Abstract
The level of polyteny of the Drosophila salivary gland chromosomes was determined throughout the chromosome region 89E1–4, the locus of the Bithorax Complex. A zone of underreplication spans the 300 kb of DNA from the Ubx to Abd-B loci. From the centromere proximal end of the complex, a 70-kb-long gradual decrease of polytenization starts with the Ubx transcription unit and, after a floor corresponding to the abd-A locus, raises gradually back to the maximum over 70 kb in the region of the Abd-B transcription unit. In flies carrying the mutation Suppressor of DNA Underreplication [Su(UR)ES], the underreplication of the Bithorax Complex is fully suppressed. In the wild type, the Bithorax Complex forms a weak point featuring thinner bands separated by clefts or constrictions. In Su(UR)ES strain in contrast, the 89E1–4 band looks like a single solid band consisting of homogenous dense material. We speculate that the wild-type Su(UR)ES protein hampers DNA replication of silenced domains and leads to their underreplication in salivary gland polytene chromosomes.
The term intercalary heterochromatin was introduced by Kaufmann (1) and follows papers describing contacts between internal regions of euchromatin arms of polytene chromosomes, and with telomeric and pericentric heterochromatin (2–6). These contacts were later named ectopic pairing (7). The 60 years of investigation on intercalary heterochromatin recently was reviewed (8).
Regions of intercalary heterochromatin in polytene chromosomes differ from typical euchromatin and rather resemble pericentric heterochromatin in a number of features: dense packing, absence of transcriptional activity, tendency to form ectopic contacts, and late replication of DNA during S-period. Chromosome breaks also are frequently observed on cytological preparations; they were named weak points or breaks by Bridges (9). Weak points could result from local DNA underreplication of intercalary heterochromatin that was already proposed in the seventies (8, 10, 11). This hypothesis was corroborated first by autoradiographic measurements of relative DNA quantities in a typical weak point, the region of intercalary heterochromatin at 11A of female X chromosome. The weak point was found 4-fold underreplicated relative to the same region of the male X chromosome where the 11A region does not break (12). Underreplication also was reported for the region of the histone genes cluster, a weak point in the 39DE region (13) and for 5′ exon of Ubx gene located in the region of weak point 89E1–4 (14, 15). Euchromatic regions relocated next to pericentric heterochromatin by chromosomal rearrangements show position effect variegation and are found to be replicated late, and in some instances to be underreplicated. They appear condensed in polytene chromosomes (16). Late replication and underreplication are likely consequences of high levels of compaction of the region that hamper the process of replication in polytene chromosomes.
The 89E1–4 band is a classical region of intercalary heterochromatin. It has focused a lot of attention recently as the nest of the Bithorax Complex. The repression of homeotic genes is inherited epigenetically, and the silencing is established early in development. Silencing is achieved and maintained with the help of complexes of proteins including those of the Polycomb group. Some of these proteins have similarities in structure and function with proteins forming silent pericentric heterochromatin (see references in refs. 17 and 18). Genomic elements in the Bithorax Complex, the Polycomb Response Elements (PREs), seem to play a central role in binding the repressive complex. For example, transgene constructs containing PREs and a miniwhite reporter gene result in mosaic inactivation of white (18–20). These data lend support to the reality of intercalary heterochromatin whose existence was questioned in several reviews (21, 22). There is ample evidence to consider that silencing in heterochromatin, whether intercalary or pericentric, is determined by similar mechanisms of fundamental importance in regulation of gene expression in eukaryotes. Recent data point to the existence of a common genetic control of underreplication in both intercalary and pericentric heterochromatin. Indeed, the Suppressor of DNA Underreplication [Su(UR)ES] mutation suppresses underreplication of DNA sequences in pericentric heterochromatin, and at the same time the weak points and underreplication in intercalary heterochromatin disappear completely in polytene chromosomes (23).
Because the Bithorax Complex is well characterized both in molecular and genetic terms, we have examined the underreplication, morphology, and other aspects of intercalary heterochromatin in the 89E1–4 region both in wild-type polytene chromosomes and chromosome carrying the Su(UR)ES mutation.
Materials and Methods
Drosophila Stocks.
Oregon R: wild type and w−/w−;Su(UR)ES/Su(UR)ES stocks (23) were raised on a standard medium at 25°C.
Quantitative Southern Blot Hybridization.
In experiments I and III and with Su(UR)ES, genomic DNA was isolated from 100 pairs of hand-dissected salivary glands of third instar larvae or from 150 fly heads as in Lamb and Laird (15) with some modifications and was digested by EcoRI, HindIII, or BamHI. DNA probes were labeled with [32P]dATP by random priming and hybridized as in Belyaeva et al. (23). Control and test clones were hybridized simultaneously in most cases. Autoradiographic signal was measured with a spectrophotometer Hitachi 557 or by using a PhosphorImage SF system (Molecular Dynamics). Quantitation of radioactivity was obtained by volume integration using imagequant software (Molecular Dynamics). Relative DNA abundance was calculated as the ratio of hybridization intensity in salivary glands to that in adult heads after normalization on control clones (rosy; clone 3144 in two experiments), which replicate fully in polytene tissue of wild type (15, 23). Experiment II was performed as described by Spierer and Spierer (24).
Electron Microscopy.
Salivary glands of third instar larvae were fixed in cold ethanol/acetic acid (3:1) for 2–4 h, squashed in 45% acetic acid, contrasted in 70% ethanol saturated with uranyl acetate for 12 h, and embedded in Araldit M epoxy resin. Thin serial sections ranging in thickness from 100 to 150 nm were examined under a JEM-100C electron microscope at 80 kV. The original magnification was in a range from ×3,000 to ×10,000. All techniques have been described in detail (25).
Results and Discussion
Polytenization Through the Bithorax Complex Region in Oregon R and Su(UR)ES Strains.
To determine the level of polytenization throughout the BX-C, we used as probes selected fragments from the chromosomal walk spanning the 300-kb-long complex (26–28). The protocols were adapted from those used by Spierer and Spierer (24) to determine the level of polyteny through the 315 kb of DNA forming the rosy-Ace chromosomal walk in 87DE. Genomic DNA was isolated from larval salivary glands (polytene tissue), flies heads (diploid cells in experiments I and III) and embryos (diploid cells in experiment II). In both experiments, genomic blots of polytene and diploid DNA were hybridized to probes spanning the Bithorax Complex. Quantitation of the relative autoradiographic signals was used to determine the relative abundance of each probe in polytene and diploid tissues (23, 24). The three determinations were performed in different laboratories, at different times, with different stocks of Oregon R, and with minor differences in methodology (see Materials and Methods).
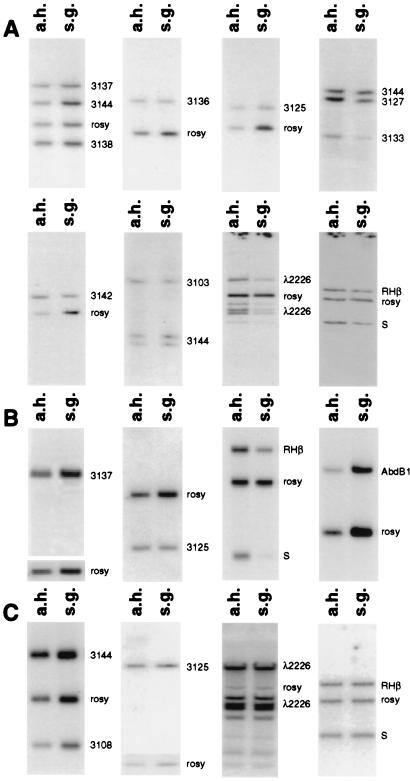
Fig. 1 shows some of the primary data of experiments I and III for both wild-type and homozygous Su(UR)ES strains. The complete results are listed in Table 1. Results of the complete analysis of all experiments are reported as a diagram in Fig. 2. In experiments I and III, the standard and internal control of full polytenization is a probe of the rosy gene (with two exceptions—clone 3144). In experiment II, the control is from the Ace locus at 87E. Both the rosy and Ace genes have been determined to fully polytenize (24). The standard level of polyteny of the rosy and 3144 clones also was confirmed independently by Lamb and Laird (15) and Belyaeva et al. (23). The precision of the method was tested by Spierer and Spierer (24). Differences of relative level of polyteny of 2-fold and more are significant.
Figure 1.
Genomic Southern blot hybridizations of polytene and diploid DNA with different probes from the BX-C. (A) Southern blots with Oregon R genomic DNA from experiment I. (B) Southern blots with Oregon R genomic DNA from experiment III. (C) Southern blots with Su(UR)ES genomic DNA. “Polytene” DNA was prepared from salivary glands (s.g.) and diploid DNA was from adult heads (a.h.). rosy or 3144 were used as an internal control.
Table 1.
Representation of DNA sequences in larval salivary gland cells compared to that in adult head cells
| Clone (map position) | Percentage of DNA polytenization
|
|||||
|---|---|---|---|---|---|---|
| Oregon R
|
Su(UR)ES
|
|||||
| Exp. I
|
Exp. III
|
|||||
| Value | Average | Value | Average | value | Average | |
| 3144-(-110; -103) | 93* | 93* | 99* | |||
| 3138-(-101; -97.5) | 84, 110 | 97 | N.D. | N.D. | ||
| 3137-(-97.5; -87) | 93, 95 | 94 | 93, 96 | 94 | N.D. | |
| 3136-(-87; -77.5) | 75, 79 | 77 | N.D. | N.D. | ||
| 3127-(-76.5; -71.5) | 68, 64 | 66 | N.D. | N.D. | ||
| 3125-(-73; -61.5) | 65, 53 | 59 | 68, 75 | 71 | 97, 95 | 96 |
| PRE G (-62.5; -60) | 58, 52 | 55 | N.D. | N.D. | ||
| 3142-(-60; -50) | 34, 17 | 25 | N.D. | N.D. | ||
| 3108-(-33; -30) | 24* | 24* | 110* | |||
| PRE F-(-15; -5) | 53, 51 | 52 | N.D. | N.D. | ||
| λ2226-(24; 40) | 58, 62 | 60 | N.D. | 103, 103 | 103 | |
| S-(116.5; 118.2) | 52, 56 | 54 | 12, 11 | 11 | 95, 99 | 97 |
| RHβ-(161; 167.5) | 80, 84 | 82 | 46, 65 | 55 | 102, 102 | 101 |
| AbdB1 (203; 205) | N.D. | 102, 108 | 110 | N.D. | ||
N.D., not determined.
Determined in previous experiments by Belyaeva et. al. (23).
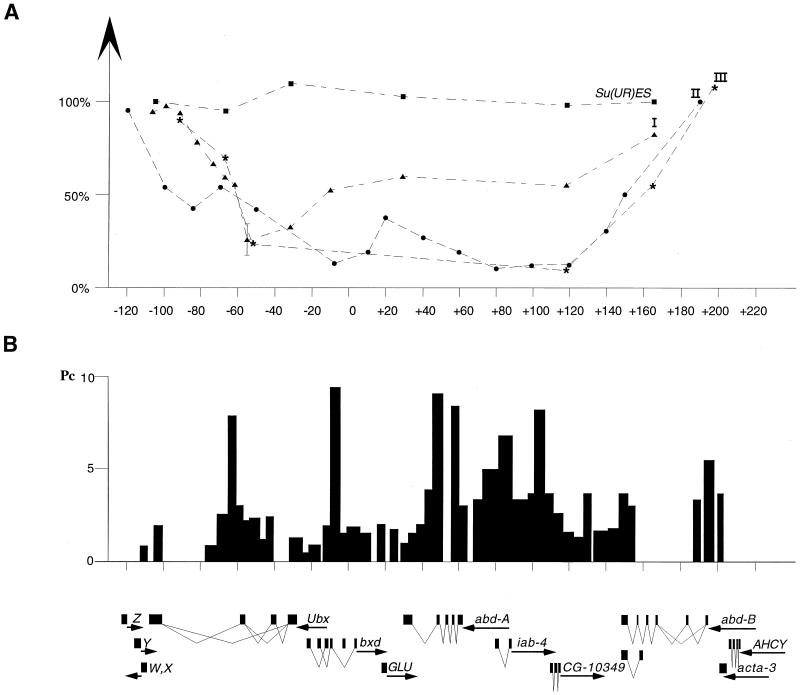
Figure 2.
Profile of underreplication and organization of chromosome region containing the Bithorax Complex. (A) Relative level of DNA replication of the BX-C in salivary gland cells in wild-type and Su(UR)ES strains. Triangles represent data obtained from wild type in experiment I (Fig. 1 and Table 1). Dots represent data of experiment II. Asterisks represent experiment III (Fig. 1 and Table 1). Squares represent data for Su(UR)ES strain (Fig. 1 and Table 1). (B) Distribution of Polycomb protein (bars) through the BX-C in the cell culture (19). Transcription units in the region taken from ref. 29 are delineated at the bottom.
From the results in the diagram of Fig. 2, we conclude that there is a wide region of underreplication colocalized with the Bithorax Complex. The most underreplicated zone is located between −60 kb and +120 kb of the BX-C map, namely a part of the Ubx gene, GLU, abd-A gene, and CG10349 (29). From its minimum level, the polyteny rises back gradually over 70 kb both in proximal (3′ region of Ubx gene) and distal (Abd-B gene) parts of BX-C (see Fig. 2). The results of all three independent experiments are comparable. There seem to be, however, significant quantitative differences between replication profiles in experiment I on the one hand and experiments II and III on the other. These differences can be related to less favorable culture conditions, resulting in a lower level of polyteny or to a less extensive dissection of fat bodies in experiment I. These conditions were carefully controlled in experiments II and III.
The maximum relative level of underreplication is about 10-fold. The level of polyteny of chromosomes in a gland is estimated at about 1,000. Therefore, even at the lowest point of polyteny, the number of DNA duplexes assuring the continuity of the chromosomes can be estimated at 100 and certainly not limited to a unique double helix.
The next experiment was to repeat these measurements in a strain homozygous for the Su(UR)ES mutation. Data in Figs. 1 and 2 and Table 1 show that the mutation completely suppresses underreplication.
As already mentioned in the Introduction, the process of underreplication might be linked to the repressed state of chromatin caused by Pc-G proteins. Fig. 2 also schematizes the data on Polycomb protein distribution obtained through the BX-C after formaldehyde crosslinking with DNA from cell culture (19). There is a correlation between the underreplication and the region of Polycomb association, with one exception over the 5′ end of the Abd-B transcript. This correlation is of potential interest, keeping in mind that the data were obtained from cell types possibly differing in expression of the BX-C genes, and that the regions flanking the BX-C were not scanned for Polycomb association.
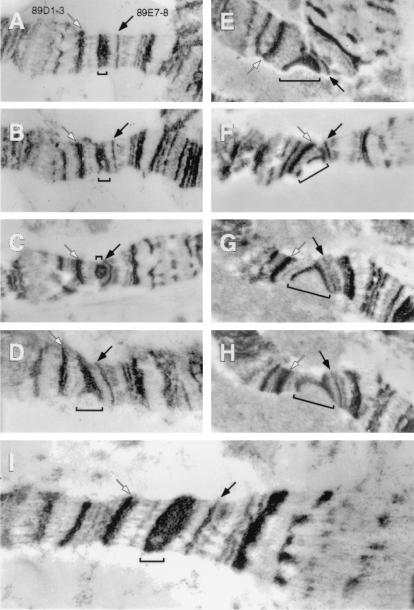
Electron Microscopy of the 89E1–4 Region in Polytene Chromosomes.
The polytene chromosome region spanning the Bithorax Complex was described by light microscopy as a constriction centered over a doublet (30). This was reexamined here by transmission electron microscopy. First, in the Su(UR)ES strain, where the BX-C locus is fully polytenized, the 89E1–4 region looks like a single massive band and its diameter does not differ from that of neighboring nonpuffing bands (Fig. 3I). In the wild type, where the Bithorax Complex is underreplicated, the morphology of the 89E1–4 region varies considerably among different salivary gland cells. The band can be solid, single, and have a normal diameter, but it is evidently thinner than in Su(UR)ES strains. Its central part is definitely looser than the borders (Fig. 3 A–H). Sometimes cavities appear between the dense borders and the band in one homologue looks like a “doublet” (Fig. 3B). Another morphological aspect is the constriction of the 89E1–4 region, when the chromosome diameter is smaller than in the neighboring regions (Fig. 3C). These three types of morphologies were not associated with partial breaks in the band. Clefts do appear in this region in wild-type strains with a frequency of 60–80% (12, 22). The fissure can be rather shallow (Fig. 3D), may run up to the middle (Fig. 3E), or to the edge of the band (Fig. 3 F and G). In this last case, the 89E1–4 region appears as two separate bands (or borders of the band 89E1–4). These “bands” can be displaced to forming a jag of irregular form (Fig. 3 F and G). In other cases, the band borders join to form a stretch consisting of elongated material (Fig. 3H). A similar variety of morphologies is characteristic also of other regions of intercalary heterochromatin (8). Strong ectopic contacts between the band boundaries flanking the underreplication zone may prevent splitting, and the weak point does not appear (Fig. 3A). This could explain why the frequency of fissures is lower in 89E1–4 than in other regions with roughly the same degree of underreplication. For example, 39E band tearing is seen in 100% of chromosomes examined. In this region spanning the histone genes cluster, the level of polyteny as determined by DNA sequence abundance is 30% of normal (15, 23).
Figure 3.
Electron micrographs of the 89E1–4 band (marked by a bracket). (A–H) Wild type (Oregon R). (I) Su(UR)ES. Bands 89D1–3 and 89D7–8 serve as flanking markers and are indicated by white and black arrows, respectively.
Thus, we infer that frequencies of weak points depend not only on the degree and extent of underreplication in the zone, but also on peculiarities of flanks limiting this zone in the band. It should be mentioned as well that the number of copies determined here is an average. We do not know whether variations among chromosomes are important. Altogether, the structural analysis reported above confirms the morphological diversity of regions of intercalary heterochromatin associated with underreplication.
Analysis by electron microscopy does not support the earlier description of the 89E1–4 band as a complex one, consisting of a heavy doublet, or even a pair of doublets, as in Bridges' map (30). The doublet in the region appears because of a splitting of its central part. In Su(UR)ES homozygotes the 89E1–4 band appears solid and continuous, without heterogeneity of packing, or any feature of loosening. Moreover, the 89E1–4 band is late replicated and binds Polycomb protein in the salivary gland chromosomes of Su(UR)ES (I.F.Z., E.S.B., and E. N. Andreeva, unpublished work). So, it looks like that the Su(UR)ES mutation does not influence silencing and late replication, but suppresses only underreplication in intercalary and pericentric heterochromatin.
Acknowledgments
We thank Welcome Bender, François Karch, Tina Kajimura, and Scott Munroe for providing us with genomic and cDNA clones from the Bithorax Complex. This work was supported by grants from the Russian Foundation for Basic Research (N 99–04-49270, N 00–15-97984), a grant from the Russian State Program “Frontiers in Genetics” (N 99–04-020), a grant from the International Association for the Promotion and Cooperation with Scientists from the New Independent States of the Former Soviet Union (N 99–1088), and grants from the Swiss National Science Foundation.
Abbreviation
- Su(UR)ES
Suppressor of DNA Underreplication.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.021353598.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.021353598
References
- 1.Kaufmann B P. Proc Natl Acad Sci USA. 1939;25:571–577. doi: 10.1073/pnas.25.11.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauer H. Proc Natl Acad Sci USA. 1936;22:216–221. doi: 10.1073/pnas.22.4.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frolova S L. Biol J. 1936;5:271–292. (in Russian). [Google Scholar]
- 4.Tinyakov G G. Biol J. 1936;5:753–802. (in Russian). [Google Scholar]
- 5.Prokofyeva–Belgovskaya A A. Bull Acad Sci URSS Ser Biol. 1937;3:719–724. (in Russian). [Google Scholar]
- 6.Prokofyeva–Belgovskaya A A. Bull Acad Sci URSS Ser Biol. 1938;4:97–103. (in Russian). [Google Scholar]
- 7.Slizynski B M. Proc R Soc Edinburgh. 1945;62:114–119. doi: 10.1017/s0080455x00009711. [DOI] [PubMed] [Google Scholar]
- 8.Zhimulev I F. Adv Genet. 1998;37:134–238. doi: 10.1016/s0065-2660(08)60341-7. [DOI] [PubMed] [Google Scholar]
- 9.Bridges P N. J Hered. 1935;26:60–64. [Google Scholar]
- 10.Barr H J, Ellison J R. Chromosoma. 1972;39:53–61. doi: 10.1007/BF00320590. [DOI] [PubMed] [Google Scholar]
- 11.Lefevre G. In: The Genetics and Biology of Drosophila. Ashburner M, Novitsky E, editors. London: Academic; 1976. pp. 1–66. [Google Scholar]
- 12.Zhimulev I F, Semeshin V F, Kulichkov V A, Belyaeva E S. Chromosoma. 1982;87:197–228. [Google Scholar]
- 13.Hammond M P, Laird C D. Chromosoma. 1985;91:279–286. doi: 10.1007/BF00328223. [DOI] [PubMed] [Google Scholar]
- 14.Lifschytz E, Hareven D. Chromosoma. 1982;86:443–445. doi: 10.1007/BF00330120. [DOI] [PubMed] [Google Scholar]
- 15.Lamb M M, Laird C D. Chromosoma. 1987;95:227–235. doi: 10.1007/BF00294779. [DOI] [PubMed] [Google Scholar]
- 16.Umbetova G H, Belyaeva E S, Baricheva E M, Zhimulev I F. Chromosoma. 1991;101:55–61. doi: 10.1007/BF00360687. [DOI] [PubMed] [Google Scholar]
- 17.Pirrotta V. Cell. 1998;93:333–336. doi: 10.1016/s0092-8674(00)81162-9. [DOI] [PubMed] [Google Scholar]
- 18.Zink D, Paro R. EMBO J. 1995;14:5660–5671. doi: 10.1002/j.1460-2075.1995.tb00253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strutt H, Cavalli G, Paro R. EMBO J. 1997;16:3621–3632. doi: 10.1093/emboj/16.12.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sigrist C J A, Pirrotta V. Genetics. 1997;147:209–221. doi: 10.1093/genetics/147.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spofford J B. In: The Genetics and Biology of Drosophila. Ashburner M, Novitsky E, editors. London: Academic; 1976. pp. 955–1018. [Google Scholar]
- 22.Gatti M, Pimpinelli S. Annu Rev Genet. 1992;26:239–275. doi: 10.1146/annurev.ge.26.120192.001323. [DOI] [PubMed] [Google Scholar]
- 23.Belyaeva E S, Zhimulev I F, Volkova E I, Alekseyenko A A, Moshkin Y M, Koryakov D E. Proc Natl Acad Sci USA. 1998;95:7532–7537. doi: 10.1073/pnas.95.13.7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spierer A, Spierer P. Nature (London) 1984;307:176–178. doi: 10.1038/307176a0. [DOI] [PubMed] [Google Scholar]
- 25.Semeshin V F, Demakov S A, Perez Alonso M, Belyaeva E S, Bonner J J, Zhimulev I F. Chromosoma. 1989;97:396–412. doi: 10.1007/BF00292767. [DOI] [PubMed] [Google Scholar]
- 26.Bender W, Akam M, Karch F, Beachy P A, Peifer M, Spierer P, Lewis E B, Hogness D S. Science. 1983;221:23–29. doi: 10.1126/science.221.4605.23. [DOI] [PubMed] [Google Scholar]
- 27.Bender W, Spierer P, Hogness D S. J Mol Biol. 1983;168:17–33. doi: 10.1016/s0022-2836(83)80320-9. [DOI] [PubMed] [Google Scholar]
- 28.Karch F, Weiffenbach B, Peifer M, Bender W, Duncan L, Celniker S, Crosby M, Lewis E B. Cell. 1985;43:81–96. doi: 10.1016/0092-8674(85)90014-5. [DOI] [PubMed] [Google Scholar]
- 29.Martin C H, Mayeda C A, Davis C A, Ericsson C L, Knafels J D, Mathog D R, Celniker S E, Lewis E B, Palazzolo M J. Proc Natl Acad Sci USA. 1995;92:8398–8402. doi: 10.1073/pnas.92.18.8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bridges P N. J Hered. 1941;32:299–300. [Google Scholar]