Abstract
A homology model of the housefly voltage-gated sodium channel was developed to predict the location of binding sites for the insecticides fenvalerate, a synthetic pyrethroid, and DDT an early generation organochlorine. The model successfully addresses the state-dependent affinity of pyrethroid insecticides, their mechanism of action and the role of mutations in the channel that are known to confer insecticide resistance. The sodium channel was modelled in an open conformation with the insecticide-binding site located in a hydrophobic cavity delimited by the domain II S4-S5 linker and the IIS5 and IIIS6 helices. The binding cavity is predicted to be accessible to the lipid bilayer and therefore to lipid-soluble insecticides. The binding of insecticides and the consequent formation of binding contacts across different channel elements could stabilize the channel when in an open state, which is consistent with the prolonged sodium tail currents induced by pyrethroids and DDT. In the closed state, the predicted alternative positioning of the domain II S4-S5 linker would result in disruption of pyrethroid-binding contacts, consistent with the observation that pyrethroids have their highest affinity for the open channel. The model also predicts a key role for the IIS5 and IIIS6 helices in insecticide binding. Some of the residues on the helices that form the putative binding contacts are not conserved between arthropod and non-arthropod species, which is consistent with their contribution to insecticide species selectivity. Additional binding contacts on the II S4-S5 linker can explain the higher potency of pyrethroid insecticides compared with DDT.
Keywords: 1,1,1-trichloro-2,2-bis(p-chlorophenyl)ethane (DDT); docking; knockdown resistance (kdr); molecular modelling; pyrethroids; sodium channel
Abbreviations: DDT, 1,1,1-trichloro-2,2-bis(p-chlorophenyl)ethane; kdr, knockdown resistance; Kv, voltage-gated potassium channel; KvAP, Aeropyrum pernix Kv channel; KcsA, Streptomyces lividans potassium channel; Mthk, Methanobacterium thermo-autotrophicum potassium channel; LD, lethal dose; para, paralytic
INTRODUCTION
The selective conductance of sodium ions across the plasma membrane by the voltage-gated sodium channel underlies the propagation of action potentials in neuronal cells of both vertebrates and invertebrates. The critical role of the sodium channel in the functioning of the nervous system has made it the target of a diverse array of toxins during evolution (reviewed in [1]). During an action potential the sodium channel undergoes transitions between closed-resting, activated and inactivated functional states, and toxins binding to specific sites on the channel either alter the equilibrium between these functional states or block the channel pore [2]. There are at least ten separate binding sites for ligands on the sodium channel, including those for local anaesthetics and anti-convulsants. The binding site for the lipid soluble DDT [1,1,1-trichloro-2,2-bis(p-chlorophenyl)ethane] and pyrethroid insecticides has been classified as site ‘7’ [1], but attempts to determine the exact location of this site using conventional techniques have been problematic [3]. The DDT and pyrethroid insecticides preferentially target the open state of the voltage-gated sodium channel [4,5], and their binding appears to stabilize the open state, thereby inhibiting the transition to the non-conducting deactivated or inactivated states [6,7]. Consequently, the inward conductance of sodium is prolonged, and this is observed as a slowly-decaying tail current during voltage-clamp experiments. The persistent depolarization of the plasma membrane resulting from the prolonged inward sodium conductance induces repetitive nerve firing and hyperexcitability, leading to paralysis and death of the insect. These channel modulators also have a greater affinity for insect over mammalian sodium channels, which coupled with differences in detoxification rates and temperature-dependent activity, make them potent and selective insecticides.
Prior to the 1970s DDT was used intensively, but following concerns over its environmental impact its use was discontinued or banned in most countries (it is, however, still the insecticide of choice for the control of malaria-transmitting mosquitoes in many countries where alternative controls have failed). Pyrethroids, with their greater selectivity and potency for insect sodium channels, low mammalian-toxicity and low persistence in the environment, became the widely favoured alternative and today still command approx. 17% of the world insecticide market. Unfortunately, resistance mechanisms selected for by continuous DDT exposure, also confer cross-resistance to the pyrethroids, decreasing the efficacy of this class of insecticides. Increasingly resistance to pyrethroids poses a serious threat to many pest-control programs.
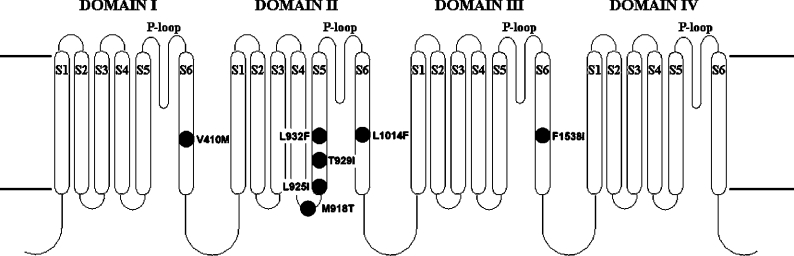
An important mechanism of resistance to DDT and pyrethroids is termed knockdown resistance or ‘kdr’ and this is known to result from alterations within the insect's (or arachnid's) voltage-gated sodium channel, rendering it less sensitive to the toxic effects of these compounds (reviewed in [8,9]). The mutations that cause resistance are most commonly found in the domain II region of the channel protein (Figure 1) where five different residues have been implicated to date: Met918 in the IIS4-IIS5 linker, Leu925, Thr929 and Leu932 in IIS5 and Leu1014 in IIS6. The most common mutation is L1014F, originally found in houseflies [10], but now reported for at least seven additional pest species [9]. This mutation is commonly referred to as the kdr mutation and is associated with moderate (10–30-fold) resistance to DDT and pyrethroids. Variant replacements of this residue, L1014H and L1014S, have also been described [11,12]. Mutations at the II S4-S5 linker and IIS5 residues are generally associated with higher levels of resistance (up to 1000-fold) and are often referred to as ‘super-kdr’ mutations. These include M918T (also first found in houseflies), L925I, T929I and T929V [10,13–16]. These mutations are often, but not always, found in combination with L1014F (for example see [16]). An interesting feature of the M918T mutation, is that when incorporated into the Drosophila para (paralytic) voltage-gated sodium channel gene and expressed in Xenopus laevis oocytes, it confers strong insensitivity to pyrethroids but does not confer cross-insensitivity to DDT [17]. This residue is also responsible (at least in part) for insect selectivity of pyrethroids; mammalian sodium channels contain isoleucine at this position and replacement of the isoleucine residue by methionine in the rat brain voltage-gated sodium channel (Nav 1.2) resulted in a 100-fold increase in its sensitivity to the pyrethroid, deltamethrin [18]. Mutations have also been reported outside of domain II, and are often located in the corresponding S4-S5, S5 and S6 segments of domains I and III (see [9]). Of these, only two, V410M in IS6 and F1538I in IIIS6 [19,20], have been shown to confer insensitivity to pyrethroids by functional assays.
Figure 1. Transmembrane topology of the voltage-gated sodium channel.
The pore-forming α-subunit consists of a single polypeptide chain with four internally homologous domains (I–IV), each having six transmembrane helices (S1–S6). The domains assemble to form a central aqueous pore, lined by the S5, S6 and S5-S6 linkers (P-loops). The identity and location of mutations associated with kdr are shown, with residues numbered according to the sequence of the housefly voltage-gated sodium channel.
The crystal structures of bacterial potassium channels have previously provided structural templates for homology modelling of the voltage-gated sodium channel pore domain (e.g. [21]). The recent publication of a 2.9 Å (1 Å=0.1 nm) resolution crystal structure of the rat brain Kv1.2 channel [22,23], a member of the Shaker family of voltage-gated potassium channels, allows the extension of this homology modelling to encompass the voltage sensor domains and S4-S5 linkers of the sodium channel. In the present study, we have used the structure of Kv1.2 as a template with which to generate a homology model for the housefly voltage-gated sodium channel in an open conformation. We have further applied a combination of protein–ligand modelling and docking procedures, in conjunction with existing experimental data on DDT/pyrethroids and sodium channel-associated insecticide resistance, to produce a hypothesis as to how these ligands interact with insect and arachnid sodium channels.
EXPERIMENTAL
Homology model generation
The S4 to S5 regions (residues 312–347) and P-loop helices (residues 362–372) from the X-ray crystal structure (PDB accession number 2A79) [22,23] of the Shaker rat-brain Kv1.2 channel and the S6 helices (residues 210–240) from the crystal structure (PDB accession number 1ORQ) [24] of KvAP (Aeropyrum pernix Kv channel) provided the structural template for a homology model of the housefly voltage-gated sodium channel in an activated state. The model was produced by superimposing the co-ordinates of an S6 helix from the crystal structure of KvAP on to each of the S6 helices of the Kv1.2 crystal structure using the SUPERIMPOSE program of the CCP4 suite [25]: the backbone atoms of residues 219–221 and 231–233 of KvAP were superimposed on to residues 397–399 and 410–412 of Kv1.2 respectively, to overlay the conserved ‘gating-hinge’ glycines, the cytoplasmic ends of the S6 helices were positioned so as to make contact with the S4-S5 linkers.
ClustalW [26] was used to align the amino acid sequences of the housefly sodium channel (Swiss-Prot accession number Q94615) with the Kv1.2 (Swiss-Prot accession number P63142) and KvAP (Swiss-Prot accession number Q9YDF8) channels. The local sequence identity of ClustalW alignments between the S4-S5 linker and S5 helical regions of Kv1.2 and the housefly sodium channel domain II was 28%, with an especially high sequence conservation in the positions for leucine and phenylalanine residues (Figure 4). The two sequences share the same pattern of amphipathicity with the S4-S5 linker and conservation of a glycine at the turn joining the S4-S5 linker and the S5 helix. Alignment of the KvAP S6 helix with housefly sodium channel S6 helices was centred on the gating-hinge glycine. The register of the sodium channel S6 helices was such that the side-chains of residues which are known to form interactions with pore-blocking local anaesthetics and batrachotoxin lined the pore.
Figure 4. Sequence alignments of the Kv1.2 channel and housefly voltage-gated sodium channel (Nav) S4-S5 linker and S5 helix, with the bottom line showing identical (*), conserved (:) and semi-conserved (.) substitutions.
Residues in the template model were changed to the housefly sodium channel sequence using the Biopolymer module of SYBYL [Version 7.0, Tripos Inc., St. Louis, MO, U.S.A.]. The final model was subjected to 500 rounds of conjugate-gradient minimization in SYBYL using the Tripos force-field [27].
Automated and manual ligand docking
The crystal structures of deltamethrin (reference code PXBVCP10) and DDT (reference code CPTCET10) were obtained from the Cambridge Structural Database (http://www.ccdc.cam.ac.uk/). The structures of fenvalerate, acrinathrin and bifenthrin were generated from the structure of deltamethrin using SYBYL to replace atoms that differed between the pyrethroids.
SYBYL was used for adding hydrogen atoms and calculating Gasteiger partial charges [28] for fenvalerate. Automated docking of fenvalerate with the model of the sodium channel was performed using the AutoDock 3.0 software package [29]. Flexible torsions of fenvalerate were defined using AUTOTORS. Grid maps with 60×60×60 points were constructed and the grid point spacing was 0.375 Å. The grids were centred on the side-chain of either Met918 or Thr929 in the housefly voltage-gated sodium channel model. Docking simulations were performed using a Lamarckian genetic algorithm [30] with the following parameters: population size=50, mutation rate=0.02, crossover rate=0.8. The number of docking runs per simulation was 50, and simulations for docking fenvalerate in the vicinity of either Met918 or Thr929 were run ten times, giving a total of 500 docking predictions. All calculations were performed on a 4-processor Compaq Alphaserver ES40. Docking predictions that had a calculated positive binding free-energy value i.e. 75 out of the 500 predictions that were run, were rejected. For the remaining 425 predictions the CCP4 program CONTACT [25] was used to screen for docking predictions where the ligand came within 4 Å of both Met918 and Thr929 (158 predictions), or atoms of the acid moiety's dimethyl group and aromatic ring came within 4 Å distance of the receptor (186 predictions). From the predictions that satisfied these conditions three docking results were chosen that: (i) had favourable binding contacts between the receptor site and the acid, ester or alcohol moieties of the pyrethroid, which is consistent with experimental data from in vivo and in vitro structure-activity studies, (ii) had binding contacts with other residues (in addition to Met918 and Thr929) known to be important in insecticide resistance, and (iii) could be incorporated into a single manually-generated conformation of fenvalerate bound with the model. Another 50 AUTODOCK predictions were subsequently run using this ‘optimal’ conformation of fenvalerate with fixed bond angles (i.e. rigid-body docking).
Manual docking of other insecticide structures with the sodium channel model and subsequent rotation about bonds in the ligand were carried out using SYBYL. A total of 500 rounds of conjugate-gradient minimization were carried out in SYBYL using the Tripos force-field after each manual docking of a ligand. The program CONTACT [25] was used to identify residues in the sodium channel model that are ≤4 Å distant from docked ligands. Figures were produced using the PyMOL molecular graphics system (DeLano Scientific, San Carlos, CA, U.S.A.).
RESULTS
Modelling of the voltage-gated sodium channel
The homology model of the housefly voltage-gated sodium channel in an open conformation is largely based on the X-ray structure of the Kv1.2 channel (PDB accession number 2A79). The Kv1.2 crystal structure is the first determination of a mammalian voltage-gated channel structure, and a key structural insight is the positioning of the S4-S5 linkers in an activated-state conformation. Each voltage sensor domain, formed by the S1 to S4 transmembrane helices, is packed loosely against the S5 helix of an adjacent monomer, and each α-helical S4-S5 linker crosses the cytoplasmic end of the S6 helix of the same domain. In addition, the first four positive charges of the S4 helices, which carry most of the gating charge in Shaker channels [31], are located on the extracellular side of the predicted membrane region, which is consistent with the outward movement of the S4 sections during activation. The pore-lining S6 helices of Kv1.2 are kinked at a conserved sequence of amino acid residues, ‘PXP’, which acts to disrupt the intra-helical hydrogen bonding, allowing the cytoplasmic ends of the S6 helices to splay apart and form a large pore opening of approx. 12 Å, which is consistent with the channel adopting an activated conformation.
Whereas the PXP hinge motif is conserved amongst members of the Shaker family of eukaryotic voltage-gated potassium channels, it has not been found in voltage-gated sodium channels or in bacterial potassium channels. C-terminal to this motif at the ‘gating-hinge’ position is a glycine residue that plays a crucial role in the activation of Shaker channels [32]. The conservation of this gating-hinge glycine in the S6 helices of KvAP (PDB accession number 1ORQ) [24] and domains I–III of the housefly sodium channel (Figure 2) gave us confidence that the S6 helix of KvAP could be superimposed on to Kv1.2 to provide an accurate template for sodium channel modelling.
Figure 2. Sequence alignments of the KvAP channel and housefly voltage-gated sodium channel (Nav) S6 helices from domains I–IV.
Glycine residues conserved in the gating-hinge position are shown in bold. Resistance-associated residues Val410, Leu1014 and Phe1538 are underlined. Residue numbers are shown on the right.
The S5-S6 linkers (P-loops) form the extracellular mouth and selectivity filter of homologous potassium, sodium and calcium channels. The section of the P-loop that forms the selectivity filter in potassium channels is preceded by an α-helix that has its C-terminus orientated towards the centre of the pore. These P-loop ‘pore-helices’ are believed to contribute a localized electronegative charge to this region of the pore due to the helix-dipole effect [33], thus reducing the energetic barrier encountered by a cation traversing the poorly-polarizable environment of a lipid bilayer [34]. Although the extended P-loop section that forms the selectivity filter in sodium channels is by necessity different from potassium channels, functional evidence and secondary structure predictions suggest that the pore-helices have been conserved during evolution [35]. Therefore only the co-ordinates of the pore-helices of the Kv1.2 P-loops were used for homology modelling of the equivalent sections in the sodium channel.
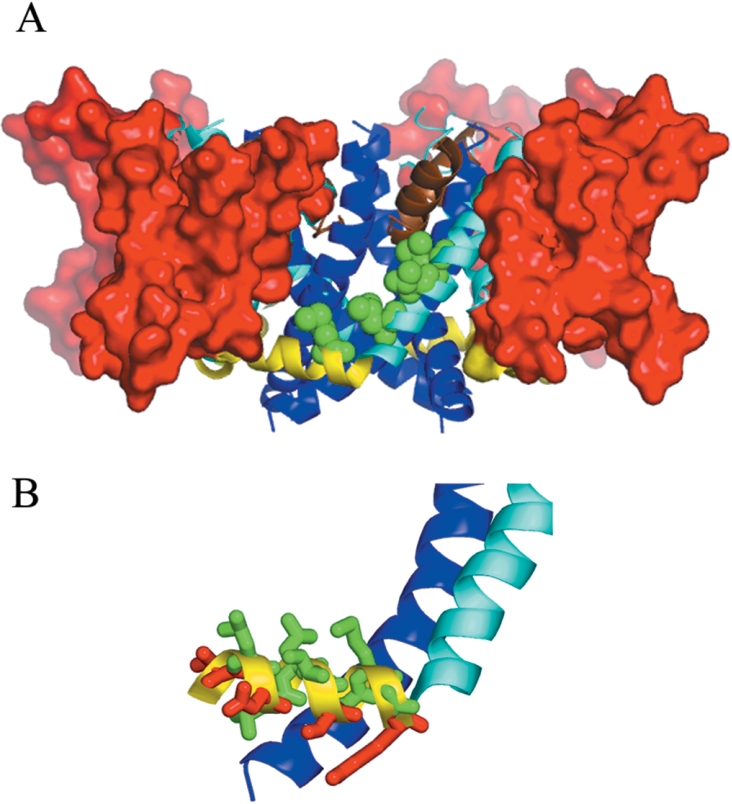
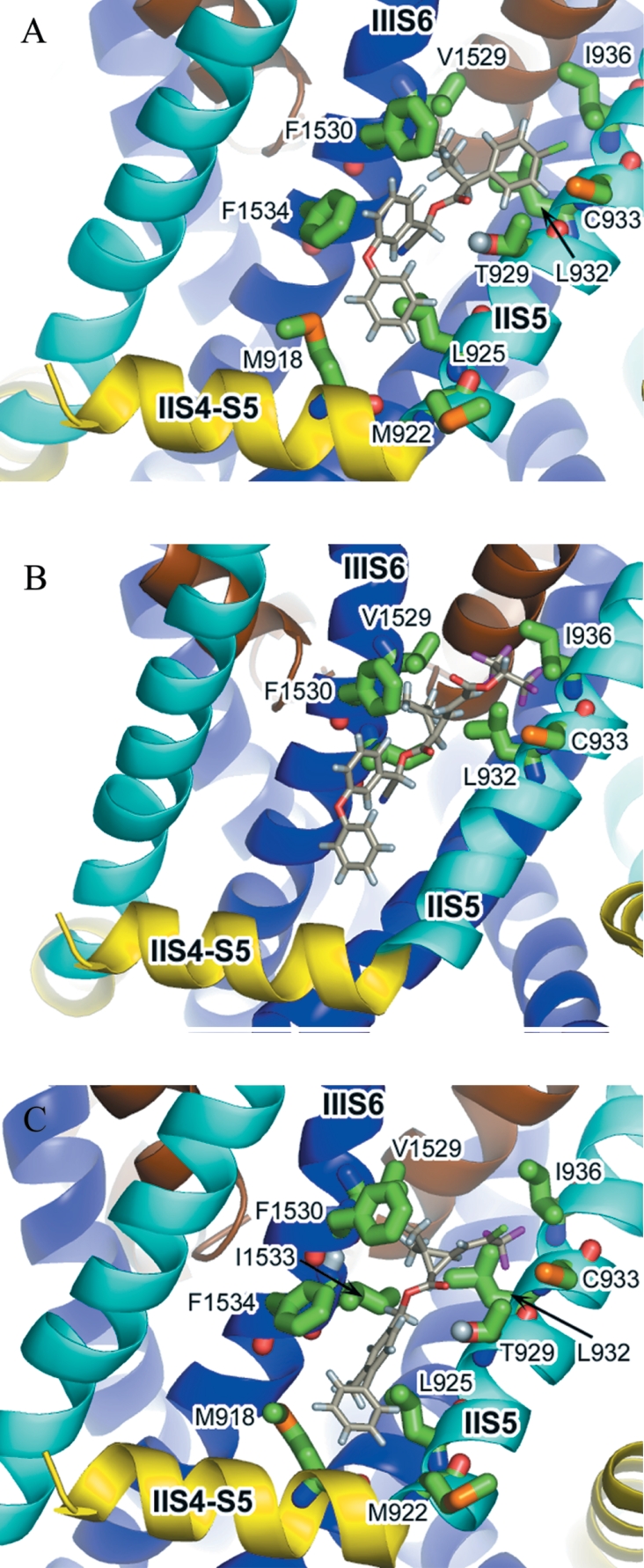
The model of the housefly voltage-gated sodium channel, incorporating the key features described above, is shown in Figure 3(A). The II S4-S5 linker of the sodium channel was modelled as an amphipathic α-helix (the sequence alignment used for modelling of this region is shown in Figure 4) with polar and charged amino acids orientated towards the predicted membrane-cytoplasm interface, whereas the opposite side of the α-helix is lined with hydrophobic residues, including Met918 (Figure 3B). The hydrophobic face of the II S4-S5 linker forms the lowest section of a hydrophobic cavity that is also delimited by the S5 helix of domain II and the S6 helix of domain III. This cavity is lined by the side-chains of key residues previously implicated in pyrethroid binding, including Met918, Leu925, Thr929 and Leu932. As the voltage sensor domains are not positioned to occlude the cavity (Figure 3A), the site is predicted to be accessible to the lipid bilayer and thus lipid-soluble insecticide molecules. Docking studies of pyrethroids and DDT were therefore undertaken to assess whether this cavity could be the insecticide-binding site.
Figure 3. Model of the activated-state of the housefly voltage-gated sodium channel.
(A) The voltage sensor domains are shown in surface-filling representation (red). The S4-S5 linkers, S5 helices, pore helices and S6 helices are shown as ribbons (yellow, cyan, brown and blue respectively). Residues Met918, Leu925, Thr929 and Leu932 are shown in space-filling representation (green). (B) The II S4-S5 linker showing its amphipathicity. The S4-S5 linker, S5 and S6 helices are coloured as described above. Residues on the II S4-S5 linker that are hydrophobic (Leu911, Leu914, Ile915, Ile917, Met918, Gly919) are shown in green, and polar or charged residues (Thr910, Asn912, Ser916, Arg920) are in shown red.
Automated docking predictions for fenvalerate
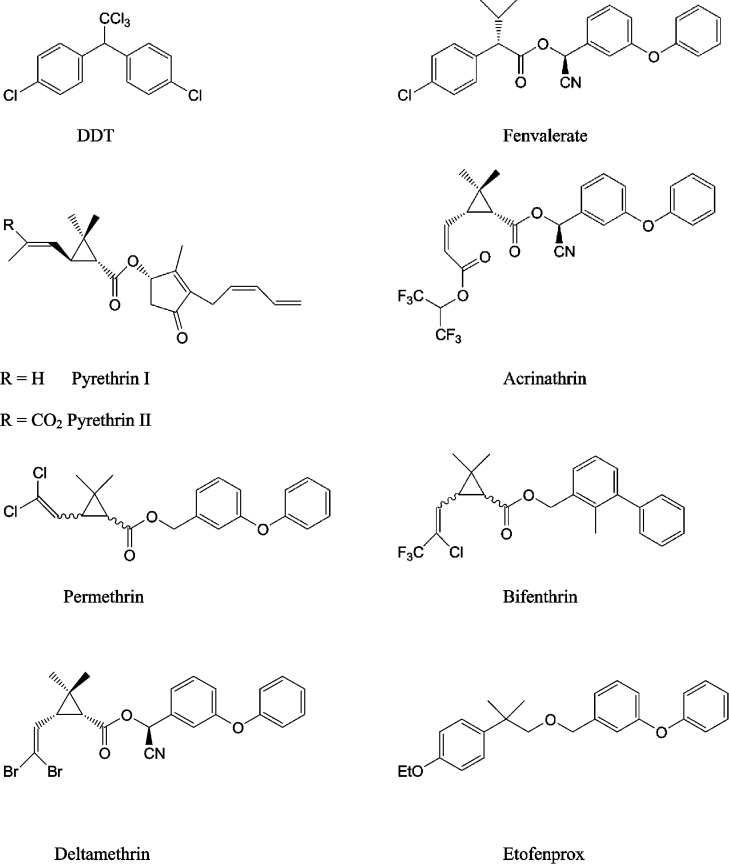
Pyrethroids have a defined structure–activity relationship that is based on the physical properties, shape and the three-dimensional configuration of the entire molecule [36]. Figure 5 illustrates that a wide variety of chemical structures have been shown to satisfy these requirements. Historically, pyrethroids have been classified as either Type I or Type II, associated with the absence (Type I) or presence (Type II) of an α-cyano group. The Type II pyrethroids are generally the more potent insecticides.
Figure 5. Chemical structures of DDT and some pyrethroid insecticides.
The programme AUTODOCK was used to generate docking predictions for the Type II pyrethroid fenvalerate in the vicinity of two key residues implicated in pyrethroid binding, namely Met918 and Thr929. Fenvalerate was chosen for these studies because of the close resemblance of its acid moiety to DDT (as described below). Energetically favourable docking predictions (i.e. those with calculated negative values for binding free-energy) were analysed to assess the binding interactions between residues in the protein model and the acid moiety, ester group and alcohol moiety of fenvalerate (Figure 6A–6C). Favourable binding interactions which were consistent with published experimental data are discussed below.
Figure 6. Docking predictions for the acid, ester and alcohol moieties of fenvalerate with the housefly voltage-gated sodium channel model.
(A) Residues ≤4 Å from the acid moiety of fenvalerate are shown in stick representation. Residues found to be non-conserved between insect and non-insect species (see Figure 7) are shown in red. The estimated free-energy of binding (ΔGb) from Autodock [29] for this docking prediction is −2.83 kcal/mol (4.184 cal≡1 J). (B) The program HBPLUS [47] was used to identify the presence of a hydrogen bond between the carbonyl group of fenvalerate and the hydroxyl group of Thr929 (ΔGb=−0.95 kcal/mol). (C) Residues ≤4 Å from the alcohol moiety of fenvalerate are shown in stick representation (ΔGb=−1.71 kcal/mol).
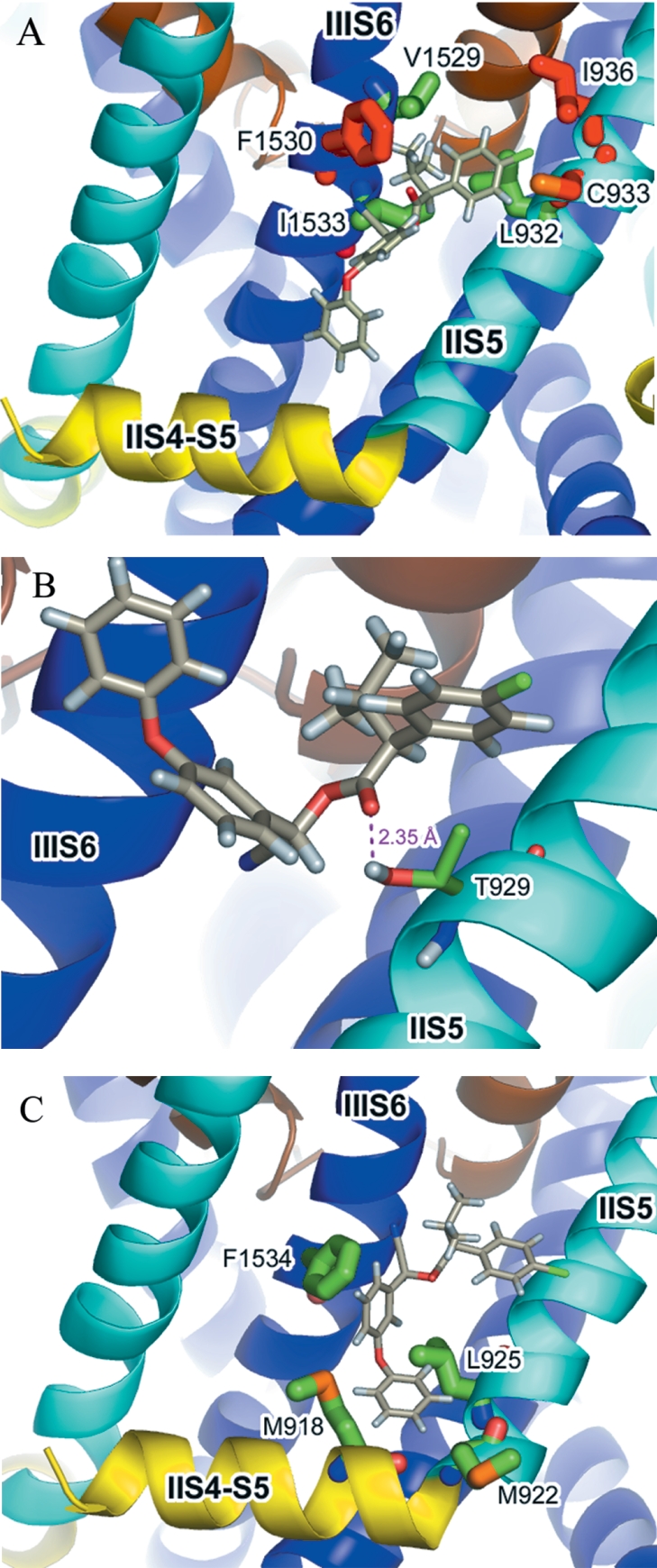
Figure 6(A) shows the acid moiety of fenvalerate docked between the IIS5 and IIIS6 helices. The dimethyl group of fenvalerate is located in a small hydrophobic pocket delimited on one side by the side-chain of Phe1530 on IIIS6; the chlorophenyl group lies within 4 Å of residues Ile936 and Cys933 on IIS5. In a region of otherwise high sequence identity, residues Ile936, Cys933 and Phe1530 are non-conserved between sodium channels in arthropods and non-arthropods (Figure 7) and may partly underlie the species-dependent binding affinity of pyrethroids. Figure 6(B) shows a docking prediction in which a hydrogen bond could be formed between the carbonyl oxygen of the pyrethroid ester group and the hydroxyl group of the resistance-associated residue, Thr929. An alternative or additional hydrogen-bonding interaction could also form between the α-proton of the fenvalerate ester group and the side-chain oxygen of Thr929 (2.42 Å in Figure 6B). A docking prediction for the 3-phenoxybenzyl alcohol moiety of fenvalerate is shown in Figure 6(C). The first aromatic ring of the alcohol moiety makes a favourable edge-to-face aromatic–aromatic contact (approx. 5 Å distance between the rings' centroids) with the side-chain of Phe1534. Such interactions, in which the partial positive charge on the edge of one ring is attracted to the partial negative charge at the face of the other ring to generate an electrostatic interaction, are widely reported in the literature [37]. A second key interaction is an edge-on interaction between the second aromatic ring of the alcohol moiety and the sulphur atom on the side-chain of Met918 (located on the II S4-S5 linker). Such an interaction has been previously reported for methionine sulphur–aromatic (protein–ligand) complexes [37]. Mutation of the Met918 residue to a non-sulphur containing threonine is known to affect dramatically the sensitivity of the sodium channel to pyrethroids, whereas a cysteine introduced at this position fully restores activity [38]. An additional resistance-associated residue that makes contact with the alcohol moiety of fenvalerate is Leu925. Although the mutation L925I is a conservative substitution, it is possible that this minor change in the side-chain volume of the amino acid might sufficiently alter the shape of the hydrophobic cavity so as to impair binding with the alcohol moiety of a pyrethroid.
Figure 7. Sequence alignment showing the location of hydrophobic residues in the postulated pyrethroid binding site that differ (in bold) between arthropod and non-arthropod species.
A model of fenvalerate binding
The structure of fenvalerate was manually docked into the model of the housefly voltage-gated sodium channel in a conformation incorporating the favourable binding interactions discussed for the acid, ester and alcohol moieties. The model of bound fenvalerate (Figure 8A) was generated using the docking predictions in Figure 6(A) and rotating single bonds in the ester and alcohol groups to form the interactions shown in Figures 6(B) and 6(C) respectively. This approach was guided by the ‘zipper’ hypothesis of Elliott [39], which proposes that one part after another of the pyrethroid molecule binds to its counterpart on the receptor site after attaining its optimum orientation. Most pyrethroids have freedom of rotation around several bonds with rotational energy barriers of less than 70 kJ mol−1, and thus are highly flexible. In the composite model, the optimally docked pyrethroid is surrounded by residues located on the II S4-S5 linker (Met918), IIS5 helix (Leu925, Thr929 and Leu932) and the cytoplasmic end of IIIS6 that have been identified as key resistance loci. The most common resistance mutation, L1014F, which gives the kdr rather than super-kdr phenotype, does not however appear to be a binding determinant, as it is located a considerable distance away from the main binding-pocket in our model. When automated docking of fenvalerate was repeated with the rigid-body conformation shown in Figure 8(A), the pyrethroid docked in the binding pocket with a calculated binding energy of −4.68 kcal/mol.
Figure 8. Optimized docking predictions for pyrethroids at the voltage-gated sodium channel.
(A) Fenvalerate docked in the postulated pyrethroid-binding site. Binding interactions between the acid moiety, ester group and alcohol moiety as shown in Figure 6(A), 6(B) and 6(C) respectively, were retained during the manual docking of fenvalerate with the housefly sodium channel model. (B) Acrinathrin docked in the postulated pyrethroid-binding site. Residues that are ≤4 Å from the acid moiety are shown in stick representation. (C) Bifenthrin docked in the postulated pyrethroid-binding site.
Structure–activity relationships and pyrethroid binding
The significance of overall molecular shape in the action of pyrethroids is most clearly demonstrated by the stringent stereospecificity required for insecticidal action. This is clearly shown in our model by attempting the docking of the inactive 1R stereoisomer of fenvalerate (which differs from the active 1S form in its stereochemistry about the chiral centre in the acid moiety) into the postulated binding site. Superimposing the inactive 1R structure on to the docked 1S conformation of fenvalerate results in steric hindrance (with the protein) when either the chlorophenyl or dimethyl groups are superimposed.
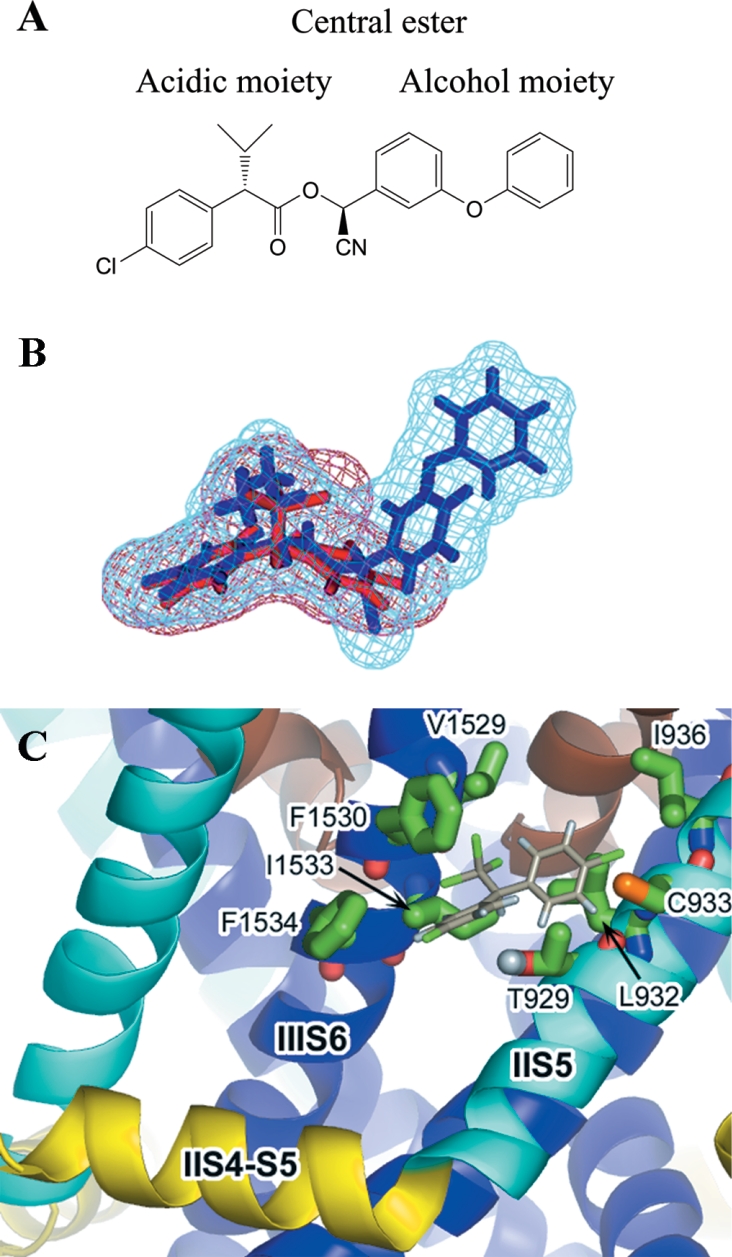
The acid moiety of a pyrethroid is tolerant to large degrees of substitution (both steric and electronic) before insecticidal activity is lost [40]. In our model, it would be possible to accommodate these types of substitutions in the narrow pocket that extends beyond where the acid moiety of fenvalerate docks, for example as shown in the docking prediction for acrinathrin (Figure 8B). The extra length of the acid moiety of this pyrethroid might explain its high insecticidal activity against mites and ticks (possibly overcoming the substitution of a glycine residue for cysteine at position 933 in these species [Figure 7]).
The stereochemical characteristics of the central region of the pyrethroid molecule (around the ester linkage) are known to be especially important for association with the binding site, since replacement of the central ester bond with other groups always leads to a substantial loss of activity unless the substituted group retains the overall configuration of the pyrethroid molecule (e.g. as in the case of the non-ester pyrethroid etofenprox).
The maximum overall dimensions for a pyrethroid's alcohol moiety are represented by two benzene rings with a bridging atom. However, both the nature and the orientation of the groups present are also critical for the pyrethroid's activity. For example, typical commercial pyrethroids have a substituted benzyl alcohol moiety. When the substituent is a cyclic group, (e.g. phenoxy or phenyl) activity is higher when attached at the meta- rather than para-position. However, introduction of an α-cyano group at the benzylic position enhances activity only in the case of a phenoxy group (or sterically equivalent groups such as benzyl and benzoyl). When the substituent is an acyclic moiety, the position of substitution (meta or para) is less critical. Furthermore, in this case introduction of an α-cyano group rarely leads to an increased activity. Non-coplanarity of the meta-substituent is seen for both the phenoxy group of docked fenvalerate (Figure 8A) and the phenyl group in bifenthrin (Figure 8C), but due to steric restriction near the side-chain of Ile1533 the α-cyano group can only be accommodated in the case of fenvalerate. The known constraints on the alcohol group are therefore seen to be consistent with the dimensions of the cleft in our model. Furthermore, a key requirement for pyrethroid binding appears to be a match in volume of the alcohol moiety with a hydrophobic cavity formed by the S4-S5 and S5 sections of the sodium channel domain II, and the optimal non-planar positioning of the alcohol's aromatic rings.
Binding predictions for DDT
Both DDT and pyrethroids exert a similar functional effect on the insect voltage-gated sodium channel, namely stabilization of the open channel state, slowing of repolarization after an action potential, and repetitive discharges. In addition, the L1014F mutation in the sodium channel confers a similar degree of kdr for both insecticides. These observations suggest that DDT and pyrethroids may share an overlapping binding site. Computer generated overlays of the crystal structure of DDT with a selection of pyrethroid compounds reveal that DDT shares a distinctive steric profile with pyrethroid acidic moieties, and this is particularly evident in the case of fenvalerate (Figures 9A and 9B). The binding prediction for DDT when this superimposition was repeated in the postulated pyrethroid binding site is shown in Figure 9(C). It can be predicted from this docking simulation that mutation of Thr929 should confer resistance to DDT. Previous studies of the T929I mutation in the diamond-back moth (Plutella xylostella) have shown that this is indeed the case, DDT being completely ineffective against larvae of the resistant Plutella strain [15]. Furthermore, it is apparent that DDT does not make contact with the Met918 residue on the II S4-S5 linker. This is consistent with experimental data which shows that mutation of Met918, a critical binding determinant for pyrethroids, has no effect on DDT affinity [17]. It is likely that the three non-conserved amino acid residues Cys933, Ile936 and Phe1530 (Figure 7), which form hydrophobic binding contacts for DDT, could provide the key to the insect selectivity of this compound.
Figure 9. Docking predictions for DDT.
(A) The acid and alcohol moieties of fenvalerate. (B) The structure of DDT superimposed on to the acid moiety of fenvalerate (based on an original overlay of deltamethrin and DDT produced by M. S. Williamson and H. Rassmussen, Rothamsted Research). The volumes of the two structures are shown in mesh format. The two dimethyl groups of fenvalerate overlap with the two chlorine atoms on the trichloro group of DDT. (C) Docking of DDT with the housefly voltage-gated sodium channel. The structure of DDT was superimposed on to the acid moiety of the fenvalerate docking simulation shown in Figure 8(A).
DISCUSSION
Proposed mechanism of insecticide activity
The X-ray crystal structures of potassium channels provide insight into how homologous members of the ion channel family are gated. Comparison of the closed-state KcsA (Streptomyces lividans potassium channel) [34] with the open-state Mthk (Methanobacterium thermo autotrophicum potassium channel) [41] and KvAP [24] channels shows that gating of the pore is associated with the pore-lining helices (S6 helices in voltage-gated channels) undergoing bending around a central glycine residue. Glycine is suitable as a gating-hinge as it can adopt a wide range of backbone dihedral angles, conferring flexibility on a polypeptide chain. A glycine also occupies the corresponding gating-hinge positions in the domain I, II and III S6 transmembrane helices of the voltage-gated sodium channel (Figure 2), thus implicating similar bending motions of the S6 helices during gating. In S6 of domain IV of the sodium channel the gating-hinge position is occupied by a serine, and the possible associated loss in helix flexibility is consistent with the observed distinctive gating kinetics of domain IV in this channel [42]. Further comparisons of the crystal structures of the closed-state KcsA and open-state MthK, KvAP and Kv1.2 channels shows that in addition to the major bending motions of the inner helices, small tilting and rotational motions of the outer (S5) helices relative to the inner (S6) helices occur during gating. As the acid moiety of the pyrethroid fenvalerate is bound between the IIS5 and IIIS6 helices in our model of the housefly voltage-gated sodium channel, the effect of the pyrethroid may be to impede the structural rearrangements in that region that are required to close the pore. The mechanism proposed for the insecticidal activity of the acid moiety of pyrethroids may also be relevant to DDT, with the IIS5 and IIIS6 helices being stabilized in their active conformation by insecticide binding. Stabilization of the IIIS6 and possibly IIS6 (through its interaction with IIS5) helices in their bent activated conformation could preserve the open conformation of the pore, which is consistent with the prolonged sodium tail currents observed with DDT- and pyrethroid-modified sodium channels. In addition, stabilization of a particular conformation of IIS6 and the III S4-S5 linker, and possibly other as yet unidentified regions of the channel, could prevent formation of the inactivation particle (i.e. the domain III-IV linker tripeptide) receptor-site [43] and thus account for the delayed inactivation of insecticide-modified channels.
The crystal structure of the mammalian Kv1.2 channel reveals how the pore domain (S5 and S6 helices) is coupled with the voltage sensor domains (S1-S4 helices). Long et al. [23] have proposed a mechanism that correlates voltage-sensor conformational changes with pore gating, whereby inward movement of the S4 helices during transition from the open- to closed-state, repositions the S4-S5 linkers closer to the cytoplasm [23]. The downward displacement of the S4-S5 linkers applies pressure on the corresponding S6 helices, straightening the helices and bringing them into close proximity to form a pore-blocking constriction at the pore's cytoplasmic end. Gating of voltage-gated channels is thus a function of the voltage-dependent positioning of the S4-S5 linkers and bending of the S6 helices. Our proposed mechanism for pyrethroid activity is that insecticide binding stabilizes the II S4-S5 linker and the IIS5 and IIIS6 helices (through interaction with the acid moiety, as discussed above) in their activated conformation. In the case of the II S4-S5 linker, the binding contacts, which include the key sulphur–aromatic interaction between Met918 and the alcohol moiety of fenvalerate, stabilize the linker conformation, allowing it to oppose inward movement of the voltage sensor. Such resistance of the II S4-S5 linker to downward displacement would maintain the IIS6 helix in a bent conformation, consistent with an open pore configuration. The change in position of Met918 when the II S4-S5 linker adopts the closed-state conformation could remove an important binding contact for pyrethroids, which may be a key determinant of their state-dependent binding affinity and their enhanced insecticidal potency compared with DDT.
Role of mutations in conferring insecticide resistance
Residues Met918, Thr929, Leu925 and Leu932 are predicted from our model to form close (≤4 Å) binding contacts with the pyrethroid, fenvalerate. These interactions are in agreement with previous experimental data obtained from resistant pest populations where mutations at these positions in the sodium channel confer resistance to pyrethroids. Furthermore, the function of these mutations in conferring resistance has been confirmed by in vitro experiments using gene expression in Xenopus oocytes. In the case of the two super-kdr substitutions M918T and T929I, this resistance may be manifested through the elimination of polar interactions with the pyrethroid. As our model predicts that changing the alcohol moiety affects the activity of a pyrethroid (e.g. a second aromatic ring might provide a close fit within the cavity and thereby enhance pyrethroid affinity), it is also possible that the close fit suggested by the model would render the pyrethroid susceptible to displacement by mutations that produce small changes in the shape of the cavity, e.g. L925I.
Val410 and Leu1014 are two loci that confer kdr-type resistance to pyrethroids and DDT [10,44]. They are located near the centre of the IS6 and IIS6 helices respectively and are located within approx. 11 Å of each other in our predicted three-dimensional structure. However, both loci are positioned relatively distant to the predicted binding cavity. Mutations at these positions lower the affinity of the open channels for pyrethroids by 10–30-fold, and decrease the availability of open channel states due to enhanced closed-state inactivation, thereby limiting the number of high-affinity binding sites available for pyrethroids [19,45]. It is not clear how these mutations exert such an attenuating effect but their close proximity to the gating-hinge position on each helix suggests they could impede the bending motions of the S6 helices necessary for channel opening, consistent with the effect of L1014F and V410M in shifting the equilibrium towards the closed state of the channel. Such a mutation (T529F) in the middle of the S6 segment of the rat rKv1.4 channel has a profound influence on channel activation and deactivation, destabilizing the open state and accelerating the rate of channel deactivation [46]. Another possibility is that mutation of Val410 or Leu1014 may sufficiently alter the packing of S5 and S6 helices to cause a displacement of residues on IIS5 and IIIS6 that are required for insecticide binding.
The mutation F1538I has been demonstrated to alter channel gating properties, producing a pyrethroid insensitive channel, and an aromatic residue at this position is critical for the action of pyrethroids [20]. Our housefly sodium channel model suggests that contact between the IIIS6 helix and II S4-S5 linker is limited to interactions between the side-chains of Phe1538, Leu914 and Met918. An F1538I substitution could disrupt these interactions and alter the relative positions of the IIIS6 helix and the II S4-S5 linker.
Key differences between Type I and Type II pyrethroids
In general, Type II pyrethroids delay the inactivation of the voltage-gated sodium channel substantially longer than do Type I compounds and their effects are less reversible, and it is these differences in the prolongation of open-channel times which probably contribute to the effectiveness of the Type II insecticides. In our model the α-cyano group on fenvalerate is located between IIS5 and IIIS6 in the vicinity of Ile1533 (on IIIS6). For pyrethroids such as fenvalerate with an α-cyano substituent, there is potential for a hydrogen bonding network between the alpha carbon proton, the ester carbonyl group and the side-chain hydroxyl group of Thr929. We postulate that positioning of the α-proton is also key for the interaction of the 3-phenoxybenzyl alcohol pyrethroids, in that it provides an additional contact point between the pyrethroid and the sodium channel only in the presence of an aromatic ring substituent in the meta position. The activity of non-cyclic substituents or substituents at the para position is not enhanced by the presence of the α-cyano group. It is further proposed that it is this additional interaction that distinguishes Type I and Type II pyrethroids, and may account for the increased effectiveness of compounds such as deltamethrin and fenvalerate. In sodium channels containing the M918T substitution, interaction with the 3-phenoxy group of the alcohol moiety is lost for both Type I and Type II pyrethroids. However, in the case of Type II pyrethroids, the additional interaction due to the α-carbon proton would also be negated, and therefore the level of insecticidal activity against the super-kdr insects is similar to that of the corresponding non-α-cyano compounds. Thus in vitro the LD50 (lethal-dose) of deltamethrin against super-kdr houseflies is similar to that of the corresponding analogue without an α-cyano group.
In summary, the modelling studies reported here have used the crystal structures of homologous ion channels and incorporated our current understanding of the conformational changes that occur during gating in order to model pyrethroid and DDT binding sites in the voltage-gated sodium channel. This has allowed us to postulate how these sites could form in a state-dependent manner and to propose a mechanism for the effect of insecticides on the channel's activity. In addition, our models are consistent with mutagenesis studies on insecticide resistance and have taken into account the structure activity relationships of pyrethroids.
Acknowledgments
This work was supported by a Biotechnology and Biological Sciences Research Council project grant (BBC5006281) to B.A.W. Rothamsted Research is supported by the Biotechnology and Biological Sciences Research Council of the United Kingdom.
References
- 1.Wang S. Y., Wang G. K. Voltage-gated sodium channels as primary targets of diverse lipid-soluble neurotoxins. Cell. Signalling. 2003;15:151–159. doi: 10.1016/s0898-6568(02)00085-2. [DOI] [PubMed] [Google Scholar]
- 2.Catterall W. A. Neurotoxins that act on voltage-sensitive sodium-channels in excitable-membranes. Annu. Rev. Pharmacol. Toxicol. 1980;20:15–43. doi: 10.1146/annurev.pa.20.040180.000311. [DOI] [PubMed] [Google Scholar]
- 3.Trainer V. L., McPhee J. C., Boutelet-Bochan H., Baker C., Scheuer T., Babin D., Demoute J. P., Guedin D., Catterall W. A. High affinity binding of pyrethroids to the α-subunit of brain sodium channels. Mol. Pharmacol. 1997;51:651–657. doi: 10.1124/mol.51.4.651. [DOI] [PubMed] [Google Scholar]
- 4.Leibowitz M. D., Schwarz J. R., Holan G., Hille B. Distinct modifications of sodium channels by alkaloids and insecticides. Biophys. J. 1986;49:A43. [Google Scholar]
- 5.Salgado V. L., Narahashi T. Immobilization of sodium-channel gating charge in crayfish giant-axons by the insecticide fenvalerate. Mol. Pharmacol. 1993;43:626–634. [PubMed] [Google Scholar]
- 6.Vijverberg H. P., van den Bercken J. Neurotoxicological effects and the mode of action of pyrethroid insecticides. Crit. Rev. Toxicol. 1990;21:105–126. doi: 10.3109/10408449009089875. [DOI] [PubMed] [Google Scholar]
- 7.Bloomquist J. R. Ion channels as targets for insecticides. Annu. Rev. Entomol. 1996;41:163–190. doi: 10.1146/annurev.en.41.010196.001115. [DOI] [PubMed] [Google Scholar]
- 8.Vais H., Williamson M. S., Devonshire A. L., Usherwood P. N. R. The molecular interactions of pyrethroid insecticides with insect and mammalian sodium channels. Pest Manag. Sci. 2001;57:877–888. doi: 10.1002/ps.392. [DOI] [PubMed] [Google Scholar]
- 9.Soderlund D. M., Knipple D. C. The molecular biology of knockdown resistance to pyrethroid insecticides. Insect Biochem. Mol. Biol. 2003;33:563–577. doi: 10.1016/s0965-1748(03)00023-7. [DOI] [PubMed] [Google Scholar]
- 10.Williamson M. S., Martinez-Torres D., Hick C. A., Devonshire A. L. Identification of mutations in the housefly para-type sodium channel gene associated with knockdown resistance (kdr) to pyrethroid insecticides. Mol. Gen. Genet. 1996;252:51–60. doi: 10.1007/BF02173204. [DOI] [PubMed] [Google Scholar]
- 11.Park Y., Taylor M. F. J. A novel mutation L1029H in sodium channel gene hscp associated with pyrethroid resistance for Heliothis virescens (Lepidoptera: Noctuidae) Insect Biochem. Mol. Biol. 1997;27:9–13. doi: 10.1016/s0965-1748(96)00077-x. [DOI] [PubMed] [Google Scholar]
- 12.Ranson H., Jensen B., Vulule J. M., Wang X., Hemingway J., Collins F. H. Identification of a point mutation in the voltage-gated sodium channel gene of Kenyan Anopheles gambiae associated with resistance to DDT and pyrethroids. Insect Mol. Biol. 2000;9:491–497. doi: 10.1046/j.1365-2583.2000.00209.x. [DOI] [PubMed] [Google Scholar]
- 13.Morin S., Williamson M. S., Goodson S. J., Brown J. K., Tabashnik B. E., Dennehy T. J. Mutations in the Bemisia tabaci para sodium channel gene associated with resistance to a pyrethroid plus organophosphate mixture. Insect Biochem. Mol. Biol. 2002;32:1781–1791. doi: 10.1016/s0965-1748(02)00137-6. [DOI] [PubMed] [Google Scholar]
- 14.Lee S. H., Yoon K. S., Williamson M. S., Goodson S. J., Takano-Lee M., Edman J. D., Devonshire A. L., Clark J. M. Molecular analysis of kdr-like resistance in permethrin-resistant strains of head lice, Pediculus capitis. Pest. Biochem. Physiol. 2000;66:130–143. [Google Scholar]
- 15.Schuler T. H., Martinez-Torres D., Thompson A. J., Denholm I., Devonshire A. L., Duce I. R., Williamson M. S. Toxicological, electrophysiological, and molecular characterisation of knockdown resistance to pyrethroid insecticides in the diamondback moth, Plutella xylostella (L.) Pest. Biochem. Physiol. 1998;59:169–182. [Google Scholar]
- 16.Bass C., Schroeder I., Turberg A., Field L. M., Williamson M. S. Identification of mutations associated with pyrethroid resistance in the para-type sodium channel of the cat flea, Ctenocephalides felis. Insect Biochem. Mol. Biol. 2004;34:1305–1313. doi: 10.1016/j.ibmb.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 17.Usherwood P. N. R., Vais H., Khambay B. P. S., Davies T. G. E., Williamson M. S. Sensitivity of the Drosophila para sodium channel to DDT is not lowered by the super-kdr mutation M918T on the IIS4-S5 linker that profoundly reduces sensitivity to permethrin and deltamethrin. FEBS Lett. 2005;579:6317–6325. doi: 10.1016/j.febslet.2005.09.096. [DOI] [PubMed] [Google Scholar]
- 18.Vais H., Atkinson S., Eldursi N., Devonshire A. L., Williamson M. S., Usherwood P. N. R. A single amino acid change makes a rat neuronal sodium channel highly sensitive to pyrethroid insecticides. FEBS Lett. 2000;470:135–138. doi: 10.1016/s0014-5793(00)01305-3. [DOI] [PubMed] [Google Scholar]
- 19.Lee S. H., Soderlund D. M. The V410M mutation associated with pyrethroid resistance in Heliothis virescens reduces the pyrethroid sensitivity of housefly sodium channels expressed in Xenopus oocytes. Insect Biochem. Mol. Biol. 2001;31:19–29. doi: 10.1016/s0965-1748(00)00089-8. [DOI] [PubMed] [Google Scholar]
- 20.Tan J. G., Liu Z. Q., Wang R. W., Huang Z. Y., Chen A. C., Gurevitz M., Dong K. Identification of amino acid residues in the insect sodium channel critical for pyrethroid binding. Mol. Pharmacol. 2005;67:513–522. doi: 10.1124/mol.104.006205. [DOI] [PubMed] [Google Scholar]
- 21.Cronin N. B., O'Reilly A., Duclohier H., Wallace B. A. Binding of the anticonvulsant drug lamotrigine and the neurotoxin batrachotoxin to voltage-gated sodium channels induces conformational changes associated with block and steady-state activation. J. Biol. Chem. 2003;278:10675–10682. doi: 10.1074/jbc.M208356200. [DOI] [PubMed] [Google Scholar]
- 22.Long S. B., Campbell E. B., MacKinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science (Washington, DC) 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 23.Long S. B., Campbell E. B., MacKinnon R. Voltage sensor of Kv1.2: Structural basis of electromechanical coupling. Science (Washington, DC) 2005;309:903–908. doi: 10.1126/science.1116270. [DOI] [PubMed] [Google Scholar]
- 24.Jiang Y. X., Lee A., Chen J. Y., Ruta V., Cadene M., Chait B. T., MacKinnon R. X-ray structure of a voltage-dependent K+ channel. Nature (London) 2003;423:33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- 25.CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 26.Thompson J. D., Higgins D. G., Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clark M., Cramer R. D., Van Opdenbosch N. Validation of the general-purpose Tripos 5.2 force-field. J. Comput. Chem. 1989;10:982–1012. [Google Scholar]
- 28.Gasteiger J., Marsili M. Iterative partial equalization of orbital electronegativity – a rapid access to atomic charges. Tetrahedron. 1980;36:3219–3228. [Google Scholar]
- 29.Goodsell D. S., Morris G. M., Olson A. J. Automated docking of flexible ligands: Applications of AutoDock. J. Mol. Recognit. 1996;9:1–5. doi: 10.1002/(sici)1099-1352(199601)9:1<1::aid-jmr241>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 30.Morris G. M., Goodsell D. S., Halliday R. S., Huey R., Hart W. E., Belew R. K., Olson A. J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998;19:1639–1662. [Google Scholar]
- 31.Aggarwal S. K., MacKinnon R. Contribution of the S4 segment to gating charge in the Shaker K+ channel. Neuron. 1996;16:1169–1177. doi: 10.1016/s0896-6273(00)80143-9. [DOI] [PubMed] [Google Scholar]
- 32.Ding S. H., Ingleby L., Ahern C. A., Horn R. Investigating the putative glycine hinge in Shaker potassium channel. J. Gen. Physiol. 2005;126:213–226. doi: 10.1085/jgp.200509287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sali D., Bycroft M., Fersht A. R. Stabilization of protein structure by interaction of alpha-helix dipole with a charged side-chain. Nature (London) 1988;335:740–743. doi: 10.1038/335740a0. [DOI] [PubMed] [Google Scholar]
- 34.Doyle D. A., Cabral J. M., Pfuetzner R. A., Kuo A. L., Gulbis J. M., Cohen S. L., Chait B. T., MacKinnon R. The structure of the potassium channel: Molecular basis of K+ conduction and selectivity. Science (Washington, DC) 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 35.Yamagishi T., Li R. A., Hsu K., Marban E., Tomaselli G. F. Molecular architecture of the voltage-dependent sodium channel: Functional evidence for α-helices in the pore. J. Gen. Physiol. 2001;118:171–181. doi: 10.1085/jgp.118.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elliott M., Farnham H., Janes N. F., Needham P. H., Pulman D. A. Insecticidally active conformations of pyrethroids. In: Kohn G. K., editor. Mechanisms of Pesticide Action. Washington DC: American Chemical Society; 1974. pp. 81–91. [Google Scholar]
- 37.Meyer E. A., Castellano R. K., Diederich F. Interactions with aromatic rings in chemical and biological recognition. Angew. Chem.-Int. Edit. 2003;42:1210–1250. doi: 10.1002/anie.200390319. [DOI] [PubMed] [Google Scholar]
- 38.Atkinson S. E. Ph.D. dissertation. U.K.: University of Nottingham; 2002. Interaction of pyrethroids with the voltage-gated sodium channel. [Google Scholar]
- 39.Elliott M. Development of synthetic pyrethroids. Pyrethroid Insecticides. Chemistry and Action. 1980. pp. 1–4. Table Ronde Rousell UCLAF.
- 40.Mullaley A., Taylor R. Conformational properties of pyrethroids. J. Comput. Aided Mol. Des. 1994;8:135–152. doi: 10.1007/BF00119864. [DOI] [PubMed] [Google Scholar]
- 41.Jiang Y. X., Lee A., Chen J. Y., Cadene M., Chait B. T., MacKinnon R. The open pore conformation of potassium channels. Nature (London) 2002;417:523–526. doi: 10.1038/417523a. [DOI] [PubMed] [Google Scholar]
- 42.Chanda B., Bezanilla F. Tracking voltage-dependent conformational changes in skeletal muscle sodium channel during activation. J. Gen. Physiol. 2002;120:629–645. doi: 10.1085/jgp.20028679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith M. R., Goldin A. L. Interaction between the sodium channel inactivation linker and domain III S4-S5. Biophys. J. 1997;73:1885–1895. doi: 10.1016/S0006-3495(97)78219-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park Y., Taylor M. F. J., Feyereisen R. A valine 421 to methionine mutation in IS6 of the hscp voltage-gated sodium channel associated with pyrethroid resistance in Heliothis virescens F. Biochem. Biophys. Res. Commun. 1997;239:688–691. doi: 10.1006/bbrc.1997.7511. [DOI] [PubMed] [Google Scholar]
- 45.Vais H., Williamson M. S., Goodson S. J., Devonshire A. L., Warmke J. W., Usherwood P. N. R., Cohen C. J. Activation of Drosophila sodium channels promotes modification by deltamethrin – reductions in affinity caused by knock-down resistance mutations. J. Gen. Physiol. 2000;115:305–318. doi: 10.1085/jgp.115.3.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou Y. Y., Jiang R., Ling S., Tseng G. N. Stabilization of a channel's open state by a hydrophobic residue in the sixth membrane-spanning segment (S6) of rKv1.4. Pflugers Arch. Eur. J. Physiol. 1998;437:114–122. doi: 10.1007/s004240050755. [DOI] [PubMed] [Google Scholar]
- 47.McDonald I. K., Thornton J. M. Satisfying hydrogen-bonding potential in proteins. J. Mol. Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]