein component of this complex as Sp1. An anti-Sp3 antibody blocked the formation of both C2 and C4, revealing that Sp3 binding to the probe formed complexes C2 and C4. Anti-EGR-1 antibody did not affect any of the DNA–protein complexes. To examine the possibility of the presence of EGR-1 in the PMA-induced DNA–protein complex (Figure 4, lane 9), supershift experiments with antibodies directed against Sp1, Sp3 or EGR-1 were performed (Figure 4, lanes 11–13). Addition of anti-EGR-1 antibody to nuclear proteins–probe mixture resulted in formation of a slow migrating supershifted band and elimination of the intense DNA–protein complex (Figure 4, lane 13). Removal of this intense band resulted in re-appearance of two faint bands that co-migrate with Sp1 and Sp3 complexes. Therefore these results demonstrate that the PMA-induced complex interacting with the hNHE3 promoter is the EGR-1 protein.
Figure 4. PMA-induced EGR-1 interacts with the downstream EGR-1/Sp1 motif.
GMSA was performed using a double-stranded oligonucleotide (bp −91 to −68) as end-labelled probe and nuclear extracts from untreated or PMA-treated C2BBe1 cells. Nuclear extracts were prepared from C2BBe1 cells at confluence. PMA-treated cells were grown in serum-reduced media at least for 24 h prior to addition of PMA (100 nM) for 2 h. A total of 5 μg of nuclear proteins was combined with 30000 c.p.m. of probe per reaction and, after 20 min incubation at room temperature, resolved on a 5% non-denaturing polyacrylamide gel and visualized by autoradiography. Competition experiments were performed in the presence of unlabelled probe (S), non-specific oligonucleotide (NS) and an oligonucleotide containing Sp1 consensus binding site (Sp1). Supershift assays were performed with anti-Sp1, -Sp2, -Sp3 and -EGR-1 antibodies (2 μg) as indicated. SS denotes the supershifted bands. + and − signs indicate the presence or absence of reaction components in the binding mixture (shown on the top). Protein components of DNA–protein complexes are shown on the right. C1, complex 1.
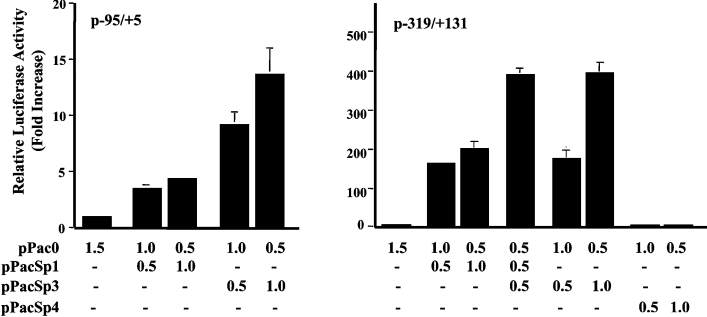
Sp1 and Sp3 co-expression activate hNHE3 promoter in Drosophila SL2 cells
The results of supershift experiments (Figure 4) indicated that at basal growth conditions, transcription factors Sp1 and Sp3 bind to the hNHE3 promoter at −91 to −68 bp promoter region. We therefore investigated whether the Sp1 family of transcription factors can functionally transactivate the hNHE3 promoter at the basal conditions. Co-transfection experiments were performed in Drosophila SL2 cells, which lack the endogenous Sp1 activity. The hNHE3 promoter-reporter construct p−95/+5 or p−319/+131 was used for co-transfection with pPacSp1, pPacSp3 or pPacSp4 carrying the Sp1, Sp3 and Sp4 cDNA respectively and the empty vector, pPac0, as a control. As shown in Figure 5, co-transfection of p−95/+5 with the empty vector resulted in very low reporter gene activity, while the presence of Sp1 expression vector led to approx. 4-fold increase in the reporter gene activity at concentrations used. The Sp3 co-transfection, on the other hand, led to approx. 10- and 15-fold increase in the NHE3 promoter activity at 0.5 and 1.0 μg of Sp3 expression vector respectively. Co-transfection of promoter construct p–319/+131 with Sp1 alone resulted in 180- and 200-fold increase in the reporter gene activity, and Sp3 led to 190- and 400-fold increase at 0.5 and 1.0 μg of the expression vector respectively. In contrast, Sp4 co-transfection had no effect on the NHE3 promoter activity in SL2 cells. The presence of both Sp1 and Sp3 at an equal concentration resulted in approx. 400-fold transactivation over the empty vector. Therefore it appears that both these transcription factors can act as activators of the NHE3 promoter activity; however, the level of induction by Sp3 alone at a higher concentration (1 μg) is 2–3-fold greater than that of Sp1, while at lower concentrations of Sp1 and Sp3, both transcription factors induce the promoter activity to the same levels. Therefore the induction by Sp1 and Sp3 seems to be concentration-dependent. Interestingly, the level of promoter activity in response to ectopic expression of Sp1 or Sp3 in SL2 cells was dramatically higher in p−319/+131 than that of p−95/+5. This was unexpected, since our transient transfection assays in C2BBe1 cells (Figure 2) did not show a significant difference in the promoter activity of p−95/+5, p−95/+131 and p−319/+131.
Figure 5. Co-transfection of Drosophila SL2 cells with p−95/+5 or p−319/+131 and Sp1 and Sp3 expression vectors.
SL2 cells were co-transfected with Sp1 (pPacSp1), Sp3 (pPacSp3) and Sp4 (pPacSp4) expression vectors individually or in combination as indicated in the bottom of the graph. Luciferase activity was normalized to the total protein concentration of the cell lysates and expressed relative to the activity of p−95/+5 co-transfected with the empty vector, pPac0. The values were obtained from three independent experiments each performed in duplicate. The results shown are the means±S.E.M. (n=3).
Due to high GC content of the DNA sequences in the NHE3 promoter region, multiple GC-box elements are present in this region. To examine whether transcription factors Sp1 and Sp3 could interact with the GC-boxes upstream from bp −95, DNA-binding activity of nuclear proteins from untreated C2BBe1 cells was tested with potential Sp1-binding sites located at bp −270 to −245 (Figure 6A, lane 1) and bp −148 to −122 (Figure 6A, lanes 3–7). These results showed that both Sp1 and Sp3 interact with Sp1-binding site at −148 to −122 and −270 to −245, as determined by supershift experiments (Figure 6A, lanes 4–6, supershift results for −270 to −245 not shown). Moreover, PMA-induced EGR-1 in nuclear proteins from PMA-treated cells binds to the probe spanning from −148 to −122 (Figure 6A, lanes 8–13), but not to the Sp1 site at bp −270 to −245 (Figure 6A, lane 2). The former probe contains an imperfect binding site for EGR-1 and two binding sites for Sp1. All three binding sites in this region overlap and are designated as upstream EGR-1/Sp1. The interaction of Sp1, Sp3 and EGR-1 with the −148 to −122 probe is shown by supershift assays (Figure 6A, lanes 11–13). Interestingly, the binding kinetics of PMA-induced EGR-1 to the upstream EGR-1/Sp1 (bp −148 to −122) is different from that of the downstream EGR-1/Sp1 (bp −91 to −68). This is shown by the simultaneous binding of Sp1, Sp3 and EGR-1 transcription factors to −148 to −122 probe (Figure 6A, lane 8), but not to the −91 to −68 probe (Figure 4, lanes 9–12). To examine the mechanism of interaction of PMA-induced EGR-1 with the upstream EGR-1/Sp1 probe, we studied the DNA-binding affinity of the EGR-1 in competition assays. In these experiments, either PMA-treated nuclear proteins (Figure 6B, lanes 1–4) were challenged with increasing concentrations of unlabelled oligonucleotides carrying the EGR-1 consensus sequence (Figure 6B, lanes 2–4), or nuclear proteins from the control cells were combined with increasing concentrations of PMA-treated nuclear proteins as a source of EGR-1 (Figure 6B, lanes 6–9) and coupled with −148 to −122 end-labelled probe. The results of these studies showed that neither inhibition of EGR-1 interaction with the probe via excess unlabelled EGR-1 consensus sequence nor increasing concentration of PMA-induced EGR-1 in binding reaction affects the formation of the Sp1 and Sp3 DNA–protein complexes. Therefore these studies suggest that EGR-1 and Sp1 family members bind to the upstream EGR-1/Sp1 motif non-competitively.
Figure 6. Sp1, Sp3 and EGR-1 interact with the upstream EGR-1/Sp1 motif simultaneously.
(A) GMSAs were performed using 32P-end-labelled probes −270 to −245 (lanes 1 and 2), −148 to −122 (lanes 3–13) with nuclear extracts prepared from the control (lanes 1 and 3–7) and PMA-treated cells (lanes 2 and 8–13). PMA-treated cells were grown in serum-reduced media at least for 24 h prior to addition of PMA (100 nM) for 2 h. The identity of the protein components of the DNA–protein complexes was determined by a competition assay with an unlabelled specific probe (lane 9) and a non-specific oligonucleotide (lane 10) and by supershift assays with anti-Sp1, -Sp3 and -EGR-1 antibodies (lanes 11–13 respectively). DNA–protein complexes were resolved on a 5% native polyacrylamide gel and visualized by autoradiography. (B) Competition assays for protein interactions were performed with −148 to −122 end-labelled probe and PMA-treated nuclear proteins (lanes 1–4) in the presence of 50-, 100- and 200-fold excess of EGR-1 consensus oligonucleotides (lanes 2–4 respectively) or the control nuclear proteins (lanes 5–9) plus increasing amounts of PMA-treated nuclear extracts at 0.5, 1.0, 1.5 and 2.0 μg (lanes 6–9 respectively). The positions of the DNA–protein complexes are shown on the right. SS denotes the supershifted bands. NS, non-specific competitor oligonucleotide or non-specific protein–DNA complex; UT, untreated.
EGR-1 overexpression leads to transactivation of the hNHE3 promoter activity
To assess directly whether EGR-1 could modulate the hNHE3 promoter expression in vivo, co-transfection experiments were performed in C2BBe1 cells as described in the Experimental section. Transfection of increasing concentrations of pAC-hEGR-1 expression vector together with either p−95/+5 or p−319/+131 NHE3 promoter-reporter constructs stimulated the NHE3 promoter activity in a dose-dependent manner. A 6-fold increase was observed with transfection of 1.0 μg of EGR-1 expression vector (results not shown). These observations, therefore, demonstrate that up-regulation of NHE3 promoter activity in response to the overexpression of EGR-1 may attribute to the EGR-1-binding site at the position −85 to −75 alone, leading to transcriptional activation of the hNHE3 promoter in both constructs.
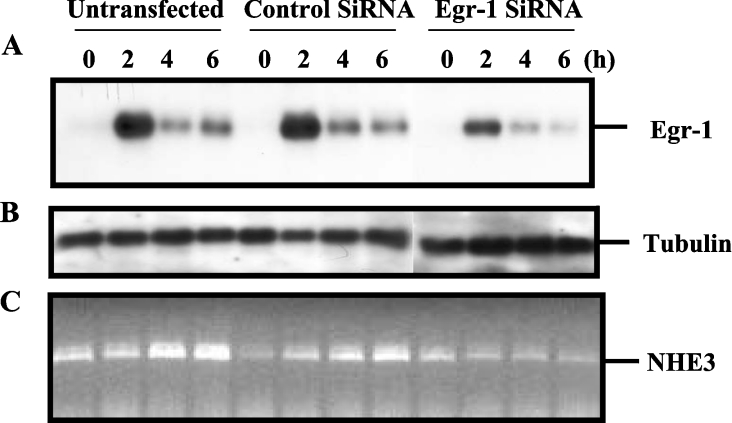
Knockdown of PMA-induced EGR-1 leads to down-regulation of PMA-induced NHE3 mRNA expression
To confirm the involvement of EGR-1 in PMA-induced endogenous NHE3 mRNA expression, we investigated the effect of EGR-1 knockdown by using EGR-1 siRNA duplex. C2BBe1 cells were transfected with either a control non-silencing siRNA or EGR-1-specific siRNA and subsequently treated with or without PMA as described in the Experimental section. Untransfected C2BBe1 cells were also used as a control. To examine the silencing effect of EGR-1 siRNA, Western-blot analyses were performed using EGR-1 antibody and total cell lysates from untransfected, control siRNA- and EGR-1 siRNA-transfected cells supplemented with or without PMA. We have shown previously that in C2BBe1 cells, EGR-1 expression is maximal at 2 h post-PMA treatment, and the protein level reduces to basal level by 16 h [21]. As shown in Figure 7(A), in untransfected cells and cells transfected with the control siRNA, EGR-1 was highly expressed at 2 h and protein levels decreased equally at 4 and 6 h of PMA treatment. However, in cells transfected with EGR-1 siRNA, the EGR-1 level was reduced in all time points compared with the controls (Figure 7A). As a control for protein loading, the same membrane was subsequently stripped and probed with tubulin antibody (Figure 7B). The effect of EGR-1 knockdown on PMA-induced NHE3 mRNA expression was determined by RT–PCR using total RNA from cells treated as described above for Western-blot analysis. As presented in Figure 7(C), transfection of C2BBe1 cells with the control siRNA did not show any inhibitory effect on PMA-induced NHE3 mRNA, while transfection with the EGR-1 siRNA led to transcriptional down-regulation of the PMA-induced NHE3 mRNA expression. These results indicate that the blockade of EGR-1 overexpression by small inhibitory RNA prevents the PMA-stimulated endogenous NHE3 mRNA expression and provides further evidence for the involvement of EGR-1 in the PMA-induced up-regulation of the hNHE3 mRNA expression.
Figure 7. EGR-1 siRNA suppresses the PMA-induced hNHE3 mRNA up-regulation.
C2BBe1 cells were transfected with a control siRNA or EGR-1-specific siRNA and treated with PMA as described in the Experimental section. Total cell proteins (15 μg/lane) were resolved in an SDS/10% polyacrylamide gel, subsequently transferred on to Immobilon-P and sequentially probed with antibodies specific to EGR-1 (A) and tubulin (B) as a control for the protein loading. The effect of EGR-1 down-regulation on PMA-induced NHE3 mRNA expression was determined by RT–PCR (C). Total RNA (3 μg) from the cells treated as above was subjected to reverse transcription using oligo(dT) as primer. One-tenth of the reverse transcription product was PCR-amplified as described in the Experimental section using NHE3-specific primers. Total proteins and RNA from untransfected cells were also used as a control in Western blotting and RT–PCR analysis.
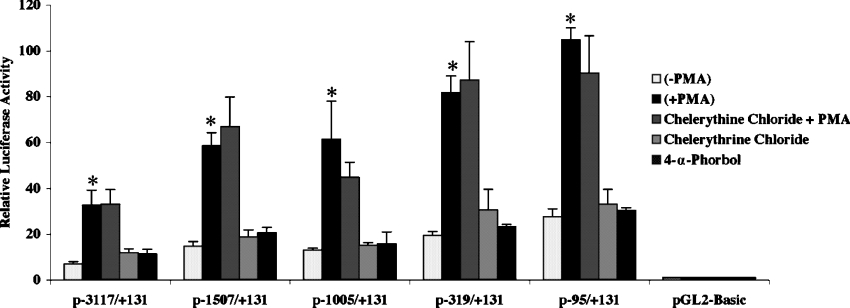
PMA-induced hNHE3 promoter activity is largely independent of PKC pathway
Activation of the PKC pathway in response to PMA plays a major role in the stimulatory effect of PMA on the expression of the target genes. To examine the contribution of PKC pathway in the PMA-induced activity of the hNHE3 promoter, C2BBe1 cells were transiently transfected with the 5′-truncated NHE3 promoter-reporter constructs and treated with PMA (100 nM) in the presence or absence of the PKC inhibitor chelerythrine chloride. Cells that were incubated simultaneously with PMA and chelerythrine chloride were pretreated with the inhibitor (2 μM) for 60 min prior to PMA addition. As a control for the specific effects of PMA on promoter activity, cells were also treated with 4α-PMA, the inactive form of PMA (Figure 8). These results revealed that the PKC inhibitor chelerythrine chloride had no effect on the PMA-induced reporter gene activity, and there was no stimulatory effect on the promoter in the presence of 4α-PMA. Thus it appears that the PMA-induced stimulation of the NHE3 promoter activity is mediated through mechanisms dependent on the mitogenic activity and protein kinases other than PKC.
Figure 8. PMA-induced hNHE3 promoter activity is PKC-independent.
C2BBe1 cells were transiently transfected with 5′-truncated NHE3 promoter-reporter constructs and incubated in serum-reduced media at least 24 h prior to treatments. Cells were either not treated or pretreated with the PKC inhibitor chelerythrine chloride (2 μM) for 60 min prior to the addition of PMA (100 nM). As control 4-α phorbol ester (100 nM) was used to serve as inactive PMA. After 16 h of PMA treatment, cells were harvested and luciferase activities were determined. Results are presented as the means±S.E.M. (n=3–5). *The treatment pair PMA and PMA+chelerythrine chloride was analysed with unpaired t test and showed no significant differences.
DISCUSSION
In the present study, we utilized a combination of transient transfection experiments, GMSAs and mutagenesis to identify the cis-elements within the hNHE3 promoter region that confer responsiveness to PMA. The expression of endogenous NHE3 mRNA was enhanced in the presence of PMA. The PMA stimulation of hNHE3 gene expression was established to be acting through the promoter and involves both transcriptional and translational events. In promoter deletion analyses, removal of the nucleotide sequences up to position −69 abolished both the basal and PMA-induced gene expressions. The region between bp −95 and −69 encompasses an 11 bp sequence motif, 5′-GCGGGGGCGGG-3′, that is identical with an overlapping EGR-1/Sp1 binding sequence in the promoter region of the PMA-responsive genes including the hNHE2 gene. By GMSAs, we showed that under the basal growth conditions Sp1 family members, and under PMA-stimulated conditions, EGR-1, bound to the probe containing the EGR-1/Sp1 motif at bp −85 to −75. Cano [24] and Kandasamy and Orlowski [25] have also noted the presence of an overlapping EGR-1/Sp1 element in the rat NHE3 promoter sequence. However, the role of this motif in regulation of the rat NHE3 promoter has not been investigated. We have shown previously that the nucleotide sequences at the proximal promoter region of both the hNHE3 and rat NHE3 share a high degree of homology [17]. An examination of the hNHE3 [17] and rat NHE3 [24,25] proximal promoter sequences revealed that the EGR-1/Sp1 motif in the hNHE3 and rat NHE3 promoters is located at the same position with respect to an atypical TATA sequence in these promoters. The high degree of conservation of the nucleotide sequences at the first 100 bp upstream from the transcription initiation site as well as a similar location of the EGR-1/Sp1 binding sites in the hNHE3 and rat NHE3 promoters underscores the importance of this motif in the regulation of the NHE3 promoter expression. Interestingly, the nucleotide sequences immediately upstream from the EGR-1/Sp1 motif diverge in these species [17].
The Sp1 transcription factor has been characterized as a ubiquitously expressed factor that regulates the basal transcription activity; however, a number of studies have shown that Sp1 binding and transactivation can be modulated by a variety of stimuli including growth factors and hormones. Sp1 is also implicated in tissue-specific [26] and developmental regulation of gene expression [27]. Sp1 family members display similar modular structures comprising three highly conserved zinc finger structures that are required for DNA binding, and as such exhibit similar binding affinity to GC-boxes. Both Sp1 and Sp3 have been reported to act as an activator or repressor of transcription, depending on the promoter context [28,29]. In addition, Sp1 can interact with Sp family members as well as other transcription factors such as the CCAAT-box-binding protein NF-Y (nuclear factor-Y) [30], Rb (retinoblastoma protein) [31] and NF-κB (nuclear factor κB) [32]. In nuclear proteins from untreated C2BBe1 cells, Sp1 and Sp3 bind to at least three positions, bp −270 to −245, bp −148 to −122 and bp −91/−68, in the hNHE3 promoter proximal region.
To confirm involvement of Sp1 family members in transactivation of the hNHE3 promoter, we performed co-transfection experiments in SL2 cell line, which is devoid of the endogenous Sp1 family members. The results of these experiments indicated that while co-expression of the empty vector showed no effect on hNHE3 promoter activity, both the Sp1 and Sp3 could act as transcriptional activators of this promoter. Surprisingly, transactivation level in p−319/+131 was dramatically higher than that of p−95/+5 in SL2 cells under the same experimental conditions. This was in contrast with our observations in C2BBe1 cells, where p−95/+5 and p−95/+131 exhibited slightly higher promoter activity than p−319/+131 [17]. This discrepancy may be explained by the presence of additional Sp1-binding sites in the DNA region upstream from position −95, and by dose effect where forced expression of high levels of Sp1 and Sp3 may result in binding of these factors to low-affinity binding sites or formation of higher order complexes that might lead to a synergistic effect on transcription from this promoter. Alternatively, SL2 cells may provide an activating cofactor that may act in concert with Sp1 family members to activate yet another upstream cis-element on the promoter. Therefore it is possible that under specific conditions, enhanced expression of Sp1 family members might play a stimulatory role in the NHE3 promoter activity by interacting with the upstream binding sites.
EGR-1 is induced by a number of diverse stimuli, including mitogenic stimuli, signals for proliferation, development, injurious insults and differentiation [33]. Competition between Sp1 and EGR-1 has been reported to result in both repression and activation of target genes. Activation of tissue factor gene expression by PMA in HeLa cells or by shear stress in epithelial or endothelial cells is mediated through induction of EGR-1 and subsequent displacement of Sp1 from the overlapping EGR-1/Sp1 site [34,35]. Likewise, Khachigian et al. [36] have demonstrated that upon cell injury, newly synthesized EGR-1 displaces the prebound Sp1 factor and occupies the overlapping Sp1/EGR-1 binding site of the promoter, leading to induction of the PDGF-B gene expression. On the other hand, a negative regulatory role for competition between EGR-1 and Sp1 has been reported for the ADA (adenosine deaminase) gene promoter, where overexpression of EGR-1 resulted in displacement of Sp1 and repression of the ADA expression [37]. In the present study, we have shown that the expression and DNA-binding affinity of the Sp1 and Sp3 transcription factors in nuclear proteins from the control and PMA-treated cells do not differ significantly (Figure 6A). However, in the presence of PMA-induced EGR-1 transcription factor, Sp1 and Sp3 do not bind to the EGR-1/Sp1 motif on −91 to −68 probe, but they interact with the upstream EGR-1/Sp1 motif non-discriminately. These differences in binding activity of two EGR-1/Sp1 motifs may explain the results of our deletion analyses and EGR-1 co-transfection results in C2BBe1 cells, which suggest that the upstream EGR-1/Sp1 motif does not play a major role in PMA-induced activation of the NHE3 promoter.
The involvement of EGR-1 in NHE3 transcriptional regulation was further confirmed by co-transfection studies with an EGR-1 expression vector. We directly demonstrated that EGR-1 overexpression leads to a dose-dependent activation of the NHE3 promoter in transiently transfected C2BBe1 cells. Moreover, the blockade of EGR-1 overexpression by EGR-1-specific small inhibitory RNA blocked the endogenous PMA-induced NHE3 mRNA up-regulation. These findings established that the PMA-induced up-regulation of the hNHE3 transcription is mediated via EGR-1 transcription factor.
Several lines of evidence suggest that multiple, independent and additive pathways may be involved in induction of EGR-1 gene expression [38,39]. NHE3 promoter activity was significantly increased by PMA treatment compared with the untreated cells. This stimulatory effect was absent in the presence of 4α-PMA, an inactive PMA analogue. PMA is a well-known inducer of the PKC pathway activation; however, the presence of the PKC inhibitor chelerythrine chloride in incubation media containing PMA had no inhibitory effect on the NHE3-reporter gene activity, suggesting that PMA stimulation of the hNHE3 is largely independent of PKC activation. It is now well established that in addition to PKC isoenzymes that bind to phorbol esters (conventional and novel PKCs), phorbol ester tumour promoters also bind to other receptors lacking kinase activity. Among these receptors are Ras-GRP (Ras guanine nucleotide releasing protein) [40], chimaerin family of receptors [41,42] and PKDs [43]. Whether these receptors are involved in mediating the PMA effect on NHE3 expression is not known at this point and needs to be defined.
Acknowledgments
These studies were supported by the NIDDK (National Institute of Diabetes and Digestive and Kidney Diseases) grants DK-33349 and DK-67990 and the U.S. Department of Veterans Affairs. We thank Ian Pearse for technical assistance.
References
- 1.Boron W. F. Intracellular pH regulation in epithelial cells. Annu. Rev. Physiol. 1986;48:377–388. doi: 10.1146/annurev.ph.48.030186.002113. [DOI] [PubMed] [Google Scholar]
- 2.Grinstein S., Rotin D., Mason M. J. Na+/H+ exchange and growth factor-induced cytosolic pH changes. Role in cellular proliferation. Biochim. Biophys. Acta. 1989;988:73–97. doi: 10.1016/0304-4157(89)90004-x. [DOI] [PubMed] [Google Scholar]
- 3.Knickelbein R. G., Aronson P. S., Dobbins J. W. Membrane distribution of sodium–hydrogen and chloride–bicarbonate exchangers in crypt and villus cell membranes from rabbit ileum. J. Clin. Invest. 1988;82:2158–2163. doi: 10.1172/JCI113838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Counillon L., Pouyssegur J. The expanding family of eucaryotic Na+/H+ exchangers. J. Biol. Chem. 2000;275:1–4. doi: 10.1074/jbc.275.1.1. [DOI] [PubMed] [Google Scholar]
- 5.Orlowski J., Grinstein S. Diversity of the mammalian sodium/proton exchanger SLC9 gene family. Pflugers Arch. 2004;447:549–565. doi: 10.1007/s00424-003-1110-3. [DOI] [PubMed] [Google Scholar]
- 6.Zachos N. C., Tse M., Donowitz M. Molecular physiology of intestinal Na+/H+ exchange. Annu. Rev. Physiol. 2005;67:411–443. doi: 10.1146/annurev.physiol.67.031103.153004. [DOI] [PubMed] [Google Scholar]
- 7.Sardet C., Counillon L., Franchi A., Pouyssegur J. Growth factors induce phosphorylation of the Na+/H+ antiporter, glycoprotein of 110 kD. Science. 1990;247:723–726. doi: 10.1126/science.2154036. [DOI] [PubMed] [Google Scholar]
- 8.Wang H., Silva N. L., Lucchesi P. A., Haworth R., Wang K., Michalak M., Pelech S., Fliegel L. Phosphorylation and regulation of the Na+/H+ exchanger through mitogen-activated protein kinase. Biochemistry. 1997;36:9151–9158. doi: 10.1021/bi970802f. [DOI] [PubMed] [Google Scholar]
- 9.Hayashi H., Szászi K., Grinstein S. Multiple modes of regulation of Na+/H+ exchangers. Ann. N.Y. Acad. Sci. 2002;976:248–258. doi: 10.1111/j.1749-6632.2002.tb04747.x. [DOI] [PubMed] [Google Scholar]
- 10.Rao G. N., Sardet C., Pouyssegur J., Berk B. C. Na+/H+ antiporter gene expression increases during retinoic acid-induced granulocytic differentiation of HL60 cells. J. Cell Physiol. 1992;151:361–366. doi: 10.1002/jcp.1041510217. [DOI] [PubMed] [Google Scholar]
- 11.Yang W., Dyck J. R., Wang H., Fliegel L. Regulation of NHE-1 promoter in mammalian myocardium. Am. J. Physiol. 1996;270:H259–H266. doi: 10.1152/ajpheart.1996.270.1.H259. [DOI] [PubMed] [Google Scholar]
- 12.Yan W., Nehrke K., Choi J., Barber D. L. The Nck-interacting kinase (NIK) phosphorylates the Na+–H+ exchanger NHE1 and regulates NHE1 activation by platelet-derived growth factor. J. Biol. Chem. 2001;276:31349–31356. doi: 10.1074/jbc.M102679200. [DOI] [PubMed] [Google Scholar]
- 13.Rocha F., Musch M. W, Lishanskiy L., Bookstein C., Sugi K., Xie Y., Chang E. B. IFN-γ downregulates expression of Na+/H+ exchangers NHE2 and NHE3 in rat intestine and human Caco-2/bbe cells. Am. J. Physiol. Cell Physiol. 2001;280:C1224–C1232. doi: 10.1152/ajpcell.2001.280.5.C1224. [DOI] [PubMed] [Google Scholar]
- 14.Kiela P. R., Hines E. R., Collins J. F., Ghishan F. K. Regulation of the rat NHE3 gene promoter by sodium butyrate. Am. J. Physiol. Gastrointest. Liver Physiol. 2001;281:G947–G956. doi: 10.1152/ajpgi.2001.281.4.G947. [DOI] [PubMed] [Google Scholar]
- 15.Yun C. C., Palmada M., Embark H. M., Fedorenko O., Feng Y., Henke G., Setiawan I., Boehmer C., Weinman E. J., Sandrasagra S., et al. The serum and glucocorticoid-inducible kinase SGK1 and the Na+/H+ exchange regulating factor NHERF2 synergize to stimulate the renal outer medullary K+ channel ROMK1. J. Am. Soc. Nephrol. 2002;13:2823–2830. doi: 10.1097/01.asn.0000035085.54451.81. [DOI] [PubMed] [Google Scholar]
- 16.Malakooti J., Dahdal R. Y., Dudeja P. K., Layden T. J., Ramaswamy K. The human Na+/H+ exchanger NHE2 gene: genomic organization and promoter characterization. Am. J. Physiol. Gastrointest. Liver Physiol. 2001;280:G763–G773. doi: 10.1152/ajpgi.2001.280.4.G763. [DOI] [PubMed] [Google Scholar]
- 17.Malakooti J., Memark V. C., Dudeja P. K., Ramaswamy K. Molecular cloning and functional analysis of the human Na+/H+ exchanger NHE3 promoter. Am. J. Physiol. Gastrointest. Liver Physiol. 2002;282:G491–G500. doi: 10.1152/ajpgi.00273.2001. [DOI] [PubMed] [Google Scholar]
- 18.Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Struhl K. (eds.) New York: Wiley and Sons; 1995. Current Protocols in Molecular Biology. [Google Scholar]
- 19.Brant S. R., Yun C. H., Donowitz M., Tse C. M. Cloning, tissue distribution, and functional analysis of the human Na+/N+ exchanger isoform, NHE3. Am. J. Physiol. 1995;269:C198–C206. doi: 10.1152/ajpcell.1995.269.1.C198. [DOI] [PubMed] [Google Scholar]
- 20.Clayburgh D. R., Rosen S., Witkowski E. D., Wang F., Blair S., Dudek S., Garcia J. G., Alverdy J. C., Turner J. R. A differentiation-dependent splice variant of myosin light chain kinase, MLCK1, regulates epithelial tight junction permeability. J. Biol. Chem. 2004;279:55506–55513. doi: 10.1074/jbc.M408822200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malakooti J., Sandoval R., Memark V. C., Dudeja P. K., Ramaswamy K. Zinc finger transcription factor EGR-1 is involved in stimulation of NHE2 gene expression by phorbol 12-myristate 13-acetate. Am. J. Physiol. Gastrointest. Liver Physiol. 2005;289:G653–G663. doi: 10.1152/ajpgi.00010.2005. [DOI] [PubMed] [Google Scholar]
- 22.Silviani V., Colombani V., Heyries L., Gerolami A., Cartouzou G., Marteau C. Role of the NHE3 isoform of the Na+/H+ exchanger in sodium absorption by the rabbit gallbladder. Pflugers Arch. 1996;432:791–796. doi: 10.1007/s004240050200. [DOI] [PubMed] [Google Scholar]
- 23.Janecki A. J., Montrose M. H., Tse C. M., de Medina F. S., Zweibaum A., Donowitz M. Development of an endogenous epithelial Na+/H+ exchanger (NHE3) in three clones of caco-2 cells. Am. J. Physiol. 1999;277:G292–G305. doi: 10.1152/ajpgi.1999.277.2.G292. [DOI] [PubMed] [Google Scholar]
- 24.Cano A. Characterization of the rat NHE3 promoter. Am. J. Physiol. 1996;271:F629–F636. doi: 10.1152/ajprenal.1996.271.3.F629. [DOI] [PubMed] [Google Scholar]
- 25.Kandasamy R. A., Orlowski J. Genomic organization and glucocorticoid transcriptional activation of the rat Na+/H+ exchanger Nhe3 gene. J. Biol. Chem. 1996;271:10551–10559. doi: 10.1074/jbc.271.18.10551. [DOI] [PubMed] [Google Scholar]
- 26.Li R. S., Abrahamsen M. S., Johnson R. R., Morris D. R. Complex interactions at a GC-rich domain regulate cell type-dependent activity of the ornithine decarboxylase promoter. J. Biol. Chem. 1994;269:7941–7949. [PubMed] [Google Scholar]
- 27.Porter W., Saville B., Hoivik D., Safe S. Functional synergy between the transcription factor Sp1 and the estrogen receptor. Mol. Endocrinol. 1997;11:1569–1580. doi: 10.1210/mend.11.11.9916. [DOI] [PubMed] [Google Scholar]
- 28.Suske G. The Sp-family of transcription factors. Gene. 1999;238:291–300. doi: 10.1016/s0378-1119(99)00357-1. [DOI] [PubMed] [Google Scholar]
- 29.McEwen D. G., Ornitz D. M. Regulation of the fibroblast growth factor receptor 3 promoter and intron I enhancer by Sp1 family transcription factors. J. Biol. Chem. 1998;273:5349–5357. doi: 10.1074/jbc.273.9.5349. [DOI] [PubMed] [Google Scholar]
- 30.Magan N., Szremska A. P., Isaacs R. J., Stowell K. M. Modulation of DNA topoisomerase IIα promoter activity by members of the Sp (specificity protein) and NF-Y (nuclear factor Y) families of transcription factors. Biochem. J. 2003;374:723–729. doi: 10.1042/BJ20030032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Udvadia A. J., Rogers K. T., Higgins P. D., Murata Y., Martin K. H., Humphrey P. A., Horowitz J. M. Sp-1 binds promoter elements regulated by the RB protein and Sp-1-mediated transcription is stimulated by RB coexpression. Proc. Natl. Acad. Sci. U.S.A. 1993;90:3265–3269. doi: 10.1073/pnas.90.8.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perkins N. D., Agranoff A. B., Pascal E., Nabel G. J. An interaction between the DNA-binding domains of RelAp65 and Sp1 mediates human immunodeficiency virus gene activation. Mol. Cell. Biol. 1994;14:6570–6583. doi: 10.1128/mcb.14.10.6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gashler A., Sukhatme V. P. Early growth response protein 1 (EGR-1): prototype of a zinc-finger family of transcription factors. Prog. Nucleic Acid Res. Mol. Biol. 1995;50:191–224. doi: 10.1016/s0079-6603(08)60815-6. [DOI] [PubMed] [Google Scholar]
- 34.Mackman N. Regulation of the tissue factor gene. FASEB J. 1995;9:883–889. doi: 10.1096/fasebj.9.10.7615158. [DOI] [PubMed] [Google Scholar]
- 35.Schwachtgen J. L., Houston P., Campbell C., Sukhatme V., Braddock M. Fluid shear stress activation of EGR-1 transcription in cultured human endothelial and epithelial cells is mediated via the extracellular signal-related kinase 1/2 mitogen-activated protein kinase pathway. J. Clin. Invest. 1998;101:2540–2549. doi: 10.1172/JCI1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khachigian L. M., Lindner V., Williams A. J., Collins T. EGR-1-induced endothelial gene expression: a common theme in vascular injury. Science. 1996;271:1427–1431. doi: 10.1126/science.271.5254.1427. [DOI] [PubMed] [Google Scholar]
- 37.Ackerman S., Minden A., Williams G., Bobonis C., Yeung C. Functional significance of an overlapping consensus binding motif for Sp1 and Zif268 in the murine adenosine deaminase gene promoter. Proc. Natl. Acad. Sci. U.S.A. 1991;88:7523–7527. doi: 10.1073/pnas.88.17.7523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arenander A. T., Lim R. W., Varnum B. C., Cole R., de Vellis J., Herschman H. R. TIS gene expression in cultured rat astrocytes: multiple pathways of induction by mitogens. J. Neurosci. Res. 1989;23:257–265. doi: 10.1002/jnr.490230303. [DOI] [PubMed] [Google Scholar]
- 39.Mechtcheriakova D., Wlachos A., Holzmuller H., Binder B. R., Hofer E. Vascular endothelial cell growth factor-induced tissue factor expression in endothelial cells is mediated by EGR-1. Blood. 1999;93:3811–3823. [PubMed] [Google Scholar]
- 40.Ebinu J. O., Bottorff D. A., Chan E. Y., Stang S. L., Dunn R. J., Stone J. C. RasGRP, a Ras guanyl nucleotide-releasing protein with calcium- and diacylglycerol-binding motifs. Science. 1998;280:1082–1086. doi: 10.1126/science.280.5366.1082. [DOI] [PubMed] [Google Scholar]
- 41.El-Shemerly M. Y., Besser D., Nagasawa M., Nagamine Y. 12-O-tetradecanoylphorbol-13-acetate activates the Ras/extracellular signal-regulated kinase (ERK) signaling pathway upstream of SOS involving serine phosphorylation of Shc in NIH3T3 cells. J. Biol. Chem. 1997;272:30599–30602. doi: 10.1074/jbc.272.49.30599. [DOI] [PubMed] [Google Scholar]
- 42.Caloca M. J., Garcia-Bermejo M. L., Blumberg P. M., Lewin N. E., Kremmer E., Mischak H., Wang S., Nacro K., Bienfait B., Marquez V. E., Kazanietz M. G. β2-Chimaerin is a novel target for diacylglycerol: binding properties and changes in subcellular localization mediated by ligand binding to its C1 domain. Proc. Natl. Acad. Sci. U.S.A. 1999;96:11854–11859. doi: 10.1073/pnas.96.21.11854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Lint J., Rykx A., Maeda Y., Vantus T., Sturany S., Malhotra V., Vandenheede J. R., Seufferlein T. Protein kinase D: an intracellular traffic regulator on the move. Trends Cell Biol. 2002;12:193–200. doi: 10.1016/s0962-8924(02)02262-6. [DOI] [PubMed] [Google Scholar]