Abstract
Interaction between Mad2 and Cdc20 (cell division cycle 20) is a key event during spindle assembly checkpoint activation. In the past, an N-terminal peptide containing amino acid residues 111–150 of Cdc20 was shown to bind Mad2 much better than the full-length Cdc20 protein. Using co-localization, co-immunoprecipitation and peptide inhibition analysis with different deletion mutants of Cdc20, we identified another Mad2-binding domain on Cdc20 from amino acids 342–355 within the WD repeat region. An intervening region between these two domains interferes with its Mad2 binding when present individually with any of these two Mad2-binding sites. We suggest that these three domains together determine the overall strength of Mad2 binding with Cdc20. Functional analysis suggests that an optimum Mad2 binding efficiency of Cdc20 is required during checkpoint arrest and release. Further, we have identified a unique polyhistidine motif with metal binding property adjacent to this second binding domain that may be important for maintaining the overall conformation of Cdc20 for its binding to Mad2.
Keywords: cell division cycle 20 (Cdc20) domain, Mad2–Cdc20 interaction, nocodazole, polyhistidine motif, spindle assembly checkpoint, WD repeat
Abbreviations: APC/C, anaphase-promoting complex/cyclosome; Bub, budding uninhibited by benomyl; BubR1, Bub related 1; C-Mad2, closed Mad2; Cdc20, cell division cycle 20; CL, clathrin; DMEM, Dulbecco's modified Eagle's medium; GFP, green fluorescent protein; Mad, mitotic arrest deficient; MCC, mitotic checkpoint component; NLS, nuclear localization signal; O-Mad2, open Mad2; RFP, red fluorescent protein; SAC, spindle assembly checkpoint
INTRODUCTION
The precise partitioning of chromosomes during mitosis is mediated by the attachment of mitotic spindles to each pair of similar chromosomes at their kinetochores [1]. Sister chromatid separation initiates after the activation of APC/C (anaphase-promoting complex/cyclosome) by Cdc20 (cell division cycle 20) during metaphase to anaphase transition [2]. Biochemically, it is achieved by ubiquitination of securin protein by activated APC/C, leading to its proteasomal degradation [3]. Degradation of securin releases the inhibition on separase, a protease, which in turn cleaves cohesin, a protein complex that holds together the two sister chromatids [3]. Destruction of cohesin allows the spindle microtubules to pull the separated chromatids to opposite poles of the cell [4]. Failure of spindle attachment to a single kinetochore activates the SAC (spindle assembly checkpoint), which arrests cells at metaphase until corrections are effected and equal distribution of chromosomes has been ensured [5–7]. A sensory mechanism initiates the ‘wait anaphase’ signal from an unattached kinetochore and triggers the accumulation of the checkpoint components that comprises the Bub (budding uninhibited by benomyl)–Mad (mitotic arrest deficient) families of proteins [8]. These proteins form complexes with Cdc20, thereby sequestering it from activating the APC/C complex [1,2].
According to the catalytic model for the generation of ‘wait anaphase’ signal, Mad1 and Bub1 are mainly resident at the kinetochore, whereas Mad2 (free of Mad1), BubR1 (Bub related 1), Bub3 (free of Bub1), Cdc20 and Mps1 dynamically exchange as part of the diffuse ‘wait anaphase’ signal [9]. Along with Mad2, other MCCs (mitotic checkpoint components) like Cdc20, Bub3 and BubR1 assemble at the unattached kinetochore and then release the signal in the form of Mad2–Cdc20 and BubR1–Bub3–Cdc20 complexes or the combined complex Mad2–Cdc20–BubR1–Bub3, which subsequently inhibit APC function. It has been shown that the phosphorylated form of Cdc20 is required for its interaction with Mad2 and BubR1–Bub3 to form these MCCs [2]. On the other hand, release from SAC is mediated by p31Comet, which competes with O-Mad2 (open Mad2) for C-Mad2 (closed Mad2) binding and preventing its interaction with Mad1–C-Mad2 or C-Mad2–Cdc20 complexes [10,11].
Although details of the function of these complexes remain to be fully understood, it is generally agreed that the distal regulators of the spindle checkpoint are Mad2 and the BubR1–Bub3 complex which sequester Cdc20 simultaneously or synergistically to inhibit the function of the APC/C [9,12–14]. In the present study, we limit our analysis to binding of Mad2 to Cdc20 and discuss the role of Mad1 in such an interaction. To understand the interplay between Mad1, Mad2 and Cdc20, it is important to note that Cdc20 and Mad1 bind to the same pocket of Mad2 [15,16] and thus Mad1 must be displaced from Mad2 before Cdc20 could bind to it. Mad2 exists in two conformational states, O-Mad2 and C-Mad2, determined by a structural change in the ‘safety belt’, the 50-residue C-terminal segment of Mad2 [15–19]. In particular, Mad2 adopts the C-Mad2 conformation when bound to Cdc20 or Mad1 [15,16,18,19], and resides predominantly in the open conformation when unbound [19]. Mad1 facilitates the interaction of Mad2 with Cdc20 by converting O-Mad2 into C-Mad2, the conformation that Mad2 adopts when bound to Cdc20 [19]. Because a large activation energy separates O-Mad2 from C-Mad2, C-Mad2 released from Mad1 is expected to bind Cdc20 faster than cytoplasmic O-Mad2 [19]. Two alternative mechanisms have been proposed to explain the role of Mad1 in converting O-Mad2 into C-Mad2 for binding with Cdc20 [11,17]. The ‘Mad2 exchange’ model predicts that Mad2 dissociated from the Mad1–Mad2 complex retains a transient active C-Mad2 conformation, compatible for binding to Cdc20 [15,19]. On the other hand the ‘Mad2 template’ model proposes that C-Mad2 bound to Mad1 acts as a template for the conversion of O-Mad2 into C-Mad2 bound to Cdc20 [17]. It has been proposed that the Cdc20–C-Mad2 complex, which can be regarded as a structural copy of Mad1–C-Mad2, may act to facilitate further binding of Mad2 to Cdc20 away from kinetochores, providing a mechanism for amplification of the checkpoint signal [15,17,20].
Cdc20 contains a Mad2-binding domain within its N-terminal region [18,21,22]. Zhang and Lees [22] reported that the N-terminal fragment of Cdc20 (amino acids 1–153) interacts more tightly with Mad2 than full-length Cdc20 protein. Moreover, increase in the fragment length up to 210 amino acids or more reduced the binding efficiency significantly [22]. These results indicate that there could be other elements in the Cdc20 protein for further modification of its Mad2 interaction. Similarly, the structural analysis of Mad2 revealed two distinct patches at the back surface of the central β-sheet that may be involved in protein–protein interaction [18]. One of the patches, formed by residues Leu154, Tyr156, Asp158, Asp160, His191 and Lys192, is in close proximity to the C-terminal tail and is required for Cdc20 binding [18]. The other conserved surface residues (Arg133, Gln134, Thr140, Phe141 and Leu142) are located at or close to helix α-C, which is far away from the C-terminal region [18]. This region was predicted to be involved in binding to other checkpoint proteins [18]. In support of the template model, it was shown that Arg133 in both O and C partner can influence O-Mad2 binding to the Mad1–C-Mad2 template but it did not confirm whether both surfaces containing Arg133 contact directly [11]. Alternatively, it may be possible that Mad2 and Cdc20 form multiple interacting surfaces and helix α-C represents another contacting site between the two [18]. One of the several Mad2-binding peptides identified by phase display assay showed very high homology with Cdc20 C-terminus region (see the Results section), indicating a possibility for Mad2–Cdc20 interaction via this domain [15].
Cdc20 has a WD40 repeat region at the C-terminal half of the protein [23–25]. WD repeats usually come in large numbers, forming a β-propeller structure that is known to be a specialized region for protein–protein interaction [23]. The interaction between Bub3 with Mad2 or Cdc20 is mediated by its WD40 repeats that serves as the platform and does not require any intact kinetochore [23]. The Cdc20 protein contains seven such WD repeats at the C-terminus but detailed structural information is not yet available and a well-defined function is not known. We were intrigued by the highly conserved nature of the C-terminus WD repeat region of Cdc20 protein and curious about the functional domains present in the C-terminus of the protein. In the present study, we have identified a second Mad2-binding domain within the WD repeats region of Cdc20, which is adjacent to a putative polyhistidine motif having metal binding property and have shown that an internal binding inhibitory domain of Cdc20 optimizes its efficiency of Mad2 binding for proper checkpoint function.
MATERIALS AND METHODS
Cell culture, transfection, mitotic arrest and synchronization
HeLa cells were cultured in DMEM (Dulbecco's modified Eagle's medium; Invitrogen, Carlsbad, CA, U.S.A.) supplemented with 10% (v/v) fetal calf serum. Transient transfection of HeLa cells with different expression plasmids was performed using Lipofectamine™ reagent (Life Technologies, Carlsbad, CA, U.S.A.) according to the manufacturer's instructions and cells were harvested after 48 h. For mitotic arrest, 300 ng/ml nocodazole (Sigma, St. Louis, MO, U.S.A.) was added to HeLa cell cultures 24–48 h (as mentioned in respective cases) before harvesting. Cells were synchronized by serum starvation for 48 h, using DMEM supplemented with 0.5% fetal calf serum as and when required.
Plasmid construction
Full-length as well as del-N and del-C mutants of CDC20 in pEGFP-N3 (Clontech, U.S.A.) were a gift from Dr J. Weinstein (Amgen, Thousand Oaks, CA, U.S.A.); and full-length MAD2 in pDs-Red1-C1 (Clontech) was a gift from Dr K. T. Jeang [National Institute of Allergy and Infectious Diseases, NIH (National Institutes of Health), Bethesda, MD, U.S.A.]. Different CDC20 deletion mutants were kindly provided by Dr J. Ruderman (Harvard Medical School, Boston, MA, U.S.A.), which were subcloned in pEGFP-C2 vector to generate pGM2-pGM6 (Clontech) constructs. CDC20 deletion constructs pGM7-pGM10 were generated by cloning respective PCR fragments into pEGFP-N3 (pGM7) or pFLAG-CMV-2 (where CMV is cytomegalovirus) vector (pGM8-pGM10; Kodak, New Haven, CT, U.S.A.). Fidelity of all these constructs was confirmed by sequencing. The other construct GFP-MAD1 used in the present study was kindly provided by Dr K. T. Jeang.
Confocal microscopy, immunofluorescence and multinuclei analysis
Asynchronous HeLa cells were transiently transfected with the GFP (green fluorescent protein)- and RFP (red fluorescent protein)-fused expression constructs individually or in combination as described in the Results section. After 48 h, cells were fixed and visualized under either a confocal (Zeiss LSM-510) or fluorescence (Olympus BX-40) microscope as required. For multinucleation analysis, full-length ormutant Cdc20 transfected HeLa cells were treated for 24 h with 300 ng/ml nocodazole followed by methanol fixation and immunostaining. To detect and count the percentage of multinuclei, fixed cells were immunostained with anti-α-tubulin antibody conjugated with FITC (Sigma) followed by counterstaining with propidium iodide. Immunostained cells were observed under a fluorescence (BX-40) or confocal (LSM-510) microscope and at least 150–200 cells were counted from each slide to determine the percentage of multinucleated cells. Cells with more than two nuclei were defined as multinucleated cells. This experiment was repeated three times and the average of these three individual values was recorded.
Co-immunoprecipitation, peptide inhibition assay and immunoblotting
For co-immunoprecipitation analysis, transfected HeLa cells were harvested after 48 h in a lysis buffer (50 mM Tris/HCl, pH 7.5, 15 mM EDTA, 150 mM NaCl and 0.1% Triton X-100) supplemented with protease inhibitor cocktail (Sigma). Cell extracts were then incubated with respective primary antibodies [anti-Cdc20-N19 and/or C-19 (Santa Cruz Biotechnology); anti-Mad2-N19 (Santa Cruz Biotechnology); anti-FLAG (M2) antibody (Sigma)] as indicated, followed by immunoprecipitation with Protein A–CL agarose (where CL is clathrin) (Bangalore Genei, India). The whole cell lysate or the immune complexes were resolved by SDS/PAGE (10–12% gel) and transferred on to a PVDF membrane (Millipore). Mad2 and different Cdc20 proteins were detected with anti-FLAG or protein-specific antibodies and visualized by a chemiluminescence kit (Amersham) after treating with horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology). The peptide inhibition assay was carried out by incubating respective cell extracts with an appropriate amount of synthetic 15-mer peptides (Ambion, Austin, TX, U.S.A.) for 2 h in ice, followed by immunoprecipitation with specific antibodies. Similarly, for EDTA inhibition, 0.01 mM EDTA was added prior to immunoprecipitation.
Ni-CL agarose binding assay
HeLa cells transfected with full-length or truncated Cdc20 expression vectors were lysed in a lysis buffer (20 mM sodium phosphate and 0.5 M NaCl, pH 7.0) by repeated freeze–thawing. Cell debris was removed by centrifugation and the supernatant containing equal amount of total protein was adsorbed to Ni-CL agarose beads (Bangalore Genei). Agarose beads were then centrifuged and the supernatant was collected (flow through). This step was repeated once with fresh Ni-CL agarose beads. Agarose beads were then pooled and washed three times with the lysis buffer by centrifugation. Ni-CL agarose bead-bound proteins were eluted in elution buffer (0.25 M imidazole, 0.05 M NaH2PO4 and 0.3 M NaCl) according to the manufacturer's instructions. Equal amount of proteins from cell lysate (input), flow through and eluted fraction were denatured in 2× SDS sample buffer (0.1 M Tris/HCl, pH 6.8, 2% SDS, 10% glycerol and 1 mg each of Bromophenol Blue and Xylene Cyanole), boiled and resolved by SDS/PAGE (12% gel), followed by immunoblotting with anti-Cdc20 (GM1 and GM2) or anti-FLAG (GM9) antibodies.
Cell-cycle analysis
Synchronized HeLa cells were transfected or co-transfected with different expression constructs and transfection efficiency was found to be at least 70–80% as determined by fluorescent cell counting under microscopic field. One set of cells was treated with nocodazole for 24 h before harvesting, and another set was left untreated. Approximately 106 cells were harvested by trypsinization and washed with PBS. Cells were fixed by resuspending in 100% ethanol and kept at 4 °C for at least 24 h. Fixed cells were again suspended in PBS containing 20 μg/ml propidium iodide (Sigma) and 200 ng/ml RNase (Sigma). Cells were then incubated at 37 °C for at least 30 min and analysed by FACS (BDLSR flow cytometer; Becton and Dickinson, U.S.A.).
In silico analysis
For homology analysis the full-length Cdc20 protein in different organisms or to find the conservation of any functional domain the pair BLAST tool of NCBI and Clustal W was used respectively. For prediction of the functional domains in the Cdc20, the domain prediction software PSTSCAN and PSORT (http://psort.ims.u-tokyo.ac.jp) were used. PSIPRED analysis of full-length Cdc20 protein was done using the web-based free software available in PSI sites (http://bioinf.cs.ucl.ac.uk/psipred/).
RESULTS
Prediction of conserved and functional domains in Cdc20 by homology search
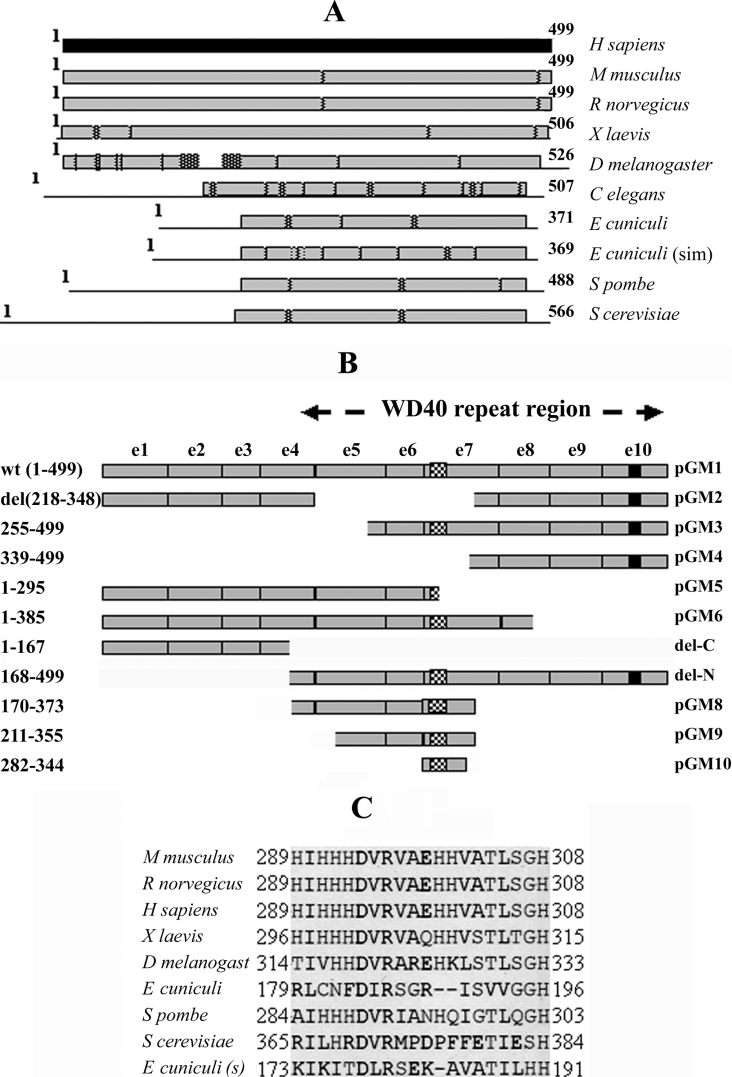
CDS (complete coding sequence) of Cdc20 homologues of different organisms (e.g. Mus musculus, Rattus norvegicus, Xenopus laevis, Drosophila melanogaster, Caenorhabditis elegans, Schizosaccharomyces pombe, Saccharomyces cerevisiae and Encephalitozoon cuniculi) available in the NCBI database were aligned pairwise with the Homo sapiens sequence and searched for conserved and variable regions. As shown in Figure 1(A), it was observed that the C-terminal half of the protein was more conserved across various species than the N-terminal half. The N-terminal region of human Cdc20 protein was either absent or exhibited poor homology (<20% similarity) in lower eukaryotes. The mouse, rat and X. laevis sequences, however, showed strong homology (91% similarity for both mouse and rat and 86% similarity for X. laevis) with human protein both at the C-terminal and N-terminal halves. Also, D. melanogaster Cdc20 showed a high level of homology (89% similarity) at both ends with an internal less similar region (<20% similarity) in the N-terminal half. Thus it appears that the C-terminal part of the Cdc20 protein is highly conserved across different species and might contain some important functional domains. It is to be noted that so far almost all characterized functional domains [22] as well as phosphorylation sites [2] have been localized to the N-terminal part of the Cdc20 protein.
Figure 1. Conserved domains of Cdc20 and the deletion mutants used in the present study.
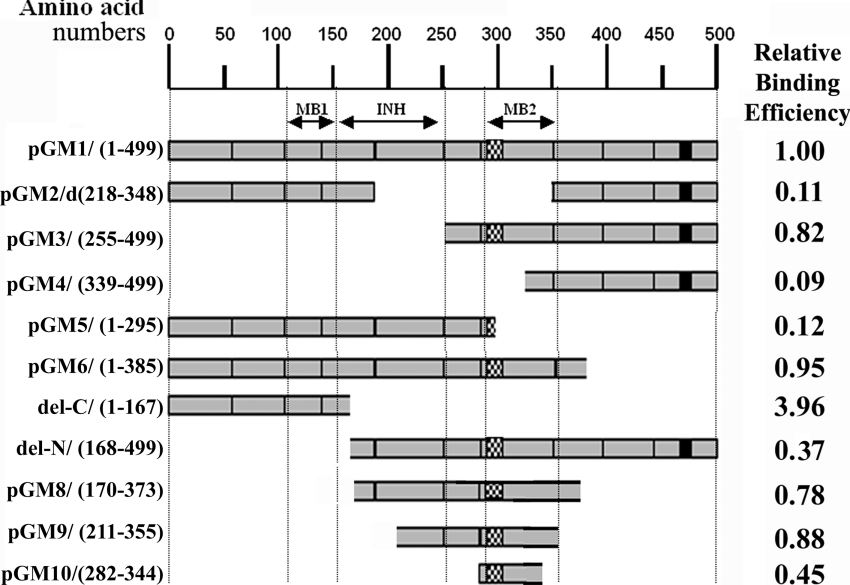
(A) Cdc20 proteins of varying length from different species are shown as horizontal bars. The light shaded bars and the lines represent regions of high (∼90% similarity) and low (<20% similarity) homology with respect to the human homologue of Cdc20 (dark shaded bar). Hatched boxes represent short interruptions of such homology. Homology analysis was done using NCBI pairwise BLAST software with default settings. The numbers at the beginning and at the end of each horizontal bar represent the first and the last amino acid number of the respective protein. (B) Map of different Cdc20 deletion mutants are shown with respect to full-length Cdc20 protein (GM1). On the left of the panel, amino acid regions present in the respective deletion constructs are shown. On the right of the panel is the name of each clone. The exons of Cdc20 cDNA are labelled as e1 to e10 and the WD repeat region is marked. The hatched box in e6 and the black box in e10 represent polyhistidine and NLS regions respectively. (C) Clustal W alignment of the predicted polyhistidine motif region in Cdc20 proteins from different organisms as mentioned at the left of the panel. The numbers at the beginning and at the end of each row represent the amino acid number at the start and end of the motifs respectively.
We next carried out the domain prediction analysis of the Cdc20 complete amino acid sequence available in NCBI database (accession no. NP_001246) using the domain prediction software PSTSCAN and PSORT (http://psort.ims.u-tokyo.ac.jp). In addition to already known seven WD40 repeats, these analyses revealed two new domains in the C-terminal half of the protein; an NLS (nuclear localization signal) at the C-terminus end and a unique polyhistidine motif overlapping third WD repeat of the protein sequence (Figure 1B; see pGM1). The polyhistidine motif region from amino acid residues 289–308 contains as much as seven histidine residues within a stretch of 20 amino acids (Figure 1C). The positions of some of the histidine moieties within this stretch were found to be similar to the consensus Cu-binding motif of catecholamine, tyrosinase or haemocyanin proteins (results not shown; [26]). It also has a cluster of four histidines in tandem like that of an artificial His6 cluster for metal-binding proteins (Figure 1C).
It is also to be noted that the Cdc20 region amino acids 344–355 (GWVPLQTFTQHQ), which is located within the WD repeat number 4 and 5, has a very high homology (six out of 12 amino acids are identical and another two are conserved) with one of the Mad2-binding peptides (GWVRLQPPPLIQ) previously identified by phage display analysis [15]. Interestingly, Clustal W alignment of the proteins revealed that lower eukaryotes like E. cuniculi and C. elegans that contain shorter Cdc20 sequences have no homology with the first Mad2-binding motif of human Cdc20 at the N-terminus as shown by Zhang and Lees [22]. These results led us to suspect that WD region of the Cdc20 protein might contain an additional Mad2-binding domain.
Physical interaction between WD region of Cdc20 and Mad2
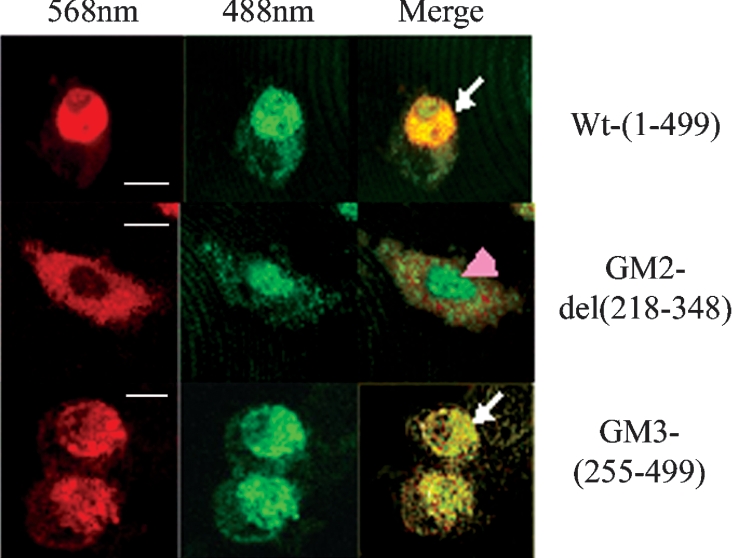
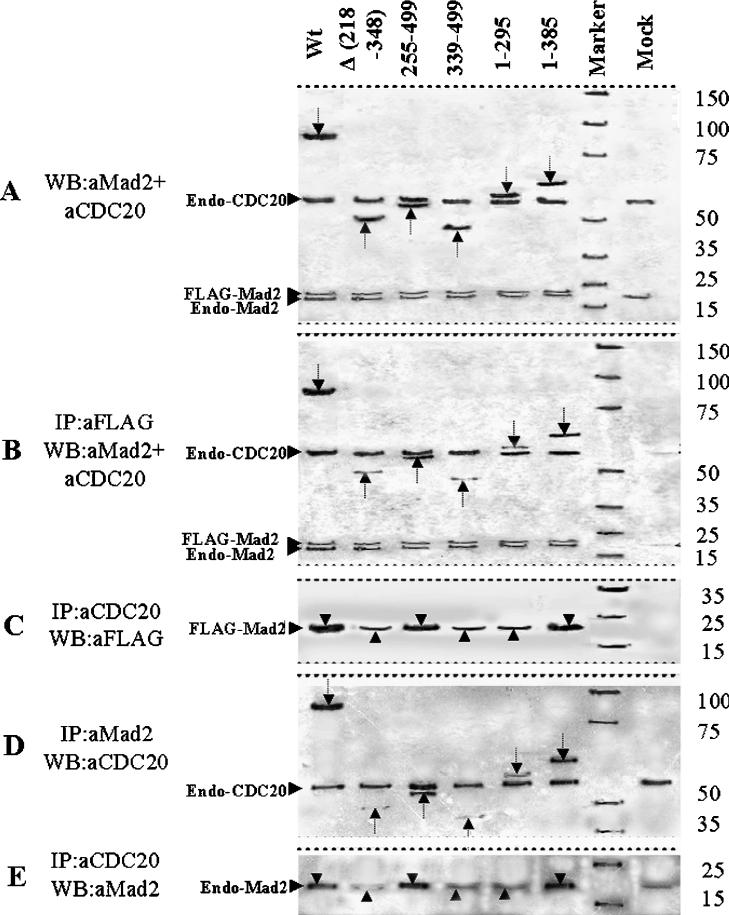
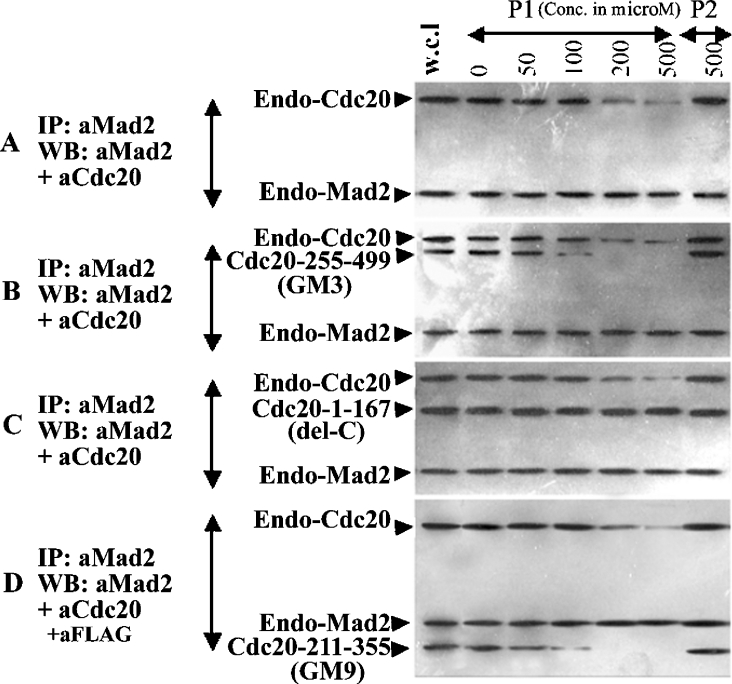
To test the hypothesis that the C-terminal WD region of Cdc20 protein interacts with Mad2, we first examined the co-localization property of two Cdc20 deletion mutants, GM2 and GM3, with Mad2 in the interphase nucleus (Figure 2). It was observed that the GFP- or RFP-fused proteins used showed similar cellular localization as well as cell cycle function similar to the endogenous proteins (results not shown). The N-terminal deletion mutant GM3 devoid of the first Mad2-binding site co-localized with Mad2 in the HeLa cell nucleus (Figure 2, panel GM3). Surprisingly, the C-terminal internal deletion mutant GM2 was unable to co-localize with Mad2 in the nucleus of the transfected HeLa cells (Figure 2, panel GM2). The full-length Cdc20 co-localized with Mad2 as expected (Figure 2, panel Wt). This initial clue suggested that the C-terminal half of Cdc20 could interact with Mad2. Next, the interaction between Mad2 and Cdc20 was examined by co-immunoprecipitation experiments. A series of deletion mutants of Cdc20 (pGM-4 to pGM-6) were generated as GFP fusion protein to functionally dissect the predicted second Mad2-binding domain in the WD region (Figure 1B). These GFP-fused Cdc20 mutants (GM2–GM6) were co-expressed with FLAG-tagged full-length Mad2 in asynchronous HeLa cells. We confirmed the co-expression of FLAG–Mad2 and various deletion mutants of GFP-fused Cdc20 in transiently transfected HeLa cell by immunoblot analysis using a combination of anti-Mad2 and anti-Cdc20 antibodies (Figure 3A). Both 81 kDa GFP-fused full-length Cdc20 and its various deletion mutants as well as exogenous FLAG–Mad2 proteins were detected in the expected size ranges (arrows in Figure 3A). Endogenous Cdc20 and Mad2 proteins were also detected, which served as an internal control in the subsequent experiments. Mock lane showed only the endogenous Cdc20 and Mad2 proteins. Immunoprecipitation of these cell lysates with anti-FLAG antibody and subsequent analysis of the immune complex by immunoblotting with a mixture of anti-Mad2 and anti-Cdc20 antibodies revealed several interesting results (Figure 3B). The internal deletion mutant GM2 (lane Δ 218–348), N-terminal deletion mutant GM4 (lane 339–499) and C-terminal deletion mutant GM5 (lane 1–295) bind to Mad2 inefficiently as indicated by the very weak signal in the co-immunoprecipitation experiment (see arrows in respective lanes). On the other hand, both N-terminal deletion mutant GM3 (lane 255–499) and C-terminal deletion mutant GM6 (lane 1–385) bound Mad2 efficiently (see arrows in respective lanes). The full-length GFP-fused Cdc20 was pulled down by anti-FLAG antibody as expected (lane Wt). Despite this differential binding of exogenously supplied mutant Cdc20s, the endogenous Cdc20 bound with Mad2 at a similar level in all cases (see endo-Cdc20 in Figure 3B). Presence of FLAG–Mad2 at almost equal levels in all lanes indicated that lack of binding was not due to failure in immunoprecipitation (see FLAG–Mad2 in Figure 3B). Interestingly, in this experiment, we also detected endogenous Mad2 in all samples probably due to formation of FLAG–Mad2–Mad2 dimers as suggested earlier [8,19].
Figure 2. Co-localization of full-length and different Cdc20 deletion mutants with Mad2.
HeLa cells co-transfected with RFP–Mad2 and GFP-fused Cdc20 proteins (full-length, GM2 and GM3 as indicated at the right side of each panel) were analysed by confocal microscopy during interphase. At 568 nm, red fluorescence from RFP–Mad2 and at 488 nm green signal from GFP–Cdc20 were observed. The yellow fluorescence in ‘merge’ panels indicates co-localization of these two proteins (arrows). Arrowhead shows the nuclear localization of Cdc20 deletion mutant (GM2) incapable of Mad2 binding. Scale bar, 10 μm.
Figure 3. Co-immunoprecipitation and immunoblot analysis of pGM1–pGM6 constructs.
HeLa cells were either co-transfected with FLAG–Mad2 and different GFP-fused Cdc20 deletion mutants (A–C) or transfected with different GFP-fused Cdc20 deletion mutants only (D, E) followed by 24 h of nocodazole treatment for metaphase arrest. Cells were harvested and the lysates with equal amount of proteins were either directly subjected to immunoblot analysis or first immunoprecipitated followed by immunoblot analysis of the immune complex. Respective antibodies used for immunopreciptation (IP) and immunoblotting (WB) are mentioned at the left-hand side of each panel. In (A), arrows indicate 81, 49, 53, 44, 58 and 67 kDa proteins expressed from Cdc20 expression constructs mentioned above each lane. Arrows in (B, D) indicate Mad2-bound full-length and truncated Cdc20 proteins. Arrowheads pointing upwards in both (C) and (E) indicate only endogenous Cdc20-bound Mad2 proteins, whereas arrowheads pointing downwards in the same panels indicate Mad2 proteins pulled down by both endogenous and ectopically expressed binding-proficient Cdc20 full-length and mutant proteins. Marker lane indicates protein molecular mass markers with sizes shown at the right-hand side of the panels. Mock lane indicates mock-transfected HeLa cell. Endo-Mad2, endogenous Mad2 protein; endo-Cdc20, endogenous Cdc20 proteins; FLAG–Mad2, FLAG-tagged Mad2 protein (see text for details).
To further validate these observations, we carried out the co-immunoprecipitation experiment in reverse order, i.e. we first immunoprecipitated with anti-Cdc20 antibody followed by immunoblotting with anti-FLAG antibody (Figure 3C). Since immunoprecipitation was done with anti-Cdc20 antibodies, it would precipitate both endogenous and exogenous Cdc20s together with bound FLAG–Mad2. Thus we expect to see FLAG–Mad2 signals in all samples due to the interaction between endogenous Cdc20 and exogenous FLAG–Mad2, irrespective of exogenous Cdc20 binding with FLAG–Mad2. However, in those cases where exogenous GFP-fused Cdc20 would bind FLAG–Mad2, a stronger signal should be observed. As expected, GM3 and GM6 showed stronger signal due to binding with exogenous FLAG–Mad2 (see arrowheads in Figure 3C) than that of other deletion mutants, indicating weak interaction of these mutants with exogenous FLAG–Mad2 (see arrowheads in Figure 3C).
Next we tested the ability of the binding of various deletion mutants of Cdc20 with endogenous Mad2 alone. Towards this end, we transiently expressed various mutant Cdc20s in metaphase arrested HeLa cell and performed the co-immunoprecipitation analysis using anti-Mad2 as precipitating antibody and anti-Cdc20 as the detecting antibody in immunoblotting of the immune complex. As shown in Figure 3(D), again mutant GM3 (lane 255–499) and GM6 (lane 1–385) exhibited strong binding with endogenous Mad2, whereas mutant GM2 (lane Δ 218–348), GM4 (lane 339–499) and GM5 (lane 1–295) failed to do so. Full-length GFP-fused Cdc20 showed strong binding as expected (lane Wt). Presence of endogenous Cdc20 in almost equal amounts in all lanes indicated that lack of binding was not due to failure in immunoprecipitation. This experiment was also validated by performing the co-immunoprecipitation experiment in reverse order as described above (Figure 3E). As expected, we observed strong signals in cases of full-length GFP-fused Cdc20 (lane Wt), GM3 (lane 255–499) and GM6 (lane 1–385) due to precipitation of endogenous Mad2 both by exogenous mutants Cdc20 and endogenous Cdc20. Endogenous Mad2 observed in other lanes were due to interaction with endogenous Cdc20. Thus these results suggest that a second Mad2-binding domain of Cdc20 may reside within the WD region spanning amino acids 255–355.
Second Mad2-binding domain of Cdc20 resides between amino acids 342 and 355 of the WD40 repeats
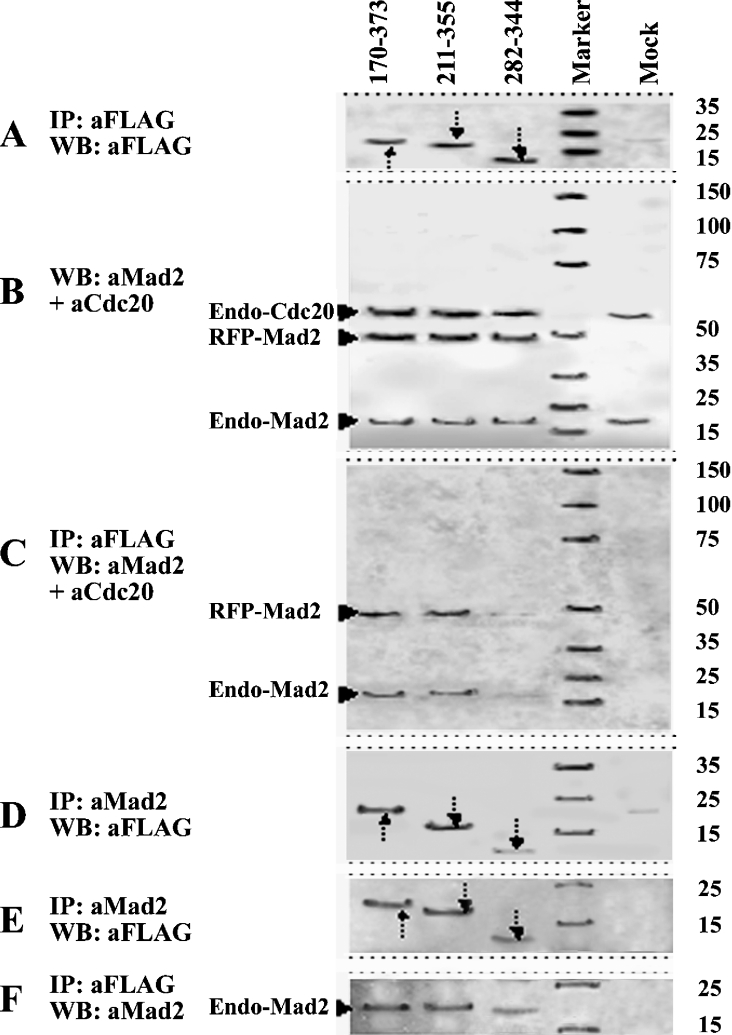
In order to precisely localize the Mad2-interacting domain within the WD40 repeats of Cdc20, we constructed three deletion mutants consisting of amino acids 170–373 (GM8), amino acids 211–355 (GM9) and amino acids 282–344 (GM10) of Cdc20 in pFLAG-CMV-2 vector to express as FLAG fusion protein (Figure 1B). Co-expression of these FLAG-fused truncated Cdc20 proteins with RFP–Mad2 resulted in physical interaction between them in a predicted manner (Figures 4A–4D). Immunoblot analysis of whole cell lysates of the transfected HeLa cells showed that these three FLAG-fused deletion mutants of Cdc20 expressed proteins of correct molecular mass (Figure 4A). A similar observation was made with RFP–Mad2, which was expressed appropriately in all co-transfected cells but absent in mock-transfected cells (Figure 4B). Both endogenous Cdc20 and Mad2 were also detected in all transfected and mock-transfected cells (Figure 4B). Immunoprecipitation of these cell lysates with anti-FLAG antibody followed by detection of the immune complex with a mixture of anti-Mad2 and anti-Cdc20 antibodies showed that both GM8 (lane 170–373) and GM9 (lane 211–355) could precipitate RFP–Mad2 efficiently but GM10 (lane 282–344) bound RFP–Mad2 very weakly (Figure 4C; see RFP–Mad2). The reverse experiment done by immunoprecipitation with anti-Mad2 antibody and detection of the immune complex with anti-FLAG antibody in immunoblots produced similar results (Figure 4D). As expected, both GM8 and GM9 were co-immunoprecipitated with RFP–Mad2 quite strongly, whereas binding of GM10 was very weak (see arrows in Figure 4D). Binding of these three internal Cdc20 fragments (GM8, GM9 and GM10) with endogenous Mad2 was also examined (Figures 4E and 4F). In concordance with the above results, both GM8 and GM9 bound endogenous Mad2 very strongly but GM10 showed very weak binding as detected by immunoprecipitation with anti-Mad2 antibody and immunoblotting by anti-FLAG antibody (Figure 4E) and vice versa (Figure 4F). So these experiments delineated amino acids 255–355 as the boundaries of the second Mad2-binding domain of Cdc20 (for further details, see Figure 10).
Figure 4. Co-immunoprecipitation and immunoblot analysis of pGM8–pGM10 constructs.
HeLa cells were either co-transfected with RFP–Mad2 and different Cdc20 deletion mutants fused with FLAG epitope (A–D) or transfected with FLAG-fused Cdc20 clones only (E, F) followed by 24 h of nocodazole treatment for metaphase arrest. Cells were harvested and the lysates with equal amounts of proteins were either directly subjected to immunoblot analysis or first immunoprecipitated followed by immunoblot analysis of the immune complex. Respective antibodies used for immunopreciptation (IP) and immunoblotting (WB) are mentioned at the left-hand side of each panel. In (A), arrows indicate 23, 17 and 7 kDa proteins from Cdc20 deletion mutants shown above the lanes. Arrows in (D, E) indicate Mad2-bound Cdc20 FLAG fusion proteins. Marker lane indicates protein molecular mass markers for which the sizes are shown at the right-hand side of each panel. Mock lane indicates mock-transfected HeLa cell. The faint band present in Mock lanes of (A, B, E) is an anti-FLAG cross-reacting protein. Endo-Mad2, endogenous Mad2 protein; endo-Cdc20, endogenous Cdc20 proteins; RFP–Mad2, RFP-fused Mad2 protein (see text for details).
Figure 10. Summary of Mad2 binding ability of Cdc20 deletion mutants.
Horizontal bars showing amino acid length of different Cdc20 deletion mutants (as mentioned at the left-hand side of each construct) in an arbitrary scale (above) of amino acid numbers. The relative Mad2 binding efficiency of each clone is shown at the right-hand side of each bar. To calculate the relative binding efficiency of each Cdc20 mutant, the ratio of band intensities in the Western blot of these mutants and endogenous Cdc20 (Figures 3B, 4C and 7) was determined. These values were then divided by the value of GM1 to obtain the relative binding efficiency of these mutants in comparison with full-length Cdc20 (GM1). Dotted vertical lines demark the approximate boundaries of first Mad2-binding (MB1), binding inhibitory (INH) and second Mad2-binding (MB2) domains of Cdc20 protein (as indicated by double-headed arrows). The hatched box and the black box represent polyhistidine and NLS regions respectively.
To find out the exact location of the second Mad2-binding site on Cdc20 protein, we performed peptide inhibition assays of Mad2–Cdc20 interaction. We used two different peptides, one containing amino acids 289–303 of the Cdc20 polyhistidine motif (P1) and the other from amino acids 342–356 (P2). HeLa cell lysates prepared from untransfected, GM3, GM9 and del-C transfected cells were incubated with either of the two peptides followed by co-immunoprecipitation analysis. Between the two peptides, P2 blocked the Mad2 binding of both endogenous Cdc20 and the Cdc20 deletion mutants GM3 and GM9 in a dose-dependent manner. The interaction of GM3 and GM9 with Mad2 was completely inhibited at 200 μM peptide concentrations, whereas inhibition of endogenous Cdc20 binding required higher amount of the P2 peptide (Figure 5). The P2 peptide was unable to inhibit the binding of del-C mutant of Cdc20 with Mad2 even at 500 μM concentrations (Figure 5). These results suggest that P2 specifically inhibits the interaction between second Mad2-binding site of Cdc20 and Mad2. The fact that P1 peptide was unable to block the interaction between Mad2 and any of Cdc20 mutants suggested that the second Mad2-binding site of Cdc20 lies within amino acids 342–355.
Figure 5. Peptide competition assay.
HeLa cells were co-transfected with FLAG–Mad2 and different Cdc20 deletion mutants [mock (first panel from the top), GM3 (second panel), del-C (third panel) and GM9 (bottom panel)] fused to GFP as mentioned at the left-hand side of each panel. Cells were harvested and the lysates with equal amount of proteins were incubated with either of the P1 or P2 peptides at various concentrations (μM) as indicated above each lane. After incubation with peptides for 2 h, immunoprecipitation and Western-blot analysis were done using anti-Mad2 and -Cdc20 antibodies as mentioned at the extreme left of each panel.
Polyhistidine motif of Cdc20 has metal binding property and indirectly influences Mad2–Cdc20 interaction
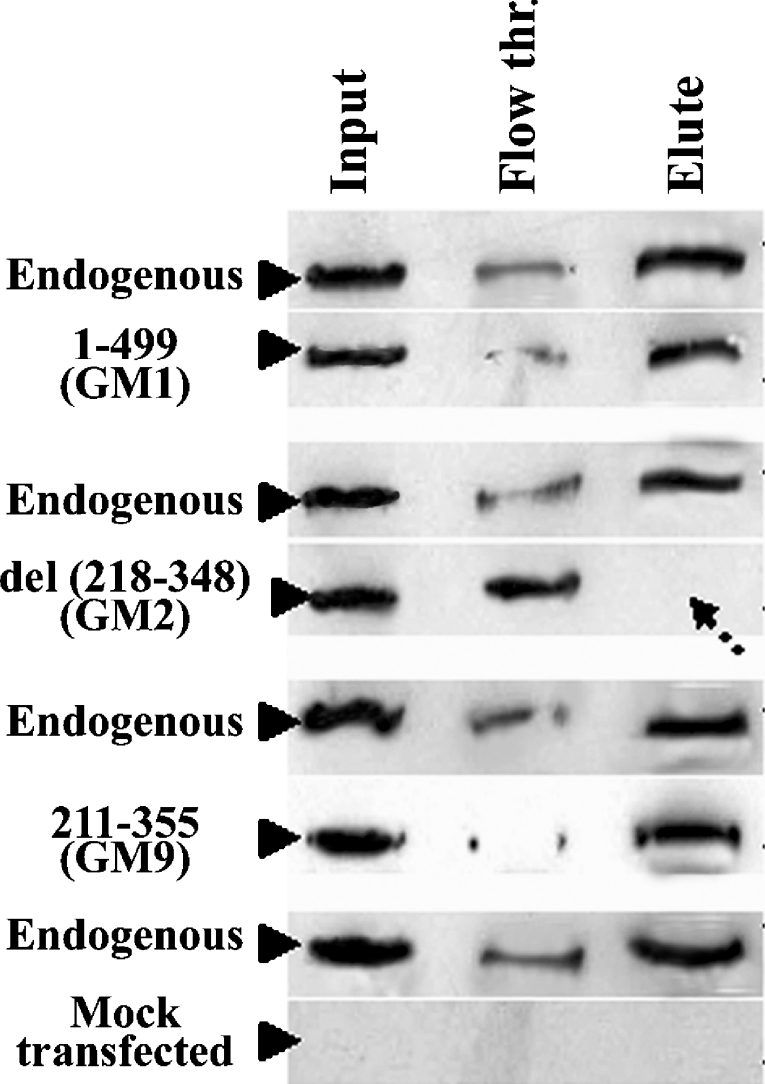
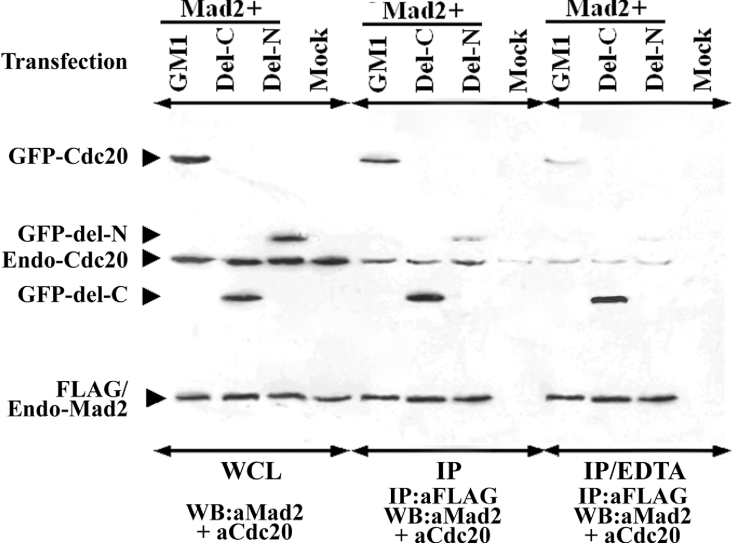
The inability of the P1 peptide to inhibit the Mad2–Cdc20 interaction suggests that the polyhistidine region of the Cdc20 protein is not directly involved in this interaction. However, to explore whether this region contributes indirectly to the Mad2–Cdc20 interaction, we first examined its metal binding capacity. HeLa cells were transfected independently with three different Cdc20 constructs: (i) GFP-fused full-length Cdc20 (GM1), (ii) a deletion mutant devoid of the polyhistidine domain (GM2), and (iii) a second deletion mutant that contains the polyhistidine domain (GM9). The binding property of the wild-type and mutant Cdc20 to Ni-CL-coated agarose beads was tested using the extracts from the respective transfected cells. As shown in Figure 6, substantial amounts of GM1 and GM9 were eluted from the Ni-columns, indicating strong binding of these exogenously expressed proteins with the Ni-agarose. However, the deletion mutant that lacks the polyhistidine motif (GM2) failed to bind to the Ni-column as evident from the absence of any signal in the lane containing the eluant. In all cases, endogenous Cdc20 bound Ni-CL agarose bead very strongly (Figure 6, panels labelled ‘endogenous’). The ‘flow through’ lane shows residual unbound proteins (Figure 6). These results suggest that polyhistidine region of Cdc20 is capable of metal binding. We repeated the co-immunoprecipitation experiment with FLAG-tagged full-length Mad2 and two Cdc20 mutants (del-N and del-C) in the presence of the metal-ion chelator EDTA. The bindings of both full-length and the del-N mutant of Cdc20 were strongly inhibited in the presence of EDTA (Figure 7, panel IP/EDTA). The interaction between Mad2 and del-C mutant of Cdc20 was not affected by the EDTA treatment (Figure 7, panel IP/EDTA). Thus disruption of the structure of the polyhistidine domain severely affected the Mad2–Cdc20 binding.
Figure 6. Ni-column binding assay.
HeLa cells were transfected with different Cdc20 constructs indicated at the left-hand side of each of the panels. Equal amounts of cell lysates were incubated with Ni-CL agarose. Ni-agarose bead-bound and unbound fractions were subjected to immunoblot analysis using anti-Cdc20 antibody for GM1, GM2 and anti-FLAG M2 antibody for GM9. Input, whole cell lysate; Flow thr., unbound supernatant; Elute, protein eluted from Ni-agarose column. Arrow indicates absence of Δ 218–348 protein. ‘Endogenous’ indicates endogenous Cdc20 protein and ‘Mock transfected’ indicates mock-transfected HeLa cell extracts.
Figure 7. Relative Mad2 binding efficiency of full-length, del-C and del-N mutants of Cdc20 and effect of EDTA on Mad2–Cdc20 interaction.
Different GFP-fused Cdc20 constructs as mentioned above each panel were co-transfected with FLAG–Mad2 and the cell lysates were subjected to Western-blot analysis, before (panel WCL) or after (IP) immunoprecipitation with respective antibodies as mentioned at the bottom of each panel. Another set of lysates was treated similarly as above but in the presence of EDTA (panel IP/EDTA).
Evidence for functionality of the second Mad2-binding domain of Cdc20
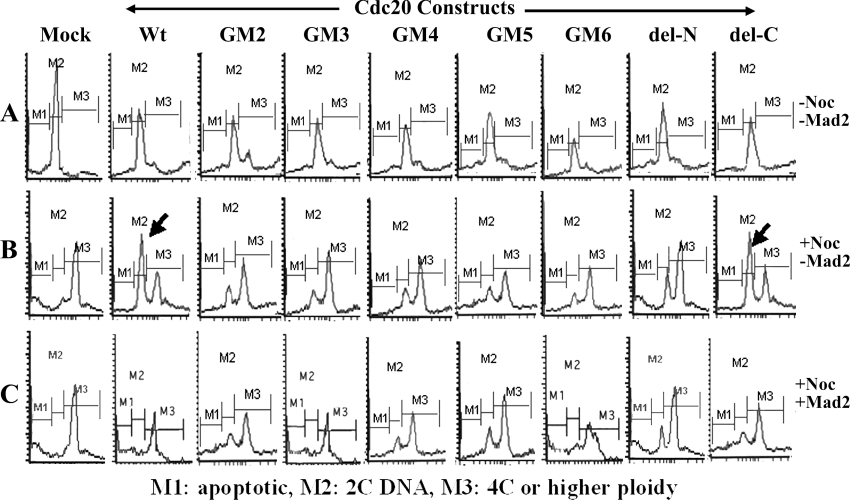
To check whether the second Mad2-binding domain of Cdc20 is required for its proper function, each of the truncated Cdc20 proteins was expressed in synchronized HeLa cells and the checkpoint function was examined by FACS analysis after nocodazole treatment (Figures 8A and 8B). One of the consequences of Cdc20 overexpression is the improper entry of the cells to the next cycle by overriding both the G1/S checkpoint and the SAC function [27,28]. In synchronized mock-transfected cells, the major DNA peak was at 2C for untreated and at 4C for the nocodazole-treated cells exhibiting normal cell cycle pattern. Interestingly, in the case of GFP-fused full-length Cdc20 (wild-type) and del-C mutant-expressed cells, although most of the untreated cells were in the 2C stage, a large fraction of the nocodazole-treated cells were also found to contain 2C DNA instead of 4C. Thus it appears that overexpression of full-length Cdc20 can partially abrogate the checkpoint function and allow cells to enter into the next cycle even in the presence of depolymerized spindle. This result indicates that overexpressed full-length Cdc20 probably titrated out the endogenous Mad2, allowing excess Cdc20 to activate APC/C complex prematurely. On the other hand, del-C mutant bound endogenous Mad2 strongly, which resulted in efficient premature anaphase by releasing endogenous Cdc20 from Mad2. The other deletion mutants of Cdc20 also exhibited such a property albeit at a lower efficiency. In the cases of GM3 and GM6, it is possible that although these two proteins bind Mad2 efficiently they might activate APC/C inefficiently. On the other hand, GM2, GM4, GM5 and del-N were inefficient in both Mad2 binding and APC/C activation. Close examination of the percentage of cells in different phases obtained by FACS analysis revealed that cells transfected with binding-proficient Cdc20 mutants have approx. 10% less cells in post-mitotic (2C) phase than the binding-deficient Cdc20 mutants (results not shown). This difference was not as much as that expected based on the Mad2 binding efficiency of the mutants, probably because of the difference in capability of APC activation, which, however, needs to be substantiated by experimental evidence. Next, we replenished Mad2 protein in different Cdc20-overexpressed cells by means of co-transfection followed by nocodazole treatment to examine whether it could attenuate the premature anaphase promotion in these cells (Figure 8C). Though the overexpression of Mad2 did not completely rescue the SAC function in del-C-expressing cells due to strong Mad2/del-C binding (Figure 8C), it restored normal SAC function efficiently in the case of full-length, GM3 and GM6 as revealed by the absence of the 2C peak. However, the Mad2 binding-deficient Cdc20 deletion mutants (GM2, GM4, GM5 and del-N) failed to rescue SAC. Thus ectopically expressed Mad2 prevented Cdc20 mutants containing a second Mad2-binding domain from promoting premature anaphase probably by sequestering them upon binding. In the case of the binding-deficient mutants, Mad2 overexpression failed to attenuate the premature anaphase probably due to lack of binding.
Figure 8. Effect of overexpression of Cdc20 deletion mutant proteins on cell cycle.
Synchronized HeLa cells were transfected with different Cdc20 constructs as mentioned at the top of each of the panels. Transfected cells were either treated with nocodazole for 24 h (B) or left untreated (A). Then cells were stained with propidium iodide and subjected to FACS analysis. (C) Synchronized HeLa cells were transfected with different Cdc20 constructs as mentioned at the top of each of the panels and Mad2, followed by nocodazole treatment for 24 h. M1, apoptotic cells; M2, cells containing 2C DNA; M3, cells containing 4C or higher amount of DNA.
Interaction between Mad2 and Cdc20 should be optimum for proper checkpoint arrest and release
We compared the ability of Mad2 binding of two Cdc20 deletion mutants, del-C (1–167 amino acids) and del-N (168–499 amino acids), with respect to the full-length Cdc20 (GM1) by co-immunoprecipitation experiments (Figure 7). Immunoblot analysis of whole cell lysate of the transfected HeLa cells showed that these two GFP-fused deletion mutants of Cdc20 and GM1 expressed proteins of expected size in equivalent amount (Figure 7, panel WCL). Interestingly, immunoblot analysis of the anti-FLAG immunoprecipitated proteins revealed that del-C bound FLAG–Mad2 more efficiently than GM1 (Figure 7, panel IP, lanes GM1 and del-C). Binding efficiency of del-N mutant was much weaker than GM1 (Figure 7, panel IP, lane del-N). Presence of equal signal intensities of endo-Cdc20 and FLAG–endo-Mad2 in all lanes indicates that differential signal with respect to Cdc20 deletion mutants was not due to either failure of immunoprecipitation or unequal amount of proteins in the cell lysate (see endo-Cdc20 and FLAG–endo-Mad2 marked bands in Figure 7). Figure 10 summarizes the efficiency of the Mad2 binding of various Cdc20 mutants with respect to full-length Cdc20 (GM1), which suggests that second Mad2-binding domain of Cdc20 confers optimal interaction of the Mad2–Cdc20 complex necessary for SAC arrest (see the Discussion section).
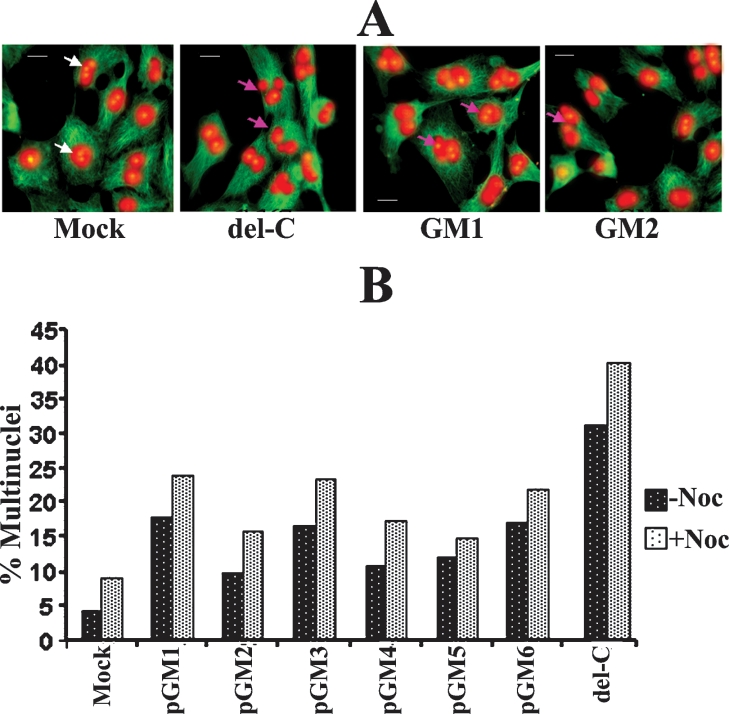
The issue of functional relevance of the second Mad2-binding domain of Cdc20 was further examined by measuring the frequency of multinuclei in HeLa cells expressing different deletion mutants of Cdc20. One of the major consequences of Cdc20 overexpression is the SAC adaptation, which is reflected by cytokinesis failure leading to multinucleation (Figure 9A) [22]. As reported earlier, we also observed high level (18% at –Noc and 24% at +Noc) of multinuclei formation in full-length Cdc20-overexpressing HeLa cells [22]. This multinucleation property was further enhanced in case of del-C-expressing cells (32% at −Noc and 41% at +Noc) as reported earlier [22] (Figure 9B). Interestingly, the Cdc20 mutants (GM3 and GM6) having Mad2 binding efficiency similar to full-length Cdc20 exhibited equivalent amount of multinuclei. On the other hand GM2, GM4 and GM5 lacking Mad2 binding capacity showed less multinucleation (Figure 9B). Thus, from these two experiments (FACS and multinuclei), we suggest that Cdc20 should bind Mad2 with optimum efficiency. Weak binding will not allow normal regulation of Cdc20 function and the MCC complex formation will be hindered, whereas strong binding will disrupt the checkpoint function probably by not allowing anaphase promotion even after reattachment and will allow cells to adapt the checkpoint followed by multinuclei formation [22,27].
Figure 9. Analysis of multinucleation frequency.
(A) HeLa cells were mock-transfected or transfected with different Cdc20 constructs as mentioned at the bottom of the panel. After 48 h, cells were stained with α-tubulin antibody (green) followed by counterstaining with propidium iodide (red) for cytoskeletal structure and nuclei respectively. White and pink arrows show normal and multinucleated cells respectively. Scale bar, 10 μm. (B) The percentage of multinucleated HeLa cells after transfection with different Cdc20 constructs as mentioned below in the presence (+Noc) or absence (−Noc) of nocodazole.
DISCUSSION
Until now, the N-terminal 1–153 amino acid region of Cdc20 was known to contain most of its functional domains such as Mad2 binding [22] and APC activation [21]. In the present paper, we provide evidence that the C-terminal part of the Cdc20 is equally important and contains at least two additional functional domains. Using co-localization, co-immunoprecipitation and peptide inhibition analysis with different deletion mutants of Cdc20, we have identified another Mad2-binding domain between amino acids 342 and 355, which is directly involved in physical interaction with Mad2. The initial clue came from the observation that instead of the N-terminal part, the C-terminal half of the Cdc20 protein sequence exhibited stronger homology with that of lower eukaryotes. Another clue was obtained from the fact that the amino acids 344–355 (GWVPLQTFTQHQ) of Cdc20 have high homology (six out of 12 amino acids are identical) with one of the ten Mad2-binding peptides (GWVRLQPPPLIQ) identified by phage display analysis [15].
Previously, it was shown that WD40 repeat regions of Cdc20 are necessary for localization to kinetochores and spindle fibres [21]. A single WD unit usually forms four β-sheets, each flanked by loops and turns and this motif can form a complete blade structure. At least four to seven such blades are necessary to make a complete propeller structure and the surface of each blade remains exposed on the surface of the propeller [29]. The PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/) analysis of full-length Cdc20 protein sequence (accession no. NP_001246) showed that the N-terminal part (amino acids 1–128) of the protein is mainly helical in nature and the other portions (amino acids 129–499), containing WD repeats, form alternate sheet-coil structures (results not shown). Cdc20 deletion mutants used in the present study pGM2, pGM3, pGM6 and pGM8 contain at least four WD repeats and thus they are expected to retain their propeller structure partially. The exact position of the WD repeats as reported in the NCBI database suggests that the small stretch of the peptide spanning amino acids 342–355, which resides in between fourth and fifth WD repeats, can form a coil structure (results not shown). From these facts, we speculate that the second Mad2-binding region may protrude as a loop on the surface of the β-propeller of full-length as well as Cdc20 deletion mutants. Thus the second Mad2-binding domain in wild-type protein will provide the required surface for the interaction with Mad2 under physiological conditions.
The contribution of the second Mad2-binding domain in Mad2–Cdc20 interaction was also evident from the fact that blocking of the second binding site (amino acids 342–355) of full-length Cdc20 by P2 peptide also destabilizes its interaction with Mad2 through its first binding site (amino acids 110–153). It was reported that Mad2 binds more tightly with short Cdc20 polypeptides such as the N-terminal 1–153 amino acids than full-length Cdc20 [22]. Moreover, increase in the fragment length up to 210 amino acids reduced the binding capacity significantly [22]. Zhang and Lees [22] did not observe any binding of Mad2 with Cdc20 deletion mutant containing amino acids 211–499, which is also consistent with our observations. It is evident from Figure 10 that whenever amino acids 167–254 of Cdc20 were present with either the first (GM5) or second (del-N) Mad2-binding sites, the efficiency of Mad2 binding decreased significantly. The Cdc20 deletion mutants devoid of this inhibitory domain either exhibited very strong binding (del-C) or equal efficiency (GM3) in comparison with full-length Cdc20 (GM1). The Cdc20 mutant GM6 that comprised both the Mad2-binding domains flanking the inhibitory domain exhibited Mad2 binding efficiency similar to GM1. Thus the first Mad2-binding site of Cdc20 alone (del-C) showed very strong Mad2 binding, which was attenuated by the presence of the inhibitory domain (GM5) and once again regained when the second Mad2-binding site was introduced (GM6). These results suggest that the second Mad2-binding domain of Cdc20 is required for optimal interaction between Mad2 and Cdc20. The amino acid region 167–254 of Cdc20 protein, on the other hand, may impart steric hindrance in Mad2 binding of both previously characterized (amino acids 1–153) and the newly characterized (amino acids 342–355) binding domains (Figure 10).
The putative polyhistidine region (amino acids 289–308), which has the metal binding property, resides near the second Mad2-binding domain of Cdc20. The smallest Cdc20 deletion mutant GM10 (amino acids 282–344) containing the polyhistidine region was still capable of binding Mad2 albeit less efficiently. Although the P1 peptide (amino acids 289–303) was unable to inhibit Mad2 binding in competition assays, the removal of metal ions from the system significantly decreased the Mad2 binding ability of the full-length (both endo- and GFP-fused) and del-N Cdc20, but not the del-C construct (Figure 7, panel IP/EDTA). Thus this result suggests that the Mad2 binding ability of the N-terminal fragment containing the first binding site is independent of metal binding, whereas the Mad2 binding of the Cdc20 deletion mutants containing only the second Mad2-binding site and the full-length Cdc20 are dependent on a specific conformation of the protein, which is maintained by the metal binding to the polyhistidine domain of Cdc20. Further studies are necessary to determine the exact contribution of the polyhistidine metal-binding domain to Mad2–Cdc20 interaction and for the structural integrity of the β-propeller [30].
The differential binding efficiency of the N-terminal peptide 1–153 amino acids and the C-terminal peptide GM3 255–499 amino acids of Cdc20 with Mad2 suggests that these two peptides may differ functionally. Our functional analysis (Figures 8 and 9) showed that an optimum Mad2–Cdc20 interaction is necessary for the sequestration of Cdc20 activity from APC/C activation. This observation is evident from the cell cycle analysis that though overexpression of any of the Cdc20 deletion mutants was capable of promoting premature anaphase, only the deletion constructs having the functional second Mad2-binding domain (GM3 and GM6) retained the capability of being inhibited by exogenous Mad2 similarly to the full-length Cdc20. However, the deletion constructs deficient of Mad2 binding (GM2, GM4, GM5 and del-N) failed to respond in SAC arrest due to their weak affinity towards Mad2. Similarly, from multinucleation analysis, it is evident that the binding-proficient deletion mutants (GM3 and GM6) of Cdc20 behave more like full-length than the binding-deficient (GM2, GM4, GM5 and del-N) mutants. Strikingly, the del-C mutant of Cdc20 showed both partial attenuation of premature anaphase promotion by exogenous Mad2 (Figure 8C, see del-C mutant) and unusually a high rate of multinucleation (Figure 9B) probably due to disruption of SAC function caused by stronger affinity towards Mad2 [22,31].
Although the template model for Mad1–Mad2 interaction could explain some issues of the exchange model, a few issues remained unaddressed [32–34]. So far, Cdc20 binding to Mad1–Mad2 complex was examined with a small N-terminal peptide (amino acids 111–153) of Cdc20 having unusually strong affinity than the full-length Cdc20 ([22] and the present study; Figure 7, lane IP). The Mad2 exchange model used a non-native peptide [19] and the template model was proposed using the Mad2-binding motif (amino acids 111–138) of Cdc20 only [11]. It is not clear whether the accessibility of the amino acid 111–153 domain in full protein is the same or not. Moreover, the template model showed involvement of Arg133 (which is one of the terminal residues in the second patch) of Mad2 from both O and C partners in the C-Mad2–O-Mad2 interaction [11]. As no evidence was found for homodimerization of O–O and C–C Mad2 [19], it is expected that the interface between the two conformations of the Mad2 will be asymmetric, which is further supported by the involvement of the C-terminal 10 amino acid residues present in the opposite surface of Mad2 in Mad2 oligomer [18]. In the light of the above discussion, we conclude that Cdc20 interacts with Mad2 through its multiple domains to confer an optimal binding between the two proteins appropriate for proper regulation of SAC. Our results also suggest that the polyhistidine metal-binding domain of Cdc20 plays an indirect role in this regulatory process which was not previously reported. Detailed structural analysis of Cdc20 with respect to its interaction with Mad2 will shed more light in understanding the intricacies of the process.
Acknowledgments
We are thankful to Dr A. Musacchio (European Institute of Oncology, Italy) and Dr K.T. Jeang for their suggestions during this work. We are also thankful to Dr P. Das (National Institute of Cholera and Enteric Diseases, Kolkata, India) and Dr C.K. Panda (Chittaranjan National Cancer Institute, Kolkata, India) for helping us with the confocal microscopy and FACS analysis respectively. We thank Dr Kunal Ray (Indian Institute of Chemical Biology, Kolkata, India) for critically reading this paper. G.M. is supported by a predoctoral fellowship from the Council of Scientific and Industrial Research (New Delhi, India). The study was partially supported by research grants from the Indian Council of Medical Research (63/147/2001-BMS) and Council of Scientific and Industrial Research, India (CMM 0003 and CMM 0016) awarded to S.R.
References
- 1.Wassmann K., Benezra R. Mitotic checkpoints: from yeast to cancer. Curr. Opin. Genet. Dev. 2001;11:83–90. doi: 10.1016/s0959-437x(00)00161-1. [DOI] [PubMed] [Google Scholar]
- 2.Chung E., Chen R. H. Phosphorylation of Cdc20 is required for its inhibition by the spindle checkpoint. Nat. Cell Biol. 2003;5:748–753. doi: 10.1038/ncb1022. [DOI] [PubMed] [Google Scholar]
- 3.Waizenegger I., Gimenez-Abian J. F., Wernic D., Peters J. M. Regulation of human separase by securin binding and autocleavage. Curr. Biol. 2002;12:1368–1378. doi: 10.1016/s0960-9822(02)01073-4. [DOI] [PubMed] [Google Scholar]
- 4.Hauf S., Waizenegger I. C., Peters J. M. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science. 2001;293:1320–1323. doi: 10.1126/science.1061376. [DOI] [PubMed] [Google Scholar]
- 5.Amon A. The spindle checkpoint. Curr. Opin. Genet. Dev. 1999;9:69–75. doi: 10.1016/s0959-437x(99)80010-0. [DOI] [PubMed] [Google Scholar]
- 6.Iwanaga Y., Kasai T., Kibler K., Jeang K. T. Characterization of regions in hsMAD1 needed for binding hsMAD2. A polymorphic change in an hsMAD1 leucine zipper affects MAD1–MAD2 interaction and spindle checkpoint function. J. Biol. Chem. 2002;277:31005–31013. doi: 10.1074/jbc.M110666200. [DOI] [PubMed] [Google Scholar]
- 7.Li Y., Benezra R. Identification of a human mitotic checkpoint gene: hsMAD2. Science. 1996;274:246–248. doi: 10.1126/science.274.5285.246. [DOI] [PubMed] [Google Scholar]
- 8.Musacchio A., Hardwick K. G. The spindle checkpoint: structural insights into dynamic signalling. Nat. Rev. Mol. Cell Biol. 2002;3:731–741. doi: 10.1038/nrm929. [DOI] [PubMed] [Google Scholar]
- 9.Howell B. J., Moree B., Farrar E. M., Stewart S., Fang G., Salmon E. D. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr. Biol. 2004;14:953–964. doi: 10.1016/j.cub.2004.05.053. [DOI] [PubMed] [Google Scholar]
- 10.Xia G., Luo X., Habu T., Rizo J., Matsumoto T., Yu H. Conformation-specific binding of p31(comet) antagonizes the function of Mad2 in the spindle checkpoint. EMBO J. 2004;23:3133–3143. doi: 10.1038/sj.emboj.7600322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeAntoni A., Sala V., Musacchio A. Explaining the oligomerization properties of the spindle assembly checkpoint protein Mad2. Philos. Trans. R. Soc. London Ser. B. 2005;360:637–647. doi: 10.1098/rstb.2004.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang G. Checkpoint protein BubR1 acts synergistically with Mad2 to inhibit anaphase-promoting complex. Mol. Biol. Cell. 2002;13:755–766. doi: 10.1091/mbc.01-09-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peters J. M. The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol. Cell. 2002;9:931–943. doi: 10.1016/s1097-2765(02)00540-3. [DOI] [PubMed] [Google Scholar]
- 14.Hoyt M. A. A new view of the spindle checkpoint. J. Cell Biol. 2001;154:909–911. doi: 10.1083/jcb.200108010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo X., Tang Z., Rizo J., Yu H. The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Mol. Cell. 2002;9:59–71. doi: 10.1016/s1097-2765(01)00435-x. [DOI] [PubMed] [Google Scholar]
- 16.Sironi L., Mapelli M., Knapp S., De Antoni A., Jeang K. T., Musacchio A. Crystal structure of the tetrameric Mad1–Mad2 core complex: implications of a ‘safety belt’ binding mechanism for the spindle checkpoint. EMBO J. 2002;21:2496–2506. doi: 10.1093/emboj/21.10.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Antoni A., Pearson C. G., Cimini D., Canman J. C., Sala V., Nezi L., Mapelli M., Sironi L., Faretta M., Salmon E. D., Musacchio A. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr. Biol. 2005;15:214–225. doi: 10.1016/j.cub.2005.01.038. [DOI] [PubMed] [Google Scholar]
- 18.Luo X., Fang G., Coldiron M., Lin Y., Yu H., Kirschner M. W., Wagner G. Structure of the Mad2 spindle assembly checkpoint protein and its interaction with Cdc20. Nat. Struct. Biol. 2000;7:224–229. doi: 10.1038/73338. [DOI] [PubMed] [Google Scholar]
- 19.Luo X., Tang Z., Xia G., Wassmann K., Matsumoto T., Rizo J., Yu H. The Mad2 spindle checkpoint protein has two distinct natively folded states. Nat. Struct. Mol. Biol. 2004;11:338–345. doi: 10.1038/nsmb748. [DOI] [PubMed] [Google Scholar]
- 20.Steensgaard P., Garre M., Muradore I., Transidico P., Nigg E. A., Kitagawa K., Earnshaw W. C., Faretta M., Musacchio A. Sgt1 is required for human kinetochore assembly. EMBO Rep. 2004;5:626–631. doi: 10.1038/sj.embor.7400154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kallio M. J., Beardmore V. A., Weinstein J., Gorbsky G. J. Rapid microtubule-independent dynamics of Cdc20 at kinetochores and centrosomes in mammalian cells. J. Cell Biol. 2002;158:841–847. doi: 10.1083/jcb.200201135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y., Lees E. Identification of an overlapping binding domain on Cdc20 for Mad2 and anaphase-promoting complex: model for spindle checkpoint regulation. Mol. Cell. Biol. 2001;21:5190–5199. doi: 10.1128/MCB.21.15.5190-5199.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fraschini R., Beretta A., Sironi L., Musacchio A., Lucchini G., Piatti S. Bub3 interaction with Mad2, Mad3 and Cdc20 is mediated by WD40 repeats and does not require intact kinetochores. EMBO J. 2001;20:6648–6659. doi: 10.1093/emboj/20.23.6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yudkovsky Y., Shteinberg M., Listovsky T., Brandeis M., Hershko A. Phosphorylation of Cdc20/fizzy negatively regulates the mammalian cyclosome/APC in the mitotic checkpoint. Biochem. Biophys. Res. Commun. 2000;271:299–304. doi: 10.1006/bbrc.2000.2622. [DOI] [PubMed] [Google Scholar]
- 25.Goto M., Eddy E. M. Speriolin is a novel spermatogenic cell-specific centrosomal protein associated with the seventh WD motif of Cdc20. J. Biol. Chem. 2004;279:42128–42138. doi: 10.1074/jbc.M403190200. [DOI] [PubMed] [Google Scholar]
- 26.Klabunde T., Eicken C., Sacchettini J. C., Krebs B. Crystal structure of a plant catechol oxidase containing a dicopper center. Nat. Struct. Biol. 1998;5:1084–1090. doi: 10.1038/4193. [DOI] [PubMed] [Google Scholar]
- 27.Lin M. L., Sakamoto K. M. p55Cdc/cdc20 overexpression promotes early G1/S transition in myeloid cells. Stem Cells. 2001;19:205–211. doi: 10.1634/stemcells.19-3-205. [DOI] [PubMed] [Google Scholar]
- 28.Shirayama M., Zachariae W., Ciosk R., Nasmyth K. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 1998;17:1336–1349. doi: 10.1093/emboj/17.5.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith T. F., Gaitatzes C., Saxena K., Neer E. J. The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci. 1999;24:181–185. doi: 10.1016/s0968-0004(99)01384-5. [DOI] [PubMed] [Google Scholar]
- 30.Wilson D. K., Cerna D., Chew E. The 1.1-angstrom structure of the spindle checkpoint protein Bub3p reveals functional regions. J. Biol. Chem. 2005;280:13944–13951. doi: 10.1074/jbc.M412919200. [DOI] [PubMed] [Google Scholar]
- 31.Lanni J. S., Jacks T. Characterization of the p53-dependent postmitotic checkpoint following spindle disruption. Mol. Cell. Biol. 1998;18:1055–1064. doi: 10.1128/mcb.18.2.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nasmyth K. How do so few control so many? Cell. 2005;120:739–746. doi: 10.1016/j.cell.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 33.Hardwick K. G. Checkpoint signalling: Mad2 conformers and signal propagation. Curr. Biol. 2005;15:R122–R124. doi: 10.1016/j.cub.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 34.Hagan R. S., Sorger P. K. Cell biology: the more MAD, the merrier. Nature (London) 2005;434:575–577. doi: 10.1038/434575a. [DOI] [PubMed] [Google Scholar]