Abstract
The S100 proteins comprise at least 25 members, forming the largest group of EF-hand signalling proteins in humans. Although the proteins are expressed in many tissues, each S100 protein has generally been shown to have a preference for expression in one particular tissue or cell type. Three-dimensional structures of several S100 family members have shown that the proteins assume a dimeric structure consisting of two EF-hand motifs per monomer. Calcium binding to these S100 proteins, with the exception of S100A10, results in an approx. 40° alteration in the position of helix III, exposing a broad hydrophobic surface that enables the S100 proteins to interact with a variety of target proteins. More than 90 potential target proteins have been documented for the S100 proteins, including the cytoskeletal proteins tubulin, glial fibrillary acidic protein and F-actin, which have been identified mostly from in vitro experiments. In the last 5 years, efforts have concentrated on quantifying the protein interactions of the S100 proteins, identifying in vivo protein partners and understanding the molecular specificity for target protein interactions. Furthermore, the S100 proteins are the only EF-hand proteins that are known to form both homo- and hetero-dimers, and efforts are underway to determine the stabilities of these complexes and structural rationales for their formation and potential differences in their biological roles. This review highlights both the calcium-dependent and -independent interactions of the S100 proteins, with a focus on the structures of the complexes, differences and similarities in the strengths of the interactions, and preferences for homo- compared with hetero-dimeric S100 protein assembly.
Keywords: calcium-binding protein interaction, cytoskeletal protein, dimerization, EF-hand, S100 protein
Abbreviations: CacyBP/SIP1, calcyclin-binding protein/Siah-1-interacting protein; CapZ, actin capping protein; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GFAP, glial fibrillary acidic protein; 5-HT1B receptor, 5-hydroxytryptamine receptor; MetAP2, methionine aminopeptidase 2; NDR kinase, nuclear Dbf2-related protein kinase; RAGE, receptor for advanced glycation end-products; SERCA2a, sarcoplasmic/endoplasmic reticulum Ca2+-ATPase 2a
INTRODUCTION
Calcium is a ubiquitous second messenger that regulates a diverse array of cellular events, including muscle contraction, neurotransmitter release, fertilization and cell growth. As a result of its pivotal role, the cellular machinery maintains tight control over the concentration of calcium, which ranges from resting levels near 100 nM to signalling levels near 1 μM. The influx of calcium from the extracellular matrix is controlled by voltage-gated or receptor-operated channels that respond to changes in membrane potential or activation via ligand binding. Calcium can also be sourced from the endoplasmic reticulum where the ion is passed to the cytoplasm by the ryanodine or inositol 1,4,5-trisphosphate receptors. Resting calcium levels are re-established by reciprocal mechanisms such as plasma membrane pumps or exchangers or through re-entry to the endoplasmic reticulum via Ca2+-ATPases [1,2]. The intermediary calcium pulse that results from competing influx and efflux of calcium stimulates a variety of cellular activities. The increased calcium level can act as a feedback inhibitor to switch the calcium import machinery off. Furthermore, the bulk of the intracellular calcium is absorbed by calcium-buffering proteins that have high capacities for calcium, tight binding affinities or unique kinetic properties that act to fine-tune the levels and availability of free cytosolic calcium. However, the most important events arising from the calcium signal are the triggering of downstream biological events modulated through binding of calcium to a large number of calcium-sensor proteins (Figure 1). By far the largest group of sensors is the EF-hand calcium-binding proteins, of which more than 600 have been identified from the human genome.
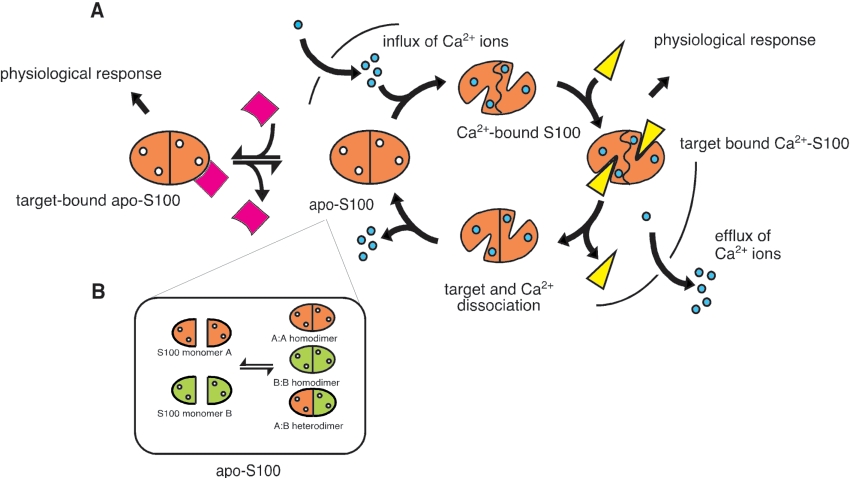
Figure 1. Calcium-dependent and -independent interactions of the S100 family.
(A) S100 proteins generate diverse physiological responses by interacting with target molecules (pink and yellow). At low calcium concentrations, S100 proteins (orange) reside in their calcium-free (apo) state. Upon influx of calcium via voltage-gated or receptor-mediated channels, the S100 protein binds calcium and undergoes a conformational change that modifies its hydrophobic surface properties. This allows the protein to interact with a wide spectrum of target proteins (yellow) to stimulate a physiological response. Release of calcium through Ca2+-ATPase activity results in the dissociation of calcium and target protein from the S100 protein, returning it to its apo state. Although the majority of S100-target interactions are calcium-dependent, some S100 members have been shown to interact with target proteins (pink) in a calcium-independent fashion. (B) Dimeric S100 proteins can exchange subunits with other members of the S100 proteins to form homo- and hetero-dimers in the same cell type as shown by in vitro and in vivo experiments. The populations of each species are dependent on the concentration of the S100 protein in the cell and the relative affinities for the S100 homo- and hetero-dimers.
EF-HAND CALCIUM-BINDING PROTEINS
Kretsinger and Nockolds [3] first identified the EF-hand motif, two α-helices with an intervening 12-residue calcium-binding loop, more than 30 years ago. Structural analysis indicates the chelating residues in the calcium-binding loop form a conserved pentagonal bipyramidal arrangement around the Ca2+ ion, utilizing the side chains at positions 1, 3, 5, 9 (via water) and 12 and backbone carbonyl group of position 7 [4]. Strong preferences exist for aspartate and glutamate in the 1 and 12 co-ordinating positions respectively, and glycine at the non-co-ordinating position 6 [5]. Functional EF-hands are found in pairs [6] and are required for the correct folding of the proteins and unique variations of calcium binding co-operativity.
The calcium signalling role for EF-hand proteins was first established for calmodulin through its activation of 3′,5′-cyclic nucleotide phosphodiesterase [7] and its ability to bind calcium [8]. Subsequently, the structural basis for calmodulin activation [9] showed a calcium-dependent rearrangement of its helices, resulting in the exposure of a hydrophobic surface used to recruit target proteins. A related calcium-sensitive mechanism exists for the muscle contractile EF-hand protein troponin-C [10,11], where calcium-binding modulates its interactions with troponin-I within the muscle complex. More recently, it has been established that the S100 proteins comprise a complex grouping of EF-hand calcium-sensors that have diverse tissue distributions and many protein interactions that result in multiple physiological responses [12] (Figure 1). Interest in the S100 proteins has been sparked by their involvement in several human diseases, such as Alzheimer's disease, cancer and rheumatoid arthritis, usually due to modified levels of expression of the S100 members [13,14].
S100 PROTEINS
The S100 proteins are small acidic proteins (10–12 kDa) that are found exclusively in vertebrates [15]. With at least 25 members found to date in humans, the S100 proteins constitute the largest subfamily of the EF-hand proteins. Of these, 21 family members (S100A1–S100A18, trichohylin, filaggrin and repetin) have genes clustered at chromosome locus 1q21, while other S100 proteins are found at chromosome loci 4p16 (S100P), 5q14 (S100Z), 21q22 (S100B) and Xp22 (S100G). First identified by Moore in 1965 [16], the S100 proteins have 25–65% identity at the amino acid level and contain two EF-hand motifs flanked by conserved hydrophobic residues and separated by a linker region [15]. The sequences of the linker region and the C-terminal extension are the most variable among the S100 proteins.
Three features are unique to the S100 proteins when compared with other EF-hand proteins. First, the two EF-hands in each monomer differ in sequence and mechanisms of calcium co-ordination. The 12-residue C-terminal EF-hand ligates calcium in a similar manner to calmodulin and troponin-C, resulting in a higher calcium affinity site with Kd≈10–50 μM [17]. The N-terminal or ‘pseudo-canonical’ EF-hand is formed by 14 residues and binds calcium mostly through main-chain carbonyl groups, except for the bidentate side chain of glutamate at the last position of the loop. This results in a weaker calcium affinity with Kd≈200–500 μM [17]. This presents an intriguing scenario for the S100 proteins in the cell whereby the C-terminal EF-hand has an affinity that would allow it to be populated during calcium influx, while the N-terminal site affinity is likely to be too weak to bind calcium at any appreciable level. The second unique characteristic of the S100 proteins is their dimeric nature. In vivo and in vitro experiments have shown that the S100 proteins can form non-covalent homo- and hetero-dimers. This indicates that dynamic exchange of the S100 subunits may occur, depending on the populations of the individual S100 protein members in a cellular compartment (Figure 1). Finally, S100 proteins are expressed in a tissue- and cell-specific fashion [12]. For example, S100A1 and S100A2 are found in the cytoplasm and nucleus respectively of smooth-muscle cells [18], whereas S100P is located in the cytoplasm of placental tissue [19,20]. Together, this results in a complex picture of calcium signalling by the S100 proteins governed by the interchange of homo- and hetero-dimeric protein species, calcium binding to the proteins, interaction with target proteins, cell specificity and regulation of biological function (Figure 1).
STRUCTURES OF S100 PROTEINS
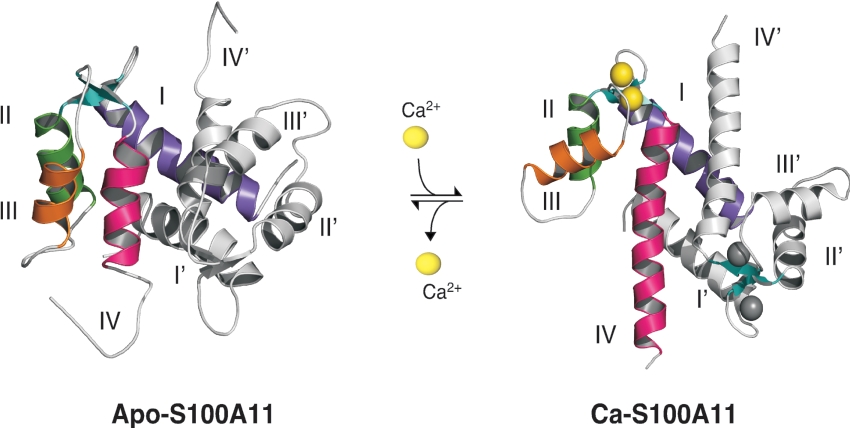
In the last 10 years, three-dimensional structures of S100 proteins have been determined in the calcium-free (apo) [21–32], calcium-bound [24,28,33–41] and target-bound [25,42–47] states. These structures have revealed that the S100 proteins undergo a significant calcium-induced conformational change that, as with calmodulin and troponin-C, results in the exposure of a hydrophobic surface allowing interaction with a target protein (Figure 2). In all S100 structures determined to date, with the exception of S100G (calbindin D9k), the S100 protein exists as a symmetric dimer, with each monomer containing two EF-hand motifs (Figure 2). The N-terminal EF-hand comprises helix I, pseudo calcium-binding site I and helix II, separated by a flexible linker from the C-terminal EF-hand that includes helix III, calcium-binding site II and helix IV. The dimer interface consists of helices I (I′) and IV (IV′) of each monomer arranged in a bicornate or X-type four-helix bundle. The interhelical relationship between these helices is strictly maintained in both the apo- and calcium-bound states. Calcium binding to site I results in only minor alterations of its backbone conformation, consistent with the site adopting a ‘calcium-ready’ state identified many years earlier for the EF-hand protein S100G (calbindin D9k) [48]. The apo-S100 proteins have a more ‘closed’ arrangement of helices III and IV. Calcium binding causes helix III to reorient and repack itself forming a more ‘open’ structure. For example, in apo-S100A11 helix III is nearly antiparallel to helix IV (154°), but opens by approx. 40° upon calcium binding with respect to both helices II and IV [21,46]. The degree of opening is similar to that observed for the C/D helices in the N-terminal domains of both troponin-C (60–70°) [49] and calmodulin (35–40°) [9], although these latter proteins have additional helix movements (i.e. helix B in troponin-C) that do not occur in the S100 proteins. Nevertheless, the calcium-induced structural change in the S100 proteins results in an exposure of residues from helices III and IV in the C-terminal EF-hand, and linker region that facilitates target protein interaction.
Figure 2. Calcium-dependent conformational change in S100 proteins.
The three-dimensional structures of calcium-free S100A11 (apo-S100A11) and calcium-bound S100A11 (Ca-S100A11) are shown to demonstrate the calcium-induced conformational change. In the symmetrical dimer, helices of one monomer (I–IV) are highlighted in different colours, while the other monomer (helices I'–IV') is coloured grey. As sensors, the S100 proteins experience a conformational change upon calcium binding (four atoms/dimer). The rearrangement of helix III (orange) exposes previously buried residues that are essential for target recognition (not shown, see Figure 3) and further biological response. This Figure was drawn using MacPyMOL (http://delsci.com/macpymol/).
BIOLOGICAL ROLES OF S100 PROTEINS
S100 proteins are proposed to have intracellular and extracellular roles in the regulation of many diverse processes such as protein phosphorylation, cell growth and motility, cell-cycle regulation, transcription, differentiation and cell survival [12] (Table 1). In the last 5 years, a wealth of information has become available, supporting these diverse biological roles and concentrating on quantifying direct interactions between an S100 protein and its biological target. Furthermore, in vivo methods such as the yeast two-hybrid assay and co-immunoprecipitation have uncovered a wide spectrum of new biological targets and roles for the S100 proteins, raising many interesting questions about the calcium-dependent and -independent functions of some S100 family members. Despite the extensive summary of S100 protein interactions, biological targets have not yet been identified for S100A3, S100A5, S100A14-S100A18, S100G (calbindin D9k), S100Z and the epidermal proteins filaggrin, trichohyalin and repetin.
Table 1. Calcium-dependent interactions of the S100 proteins.
Support refers to the techniques used to study interactions. Abbreviations used: AB, Ab epitope mapping; AC, affinity chromatography; C, competition assays; CC, chemical cross-linking; CD, circular dichroism; CE, co-expression; CI, co-immunoprecipitation; CL, co-localization; CP, co-purification; CS, co-sedimentation; E, native gel electrophoresis; F, fluorescence; FT, fluorescence resonance energy transfer; GO, gel overlay; IF, isoelectric focusing; M, mutagenesis; MS, mass spectrometry; NMR, nuclear magnetic resonance; O, others; OB, optical biosensor; Ph, phage display; X-Ray, X-ray crystallography; Y2H, yeast two-hybrid. The regions involved in protein interaction are shaded in black or have a heavier line in the schematic of EF-hand motifs in S100 members (N-terminus–HelixI–Site1–HelixII–Linker–HelixIII–Site2–HelixIV–C-terminus). Kd is the dissociation constant for S100–target interaction. Ranges of Kd reflect multiple measurement reports. AHNAK, giant phosphoprotein, identical with desmoyokin; CCN3, cysteine-rich 61/connective tissue growth factor/nephroblastoma overexpressed; EC, excitation–contraction; IQGAP1, IQ motif GTPase-activating protein; MAG, myelin-associated glycoprotein; MRP, myeloid-related protein; MyOD, myogenic determination gene; PLB, phospholamban; S100PBPR, S100P-binding protein.
| Protein | Target | Support | Region | Kd (nM) | Function | Reference(s) |
|---|---|---|---|---|---|---|
| S100A1 | Aldolase C | GO | Stimulation of aldolase C activity | [60] | ||
| Annexin A5 | F, CC | [73] | ||||
| Annexin A6 | CL, CI, F, CC |  |
<1000 | Regulation of calcium flux and IF assembly | [72,73,116] | |
| CacyBP/SIP1 | GO, AC, F, CI | Regulation of CacyBP/SIP | [79] | |||
| Caldesmon | E, F, CC | 170 | Decrease in inhibition of actomyosin by caldesmon | [55] | ||
| CapZ (TRTK-12) | GO, M, CC, F, CS, O |  |
5550 | Modulation of actin organization | [83,117–119] | |
| Desmin | F, O | Inhibition of desmin intermediate filament assembly | [120] | |||
| F-actin | CL, CS | Regulation of actin filament polymerization | [18] | |||
| GFAP | F, CC, CS |  |
500 | Inhibition of GFAP assembly | [88,121] | |
| Microtubules/tubulin | F, CC, O |  |
6000 | Regulation of microtubule dynamics | [122,123] | |
| MyOD | AC, CC, E, F, CI | 2 | Inhibition of MyoD phosphorylation and DNA binding | [124,125] | ||
| NDR | O | Activation of NDR kinase activity | [57] | |||
| p53 | F, CS |  |
Disruption of tubulin–S100A1 complex formation | [117] | ||
| Phosphoglucomutase | GO, AC, O | Inhibition of phosphoglucomutase activity | [61] | |||
| RAGE | O | Promotion of cell survival | [68] | |||
| Ryanodine receptor | CI, OB, AC, O |  |
214 | Regulation of ryanodine and cardiac contractility | [70,71] | |
| SERCA2a and PLB | AC, CI, CL, CE | Regulation of EC-coupling in the heart | [126] | |||
| Synapsin I | CL, C, AC, OB | 245 | Regulation of catalytic activity of synapsins | [127,128] | ||
| Titin (PEVK domain) | GO, OB, O | Inhibition of the actin– or nebulin–PEVK interaction | [129,130] | |||
| Twitchin kinase | OB, O | Activation of twitchin kinase | [131,132] | |||
| S100A2 | ΔNp63 | O | Downstream mediation of ΔNp63 | [133] | ||
| p53 | CL, CI, AC, O | Activation of p53 transcriptional activity | [53] | |||
| Tropomyosin | CC, CI, CL, AC, O | Modulation of the actin cytoskeleton organization | [134] | |||
| S100A4 | CCN3 | Y2H, AC | Modulation of S100A4 affinity to calcium and function | [50,135] | ||
| F-actin | CS | 34000 | [85,136] | |||
| MetAP2 | AC, CL, CI, Y2H | Regulation of MetAP2 | [51] | |||
| Myosin heavy chain II-A | AC, M, CI, FT, GO, CS, CL, C, OB, O |  |
220–600 | Regulation of cytoskeletal dynamics | [77,77a,80,85,92–96] | |
| Myosin heavy chain II-B | CS, O | 23100 | [85,94,138] | |||
| Tropomyosin isoform2 | AC, CL | Regulation of tropomyosin–actin association | [86] | |||
| p37 | F, AC | Increase affinity of S100A4 to calcium | [139] | |||
| p53 | OB, CI, CE, AC, F, O | 9–14 | Enhancement of p53-dependent apoptosis in tumours | [75–77] | ||
| Sept2, Sept6 and Sept7 | AC | [140] | ||||
| S100A6 | Annexin A11 | AC, GO, CC, CI, CL, M, O |  |
1610 | Regulation of annexin A11 function | [141–146] |
| Annexin A2 | AC, GO | Very weak | [74,89,145] | |||
| Annexin A5 | GO | Very weak | [145] | |||
| Annexin A6 | AC, O | [74] | ||||
| CacyBP | F, M, NMR, AC, GO, O | 960 | [141,147] | |||
| CacyBP/SIP | AC, CI, O | Regulation of CacyBP/SIP ubiquitinylation complex | [79] | |||
| Caldesmon | F | [148] | ||||
| Fetuin (biotinylated) | O | [149] | ||||
| GADPH | AC, GO, F, CS, O |  |
100 | [74,89,150] | ||
| Lysozyme | GO | [151] | ||||
| Tropomyosin | F, O | 0.3–1 | Regulation of smooth muscle contraction | [152] | ||
| Sgt1 | AC, CC, CI | Regulation of protein ubiquitination via Stg1 | [59] | |||
| S100A8/S100A9 | Cytochrome b558 | O | Activation of cytochrome b558 | [153] | ||
| Glycosaminoglycans | O | 6 | Localization of MRPs to endothelium | [154] | ||
| S100A8/S100A9 (tetramer) | MS, O | Increasing affinity for calcium | [155,156] | |||
| S100A11 | Actin | CS | 1000 | Regulation of actin activated myosin ATPase | [157] | |
| Annexin A1 | X-Ray, F, M, CL, AC, CI, CS, O |  |
15000 | Targeting and membrane cross linking | [46,66,158,159] | |
| S100A12 | Annexin A5 | AC, OB | 621 | [160] | ||
| CacyBP/SIP | AC, O | Degradation of α/β-catenin | [79] | |||
| Paramyosin | E, O | Development of keratitis | [161] | |||
| RAGE | CD, E, O | Inflammatory processes | [67,162] | |||
| S100A12 (hexamer) | X-Ray | Receptor signaling | [163] | |||
| S100B | AHNAK | AC, CI, OB | 50 | Regulation of calcium homoeostasis | [164] | |
| Aldolase C | GO | Stimulation of aldolase C activity | [60] | |||
| Annexin A6 | CL, CI, F, CC |  |
<1000 | Regulation of calcium flux and IF assembly | [72,73,116] | |
| CacyBP/SIP | GO, AC, CI, F | Regulation of ubiquitination | [55] | |||
| Caldesmon | E, F, CC | 500 | Decrease in inhibition of actomyosin by caldesmon | [79] | ||
| CapZ (TRTK-12) | NMR, F, AC, CC, O |  |
150–1000 | Regulation of actin filament extension | [43,45,65,84,165] | |
| FtsZ | CL, AC |  |
[64] | |||
| GFAP | O | Assembly of intermediate filaments | [63] | |||
| Guanylate cyclase | O | 2000 | Activation of guanylate cyclase | [166] | ||
| Intermediate filaments | CC, CL, O | Regulation of IF assembly and disassembly | [167] | |||
| IQGAP1 | CL, CI, AC | Membrane rearrangement | [168] | |||
| MAG | AC, CC, O | 7000 | Regulation of glial cell cytoskeleton | [169] | ||
| Microtubules | Cl, O |  |
Regulation of microtubule dynamics | [62] | ||
| NDR | NMR, AC, O |  |
500 | Modulation of NDR kinase activity | [42,57] | |
| Neuromodulin | CC | Inhibition of neuromodulin phosphorylation by PKC | [170] | |||
| p53 | NMR, AC, F, OB, O |  |
24–23500 | Inhibition of p53 function | [47,78,110,171–173] | |
| Phosphoglucomutase | GO, AC, M, O |  |
Stimulation of phosphoglucomutase activity | [61,174] | ||
| RAGE | O | Promotion of cell survival | [68] | |||
| Tau | AC, CC |  |
100–1000 | Inhibition of tau phosphorylation by protein kinase II | [56,175] | |
| S100P | CacyBP | O | [79] | |||
| Dormant ezrin | AC, CL | Regulation of ezrin ability to bind actin | [176] | |||
| Melittin | F, CD, O | 5000 | [97] | |||
| RAGE | CI | Stimulation of cell proliferation and survival via RAGE | [69,177] | |||
| S100PBPR | AC, CI, O | Involvement in early pancreatic cancer | [178] | |||
| Sgt1 | AC | [59] |
Calcium-dependent protein interactions
The calcium-dependent signalling roles of the S100 proteins arise because their affinities for calcium are comparable with the free calcium concentration in the cytoplasm during a calcium wave (∼1 μM). Thus the binding of S100 proteins to their targets in the presence of calcium and their release, or lack of binding with EDTA, have been used to provide evidence of a calcium-dependent interaction using in vitro techniques such as affinity chromatography, optical biosensing and gel overlay, among others. Despite more than 90 potential protein complexes for the S100 proteins (Table 1), only two examples are available that use a yeast two-hybrid screen to identify calcium-dependent interactions (S100A4 with the cell growth regulator protein CCN3 [50], and methionine aminopeptidase 2, MetAP2 [51]). The lack of calcium-dependent interactions observed in yeast is likely to be a function of their tightly controlled intracellular calcium levels (∼200 nM) [52] that are well below the calcium Kd values of most S100 proteins (∼10–50 μM) [17] and resulting in very low populations of the calcium-bound S100 protein during these experiments. S100 proteins have been found to interact with biological targets using co-immunoprecipitation or co-localization experiments. For instance, S100A2 and its interacting partner p53 co-localize in the nucleus and the cytoplasm in oral carcinoma cell lines, suggesting a relevant interaction of these two proteins [53]. Furthermore, binding of p53 to S100A2 was confirmed using pull-down assays and electrophoretic mobility-shift assays [53]. In contrast, S100B interactions with p53 [47], MARCKS (myristoylated alanine-rich C-kinase substrate) [54] and caldesmon [55] have been observed in vitro, but no evidence of co-localization has been demonstrated [12]. This lack of co-localization evidence not only for S100B, but also for other S100 proteins and their targets, has generated some confusion regarding the biological relevance of some in vitro findings.
In general, the calcium-dependent roles of the S100 proteins can be divided into five major functional groupings: (i) regulation of phosphorylation mediated by protein kinases, (ii) modulation of enzymatic activity, (iii) maintenance of cell shape and motility, (iv) influence of some signal-transduction pathways, and (v) promotion of calcium homoeostasis. For example, S100B has been implicated in the phosphorylation of tau protein [56], and the modulation of kinase activity by NDR kinase (nuclear Dbf2-related protein kinase) and protein kinase II [42,56–58]. The mechanism of this inhibition is through direct interaction with the kinase rather than recruitment of substrate. The regulation of the yeast ubiquitination pathway protein Sgt1 by S100A6 appears to operate by a similar mechanism [59]. The enzymatic activity of aldolase (isoforms A and C) and phosphoglucomutase, occurs via interactions with S100A1 [60,61] or S100B [60,61], whereas MetAP2 interacts with S100A4 [51]. It is surprising that reversed biological function can be promoted by different S100 proteins. For example, the interaction of S100A1 with phosphoglucomutase inhibits enzyme activity, whereas S100B interaction seems to stimulate this enzyme's function [61]. Furthermore, S100A1 binding to aldolase C requires calcium, whereas the interaction with aldolase A is calcium-insensitive [60].
The most significant number of protein interactions for the S100 proteins are with components of the cytoskeleton, including the tubulins, intermediate filaments, actin, myosin and tropomyosin. For example, S100B controls the assembly of microtubules [62] and GFAP (glial fibrillary acidic protein) [63] interacts with the tubulin homologue FtsZ [64] and is proposed to regulate actin filament extension through interaction with CapZ (actin capping protein) [65]. Other S100 proteins, including S100A1 S100A2, S100A4, S100A6 and S100A11, have been shown to affect similar components of the cytoskeleton (Table 1). In particular, elegant mechanisms for calcium-dependent membrane aggregation, important for cell vesiculation, have been proposed for the interaction of S100A11 with annexin A1 [66]. Several S100 proteins have been implicated in a variety of signal-transduction pathways. For example, S100B, S100A1, S100A12 and S100P can bind to the RAGE (receptor for advanced glycation end-products) [67–69], activating an intracellular signal cascade that contributes to cell proliferation and survival. Finally, the involvement of the S100 proteins on calcium homoeostasis has been suggested through interactions of S100A1 with the ryanodine receptor [70,71].
A few general observations that can be made from Table 1. Clearly, some S100 members are proposed to regulate the activity of the same target molecule. For example, S100A1, S100A6 and S100B bind annexin A6 [72–74]; S100A1, S100A2, S100A4 and S100B interact with the tumour suppressor p53 ([47,53,75–77], but see [77a], [78]); and S100B, S100A6, S100A12 and S100A1 form complexes with the ubiquitination protein CacyBP/SIP (calcyclin-binding protein/Siah-1-interacting protein) [55,79]. This is perhaps not surprising, given the significant sequence similarities between many of the S100 proteins. The apparent multi-S100 protein interactions could result from differential expression in tissues such that two different S100 proteins might control similar processes but in different tissues. It will be important to link these interactions with the expression and availability of both S100 protein and target in the same cell type and tissue. This has been elegantly shown for S100A4 and the non-muscle myosin A heavy chain that is thought to have a role in metastasis [80].
For most of the S100-target protein complexes, the dissociation constant of the S100 member for the target is approx. 1 μM (Table 1). This affinity is similar to that obtained for other calcium-dependent EF-hand protein complexes, such as troponin-C with troponin-I (0.1–30 μM) [81,82]. In some cases, affinity measurements may provide evidence for target selectivity amongst the S100 proteins. For example, the CapZ peptide TRTK-12 (TRTKIDWNKILS) binds 10-fold more tightly to S100B than to S100A1, indicative of a strong preference for S100B [83,84]. It will be important to extend these studies to off-rate measurements to determine whether the release rates are consistent with the lifetimes of the biological function as shown recently for S100A4 [80]. Table 1 also reveals that there are some disagreements regarding some S100 complexes. For example, co-sedimentation experiments show an interaction of F-actin with S100A4 [85], but this binding could not be observed using optical biosensor assays ([77], but see [77a]). In the same manner, the S100A4–tropomyosin (TM2) complex was observed using affinity chromatography [86], but not using surface plasmon resonance detection ([77], but see [77a]) and the binding of annexin A2 to S100A6 was reported using affinity-chromatographic techniques, but was not detected by fluorescence or chemical cross-linking [74]. These results emphasize the need to use several methods and/or sample conditions to monitor the in vitro S100–target interactions.
Several three-dimensional structures of calcium-bound S100 proteins, bound to target molecules, have been solved using either NMR or X-ray-crystallographic methods. The structures of S100A11 bound to the N-terminal peptide of annexin A1 [46], and S100B in complex with peptides derived from CapZ (TRTK-12) [43,45], p53 [47] and NDR kinase [42] provide atomic details about the protein–protein interface that can be used to rationalize the specificity of the interactions. The first structure of a calcium-bound S100 protein in complex with a region of its biological target was the crystal structure of S100A11 bound to the N-terminal 14 residues of annexin A1 [46]. This structure reveals that the S100A11–annexin A1 complex remains symmetrical, binding two annexin molecules per S100 dimer on the periphery of the protein. The annexin peptide lies across the linker, helix III and helix IV of one monomer and contacts helix I of the partner monomer, thus bridging the two S100 subunits (Figure 3). Residues near the C-terminus of helix IV in S100A11 are necessary for the annexin interaction. This region is not helical in apo-S100A11 (Figure 2), indicating that induction of an α-helix is important for target protein binding. The annexin peptide forms an amphipathic α-helix with its hydrophobic surface (Ala1, Met2, Val3, Phe6, Leu7, Ala10, Trp11) facing the S100 protein, thereby identifying some of the amino acid determinants for the S100 specificity.
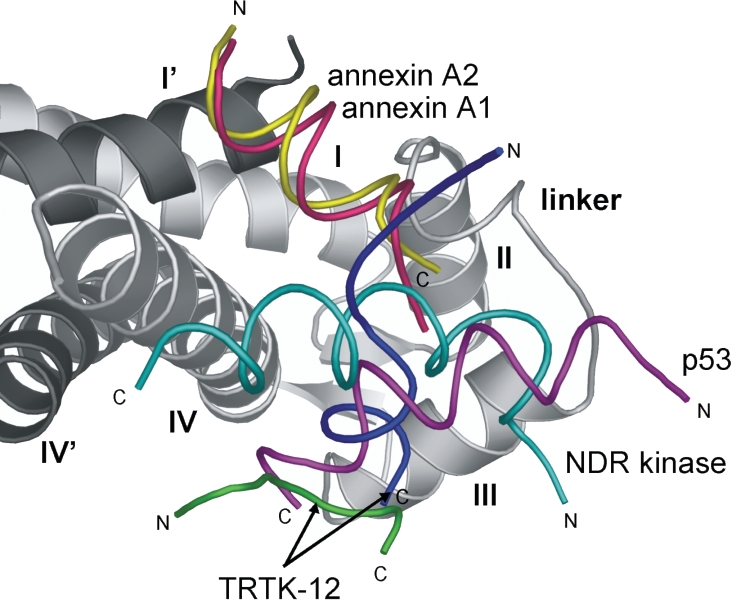
Figure 3. Target protein orientation for S100 proteins.
A ribbon representation of calcium-bound S100A11 is presented, showing one of the monomers in light grey (helices labelled as I, II, III and IV) and the other monomer in dark grey (helices labelled as I' and IV'). The S100 portion from the three dimensional complexes of S100A10–annexin A2, S100B–p53, S100B–TRTK and S100B–NDR kinase was superimposed with S100A11 to give the relative orientations of each of the target peptides. Annexin A1 (residues 1–11) is shown in pink, annexin A2 (residues 1–11) in yellow, NDR kinase (residues 71–87) in cyan, p53 (residues 99–112) in purple and TRTK-12 (residues 1–12) in both green and blue. The N- and C-termini of each of the peptides are indicated with either an N or C respectively. The S100A11 and S100B complexes are in the calcium-bound state, although no calcium ions are shown in the Figure. S100A10 is in the calcium-free state. This Figure was drawn using MacPyMOL (http://delsci.com/macpymol/).
Three-dimensional structures of calcium-bound S100B with peptides derived from p53, CapZ (TRTK-12) and NDR kinase have been determined by NMR spectroscopy (Figure 3). These structures provide initial details for the specificity of different targets for the same S100 protein. All four of these structures utilize a similar binding region of the S100B protein, namely the surface formed by the linker and helices III and IV. The p53 and NDR targets adopt α-helical conformations that are 12–14 residues in length. Both peptides have their N-termini positioned towards the N-terminus of helix III, but the orientation of the two peptides diverges by more than 8 Å (1 Å=0.1 nm) as they approach helix IV. Structures of calcium-bound S100B with TRTK-12 show these peptides have little regular secondary structure and are arranged roughly at 90° to the orientations of the p53 or NDR peptides. These four structures point towards a malleable flat binding surface in S100B that could accommodate many different target proteins, and is shallower than that found in calmodulin [42]. This is consistent with observations in Table 1 showing that S100B can interact with at least 20 different proteins in a calcium-dependent manner. The possibility does exist that the peptide segments being used to map S100 target interactions are void of secondary, or supplementary, binding sites. Evidence for this arises from the calcium-dependent interaction of S100B with p53, where peptides comprising residues 319–393 possess very tight binding to S100B (Kd=24 nM) [87], but this is weakened more than 100-fold with shorter constructs (367–381 or 367–393) [75]. In order to define further the binding surfaces on the S100 proteins with specific binding partners it will be important to expand the number of structures of calcium-bound S100 family members with targets, including interactions with intact target proteins or much larger peptide sequences. Furthermore, the precision of the structures may need to be improved, especially for side-chain interactions involving the bound peptides, in order to develop better rationales for binding specificity.
All of the calcium-bound S100A11 and S100B complexes utilize helix IV for their target protein interaction. The TRTK-12 and p53 sequences interact with regions near Val80–Cys84, whereas the NDR peptide interacts mostly downstream of this (Cys84–Phe88). In contrast, annexin A1 utilizes the opposite face of helix IV in S100A11 (Figure 3). This shows the central adaptive role that helix IV plays to modify S100 target protein interactions. Examination of Table 1 corroborates this idea, as more than 25 calcium-dependent binding partners with different S100 proteins utilize helix IV or the subsequent C-terminus. For example, deletion of the S100A1 C-terminus abolishes its interaction with TRTK-12, GFAP, p53, SERCA2a (sarcoplasmic/endoplasmic reticulum Ca2+-ATPase 2a) and phospholamban [88]. However, not all S100 target proteins follow this trend. For instance, the N-terminal region of S100A6 is essential for its interaction with GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and annexin A2 [89]. These observations and the variations in the three-dimensional structures of calcium-bound S100–target complexes provide some insight into the level of target specificity among the S100 family members. It is clear that, although several target proteins appear to ‘cross-react’ with more than one S100 protein and most S100 proteins interact with several targets, there remains a fine level of discrimination that does not allow random interaction of targets with all S100 proteins. The broad selection of the S100 targets such as TRTK-12, NDR and p53 is likely to be a reflection of the moderate hydrophobicity in the short target peptide sequence and the extensive hydrophobic patch that exists on the S100 structures. Nevertheless, this exposed region comprises the most divergent portions of the S100 sequences (the linker and C-terminal regions) and this must be enough to discriminate against different target proteins.
Calcium-independent protein interactions
Although the majority of S100 protein interactions are calcium-dependent, several calcium-independent interactions have been reported (Table 2). The most common binding partners for the apo-S100 proteins are enzymes. For example, S100B and S100A1 bind with glycogen phosphorylase [90], whereas S100A10 and S100A11 show interactions with transglutaminase [91]. Some discrepancies for the calcium-sensitivity of S100-target interactions are encountered. For example, optical biosensor techniques show a calcium-insensitive complex between S100A4 and non-muscle myosin (heavy chain II, isoform A), and between S100A4 and p53 ([77], but see [77a]), whereas strict calcium-dependence was observed using other techniques ([77], but see [77a], [80,85,92–96]). In some cases, such as the calcium-insensitive interaction of S100A4 with F-actin (filamentous actin), the dissociation constant is very weak (Kd≈500 μM), but strengthens significantly upon calcium binding to the S100 protein (Kd≈30 μM) [85]. Similar observations have been noted for the interaction of melittin with S100P [97]. These poor binding affinities in the absence of calcium probably indicate that the in vivo interaction is insignificant in the absence of calcium.
Table 2. Calcium-independent interactions of the S100 proteins.
For details see the legend to Table 1. Homodimer proteins are listed only if the Kd has been reported. E-FABP, epidermal-type fatty-acid-binding protein; FGF, fibroblast growth factor; HBV pol, hepatitis B virus polymerase; TASK-1, TWIK (two-pore domain weak inwardly rectifying K+ channel)-related acid-sensitive K+ channel; TRPV, transient receptor potential channel (subfamily V).
| (a) Interactions with other proteins | ||||||
|---|---|---|---|---|---|---|
| Protein | Target | Support | Region | Kd (nM) | Function | Reference(s) |
| S100A1 | Adenylate cyclase | O | 200 | Stimulation of adenylate cyclase | [179] | |
| Aldolase A | O | [83] | ||||
| Glycogen phosphorylase | GO, AC, CL | Inhibition of glycogen phosphorylase activity | [90] | |||
| Microtubules | O | Microtubule assembly | [180] | |||
| S100A4 | Annexin A2 | CI, NMR, O | Mediation of plasmin production from plasminogen | [181] | ||
| F-actin | OB | 543000 | [85] | |||
| Liprin β1 | CI, CL, MS, O |  |
Inhibition of liprin β1 phosphorylation | [182] | ||
| Myosin heavy chain II-A | OB | 220–600 | Regulation of cytoskeletal dynamics | [77] | ||
| p53 | OB | 9–14 | Enhancement of p53-dependent apoptosis in tumours | [77] | ||
| S100A4 (tetramer) | MS | Stimulation of angiogenesis and neurite growth | [183] | |||
| S100A7 | E-FABP | CL, CI, CP, GO | Shuttle E-FABP to membrane or ligands | [184–186] | ||
| Jab-1 | Y2H, CI, O |  |
Pro-survival pathway | [187,188] | ||
| RanBPM | Y2H, CI | Cytoskeletal functions, adhesion and migration | [189] | |||
| Transglutaminase | O |  |
Regulation of S100A7 function | [91] | ||
| S100A8/S100A9 | Carboxylated glycans | AC | Inflammation | [190] | ||
| CD36 | AC | Fatty acid uptake | [191] | |||
| Phox proteins | CP, CI | Scaffold for phox and NADPH oxidase activation | [153,192] | |||
| S100A10 | 5-HT1B receptor | Y2H, CI, CL, O | Localization of 5-HT1B receptors to the cell surface | [99] | ||
| Annexin A2 | X-Ray, CD, F, M, CC, CL, O |  |
∼30 | Signal transduction | [98,193–203] | |
| NS3 | Y2H | Mediation of virus release | [100] | |||
| Connexin 31 | Y2H | [204] | ||||
| Phospholipase A2 | CI, O | Regulation of phospholipase A2 activation | [205] | |||
| HBV Pol | Y2H, AC, CI, CL, O | Inhibition of the DNA polymerase activity of HBV pol | [101] | |||
| TASK-1 K+ channel | Y2H, AC, CI, M, O | Trafficking of TASK-1 to the plasma membrane | [206] | |||
| Plasminogen activator | AC, O |  |
[207] | |||
| Sodium channel Nav1.8 | Y2H, AC, O |  |
Translocation of Nav1.8 to the plasma membrane | [102,208,209] | ||
| Transglutaminase | O |  |
Regulation of S100A10 function | [91] | ||
| TRPV5/TRPV6 | Y2H, AC, CI |  |
Translocation TRPV5/TRPV6 to the plasma membrane | [210] | ||
| S100A11 | Transglutaminase | O | Regulation of S100A11 function | [91] | ||
| Isocitrate dehydrogenase | AC, OB | [160] | ||||
| Aldolase A | [160] | |||||
| GAPDH | AC, OB | 83 | [160] | |||
| S100A13 | FGF-1, p40 Syt-1 | CP, E, CL, O | Regulation of FGF-1 release | [211–213] | ||
| Interleukin-1α | CL, CI |  |
Stress induced release of interleukin-1α | [214] | ||
| S100B | Aldolase A | O | Stimulation of aldolase A activity | [60] | ||
| Glycogen phosphorylase | GO, AC | [90] | ||||
| S100P | Melittin | F, CD, O | 200000 | [97] | ||
| (b) Observed heterodimers of the S100 proteins | ||||||
| Monomer I | Monomer II | Support | Region | Kd (nM) | Function | Reference(s) |
| S100A1 | S100A4 | Y2H, F, FT, MS, OB, AC, GO, CP, CE, M |  |
300–500 (+Ca2+) | Modulation of metastasis in cancer cells | [107,112,114] |
| S100B | Y2H, O | [108,113,215] | ||||
| S100P | Y2H, OB, FT, CE, M |  |
1100–1800 (apo); 10–20 (+Ca2+) | Target binding and function regulation | [115] | |
| S100A4 | S100A4 | Y2H, OB, NMR, O |  |
4000 (apo); 670–1000 (+Ca2+) | [107,111,112,114] | |
| S100A6 | S100B | Y2H, CI, CL | Melanoma cell growth | [108,113] | ||
| S100A7 | S100A10 | MS, CI, O | [216] | |||
| S100A8 | S100A9 | Y2H, MS, AC, M, Ph, AB, CC, CD, F, NMR, X-Ray, E, IF |  |
Inflammatory processes | [106,217–220] | |
| S100A10 | MS, CI, O | [216] | ||||
| S100A9 | S100A12 | AC, CI, OB, M, O |  |
1520 (apo) | [160] | |
| S100A11 | S100B | Y2H, CL, CI |  |
Modulation of target binding | [113,158] | |
| S100A12 | S100A12 | X-Ray, OB | 4 (+Ca2+) | [160] | ||
| S100B | S100B | NMR, O | <0.5 (apo); <0.5 (+Ca2+) | [103] | ||
| S100P | S100P | Y2H, X-Ray, OB | 1400–2500 (apo); 40–120 (+Ca2+) | Involved in various diseases | [41,115,221] | |
| S100Z | Y2H, IF, E | [222] | ||||
Although questions remain about the significance of some apo-S100–protein interactions, there is overwhelming support for the notion that S100A10 has multiple binding partners in the calcium-free state. The calcium-insensitivity of S100A10 results from its inability to bind calcium owing to mutations in both calcium-binding sites. Early evidence showed that S100A10 was co-purified as a heterotetramer with annexin A2 [98]. More recently, an important interaction between S100A10 and the 5-hydroxytryptamine (serotonin) receptor 5-HT1B has been identified using a yeast two-hybrid screen. Further experiments have provided strong evidence that this S100A10–5-hydroxytryptamine complex may have a role in the onset of depression [99]. Other complexes with S100A10, including those with the viral proteins NS3 and hepatitis virus B polymerase, have been identified using yeast two-hybrid assays [100,101] (Table 2), as these protein interactions are very strong (Kd≈30 nM for annexin A2) [98]. The S100A10–annexin A2 complex was the first three-dimensional structure of an S100 complex to be determined [25]. As with S100A11, the S100A10 protein interacts with the N-terminus of the annexin molecule. The fold of the S100A10 protein is nearly identical with that of calcium-bound S100A11 (Figure 2), adopting the more ‘open’ conformation that provides a hydrophobic surface for protein binding, even in the absence of calcium. The location and interactions of the annexin A2 peptide with apo-S100A10 are remarkably similar to those identified for S100A11 (root mean square deviation 0.87 Å) [46], interacting with residues from helices III and IV, and the linker of one monomer, and helix I of the other monomer (Figure 3). Other protein partners, including tissue-type plasminogen activator, appear to utilize the same helix IV region (Table 2a), while targets such as the sodium channel NaV1.8 interact at different sites [102].
The most important calcium-independent interactions of the S100 proteins are their abilities to form homo- and hetero-dimers, as well as some higher-order complexes (Table 2b). This property results in a dynamic interplay between formation of the dimeric species, calcium binding and interactions with a biological target protein (Figure 1). Traditionally, homodimeric interactions have been the focus of the S100 proteins; however, the use of yeast two-hybrid experiments has uncovered a large number of heterodimeric complexes. Furthermore, optical biosensor experiments have been used to quantify the strength of the homo- and hetero-dimeric interactions in vitro, so that the importance of these can be assessed in vivo.
The association of several homodimeric S100 proteins has been quantified, providing an indication of the overall stability of S100B, S100A4, S100A12 and S100P. These studies show a wide range of Kd values for the monomer–dimer equilibrium. For example, S100B forms the tightest dimer (Kd<500 pM) in the calcium-free state [103] and, remarkably, is nearly 5000-fold more stable than either the S100A4 or S100P homodimers (Kd≈1–2 μM). In some cases (S100P and S100A12), dimerization is enhanced by more than 100-fold in the presence of calcium. In other cases (S100B, S100A4), calcium binding has a negligible effect. In vivo, the extent of homodimerization will be dependent on the concentration of the S100 protein and the Kd for dimer formation. For example, S100B has been found at high concentrations (nearly 10 μM) in glial cells [104], indicating that this protein would be completely in the dimeric form. On the other hand, proteins such as S100A4 and S100P, if found in similar or lower concentrations than S100B, would have a significant population of monomeric protein in the cell. This would facilitate formation of heterodimers with other S100 proteins (Figure 1).
The observation that dynamic exchange occurs for the S100 subunits indicates that heterodimeric proteins probably exist in vivo. Consistent with this, at least ten different heterodimeric S100 species have been identified. The S100A8–S100A9 heterodimer is probably the most well characterized of these. Originally isolated from synovial fluid [105], X-ray structures are available for the S100A8 and S100A9 homodimers [36,44]. However, other experiments have shown there is a strong preference for the S100A8–S100A9 heterodimer, especially in the presence of calcium, and it has been suggested that the S100A8–S100A9 heterodimeric species is the only relevant biological species [106]. A similar calcium stabilization of the heterodimer has been observed for S100B–S100A6 and S100B–S100A11. In the case of the other S100 heterodimers, there are several intriguing possibilities. The first, of course, is that the relevant S100 proteins must be found in the same cell type to substantiate heterodimer formation. This has been confirmed for several species, including S100A1–S100A4 found in several mammary cell lines, and S100B–S100A6, found in some human melanoma cells [107,108]. Furthermore, the formation of the S100 heterodimeric proteins will be governed by the thermodynamics of the equilibria involved, shown previously for homo- and hetero-dimeric calcium-binding peptides [109], and for tropomyosin [110]. Using the example of S100A1 and S100A4, the S100A1–S100A4 heterodimer will be preferentially formed when ΔGS100A1–S100A4<1/2[ΔGS100A1+ΔGS100A4]+R·T·ln 2. On the basis of Table 2(b), the stability of S100A1–S100A4 (Kd≈300 nM) is nearly 10-fold that of the S100A4 homodimer (Kd≈1–2 μM), as determined using yeast two-hybrid, optical biosensor and analytical ultracentrifugation experiments [107,111,112]. Furthermore, yeast two-hybrid studies have shown the S100A1 homodimer probably has a similar stability as the S100A1–S100A4 heterodimer. On the basis of these observations and the above inequality, the S100A1–S100A4 heterodimer would be favoured thermodynamically by approx. 3 kJ/mol. More complicated situations may arise in the cell, including interactions with other S100 proteins or binding partners, or large differences in cellular concentrations of the S100 protein. However, the thermodynamic point of view would indicate that the S100A1–S100A4 heterodimer is the dominant in vivo species. For other heterodimers, this approach indicates that the homodimer is the major species. For example, the extremely tight dimer association of S100B indicates that heterodimers such as S100B–S100A11 and S100B–S100A6 would be poorly formed in vivo. Although Kd values are not available for these two heterodimers, two-hybrid experiments show an approx. 2-fold poorer β-galactosidase activity for the heterodimers [108,113]. Furthermore, experiments will be needed for other S100 homo- and hetero-dimers to establish their strengths of interaction and relative populations in different cell types.
Some important differences regarding the specificity of interaction at the dimer interface have been noted for S100 homo- and hetero-dimers using site-directed mutagenesis and yeast two-hybrid experiments. For example, deletion of the C-terminal eight residues for S100B abolishes heterodimer formation with both S100A6 and S100A11, whereas S100B homodimer formation is unaffected [113]. Within this region, Phe87 and Phe88 are particularly important. These residues are in an unstructured region following helix IV and have very few intersubunit contacts in the apo-S100B structure. In order to contribute substantially to the heterodimer complex, these interactions must be significantly altered in the heterodimeric structures. Similar observations have been made for S100A1–S100A4 and S100A1–S100P where Cys76 and Cys81 in S100A4 [114] and Val76 in S100P [115] are required for heterodimerization with S100A1, but have little affect on the respective homodimer formation. More recently, the first X-ray structure of a heterodimeric S100 protein, S100A8–S100A9 shows that the heterodimer might be energetically driven by a slightly more extensive burial of solvent-accessible surface area compared with either the S100A8 or S100A9 homodimers (I. Korndoefer and A. Skerra, personal communication). These experiments will form an excellent initial framework for assessing homo- and hetero-dimer formation.
FUTURE INSIGHTS
The S100 proteins are a unique family of EF-hand calcium-binding proteins involved in a large network of calcium-dependent, and independent protein–protein interactions. Although structures exist for the S100 proteins in the apo, calcium-bound and target-bound states, there are still many unresolved questions about the in vivo biological functions of the proteins. The dimeric structure of S100 family members suggests that these proteins, in principle, interact with either two identical, or two non-identical, target proteins in the case of homodimers and heterodimers respectively. For some S100–protein complexes, such as those with the oligomeric targets GFAP, tubulin and p53, this presents an attractive mechanism whereby the S100 can modulate the assembly/disassembly of the target protein complex. In vitro experiments support this to some extent, whereas in vivo evidence is not as conclusive. Other hypotheses exist such as those for S100A10 and S100A11 where the protein acts to bridge adjacent phospholipid membrane surfaces en route to vesiculation, further providing a rationale for the S100 dimeric structure.
There is a great deal of excitement in the calcium signalling field as the S100 protein story unfolds. Clearly, some inroads have been made towards the structural recognition of biological target proteins through recent three-dimensional structures. As with calmodulin and troponin-C, further quantification and structural information will undoubtedly allow researchers to understand the specificity of the S100 proteins for target recognition. Furthermore, the formation of the homo- and hetero-dimers presents a seemingly unending variation in S100 dimer composition. Is it possible that the monomeric S100 form has a biological role in some cells? This, and other questions, will provide researchers with many challenges in the future.
Acknowledgments
We thank Dr Arne Skerra (Lehrstuhl für Biologische Chemie, Technische Universität München, Freising-Weihenstephan, Germany) for pre-submission discussions regarding the S100A8–S100A9 heterodimer structure. We thank Kathryn Barber (of this department) for her critical reading and invaluable assistance during the final stages of writing this review. This work was supported by the Canadian Institutes of Health Research, and Canada Research Chairs Program (G.S.S.). We are also grateful for graduate scholarship support from the Canadian Institutes of Health Research (A.R.D.) and the Ontario Graduate Scholarship in Science and Technology (L.S.K.).
References
- 1.Berridge M. J., Lipp P., Bootman M. D. The versatility and universality of calcium signalling. Nat. Rev. Mol. Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 2.Berridge M. J., Bootman M. D., Roderick H. L. Calcium signalling: dynamics, homeostasis and remodelling. Nat. Rev. Mol. Cell Biol. 2003;4:517–529. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- 3.Kretsinger R. H., Nockolds C. E. Carp muscle calcium-binding protein. II. Structure determination and general description. J. Biol. Chem. 1973;248:3313–3326. [PubMed] [Google Scholar]
- 4.Strynadka N. C. J., James M. N. G. Crystal structures of the helix–loop–helix calcium-binding proteins. Annu. Rev. Biochem. 1989;58:951–998. doi: 10.1146/annurev.bi.58.070189.004511. [DOI] [PubMed] [Google Scholar]
- 5.Marsden B. J., Shaw G. S., Sykes B. D. Calcium binding proteins: elucidating the contributions to calcium affinity from analysis of species variants and peptide fragments. Biochem. Cell Biol. 1990;68:587–601. doi: 10.1139/o90-084. [DOI] [PubMed] [Google Scholar]
- 6.Shaw G. S., Hodges R. S., Sykes B. D. Calcium-induced peptide association to form an intact protein domain: 1H NMR structural evidence. Science. 1990;249:280–283. doi: 10.1126/science.2374927. [DOI] [PubMed] [Google Scholar]
- 7.Cheung W. Y. Cyclic 3′,5′-nucleotide phosphodiesterase: demonstration of an activator. Biochem. Biophys. Res. Commun. 1970;38:533–538. doi: 10.1016/0006-291x(70)90747-3. [DOI] [PubMed] [Google Scholar]
- 8.Teo T. S., Wang J. H. Mechanism of activation of a cyclic adenosine 3′:5′-monophosphate phosphodiesterase from bovine heart by calcium ions: identification of the protein activator as a Ca2+ binding protein. J. Biol. Chem. 1973;248:5950–5955. [PubMed] [Google Scholar]
- 9.Ikura M., Clore G. M., Gronenborn A. M., Zhu G., Klee C. B., Bax A. Solution structure of a calmodulin-target peptide complex by multidimensional NMR. Science. 1992;256:632–638. doi: 10.1126/science.1585175. [DOI] [PubMed] [Google Scholar]
- 10.Gagne S. M., Tsuda S., Li M. X., Smillie L. B., Sykes B. D. Structures of the troponin C regulatory domains in the apo and calcium-saturated states. Nat. Struct. Biol. 1995;2:784–789. doi: 10.1038/nsb0995-784. [DOI] [PubMed] [Google Scholar]
- 11.Strynadka N. C., Cherney M., Sielicki A. R., Li M. X., Smillie L. B., James M. N. G. Structural details of a calcium-induced molecular switch: X-ray crystallographic analysis of the calcium-saturated N-terminal domain of troponin-C at 1.75 Å resolution. J. Mol. Biol. 1997;273:238–255. doi: 10.1006/jmbi.1997.1257. [DOI] [PubMed] [Google Scholar]
- 12.Donato R. S100: a multigenic family of calcium-modulated proteins of the EF-hand type with intracellular and extracellular functional roles. Int. J. Biochem. Cell Biol. 2001;33:637–668. doi: 10.1016/s1357-2725(01)00046-2. [DOI] [PubMed] [Google Scholar]
- 13.Odink K., Cerletti N., Bruggen J., Clerc R. G., Tarcsay L., Zwadlo G., Gerhards G., Schlegel R., Sorg C. Two calcium-binding proteins in infiltrate macrophages of rheumatoid arthritis. Nature (London) 1987;330:80–82. doi: 10.1038/330080a0. [DOI] [PubMed] [Google Scholar]
- 14.Van Eldik L. J., Griffin W. S. T. S100b expression in Alzheimer's disease: relation to neuropathology in brain regions. Biochim. Biophys. Acta. 1994;1223:398–403. doi: 10.1016/0167-4889(94)90101-5. [DOI] [PubMed] [Google Scholar]
- 15.Schafer B. W., Heizmann C. W. The S100 family of EF-hand calcium-binding proteins: functions and pathology. Trends Biochem. Sci. 1996;21:134–140. doi: 10.1016/s0968-0004(96)80167-8. [DOI] [PubMed] [Google Scholar]
- 16.Moore B. W. A soluble protein characteristic of the nervous system. Biochem. Biophys. Res. Commun. 1965;19:739–744. doi: 10.1016/0006-291x(65)90320-7. [DOI] [PubMed] [Google Scholar]
- 17.Donato R. S-100 proteins. Cell Calcium. 1986;7:123–145. doi: 10.1016/0143-4160(86)90017-5. [DOI] [PubMed] [Google Scholar]
- 18.Mandinova A., Atar D., Schafer B. W., Spiess M., Aebi U., Heizmann C. W. Distinct subcellular localization of calcium binding S100 proteins in human smooth muscle cells and their relocation in response to rises in intracellular calcium. J. Cell Sci. 1998;111:2043–2054. doi: 10.1242/jcs.111.14.2043. [DOI] [PubMed] [Google Scholar]
- 19.Emoto Y., Kobayashi R., Akatsuka H., Hidaka H. Purification and characterization of a new member of the S-100 protein family from human placenta. Biochem. Biophys. Res. Commun. 1992;182:1246–1253. doi: 10.1016/0006-291x(92)91865-n. [DOI] [PubMed] [Google Scholar]
- 20.Becker T., Gerke V., Kube E., Weber K. S100P, a novel Ca2+-binding protein from human placenta. cDNA cloning, recombinant protein expression and Ca2+ binding properties. Eur. J. Biochem. 1992;207:541–547. doi: 10.1111/j.1432-1033.1992.tb17080.x. [DOI] [PubMed] [Google Scholar]
- 21.Dempsey A. C., Walsh M. P., Shaw G. S. Unmasking the annexin I interaction from the structure of Apo-S100A11. Structure. 2003;11:887–897. doi: 10.1016/s0969-2126(03)00126-6. [DOI] [PubMed] [Google Scholar]
- 22.Drohat A. C., Tjandra N., Baldisseri D. M., Weber D. J. The use of dipolar couplings for determining the solution structure of rat apo-S100B(ββ) Protein Sci. 1999;8:800–809. doi: 10.1110/ps.8.4.800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kilby P. M., Van Eldik L. J., Roberts G. C. The solution structure of the bovine S100B protein dimer in the calcium-free state. Structure. 1996;4:1041–1052. doi: 10.1016/s0969-2126(96)00111-6. [DOI] [PubMed] [Google Scholar]
- 24.Otterbein L. R., Kordowska J., Witte-Hoffmann C., Wang C.-L., Dominguez R. Crystal structures of S100A6 in the Ca2+-free and Ca2+-bound states: the calcium sensor mechanism of S100 proteins revealed at atomic resolution. Structure. 2002;10:557–567. doi: 10.1016/s0969-2126(02)00740-2. [DOI] [PubMed] [Google Scholar]
- 25.Rety S., Sopkova J., Renouard M., Osterloh D., Gerke V., Tabaries S., Russo-Marie F., Lewit-Bentley A. The crystal structure of a complex of p11 with the annexin II N-terminal peptide. Nat. Struct. Biol. 1999;6:89–95. doi: 10.1038/4965. [DOI] [PubMed] [Google Scholar]
- 26.Rustandi R. R., Baldisseri D. M., Inman K. G., Nizner P., Hamilton S. M., Landar A., Landar A., Zimmer D. B., Weber D. J. Three-dimensional solution structure of the calcium-signaling protein apo S100A1 as determined by NMR. Biochemistry. 2002;41:788–796. doi: 10.1021/bi0118308. [DOI] [PubMed] [Google Scholar]
- 27.Vallely K. M., Rustandi R. R., Ellis K. C., Varlamova O., Bresnick A. R., Weber D. J. Solution structure of human Mts 1 (S100A4) as determined by NMR spectroscopy. Biochemistry. 2002;41:12670–12680. doi: 10.1021/bi020365r. [DOI] [PubMed] [Google Scholar]
- 28.Arnesano F., Banci L., Bertini I., Fantoni A., Tenori L., Viezzoli M. S. Structural interplay between calcium(II) and copper(II) binding to S100A13 protein. Angew. Chem. Int. Ed. Engl. 2005;44:6341–6344. doi: 10.1002/anie.200500540. [DOI] [PubMed] [Google Scholar]
- 29.Lee Y. C., Volk D. E., Thiviyanathan V., Kleerekoper Q., Gribenko A. V., Zhang S., Gorenstein D. G., Makhatadze G. I., Luxon B. A. NMR structure of the Apo-S100P protein. J. Biomol. NMR. 2004;29:399–402. doi: 10.1023/B:JNMR.0000032617.88899.4b. [DOI] [PubMed] [Google Scholar]
- 30.Maler L., Potts B. C., Chazin W. J. High resolution solution structure of apo calcyclin and structural variations in the S100 family of calcium-binding proteins. J. Biomol. NMR. 1999;13:233–247. doi: 10.1023/a:1008315517955. [DOI] [PubMed] [Google Scholar]
- 31.Mittl P. R. E., Fritz G., Sargent D. F., Richmond T. J., Heizmann C. W., Grutter M. G. Metal-free MIRAS phasing: structure of apo-S100A3. Acta Crystallogr. Sect. D Biol. Crystallogr. 2002;58:1255–1261. doi: 10.1107/s0907444902008430. [DOI] [PubMed] [Google Scholar]
- 32.Potts B. C. M., Smith J., Akke M., Macke T. J., Okazaki K., Hidaka H., Case D. A., Chazin W. J. The structure of calcyclin reveals a novel homodimeric fold for S100 Ca2+-binding proteins. Nat. Struct. Biol. 1995;2:790–796. doi: 10.1038/nsb0995-790. [DOI] [PubMed] [Google Scholar]
- 33.Brodersen D. E., Etzerodt M., Madsen P., Celis J. E., Thogersen H. C., Nyborg J., Kjeldgaard M. EF-hands at atomic resolution: the structure of human psoriasin (S100A7) solved by MAD phasing. Structure. 1998;6:477–489. doi: 10.1016/s0969-2126(98)00049-5. [DOI] [PubMed] [Google Scholar]
- 34.Brodersen D. E., Nyborg J., Kjeldgaard M. Zinc-binding site of an S100 protein revealed. Two crystal structure of Ca2+-bound human psoriasin (S100A7) in the Zn2+-loaded and Zn2+-free states. Biochemistry. 1999;38:1695–1704. doi: 10.1021/bi982483d. [DOI] [PubMed] [Google Scholar]
- 35.Drohat A. C., Baldisseri D. M., Rustandi R. R., Weber D. J. Solution structure of calcium-bound rat S100B (ββ) as determined by nuclear magnetic resonance spectroscopy. Biochemistry. 1998;37:2729–2740. doi: 10.1021/bi972635p. [DOI] [PubMed] [Google Scholar]
- 36.Ishikawa K., Nakagawa A., Tanaka I., Suzuki M., Nishihira J. The structure of human MRP8, a member of the S100 calcium-binding protein family, by MAD phasing at 1.9 Å resolution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2000;56:559–566. doi: 10.1107/s0907444900002833. [DOI] [PubMed] [Google Scholar]
- 37.Maler L., Sastry M., Chazin W. J. A structural basis for S100 protein specificity derived from comparative analysis of apo and Ca2+-calcyclin. J. Mol. Biol. 2002;317:279–290. doi: 10.1006/jmbi.2002.5421. [DOI] [PubMed] [Google Scholar]
- 38.Moroz O. V., Antson A. A., Murshudov G. N., Maitland N. J., Dodson G. G., Wilson K. S., Skibshoj I., Lukanidin E. M., Bronstein I. B. The three-dimensional structure of human S100A12. Acta Crystallogr. Sect. D Biol. Crystallogr. 2001;57:20–29. doi: 10.1107/s090744490001458x. [DOI] [PubMed] [Google Scholar]
- 39.Sastry M., Ketchem R. R., Crescenzi O., Weber C., Lubienski M. J., Hidaka H., Chazin W. J. The three-dimensional structure of Ca2+-bound calcyclin: implications for Ca2+-signal transduction by S100 proteins. Structure. 1998;6:223–231. doi: 10.1016/s0969-2126(98)00023-9. [DOI] [PubMed] [Google Scholar]
- 40.Smith S. P., Shaw G. S. A novel calcium-sensitive switch revealed by the structure of human S100B in the calcium-bound form. Structure. 1998;6:211–222. doi: 10.1016/s0969-2126(98)00022-7. [DOI] [PubMed] [Google Scholar]
- 41.Zhang H., Wang G., Ding Y., Wang Z., Barraclough R., Rudland P. S., Fernig D. G., Rao Z. The crystal structure at 2Å resolution of the Ca2+-binding protein S100P. J. Mol. Biol. 2003;325:785–794. doi: 10.1016/s0022-2836(02)01278-0. [DOI] [PubMed] [Google Scholar]
- 42.Bhattacharya S., Large E., Heizmann C. W., Hemmings B., Chazin W. J. Structure of the Ca2+/S100B/NDR kinase peptide complex: insights into S100 target specificity and activation of the kinase. Biochemistry. 2003;42:14416–14426. doi: 10.1021/bi035089a. [DOI] [PubMed] [Google Scholar]
- 43.Inman K. G., Yang R., Rustandi R. R., Miller K. E., Baldisseri D. M., Weber D. J. Solution NMR structure of S100B bound to the high-affinity target peptide TRTK-12. J. Mol. Biol. 2002;324:1003–1014. doi: 10.1016/s0022-2836(02)01152-x. [DOI] [PubMed] [Google Scholar]
- 44.Itou H., Yao M., Fujita I., Watanabe N., Suzuki M., Nishihira J., Tanaka I. The crystal structure of human MRP14 (S100A9), a Ca2+-dependent regulator protein in inflammatory process. J. Mol. Biol. 2002;316:265–276. doi: 10.1006/jmbi.2001.5340. [DOI] [PubMed] [Google Scholar]
- 45.McClintock K. A., Shaw G. S. A novel S100 target conformation is revealed by the solution structure of the Ca2+–S100B–TRTK-12 complex. J. Biol. Chem. 2003;278:6251–6257. doi: 10.1074/jbc.M210622200. [DOI] [PubMed] [Google Scholar]
- 46.Rety S., Osterloh D., Arie J. P., Tabaries S., Seeman J., Russo-Marie F., Gerke V., Lewit-Bentley A. Structural basis of the Ca2+-dependent association between S100C (S100A11) and its target, the N-terminal part of annexin I. Structure. 2000;8:175–184. doi: 10.1016/s0969-2126(00)00093-9. [DOI] [PubMed] [Google Scholar]
- 47.Rustandi R. R., Baldisseri D. M., Weber D. J. Structure of the negative regulatory domain of p53 bound to S100B(ββ) Nat. Struct. Biol. 2000;7:570–574. doi: 10.1038/76797. [DOI] [PubMed] [Google Scholar]
- 48.Skelton N. J., Kordel J., Akke M., Forsen S., Chazin W. J. Signal transduction versus buffering activity in Ca2+-binding proteins. Struct. Biol. 1994;1:239–245. doi: 10.1038/nsb0494-239. [DOI] [PubMed] [Google Scholar]
- 49.Gagne S. M., Li M. X., McKay R. T., Sykes B. D. The NMR angle on troponin C. Biochem. Cell Biol. 1998;76:302–312. doi: 10.1139/bcb-76-2-3-302. [DOI] [PubMed] [Google Scholar]
- 50.Li C. L., Martinez V., He B., Lombet A., Perbal B. A role for CCN3 (NOV) in calcium signalling. Mol. Pathol. 2002;55:250–261. doi: 10.1136/mp.55.4.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Endo H., Takenaga K., Kanno T., Satoh H., Mori S. Methionine aminopeptidase 2 is a new target for the metastasis-associated protein, S100A4. J. Biol. Chem. 2002;277:26396–26402. doi: 10.1074/jbc.M202244200. [DOI] [PubMed] [Google Scholar]
- 52.Halachmi D., Eilam Y. Calcium homeostasis in yeast cells exposed to high concentrations of calcium: roles of vacuolar H+-ATPase and cellular ATP. FEBS Lett. 1993;316:73–78. doi: 10.1016/0014-5793(93)81739-m. [DOI] [PubMed] [Google Scholar]
- 53.Mueller A., Schafer B. W., Ferrari S., Weibel M., Makek M., Hochli M., Heizmann C. W. The calcium-binding protein S100A2 interacts with p53 and modulates its transcriptional activity. J. Biol. Chem. 2005;280:29186–29193. doi: 10.1074/jbc.M505000200. [DOI] [PubMed] [Google Scholar]
- 54.Sheu F.-S., Huang F. L., Huang K.-P. Differential responses of protein kinase C substrates (MARCKS, neuromodulin and neurogranin) phosphorylation to calmodulin and S100. Arch. Biochem. Biophys. 1995;316:335–342. doi: 10.1006/abbi.1995.1045. [DOI] [PubMed] [Google Scholar]
- 55.Polyakov A. A., Huber P. A., Marston S. B., Gusev N. B. Interaction of isoforms of S100 protein with smooth muscle caldesmon. FEBS Lett. 1998;422:235–239. doi: 10.1016/s0014-5793(98)00014-3. [DOI] [PubMed] [Google Scholar]
- 56.Baudier J., Cole R. D. Interactions between the microtubule-associated τ proteins and S100b regulate τ protein phosphorylation by the Ca2+-calmodulin-dependent protein kinase II. J. Biol. Chem. 1988;263:5876–5883. [PubMed] [Google Scholar]
- 57.Millward T. A., Heizmann C. W., Schafer B. W., Hemmings B. A. Calcium regulation of Ndr protein kinase mediated by S100 calcium-binding proteins. EMBO J. 1998;17:5913–5922. doi: 10.1093/emboj/17.20.5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stegert M. R., Tamaskovic R., Bichsel S. J., Hergovich A., Hemmings B. A. Regulation of NDR2 protein kinase by multi-site phosphorylation and the S100B calcium-binding protein. J. Biol. Chem. 2004;279:23806–23812. doi: 10.1074/jbc.M402472200. [DOI] [PubMed] [Google Scholar]
- 59.Nowotny M., Spiechowicz M., Jastrzebska B., Filipek A., Kitagawa K., Kuznicki J. Calcium-regulated interaction of Sgt1 with S100A6 (calcyclin) and other S100 proteins. J. Biol. Chem. 2003;278:26923–26928. doi: 10.1074/jbc.M211518200. [DOI] [PubMed] [Google Scholar]
- 60.Zimmer D. B., Van Eldik L. J. Identification of a molecular target for the calcium-modulated protein S100: fructose-1,6-bisphosphate aldolase. J. Biol. Chem. 1986;261:11424–11428. [PubMed] [Google Scholar]
- 61.Landar A., Caddell G., Chessher J., Zimmer D. B. Identification of an S100A1/S100B target protein: phosphoglucomutase. Cell Calcium. 1996;20:279–285. doi: 10.1016/s0143-4160(96)90033-0. [DOI] [PubMed] [Google Scholar]
- 62.Sorci G., Agneletti A. L., Bianchi R., Donato R. Association of S100B with intermediate filaments and microtubules in glial cells. Biochim. Biophys. Acta. 1998;1448:277–289. doi: 10.1016/s0167-4889(98)00134-7. [DOI] [PubMed] [Google Scholar]
- 63.Frizzo J. K., Tramontina F., Bortoli E., Gottfried C., Leal R. B., Lengyel I., Donato R., Dunkley P. R., Goncalves C. A. S100B-mediated inhibition of the phosphorylation of GFAP is prevented by TRTK-12. Neurochem. Res. 2004;29:735–740. doi: 10.1023/b:nere.0000018844.51009.40. [DOI] [PubMed] [Google Scholar]
- 64.Ferguson P. L., Shaw G. S. Human S100B protein interacts with the Escherichia coli division protein FtsZ in a calcium-sensitive manner. J. Biol. Chem. 2004;279:18806–18813. doi: 10.1074/jbc.M313948200. [DOI] [PubMed] [Google Scholar]
- 65.Ivanenkov V. V., Jamieson G. A., Jr, Gruenstein E., Dimlich R. V. W. Characterization of S-100b binding epitopes: identification of a novel target, the actin capping protein CapZ. J. Biol. Chem. 1995;270:14651–14658. doi: 10.1074/jbc.270.24.14651. [DOI] [PubMed] [Google Scholar]
- 66.Seemann J., Weber K., Gerke V. Annexin I targets S100C to early endosomes. FEBS Lett. 1997;413:185–190. doi: 10.1016/s0014-5793(97)00911-3. [DOI] [PubMed] [Google Scholar]
- 67.Hofmann M. A., Drury S., Fu C., Qu W., Taguchi A., Lu Y., Avila C., Kambham N., Bierhaus A., Nawroth P., et al. RAGE mediates a novel proinflammatory axis: a central cell surface receptor for S100/calgranulin polypeptides. Cell. 1999;97:889–901. doi: 10.1016/s0092-8674(00)80801-6. [DOI] [PubMed] [Google Scholar]
- 68.Huttunen H. J., Kuja-Panula J., Sorci G., Agneletti A. L., Donato R., Rauvala H. Coregulation of neurite outgrowth and cell survival by amphoterin and S100 proteins through receptor for advanced glycation end products (RAGE) activation. J. Biol. Chem. 2000;275:40096–40105. doi: 10.1074/jbc.M006993200. [DOI] [PubMed] [Google Scholar]
- 69.Arumugam T., Simeone D. M., Schmidt A. M., Logsdon C. D. S100P stimulates cell proliferation and survival via receptor for activated glycation end products (RAGE) J. Biol. Chem. 2004;279:5059–5065. doi: 10.1074/jbc.M310124200. [DOI] [PubMed] [Google Scholar]
- 70.Most P., Remppis A., Weber C., Bernotat J., Ehlermann P., Pleger S. T., Kirsch W., Weber M., Uttenweiler D., Smith G. L., et al. The C terminus (amino acids 75–94) and the linker region (amino acids 42–54) of the Ca2+-binding protein S100A1 differentially enhance sarcoplasmic Ca2+ release in murine skinned skeletal muscle fibers. J. Biol. Chem. 2003;278:26356–26364. doi: 10.1074/jbc.M303338200. [DOI] [PubMed] [Google Scholar]
- 71.Most P., Remppis A., Pleger S. T., Loffler E., Ehlermann P., Bernotat J., Kleuss C., Heierhorst J., Ruiz P., Witt H., et al. Transgenic overexpression of the Ca2+-binding protein S100A1 in the heart leads to increased in vivo myocardial contractile performance. J. Biol. Chem. 2003;278:33809–33817. doi: 10.1074/jbc.M301788200. [DOI] [PubMed] [Google Scholar]
- 72.Arcuri C., Giambanco I., Bianchi R., Donato R. Annexin V, annexin VI, S100A1 and S100B in developing and adult avian skeletal muscles. Neuroscience. 2002;109:371–388. doi: 10.1016/s0306-4522(01)00330-x. [DOI] [PubMed] [Google Scholar]
- 73.Garbuglia M., Verzini M., Donato R. Annexin VI binds S100A1 and S100B and blocks the ability of S100A1 and S100B to inhibit desmin and GFAP assemblies into intermediate filaments. Cell Calcium. 1998;24:177–191. doi: 10.1016/s0143-4160(98)90127-0. [DOI] [PubMed] [Google Scholar]
- 74.Zeng F. Y., Gerke V., Gabius H. J. Identification of annexin II, annexin VI and glyceraldehyde-3-phosphate dehydrogenase as calcyclin-binding proteins in bovine heart. Int. J. Biochem. 1993;25:1019–1027. doi: 10.1016/0020-711x(93)90116-v. [DOI] [PubMed] [Google Scholar]
- 75.Fernandez-Fernandez M. R., Veprintsev D. B., Fersht A. R. Proteins of the S100 family regulate the oligomerization of p53 tumor suppressor. Proc. Natl. Acad. Sci. U.S.A. 2005;102:4735–4740. doi: 10.1073/pnas.0501459102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Grigorian M., Andressen S., Tulchinsky E., Kriajevska M., Carlberg C., Kruse C., Cohn M., Ambartsumian N., Christensen A., Selivanova G., Lukanidin E. Tumor suppressor p53 protein is a new target for the metastasis-associated mts1/S100A4 protein. J. Biol. Chem. 2001;276:22699–22708. doi: 10.1074/jbc.M010231200. [DOI] [PubMed] [Google Scholar]
- 77.Chen H., Fernig D. G., Rudland P. S., Sparks A., Wilkinson M. C., Barraclough R. Binding to intracellular targets of the metastasis-inducing protein, S100A4 (p9Ka) Biochem. Biophys. Res. Commun. 2001;286:1212–1217. doi: 10.1006/bbrc.2001.5517. [DOI] [PubMed] [Google Scholar]
- 77a.Erratum. Biochem. Biophys. Res. Commun. 2003. p. 408.
- 78.Baudier J., Delphin C., Grunwald D., Khochbin S., Lawrence J. J. Characterization of the tumor suppressor protein p53 as a protein kinase C substrate and a S100b-binding protein. Proc. Natl. Acad. Sci. U.S.A. 1992;89:11627–11631. doi: 10.1073/pnas.89.23.11627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Filipek A., Jastrzebska B., Nowotny M., Kuznicki J. CacyBP/SIP, a calcyclin and Siah-1-interacting protein, binds EF-hand proteins of the S100 family. J. Biol. Chem. 2002;277:28848–28852. doi: 10.1074/jbc.M203602200. [DOI] [PubMed] [Google Scholar]
- 80.Zhang S., Wang G., Fernig D. G., Rudland P. S., Webb S. E., Barraclough R., Martin-Fernandez M. Interaction of metastasis-inducing S100A4 protein in vivo by fluorescence lifetime imaging microscopy. Eur. Biophys. J. 2005;34:19–27. doi: 10.1007/s00249-004-0428-x. [DOI] [PubMed] [Google Scholar]
- 81.Pearlstone J. R., Sykes B. D., Smillie L. B. Interactions of structural C and regulatory N domains of troponin C with repeated sequence motifs in troponin I. Biochemistry. 1997;36:7601–7606. doi: 10.1021/bi970200w. [DOI] [PubMed] [Google Scholar]
- 82.McKay R. T., Tripet B. P., Hodges R. S., Sykes B. D. Interaction of the second binding region of troponin I with the regulatory domain of skeletal muscle troponin C as determined by NMR spectroscopy. J. Biol. Chem. 1997;272:28494–28500. doi: 10.1074/jbc.272.45.28494. [DOI] [PubMed] [Google Scholar]
- 83.Landar A., Rustandi R. R., Weber D. J., Zimmer D. B. S100A1 utilizes different mechanisms for interacting with calcium-dependent and calcium-independent target proteins. Biochemistry. 1998;37:17429–17438. doi: 10.1021/bi9817921. [DOI] [PubMed] [Google Scholar]
- 84.Barber K. R., McClintock K. A., Jamieson G. A., Jr, Dimlich R. V., Shaw G. S. Specificity and Zn2+ enhancement of the S100B binding epitope TRTK-12. J. Biol. Chem. 1999;274:1502–1508. doi: 10.1074/jbc.274.3.1502. [DOI] [PubMed] [Google Scholar]
- 85.Li Z. H., Spektor A., Varlamova O., Bresnick A. R. Mts1 regulates the assembly of nonmuscle myosin-IIA. Biochemistry. 2003;42:14258–14266. doi: 10.1021/bi0354379. [DOI] [PubMed] [Google Scholar]
- 86.Takenaga K., Nakamura Y., Sakiyama S., Hasegawa Y., Sato K., Endo H. Binding of pEL98 protein, an S100-related calcium-binding protein, to nonmuscle tropomyosin. J. Cell. Biol. 1994;124:757–768. doi: 10.1083/jcb.124.5.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Delphin C., Ronjat M., Deloulme J. C., Garin G., Debussche L., Higashimoto Y., Sakaguchi K., Baudier J. Calcium-dependent interaction of S100B with the C-terminal domain of the tumor suppressor p53. J. Biol. Chem. 1999;274:10539–10544. doi: 10.1074/jbc.274.15.10539. [DOI] [PubMed] [Google Scholar]
- 88.Garbuglia M., Verzini M., Sorci G., Bianchi R., Giambanco I., Agneletti A. L., Donato R. The calcium-modulated proteins, S100A1 and S100B, as potential regulators of the dynamics of type III intermediate filaments. Braz. J. Med. Biol. Res. 1999;32:1177–1185. doi: 10.1590/s0100-879x1999001000001. [DOI] [PubMed] [Google Scholar]
- 89.Filipek A., Wojda U., Lesniak W. Interaction of calcyclin and its cyanogen bromide fragments with annexin II and glyceraldehyde 3-phosphate dehydrogenase. Int. J. Biochem. Cell Biol. 1995;27:1123–1131. doi: 10.1016/1357-2725(95)00096-8. [DOI] [PubMed] [Google Scholar]
- 90.Zimmer D. B., Dubuisson J. G. Identification of an S100 target protein: glycogen phosphorylase. Cell Calcium. 1993;14:323–332. doi: 10.1016/0143-4160(93)90053-9. [DOI] [PubMed] [Google Scholar]
- 91.Ruse M., Lambert A., Robinson N. A., Ryan D., Shon K., Eckert R. L. S100A7, S100A10, and S100A11 are transglutaminase substrates. Biochemistry. 2001;40:3167–3173. doi: 10.1021/bi0019747. [DOI] [PubMed] [Google Scholar]
- 92.Zhang S., Wang G., Liu D., Bao Z., Fernig D. G., Rudland P. S., Barraclough R. The C-terminal region of S100A4 is important for its metastasis-inducing properties. Oncogene. 2005;24:4401–4411. doi: 10.1038/sj.onc.1208663. [DOI] [PubMed] [Google Scholar]
- 93.Kriajevska M. V., Cardenas M. N., Grigorian M. S., Ambartsumian N. S., Georgiev G. P., Lukanidin E. M. Non-muscle myosin heavy chain as a possible target for protein encoded by metastasis-related mts-1 gene. J. Biol. Chem. 1994;269:19679–19682. [PubMed] [Google Scholar]
- 94.Ford H. L., Silver D. L., Kachar B., Sellers J. R., Zain S. B. Effect of Mts1 on the structure and activity of nonmuscle myosin II. Biochemistry. 1997;36:16321–16327. doi: 10.1021/bi971182l. [DOI] [PubMed] [Google Scholar]
- 95.Ford H. L., Zain S. B. Interaction of metastasis associated Mts1 protein with nonmuscle myosin. Oncogene. 1995;10:1597–1605. [PubMed] [Google Scholar]
- 96.Kim E. J., Helfman D. M. Characterization of the metastasis-associated protein, S100A4: roles of calcium binding and dimerization in cellular localization and interaction with myosin. J. Biol. Chem. 2003;278:30063–30073. doi: 10.1074/jbc.M304909200. [DOI] [PubMed] [Google Scholar]
- 97.Gribenko A. V., Guzman-Casado M., Lopez M. M., Makhatadze G. I. Conformational and thermodynamic properties of peptide binding to the human S100P protein. Protein Sci. 2002;11:1367–1375. doi: 10.1110/ps.0202202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Johnsson N., Marriott G., Weber K. p36, the major cytoplasmic substrate of src tyrosine protein kinase, binds to its p11 regulatory subunit via a short amino-terminal amphipathic helix. EMBO J. 1988;7:2435–2442. doi: 10.1002/j.1460-2075.1988.tb03089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Svenningsson P., Chergui K., Rachleff I., Flajolet M., Zhang X., El Yacoubi M., Vaugeois J. M., Nomikos G. G., Greengard P. Alterations in 5-HT1B receptor function by p11 in depression-like states. Science. 2006;311:77–80. doi: 10.1126/science.1117571. [DOI] [PubMed] [Google Scholar]
- 100.Beaton A. R., Rodriguez J., Reddy Y. K., Roy P. The membrane trafficking protein calpactin forms a complex with bluetongue virus protein NS3 and mediates virus release. Proc. Natl. Acad. Sci. U.S.A. 2002;99:13154–13159. doi: 10.1073/pnas.192432299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Choi J., Chang J. S., Song M. S., Ahn B. Y., Park Y., Lim D. S., Han Y. S. Association of hepatitis B virus polymerase with promyelocytic leukemia nuclear bodies mediated by the S100 family protein p11. Biochem. Biophys. Res. Commun. 2003;305:1049–1056. doi: 10.1016/s0006-291x(03)00881-7. [DOI] [PubMed] [Google Scholar]
- 102.Poon W. Y., Malik-Hall M., Wood J. N., Okuse K. Identification of binding domains in the sodium channel NaV1.8 intracellular N-terminal region and annexin II light chain p11. FEBS Lett. 2004;558:114–118. doi: 10.1016/S0014-5793(03)01512-6. [DOI] [PubMed] [Google Scholar]
- 103.Drohat A. C., Nenortas E., Beckett D., Weber D. J. Oligomerization state of S100B at nanomolar concentration determined by large-zone analytical gel filtration chromatography. Protein Sci. 1997;6:1577–1582. doi: 10.1002/pro.5560060721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fano G., Biocca S., Fulle S., Mariggio M., Belia S., Calissano P. The S100: a protein family in search of a function. Prog. Neurobiol. 1995;46:71–82. doi: 10.1016/0301-0082(94)00062-m. [DOI] [PubMed] [Google Scholar]
- 105.Odink K., Cerletti N., Bruggen J., Clerc R. G., Tarcsay L., Zwadlo G., Gerhards G., Schlegel R., Sorg C. Two calcium-binding proteins in infiltrate macrophages of rheumatoid arthritis. Nature (London) 1987;330:80–82. doi: 10.1038/330080a0. [DOI] [PubMed] [Google Scholar]
- 106.Propper C., Huang X., Roth J., Sorg C., Nacken W. Analysis of the MRP8–MRP14 protein–protein interaction by the two-hybrid system suggests a prominent role of the C-terminal domain of S100 proteins in dimer formation. J. Biol. Chem. 1999;274:183–188. doi: 10.1074/jbc.274.1.183. [DOI] [PubMed] [Google Scholar]
- 107.Wang G., Rudland P. S., White M. R., Barraclough R. Interaction in vivo and in vitro of the metastasis-inducing S100 protein, S100A4 (p9Ka) with S100A1. J. Biol. Chem. 2000;275:11141–11146. doi: 10.1074/jbc.275.15.11141. [DOI] [PubMed] [Google Scholar]
- 108.Yang Q., O'Hanlon D., Heizmann C. W., Marks A. Demonstration of heterodimer formation between S100B and S100A6 in the yeast two-hybrid system and human melanoma. Exp. Cell Res. 1999;246:501–509. doi: 10.1006/excr.1998.4314. [DOI] [PubMed] [Google Scholar]
- 109.Shaw G. S., Hodges R. S., Kay C. M., Sykes B. D. Relative stabilities of synthetic peptide homo- and heterodimeric troponin-C domains. Protein Sci. 1994;3:1010–1019. doi: 10.1002/pro.5560030703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lehrer S. S., Qian Y. Unfolding/refolding studies of smooth muscle tropomyosin: evidence for a chain exchange mechanism in the preferential assembly of the native heterodimer. J. Biol. Chem. 1990;265:1134–1138. [PubMed] [Google Scholar]
- 111.Tarabykina S., Scott D. J., Herzyk P., Hill T. J., Tame J. R., Kriajevska M., Lafitte D., Derrick P. J., Dodson G. G., Maitland N. J., et al. The dimerization interface of the metastasis-associated protein S100A4 (Mts1): in vivo and in vitro studies. J. Biol. Chem. 2001;276:24212–24222. doi: 10.1074/jbc.M009477200. [DOI] [PubMed] [Google Scholar]
- 112.Wang G., Zhang S., Fernig D. G., Martin-Fernandez M., Rudland P. S., Barraclough R. Mutually antagonistic actions of S100A4 and S100A1 on normal and metastatic phenotypes. Oncogene. 2005;24:1445–1454. doi: 10.1038/sj.onc.1208291. [DOI] [PubMed] [Google Scholar]
- 113.Deloulme J. C., Assard N., Mbele G. O., Mangin C., Kuwano R., Baudier J. S100A6 and S100A11 are specific targets of the calcium- and zinc-binding S100B protein in vivo. J. Biol. Chem. 2000;275:35302–35310. doi: 10.1074/jbc.M003943200. [DOI] [PubMed] [Google Scholar]
- 114.Tarabykina S., Kriajevska M., Scott D. J., Hill T. J., Lafitte D., Derrick P. J., Dodson G. G., Lukanidin E., Bronstein I. Heterocomplex formation between metastasis-related protein S100A4 (Mts1) and S100A1 as revealed by the yeast two-hybrid system. FEBS Lett. 2000;475:187–191. doi: 10.1016/s0014-5793(00)01652-5. [DOI] [PubMed] [Google Scholar]
- 115.Wang G., Zhang S., Fernig D. G., Spiller D., Martin-Fernandez M., Zhang H., Ding Y., Rao Z., Rudland P. S., Barraclough R. Heterodimeric interaction and interfaces of S100A1 and S100P. Biochem. J. 2004;382:375–383. doi: 10.1042/BJ20040142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Garbuglia M., Verzini M., Hofmann A., Huber R., Donato R. S100A1 and S100B interactions with annexins. Biochim. Biophys. Acta. 2000;1498:192–206. doi: 10.1016/s0167-4889(00)00096-3. [DOI] [PubMed] [Google Scholar]
- 117.Garbuglia M., Verzini M., Rustandi R. R., Osterloh D., Weber D. J., Gerke V., Donato R. Role of the C-terminal extension in the interaction of S100A1 with GFAP, tubulin, the S100A1- and S100B-inhibitory peptide, TRTK-12, and a peptide derived from p53, and the S100A1 inhibitory effect on GFAP polymerization. Biochem. Biophys. Res. Commun. 1999;254:36–41. doi: 10.1006/bbrc.1998.9881. [DOI] [PubMed] [Google Scholar]
- 118.Ivanenkov V. V., Dimlich R. V. W., Jamieson G. A., Jr Interaction of S100a0 protein with the actin capping protein, CapZ: characterization of a putative S100a0 binding site in CapZα-subunit. Biochem. Biophys. Res. Commun. 1996;221:46–50. doi: 10.1006/bbrc.1996.0542. [DOI] [PubMed] [Google Scholar]
- 119.Osterloh D., Ivanenkov V. V., Gerke V. Hydrophobic residues in the C-terminal region of S100A1 are essential for target protein binding but not for dimerization. Cell Calcium. 1998;24:137–151. doi: 10.1016/s0143-4160(98)90081-1. [DOI] [PubMed] [Google Scholar]
- 120.Garbuglia M., Verzini M., Giambanco I., Spreca A., Donato R. Effects of calcium-binding proteins (S-100a0, S-100a, S-100b) on desmin assembly in vitro. FASEB. 1996;10:317–324. doi: 10.1096/fasebj.10.2.8641565. [DOI] [PubMed] [Google Scholar]
- 121.Bianchi R., Giambanco I., Donato R. S-100 protein, but not calmodulin binds to the glial fibrillary acid protein and inhibits its polymerization in a Ca2+-dependent manner. J. Biol. Chem. 1993;268:12669–12674. [PubMed] [Google Scholar]
- 122.Sorci G., Agneletti A. L., Donato R. Effects of S100A1 and S100B on microtubule stability: an in vitro study using triton-cytoskeletons from astrocyte and myoblast cell lines. Neurosci. 2000;99:773–783. doi: 10.1016/s0306-4522(00)00238-4. [DOI] [PubMed] [Google Scholar]
- 123.Garbuglia M., Verzini M., Sorci G., Bianchi R., Giambanco I., Agneletti A. L., Donato R. The calcium-modulated proteins, S100A1 and S100B, as potential regulators of the dynamics of type III intermediate filaments. Braz. J. Med. Biol. Res. 1999;32:1177–1185. doi: 10.1590/s0100-879x1999001000001. [DOI] [PubMed] [Google Scholar]
- 124.Baudier J., Bergeret E., Bertacchi N., Weintraub H., Gagnon J., Garin J. Interactions of myogenic bHLH transcription factors with calcium-binding calmodulin and S100a (αα) proteins. Biochemistry. 1995;34:7834–7846. doi: 10.1021/bi00024a007. [DOI] [PubMed] [Google Scholar]
- 125.Onions J., Hermann S., Grundstrom T. Basic helix–loop–helix protein sequences determining differential inhibition by calmodulin and S100 proteins. J. Biol. Chem. 1997;272:23930–23937. doi: 10.1074/jbc.272.38.23930. [DOI] [PubMed] [Google Scholar]
- 126.Kiewitz R., Acklin C., Schafer B. W., Maco B., Uhrik B., Wuytack F., Erne P., Heizmann C. W. Ca2+-dependent interaction of S100A1 with the sarcoplasmic reticulum Ca2+-ATPase 2a and phospholamban in the human heart. Biochem. Biophys. Res. Commun. 2003;306:550–557. doi: 10.1016/s0006-291x(03)00987-2. [DOI] [PubMed] [Google Scholar]
- 127.Benfenati F., Ferrari R., Onofri F., Arcuri C., Giambanco I., Donato R. S100A1 codistributes with synapsin I in discrete brain areas and inhibits the F-actin-bundling activity of synapsin I. J. Neurochem. 2004;89:1260–1270. doi: 10.1111/j.1471-4159.2004.02419.x. [DOI] [PubMed] [Google Scholar]
- 128.Heierhorst J., Mitchelhill K. I., Mann R. J., Tiganis T., Czernik A. J., Greengard P., Kemp B. E. Synapsins as major neuronal Ca2+/S100A1-interacting proteins. Biochemistry. 1999;344:577–583. [PMC free article] [PubMed] [Google Scholar]
- 129.Gutierrez-Cruz G., Van Heerden A. H., Wang K. Modular motif, structural folds and affinity profiles of the PEVK segment of human fetal skeletal muscle titin. J. Biol. Chem. 2001;276:7442–7449. doi: 10.1074/jbc.M008851200. [DOI] [PubMed] [Google Scholar]
- 130.Yamasaki R., Berri M., Wu Y., Trombitas K., McNabb M., Kellermayer M. S., Witt C., Labeit D., Labeit S., Greaser M., Granzier H. Titin–actin interaction in mouse myocardium: passive tension modulation and its regulation by calcium/S100A1. Biophys. J. 2001;81:2297–2313. doi: 10.1016/S0006-3495(01)75876-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Heierhorst J., Mann R. J., Kemp B. E. Interaction of the recombinant S100A1 protein with twitchin kinase, and comparison with other Ca2+-binding proteins. Eur. J. Biochem. 1997;249:127–133. doi: 10.1111/j.1432-1033.1997.00127.x. [DOI] [PubMed] [Google Scholar]
- 132.Heierhorst J., Kobe B., Feil S. C., Parker M. W., Benian G. M., Weiss K. R., Kemp B. E. Ca2+/S100 regulation of giant protein kinases. Nature (London) 1996;380:636–639. doi: 10.1038/380636a0. [DOI] [PubMed] [Google Scholar]
- 133.Hibi K., Fujitake S., Takase T., Kodera Y., Ito K., Akiyama S., Shirane M., Nakao A. Identification of S100A2 as a target of the ΔNp63 oncogenic pathway. Clin. Cancer Res. 2003;9:4282–4285. [PubMed] [Google Scholar]
- 134.Gimona M., Lando Z., Dolginov Y., Vandekerckhove J., Kobayashi R., Sobieszek A., Helfman D. M. Ca2+-dependent interaction of S100A2 with muscle and nonmuscle tropomyosins. J. Cell Sci. 1997;110:611–621. doi: 10.1242/jcs.110.5.611. [DOI] [PubMed] [Google Scholar]
- 135.Lombet A., Planque N., Bleau A. M., Li C. L., Perbal B. CCN3 and calcium signaling. Cell Commun. Signaling. 2003;1:1. doi: 10.1186/1478-811X-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Watanabe Y., Usada N., Minami H., Morita T., Tsugane S., Ishikawa R., Kohama K., Tomida Y., Hidaka H. Calvasculin, as a factor affecting the microfilament assemblies in rat fibroblasts transfected by src gene. FEBS Lett. 1993;324:51–55. doi: 10.1016/0014-5793(93)81530-d. [DOI] [PubMed] [Google Scholar]
- 137. Reference deleted.
- 138.Kriajevska M., Tarabykina S., Bronstein I., Maitland N., Lomonosov M., Hansen K., Georgiev G., Lukanidin E. Metastasis-associated Mts1 (S100A4) protein modulates protein kinase C phosphorylation of the heavy chain of nonmuscle myosin. J. Biol. Chem. 1998;273:9852–9856. doi: 10.1074/jbc.273.16.9852. [DOI] [PubMed] [Google Scholar]
- 139.Dukhanina E. A., Dukhanin A. S., Lomonosov M. Y., Lukanidin E. M., Georgiev G. P. Spectral studies on the calcium-binding properties of Mts1 protein and its interaction with target protein. FEBS Lett. 1997;410:403–406. doi: 10.1016/s0014-5793(97)00576-0. [DOI] [PubMed] [Google Scholar]
- 140.Koshelev Y. A., Kiselev S. L., Georgiev G. P. Interaction of the S100A4 (Mts1) protein with septins Sept2, Sept6, and Sept7 in vitro. Dokl. Biochem. Biophys. 2003;391:195–197. doi: 10.1023/a:1025149005902. [DOI] [PubMed] [Google Scholar]
- 141.Filipek A., Wojda U. p30, a novel protein target of mouse calcyclin (S100A6) Biochem. J. 1996;320:585–587. doi: 10.1042/bj3200585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Watanabe M., Ando Y., Tokumitsu H., Hidaka H. Binding site of annexin XI on the calcyclin molecule. Biochem. Biophys. Res. Commun. 1993;196:1376–1382. doi: 10.1006/bbrc.1993.2405. [DOI] [PubMed] [Google Scholar]
- 143.tMinami H., Tokumitsu H., Mizutani A., Watanabe Y., Watanabe M., Hidaka H. Specific binding of CAP-50 to calcyclin. FEBS Lett. 1992;305:217–219. doi: 10.1016/0014-5793(92)80671-3. [DOI] [PubMed] [Google Scholar]
- 144.Tomas A., Moss S. E. Calcium- and cell cycle-dependent association of annexin 11 with the nuclear envelope. J. Biol. Chem. 2003;278:20210–20216. doi: 10.1074/jbc.M212669200. [DOI] [PubMed] [Google Scholar]
- 145.Tokumitsu H., Mizutani A., Minami H., Kobayashi R., Hidaka H. A calcyclin-associated protein is a newly identified member of the Ca2+/phospholipid binding proteins, annexin family. J. Biol. Chem. 1992;267:8919–8924. [PubMed] [Google Scholar]
- 146.Williams L. H., McClive P. J., Van Den Bergen J. A., Sinclair A. H. Annexin XI co-localises with calcyclin in proliferating cells of the embryonic mouse testis. Dev. Dyn. 2005;234:432–437. doi: 10.1002/dvdy.20548. [DOI] [PubMed] [Google Scholar]
- 147.Nowotny M., Bhattacharya S., Filipek A., Krezel A. M., Chazin W., Kuznicki J. Characterization of the interaction of calcyclin (S100A6) and calcyclin-binding protein. J. Biol. Chem. 2000;275:31178–31182. doi: 10.1074/jbc.M001622200. [DOI] [PubMed] [Google Scholar]
- 148.Filipek A., Zasada A., Wojda U., Makuch R., Dabrowska R. Characterization of chicken gizzard calcyclin and examination of its interaction with caldesmon. Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 1996;113:745–752. doi: 10.1016/0305-0491(95)02095-0. [DOI] [PubMed] [Google Scholar]
- 149.Zeng F. Y., Gabius H. J. Carbohydrate-binding specificity of calcyclin and its expression in human tissues and leukemic cells. Arch. Biochem. Biophys. 1991;289:137–144. doi: 10.1016/0003-9861(91)90453-p. [DOI] [PubMed] [Google Scholar]
- 150.Filipek A., Gerke V., Weber K., Kuznicki J. Characterization of the cell-cycle-regulated protein calcyclin from Ehrlich ascites tumor cells: identification of two binding proteins obtained by Ca2+-dependent affinity chromatography. Eur. J. Biochem. 1991;195:795–800. doi: 10.1111/j.1432-1033.1991.tb15768.x. [DOI] [PubMed] [Google Scholar]
- 151.Filipek A., Wojda U. Chicken gizzard calcyclin: distribution and potential target proteins. Biochem. Biophys. Res. Commun. 1996;225:151–154. doi: 10.1006/bbrc.1996.1145. [DOI] [PubMed] [Google Scholar]
- 152.Golitsina N. L., Kordowska J., Wang C. L., Lehrer S. S. Ca2+-dependent binding of calcyclin to muscle tropomyosin. Biochem. Biophys. Res. Commun. 1996;220:360–365. doi: 10.1006/bbrc.1996.0410. [DOI] [PubMed] [Google Scholar]
- 153.Berthier S., Paclet M. H., Lerouge S., Roux F., Vergnaud S., Coleman A. W., Morel F. Changing the conformation state of cytochrome b558 initiates NADPH oxidase activation: MRP8/MRP14 regulation. J. Biol. Chem. 2003;278:25499–25508. doi: 10.1074/jbc.M209755200. [DOI] [PubMed] [Google Scholar]
- 154.Robinson M. J., Tessier P., Poulsom R., Hogg N. The S100 family heterodimer, MRP-8/14, binds with high affinity to heparin and heparan sulfate glycosaminoglycans on endothelial cells. J. Biol. Chem. 2002;277:3658–3665. doi: 10.1074/jbc.M102950200. [DOI] [PubMed] [Google Scholar]
- 155.Strupat K., Rogniaux H., Van Dorsselaer A., Roth J., Vogl T. Calcium-induced noncovalently linked tetramers of MRP8 and MRP14 are confirmed by electrospray ionization-mass analysis. J. Am. Soc. Mass. Spectrom. 2000;11:780–788. doi: 10.1016/S1044-0305(00)00150-1. [DOI] [PubMed] [Google Scholar]
- 156.Vogl T., Roth J., Sorg C., Hillenkamp F., Strupat K. Calcium-induced noncovalently linked tetramers of MRP8 and MRP14 detected by ultraviolet matrix-assisted laser desorption/ionization mass spectrometry. J. Am. Soc. Mass. Spectrom. 1999;10:1124–1130. doi: 10.1016/s1044-0305(99)00085-9. [DOI] [PubMed] [Google Scholar]
- 157.Zhao X. Q., Naka M., Muneyuki M., Tanaka T. Ca2+-dependent inhibition of actin-activated myosin ATPase activity by S100C (S100A11), a novel member of the S100 protein family. Biochem. Biophys. Res. Commun. 2000;267:77–79. doi: 10.1006/bbrc.1999.1918. [DOI] [PubMed] [Google Scholar]
- 158.Bianchi R., Giambanco I., Arcuri C., Donato R. Subcellular localization of S100A11 (S100C) in LLC-PK1 renal cells: calcium- and protein kinase C-dependent association of S100A11 with S100B and vimentin intermediate filaments. Microsc. Res. Tech. 2003;60:639–651. doi: 10.1002/jemt.10305. [DOI] [PubMed] [Google Scholar]
- 159.Seemann J., Weber K., Gerke V. Structural requirements for annexin I–S100C complex-formation. Biochem. J. 1996;319:123–129. doi: 10.1042/bj3190123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hatakeyama T., Okada M., Shimamoto S., Kubota Y., Kobayashi R. Identification of intracellular target proteins of the calcium-signaling protein S100A12. Eur. J. Biochem. 2004;271:3765–3775. doi: 10.1111/j.1432-1033.2004.04318.x. [DOI] [PubMed] [Google Scholar]
- 161.Akpek E. K., Liu S. H., Thompson R., Gottsch J. D. Identification of paramyosin as a binding protein for calgranulin C in experimental helminthic keratitis. Invest. Ophthalmol. Vis. Sci. 2002;43:2677–2684. [PubMed] [Google Scholar]
- 162.Huttunen H. J., Fages C., Rauvala H. Receptor for advanced glycation end products (RAGE)-mediated neurite outgrowth and activation of NF-κB require the cytoplasmic domain of the receptor but different downstream signaling pathways. J. Biol. Chem. 1999;274:19919–19924. doi: 10.1074/jbc.274.28.19919. [DOI] [PubMed] [Google Scholar]
- 163.Moroz O. V., Antson A. A., Dodson E. J., Burrell H. J., Grist S. J., Lloyd R. M., Maitland N. J., Dodson G. G., Wilson K. S., Lukanidin E., Bronstein I. B. The structure of S100A12 in a hexameric form and its proposed role in receptor signalling. Acta Crystallogr. Sect. D Biol. Crystallogr. 2002;58:407–413. doi: 10.1107/s0907444901021278. [DOI] [PubMed] [Google Scholar]
- 164.Gentil B. J., Delphin C., Mbele G. O., Deloulme J. C., Ferro M., Garin J., Baudier J. The giant protein AHNAK is a specific target for the calcium- and zinc-binding S100B protein. J. Biol. Chem. 2001;276:23253–23261. doi: 10.1074/jbc.M010655200. [DOI] [PubMed] [Google Scholar]
- 165.McClintock K. A., Van Eldik L. J., Shaw G. S. The C-terminus and linker region of S100B exert dual control on protein–protein interactions with TRTK-12. Biochemistry. 2002;41:5421–5428. doi: 10.1021/bi011732m. [DOI] [PubMed] [Google Scholar]
- 166.Margulis A., Pozdnyakov N., Sitaramayya A. Activation of bovine photoreceptor guanylate cyclase by S100 proteins. Biochem. Biophys. Res. Commun. 1996;218:243–247. doi: 10.1006/bbrc.1996.0043. [DOI] [PubMed] [Google Scholar]
- 167.Garbuglia M., Verzini M., Dimlich R. V. W., Jamieson G. A., Jr, Donato R. Characterization of type III intermediate filament regulatory protein target epitopes: S100 (β and/or α) binds the N-terminal head domain; annexin II2-p112 binds rod domain. Biochim. Biophys. Acta. 1996;1313:268–276. doi: 10.1016/0167-4889(96)00099-7. [DOI] [PubMed] [Google Scholar]
- 168.Mbele G. O., Deloulme J. C., Gentil B. J., C. D., Ferro M., Garin J., Takahashi M., Baudier J. The zinc- and calcium-binding S100B interacts and co-localizes with IQGAP1 during dynamic rearrangement of cell membranes. J. Biol. Chem. 2002;277:49998–50007. doi: 10.1074/jbc.M205363200. [DOI] [PubMed] [Google Scholar]
- 169.Kursula P., Tikkanen G., Lehto V. P., Nishikimi M., Heape A. M. Calcium-dependent interaction between the large myelin-associated glycoprotein and S100β. J. Neurochem. 1999;73:1724–1732. doi: 10.1046/j.1471-4159.1999.731724.x. [DOI] [PubMed] [Google Scholar]
- 170.Baudier J., Bronner C., Kligman D., Cole R. D. Protein kinase C substrates from bovine brain. J. Biol. Chem. 1989;264:1824–1828. [PubMed] [Google Scholar]
- 171.Lin J., Blake M., Tang C., Zimmer D., Rustandi R. R., Weber D. J., Carrier F. Inhibition of p53 transcriptional activity by the S100B calcium-binding protein. J. Biol. Chem. 2001;276:35037–35041. doi: 10.1074/jbc.M104379200. [DOI] [PubMed] [Google Scholar]
- 172.Rustandi R. R., Baldisseri D. M., Drohat A. C., Weber D. J. Structural changes in the C-terminus of Ca2+-bound rat S100B (ββ) upon binding to a peptide derived from the C-terminal regulatory domain of p53. Protein Sci. 1999;8:1743–1751. doi: 10.1110/ps.8.9.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Rustandi R. R., Drohat A. C., Baldisseri D. M., Wilder P. T., Weber D. J. The Ca2+-dependent interaction of S100B(ββ) with a peptide derived from p53. Biochemistry. 1998;37:1951–1960. doi: 10.1021/bi972701n. [DOI] [PubMed] [Google Scholar]
- 174.Landar A., Hall T. L., Cornwall E. H., Correia J. J., Drohat A. C., Weber D. J., Zimmer D. B. The role of cysteine residues in S100B dimerization and regulation of target protein activity. Biochim. Biophys. Acta. 1997;1343:117–129. doi: 10.1016/s0167-4838(97)00126-x. [DOI] [PubMed] [Google Scholar]
- 175.Baudier J., Mochly-Rosen D., Newton A., Lee S. H., Koshland D. E., Jr, Cole R. D. Comparison of S100b protein with calmodulin: interactions with melittin and microtubule-associated tau proteins and inhibition of phosphorylation of tau proteins by protein kinase C. Biochemistry. 1987;26:2886–2893. doi: 10.1021/bi00384a033. [DOI] [PubMed] [Google Scholar]
- 176.Koltzscher M., Neumann C., Konig S., Gerke V. Ca2+-dependent binding and activation of dormant ezrin by dimeric S100P. Mol. Biol. Cell. 2003;14:2372–2384. doi: 10.1091/mbc.E02-09-0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Arumugam T., Simeone D. M., Van Golen K., Logsdon C. D. S100P promotes pancreatic cancer growth, survival, and invasion. Clin. Cancer Res. 2005;11:5356–5364. doi: 10.1158/1078-0432.CCR-05-0092. [DOI] [PubMed] [Google Scholar]
- 178.Dowen S. E., Crnogorac-Jurcevic T., Gangeswaran R., Hansen M., Eloranta J. J., Bhakta V., Brentnall T. A., Luttges J., Kloppel G., Lemoine N. R. Expression of S100P and its novel binding partner S100PBPR in early pancreatic cancer. Am. J. Pathol. 2005;166:81–92. doi: 10.1016/S0002-9440(10)62234-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Fano G., Angelella P., Mariggio D., Aisa M. C., Giambanco I., Donato R. S-100a0 protein stimulates the basal (Mg2+-activated) adenylate cyclase activity associated with skeletal muscle membranes. FEBS Lett. 1989;248:9–12. doi: 10.1016/0014-5793(89)80421-1. [DOI] [PubMed] [Google Scholar]
- 180.Donato Calcium-independent, pH-regulated effects of S-100 proteins on assembly–disassembly of brain microtubule protein in vitro. J. Biol. Chem. 1988;263:106–110. [PubMed] [Google Scholar]
- 181.Semov A., Moreno M. J., Onichtchenko A., Abulrob A., Ball M., Ekiel I., Pietrzynski G., Stanimirovic D., Alakhov V. Metastasis-associated protein S100A4 induces angiogenesis through interaction with annexin II and accelerated plasmin formation. J. Biol. Chem. 2005;280:20833–20841. doi: 10.1074/jbc.M412653200. [DOI] [PubMed] [Google Scholar]
- 182.Kriajevska M., Fischer-Larsen M., Moertz E., Vorm O., Tulchinsky E., Grigorian M., Ambartsumian N., Lukanidin E. Liprin β1, a member of the family of LAR transmembrane tyrosine phosphatase-interacting proteins, is a new target for the metastasis-associated protein S100A4 (Mts1) J. Biol. Chem. 2002;277:5229–5235. doi: 10.1074/jbc.M110976200. [DOI] [PubMed] [Google Scholar]
- 183.Burkitt W. I., Derrick P. J., Lafitte D., Bronstein I. Protein–ligand and protein–protein interactions studied by electrospray ionization and mass spectrometry. Biochem. Soc. Trans. 2003;31:985–989. doi: 10.1042/bst0310985. [DOI] [PubMed] [Google Scholar]
- 184.Hagens G., Roulin K., Hotz R., Saurat J. H., Hellman U., Siegenthaler G. Probable interaction between S100A7 and E-FABP in the cytosol of human keratinocytes from psoriatic scales. Mol. Cell. Biochem. 1999;192:123–128. [PubMed] [Google Scholar]
- 185.Hagens G., Masouye I., Augsburger E., Hotz R., Saurat J. H., Siegenthaler G. Calcium-binding protein S100A7 and epidermal-type fatty acid-binding protein are associated in the cytosol of human keratinocytes. Biochem. J. 1999;339:419–427. [PMC free article] [PubMed] [Google Scholar]
- 186.Ruse M., Broome A. M., Eckert R. L. S100A7 (psoriasin) interacts with epidermal fatty acid binding protein and localizes in focal adhesion-like structures in cultured keratinocytes. J. Invest. Dermatol. 2003;121:132–141. doi: 10.1046/j.1523-1747.2003.12309.x. [DOI] [PubMed] [Google Scholar]
- 187.Emberley E. D., Niu Y., Curtis L., Troup S., Mandal S. K., Myers J. N., Gibson S. B., Murphy L. C., Watson P. H. The S100A7–c-Jun activation domain binding protein 1 pathway enhances prosurvival pathways in breast cancer. Cancer Res. 2005;65:5696–5702. doi: 10.1158/0008-5472.CAN-04-3927. [DOI] [PubMed] [Google Scholar]
- 188.Emberley E. D., Niu Y., Leygue E., Tomes L., Gietz R. D., Murphy L. C., Watson P. H. Psoriasin interacts with Jab1 and influences breast cancer progression. Cancer Res. 2003;63:1954–1961. [PubMed] [Google Scholar]
- 189.Emberley E. D., Gietz R. D., Campbell J. D., HayGlass K. T., Murphy L. C., Watson P. H. RanBPM interacts with psoriasin in vitro and their expression correlates with specific clinical features in vivo in breast cancer. BMC Cancer. 2002;2:28. doi: 10.1186/1471-2407-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Srikrishna G., Turovskaya O., Shaikh R., Newlin R., Foell D., Murch S., Kronenberg M., Freeze H. H. Carboxylated glycans mediate colitis through activation of NF-κB. J. Immunol. 2005;175:5412–5422. doi: 10.4049/jimmunol.175.8.5412. [DOI] [PubMed] [Google Scholar]
- 191.Kerkhoff C., Sorg C., Tandon N. N., Nacken W. Interaction of S100A8/S100A9–arachidonic acid complexes with the scavenger receptor CD36 may facilitate fatty acid uptake by endothelial cells. Biochemistry. 2001;40:241–248. doi: 10.1021/bi001791k. [DOI] [PubMed] [Google Scholar]
- 192.Doussiere J., Bouzidi F., Vignais P. V. The S100A8/A9 protein as a partner for the cytosolic factors of NADPH oxidase activation in neutrophils. Eur. J. Biochem. 2002;269:3246–3255. doi: 10.1046/j.1432-1033.2002.03002.x. [DOI] [PubMed] [Google Scholar]
- 193.Osborn M., Johnsson N., Wehland J., Weber K. The submembranous location of p11 and its interaction with the p36 substrate of pp60 src kinase in situ. Exp. Cell Res. 1988;175:81–96. doi: 10.1016/0014-4827(88)90257-1. [DOI] [PubMed] [Google Scholar]
- 194.Bianchi R., Pula G., Ceccarelli P., Giambanco I., Donato R. S-100 protein binds to annexin II and p11, the heavy and light chains of calpactin I. Biochim. Biophys. Acta. 1992;1160:67–75. doi: 10.1016/0167-4838(92)90039-g. [DOI] [PubMed] [Google Scholar]
- 195.Johnsson N., Weber K. Alkylation of cysteine 82 of p11 abolishes the complex formation with the tyrosine-protein kinase substrate p36 (annexin 2, calpactin 1, lipocortin 2) J. Biol. Chem. 1990;265:14464–14468. [PubMed] [Google Scholar]
- 196.Johnsson N., Weber K. Structural analysis of p36, a Ca2+/lipid-binding protein of the annexin family, by proteolysis and chemical fragmentation. Eur. J. Biochem. 1990;188:1–7. doi: 10.1111/j.1432-1033.1990.tb15363.x. [DOI] [PubMed] [Google Scholar]
- 197.Harder T., Gerke V. The annexin II2p11(2) complex is the major protein component of the triton X-100-insoluble low-density fraction prepared from MDCK cells in the presence of Ca2+ Biochim. Biophys. Acta. 1994;1223:375–382. doi: 10.1016/0167-4889(94)90098-1. [DOI] [PubMed] [Google Scholar]
- 198.Rety S., Sopkova J., Renouard M., Osterloh D., Gerke V., Tabaries S., Russo-Marie R., Lewit-Bentley A. The crystal structure of a complex of p11 with the annexin II N-terminal peptide. Nat. Struct. Biol. 1999;6:89–95. doi: 10.1038/4965. [DOI] [PubMed] [Google Scholar]
- 199.Zobiack N., Gerke V., Rescher U. Complex formation and submembranous localization of annexin 2 and S100A10 in live HepG2 cells. FEBS Lett. 2001;500:137–140. doi: 10.1016/s0014-5793(01)02604-7. [DOI] [PubMed] [Google Scholar]
- 200.Becker T., Weber K., Johnsson N. Protein-protein recognition via short amphiphilic helices: a mutational analysis of the binding site of annexin II for p11. EMBO J. 1990;9:4207–4213. doi: 10.1002/j.1460-2075.1990.tb07868.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Kube E., Weber K., Gerke V. Primary structure of human, chicken, and Xenopus laevis p11, a cellular ligand of the Src-kinase substrate, annexin II. Gene. 1991;102:255–259. doi: 10.1016/0378-1119(91)90086-q. [DOI] [PubMed] [Google Scholar]
- 202.Kube E., Becker T., Weber K., Gerke V. Protein–protein interaction studied by site-directed mutagenesis: characterization of the annexin II binding site on p11, a member of the S100 protein family. J. Biol. Chem. 1992;267:14175–14182. [PubMed] [Google Scholar]
- 203.Zokas L., Glenney J. R., Jr The calpactin light chain is tightly linked to the cytoskeletal form of calpactin I: studies using monoclonal antibodies to calpactin subunits. J. Cell Biol. 1987;105:2111–2121. doi: 10.1083/jcb.105.5.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Huang X. L., Pawliczak R., Cowan M. J., Gladwin M. T., Madara P., Logun C., Shelhamer J. H. Epidermal growth factor induces p11 gene and protein expression and down-regulates calcium ionophore-induced arachidonic acid release in human epithelial cells. J. Biol. Chem. 2002;277:38431–38440. doi: 10.1074/jbc.M207406200. [DOI] [PubMed] [Google Scholar]
- 205.Bailleux A., Wendum D., Audubert F., Jouniaux A. M., Koumanov K., Trugnan G., Masliah J. Cytosolic phospholipase A2–p11 interaction controls arachidonic acid release as a function of epithelial cell confluence. Biochem. J. 2004;378:307–315. doi: 10.1042/BJ20031014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Girard C., Tinel N., Terrenoire C., Romey G., Lazdunski M., Borsotto M. p11, an annexin II subunit, an auxiliary protein associated with the background K+ channel, TASK-1. EMBO J. 2002;21:4439–4448. doi: 10.1093/emboj/cdf469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Kassam G., Le B. H., Choi K. S., Kang H. M., Fitzpatrick S. L., Louie P., Waisman D. M. The p11 subunit of the annexin II tetramer plays a key role in the stimulation of t-PA-dependent plasminogen activation. Biochemistry. 1998;37:16958–16966. doi: 10.1021/bi981713l. [DOI] [PubMed] [Google Scholar]
- 208.Malik-Hall M., Poon W. Y., Baker M. D., Wood J. N., Okuse K. Sensory neuron proteins interact with the intracellular domains of sodium channel NaV 1.8. Mol. Brain Res. 2003;110:298–304. doi: 10.1016/s0169-328x(02)00661-7. [DOI] [PubMed] [Google Scholar]
- 209.Okuse K., Malik-Hall M., Baker M. D., Poon W. Y., Kong H., Chao M. V., Wood J. N. Annexin II light chain regulates sensory neuron-specific sodium channel expression. Nature (London) 2002;417:653–656. doi: 10.1038/nature00781. [DOI] [PubMed] [Google Scholar]
- 210.Van de Graaf S. F. J., Hoenderop J. G. J., Gkika D., Lamers D., Prenen J., Rescher U., Gerke V., Staub O., Nilius B., Bindels R. J. M. Functional expression of the epithelial Ca2+ channels (TRPV5 and TRPV6) requires association of the S100A10–annexin 2 complex. EMBO J. 2003;22:1478–1487. doi: 10.1093/emboj/cdg162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Landriscina M., Bagala C., Mandinova A., Soldi R., Micucci I., Bellum S., Prudovsky I., Maciag T. Copper induces the assembly of a multiprotein aggregate implicated in the release of fibroblast growth factor 1 in response to stress. J. Biol. Chem. 2001;276:25549–25557. doi: 10.1074/jbc.M102925200. [DOI] [PubMed] [Google Scholar]
- 212.Mouta Carreira C., LaVallee T. M., Tarantini F., Jackson A., Lathrop J. T., Hampton B., Burgess W. H., Maciag T. S100A13 is involved in the regulation of fibroblast growth factor-1 and p40 synaptotagmin-1 release in vitro. J. Biol. Chem. 1998;273:22224–22231. doi: 10.1074/jbc.273.35.22224. [DOI] [PubMed] [Google Scholar]
- 213.Prudovsky I., Bagala C., Tarantini F., Mandinova A., Soldi R., Bellum S., Maciag T. The intracellular translocation of the components of the fibroblast growth factor 1 release complex precedes their assembly prior to export. J. Cell Biol. 2002;158:201–208. doi: 10.1083/jcb.200203084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Mandinova A., Soldi R., Graziani I., Bagala C., Bellum S., Landriscina M., Tarantini F., Prudovsky I., Maciag T. S100A13 mediates the copper-dependent stress-induced release of IL-1α from both human U937 and murine NIH 3T3 cells. J. Cell Sci. 2003;116:2687–2696. doi: 10.1242/jcs.00471. [DOI] [PubMed] [Google Scholar]
- 215.Isobe T., Ishioka N., Masuda T., Takahashi Y., Ganno S., Okuyama T. A rapid separation of S100 subunits by high performance liquid chromatography: the subunit compositions of S100 proteins. Biochem. Int. 1983;6:419–426. [PubMed] [Google Scholar]
- 216.Lehmann R., Melle C., Escher N., von Eggeling F. Detection and Identification of protein interactions of S100 proteins by ProteinChip technology. J. Proteome Res. 2005;4:1717–1721. doi: 10.1021/pr050163s. [DOI] [PubMed] [Google Scholar]
- 217.Nacken W., Sopalla C., Propper C., Sorg C., Kerkhoff C. Biochemical characterization of the murine S100A9 (MRP14) protein suggests that it is functionally equivalent to its human counterpart despite its low degree of sequence homology. Eur. J. Biochem. 2000;267:560–565. doi: 10.1046/j.1432-1327.2000.01040.x. [DOI] [PubMed] [Google Scholar]
- 218.Dell'Angelica E. C., Schleicher C. H., Simpson R. J., Santome J. A. Complex assembly of calgranulins A and B, two S100-like calcium-binding proteins from pig granulocytes. Int. J. Biochem. Cell Biol. 1996;28:53–62. doi: 10.1016/1357-2725(95)00115-8. [DOI] [PubMed] [Google Scholar]
- 219.Teigelkamp S., Bhardwaj R. S., Roth J., Meinardus-Hager G., Karas M., Sorg C. Calcium-dependent complex assembly of the myeloic differentiation proteins MRP-8 and MRP-14. J. Biol. Chem. 1991;266:13462–13467. [PubMed] [Google Scholar]
- 220.Leukert N., Sorg C., Roth J. Molecular basis of the complex formation between the two calcium-binding proteins S100A8 (MRP8) and S100A9 (MRP14) Biol. Chem. 2005;386:429–434. doi: 10.1515/BC.2005.051. [DOI] [PubMed] [Google Scholar]
- 221.Koltzscher M., Gerke V. Identification of hydrophobic amino acid residues involved in the formation of S100P homodimers in vivo. Biochemistry. 2000;39:9533–9539. doi: 10.1021/bi000257+. [DOI] [PubMed] [Google Scholar]
- 222.Gribenko A. V., Hopper J. E., Makhatadze G. I. Molecular characterization and tissue distribution of a novel member of the S100 family of EF-hand proteins. Biochemistry. 2001;40:15538–15548. doi: 10.1021/bi0114731. [DOI] [PubMed] [Google Scholar]