Abstract
Tandemly repeated DNA sequences are highly dynamic components of genomes1. Most repeats are in intergenic regions, but some are found within coding sequences or pseudogenes2. In humans, expansion of intragenic triplet repeats is associated with various diseases, including Huntington’s chorea and fragile X syndrome3,4. The persistence of intragenic repeats in genomes argues in favor of a compensating benefit. Here we show that in the Saccharomyces genome, the majority of the genes containing intragenic repeats encode cell wall proteins. The repeats trigger frequent recombination events within the gene or between the gene and a pseudogene, causing expansion and contraction in the gene size. This size variation creates quantitative alterations in phenotypes (e.g. adhesion, flocculation, biofilm formation). We propose that variation in intragenic repeat number provides the functional diversity of cell surface antigens that, in fungi and other pathogens, allows rapid adaptation to the environment and/or elusion of the host immune system.
The sequenced and annotated genome of Saccharomyces cerevisiae provides a unique opportunity to determine the function of intragenic repeat sequences. To identify the S. cerevisiae open reading frames (ORFs) that contain intragenic tandem repeats, we scanned all 6591 open reading frames for the presence of long (>40 nt) or short (3-39 nt) repeats (see Methods for details). The search yielded 44 ORFs: 29 ORFs with repeats longer than 40 nt (Fig. 1a) and 15 ORFs with small repeats (Fig. 1b). These 44 genes showed unexpected functional similarities. Eighteen of the 29 ORFs (62 %) with conserved long repeats encode cell wall proteins. By comparison, only 1.3 % of all S. cerevisiae open reading frames are cell surface proteins (88 out of 6591). An additional 4 genes (CTR1, MNN4, MSB2, HKR1) are plasma membrane proteins with extracellular domains. Hence, more than 75% (22/29) of all genes with long intragenic tandem repeats encode cell-surface proteins. The group of 15 genes with short repeats contains only one cell wall gene (SCW11). However, several genes in this group encode regulators of cell wall synthesis and maintenance, such as MSS11 (regulator of adhesion), WSC3 (regulator of cell wall integrity) and CHS5 (regulator of chitin biosynthesis).
Fig. 1. S. cerevisiae genes containing conserved intragenic repeats.
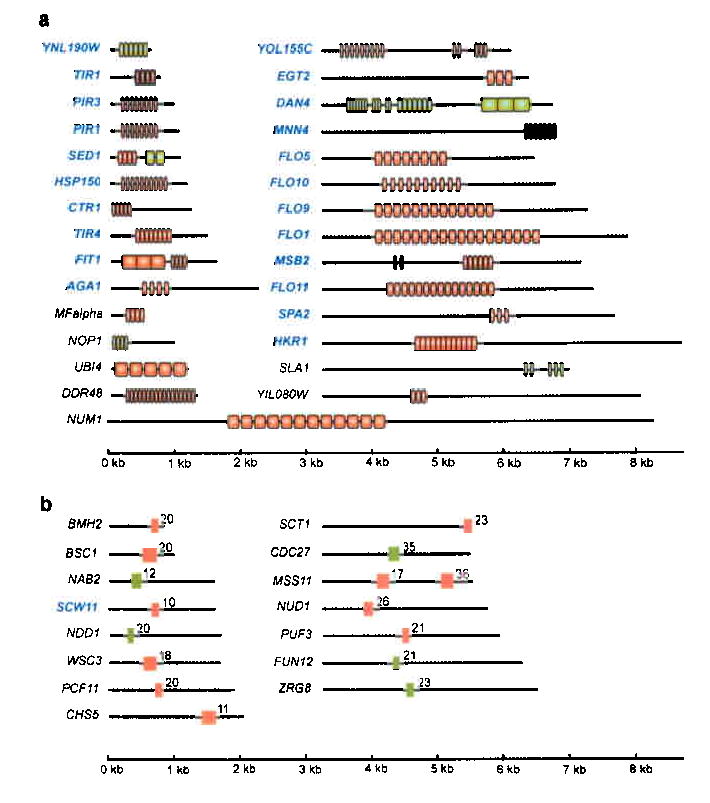
A screen of all open reading frames in the S. cerevisiae genome for those containing conserved intragenic tandem repeats identified 29 genes with large (≥ 40 nt) repeats (panel a) and 15 genes with short (<40 nt) repeats (panel b). Repeats (vertical boxes) that vary in size among 6 different S. cerevisiae strains are colored red (see text and Supplementary Fig. 1-2 online); repeats that do not show size variation among these strains are colored green. Cell surface genes are indicated in blue. The numbers in panel b indicate the number of repeats. More information about the repeats (consensus sequence, conservation…) is given in Supplementary Table 1 online. The repeat units in most genes are distinct from those in others except in FLO1, FLO5 and FLO9, which share the same repeat unit, and in PIR1, PIR3 and HSP150, which share the same repeat unit. SED1, FIT1, SLA1 and MSB2 contain two intragenic repeat regions with different repeat sequences. YOL155C and DAN4 contain three distinct repeat regions.
Remarkably, all repeats were found to be in-frame, so that deletion or addition of repeat units would not alter the reading frame. In order to verify that the intragenic repeat regions show size variations between yeast strains due to expansion or contraction of the repeats, we amplified each of the identified repetitive regions by PCR and compared the sizes for six different S. cerevisiae strains. The length of the repeat region in 35 of the 44 genes with intragenic repeats varies from strain to strain (Fig. 2 and Supplementary Fig. 1, 2 online). Virtually all cell surface genes with conserved repeats showed size variation. Moreover, strains that have a ploidy greater than haploid often harbor several different alleles of the same gene. The size difference between the genes in different S. cerevisiae strains is remarkable, as the size of most genes has been conserved over millions of years in different yeast species5. To confirm that genes in these six strains do not generally vary in size, we analyzed 16 genes without repeats: 8 cell surface genes, 4 long genes (> 3kb) and 4 genes encoding various enzymes. None of these 16 genes lacking repeats show any length differences among the six S. cerevisiae strains (Supplementary Fig. 3 online).
Fig. 2. Intragenic repetitive domains vary in size.
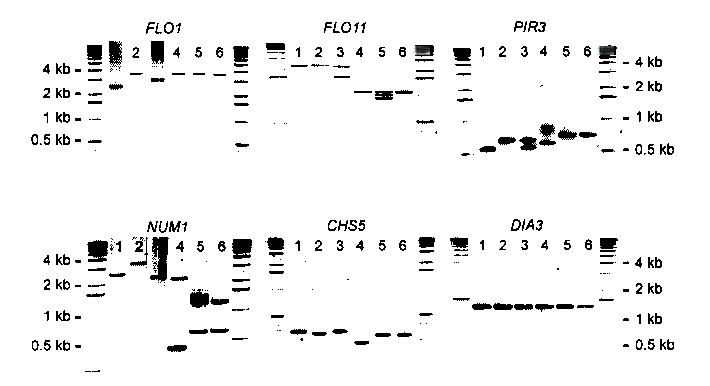
The repetitive domains of all 44 genes carrying intragenic repeats (Fig. 1) and the ORFs of 16 control genes without repeats were amplified by PCR for six different S. cerevisiae strains. This figure shows the results for 5 genes with repeats (FLO1, FLO11, PIR3, NUM1 and CSH5) and one gene without repeats (DIA3). Lane 1: S288C (haploid); Lane 2: Sigma1278b (haploid); Lane 3: EM93 (diploid); Lane 4: CMBS355 (polyploid); Lane 5: CMBS DL16 (polyploid); Lane 6: CMBS33 (polyploid). The analysis of other genes is in Supplementary Figures 1-3 online. The variability in some cell surface genes has already been used for the genotyping of wine yeasts30.
To characterize the events leading to expansion and contraction of intragenic repeats, we designed a system that permitted us to detect events occurring within the repeat region in one of the genes with repeats (FLO1). FLO1 is a homologue of the human mucin genes and encodes a cell surface adhesin, a mannoprotein responsible for adherence to other yeast cells (flocculation) as well as certain surfaces2,6. A single copy of the URA3 gene was inserted among the repeats of the genomic copy of FLO1 in the S288C strain (Fig 3a). In this strain, the FLO1 gene is 4.6 kb long and contains 18 repeats of about 100 nt, separated by a less conserved 45-nt sequence. The FLO1::URA3 strains were grown on medium without uracil and then spread on plates containing 5 fluoro-orotic acid (5-FOA), which selects for mitotic segregants that have lost the URA3 marker (Fig. 3b).
Fig 3. Intragenic repeats are hot-spots for recombination.
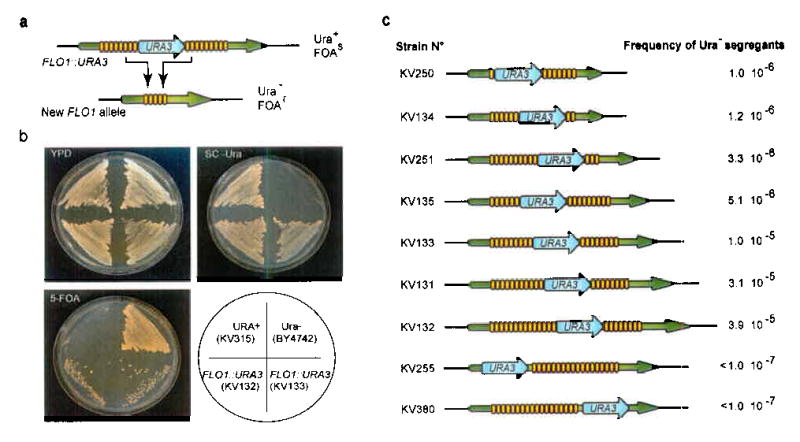
(a). In order to monitor recombination between intragenic repeats in the FLO1 gene, a URA3 expression cassette was integrated at various positions in the FLO1 repeats. As a consequence of the numerous recombination events within the repeats, the URA3 marker is lost at exceptionally high frequencies, resulting in a 5-FOA-resistant (Ura−) strain containing a new FLO1 allele. (b). Assay for loss of the URA3 marker. Ura+ strains (KV315, URA3 integrated at its native locus in the genome) grow on minimal medium (SC -Ura), but not on 5 fluoro-orotic acid (5-FOA) medium. Ura− strains (BY4742) grow on 5-FOA but not on minimal medium. Strains KV132 and KV133, with a URA3 cassette in the FLO1 repeats (FLO1::URA3), grow on minimal medium. Due to recombination events within the repeats, the URA3 cassette is looped out at high frequencies, yielding many 5-FOA-resistant segregants. (c). S. cerevisiae strains with the URA3 cassette integrated into various positions in the genomic FLO1 repeats (FLO1::URA3) were grown on medium lacking uracil and subsequently plated onto 5-FOA medium. The numbers indicate the frequency of Ura− segregants.
The FLO1::URA3 strains give rise to Ura− segregants at a high frequency (≈ 1 × 10−5; Fig. 3b,c). Moreover, the frequency of segregants gradually increases with increasing numbers of repeated DNA motifs surrounding the URA3 marker (Fig. 3c). The Ura− segregants have alterations in the number of repeats as compared with the starting strain. Most of the new FLO1 alleles obtained after loss of the URA3 marker have fewer repeat units than the wild-type FLO1 allele. However, about 15 percent (7/50) of the alleles gained extra repeats, causing increases in the gene size of up to 1 kb. This remarkable result indicates that the URA3 marker is not just “looped out” by unequal crossover between repeat units surrounding the marker (see further). Moreover, the wide range of different alleles generated in this procedure indicates that different repeat units can freely recombine with each other. Sequence analysis of the wild-type FLO1 allele and three of the novel short FLO1 alleles confirmed that each of these short alleles had lost several repeat units. Moreover, since all different repeats in the FLO genes show slight sequence differences2, it is possible to determine which repeat units were lost by aligning the sequences of the new alleles to that of the wild-type FLO1 sequence (Supplementary Fig. 4 online). This analysis shows that, in all cases, an upstream repeat unit had fused with one of the downstream units, thereby removing several repeat units in between while preserving the open reading frame.
Both the PIR and the FLO gene families have pseudogenes containing repeats that are similar to those in the functional copies2,7. These pseudogenes may provide additional genetic information that could be incorporated by a recombination event. In fact, two independent strains contain a novel FLO1 allele formed by the fusion of the first repeat unit of FLO1 with a repeat unit similar to those in FLO1 found in the FLO1 pseudogene YAR062W. This pseudogene is located approximately 12 kb downstream from the FLO1 termination codon2 (Fig. 4a). Sequence analysis of the FLO1-YAR062W fusion shows that the first FLO1 repeat had recombined with the repeat in the pseudogene, thereby looping out the complete 12 kb between the repeats in FLO1 and the pseudogene. Southern blotting and CHEF chromosome analysis confirmed the loss of about 12 kb between FLO1 and the pseudogene on chromosome I (Fig. 4b,c).
Fig 4. Repeats in pseudogenes provide an additional source of variability.
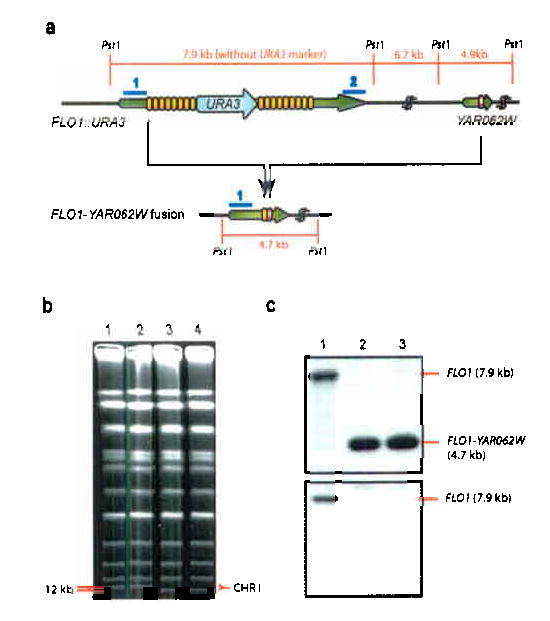
(a). Most of the recombination events between FLO1 repeats are strictly intragenic, i.e. only repeats within FLO1 recombine with each other. However, in some cases, the repeats in FLO1 recombine with a similar repeat unit found in the FLO1 pseudogene YAR062W, which is located about 12 kb downstream of FLO1. Fusion of a repeat in FLO1 with that in YAR062W results in deletion of the 3′ end of FLO1 and the entire 12 kb of DNA separating both open reading frames. (b). The FLO1-YAR062W deletion/fusion results in altered mobility of chromosome I (231 kbp) using clamped homogeneous electrical field (CHEF) electrophoresis. Lane 1: wild-type S. cerevisiae S288C; Lanes 2: control Ura− segregant (KV291) that has lost only intragenic FLO1 repeats. Lanes 3-4: FLO1-YAR062W fusion strains (KV360 and KV361). (c). Southern analysis confirms the deletion of the 12 kb region in between FLO1 and YAR062W. Genomic DNA of wild-type cells (lane 1) and the FLO1-YAR062W fusion strains (lanes 2-3) was cut with Pst1 and used for Southern blotting with probes that bind to the 5′ portion of FLO1 (probe 1, top) and the 3′ portion of FLO1 (probe 2, bottom). Other probes were used to further confirm the fusion (not shown).
To determine the functional consequence of continued variation of cell wall genes carrying intragenic repeats, we compared eight newly generated FLO1 alleles (2.9 kb to 5.4 kb, Fig. 5a,b) for their effects on various adhesion phenotypes associated with the FLO1 gene6. Each FLO1 size variant was fused to the inducible GAL1 promoter in the S288C background. In S288C, all 5 FLO genes are transcriptionally silent2,8, so the ectopic expression of these GAL1P-FLO1 constructs permits evaluation of the contribution of the particular FLO1 allele. As expected, none of the strains displayed any adhesion phenotypes on glucose medium. However, when these strains carrying the GAL1P-FLO1 fusion were grown on galactose medium (YPGal), there was a striking, linear correlation between gene size and the extent of adhesion: as the FLO1 proteins become longer (carrying more repeats), the adhesion properties gradually become stronger (Fig. 5c,d). Flocculation (i.e. adhesion to other yeast cells) shows the same quantitative relationship to the repeat number: the more repeats, the greater the fraction of flocculating cells (Fig. 5e). The observed correlation between the number of repeats and gain-of-function of Flo1 relies on the specific amino acids encoded by the repeats because insertion of URA3 in the FLO1 repeat region totally abolished adhesion (not shown).
Fig. 5. Instability of the FLO1 repeats generates functional variability.
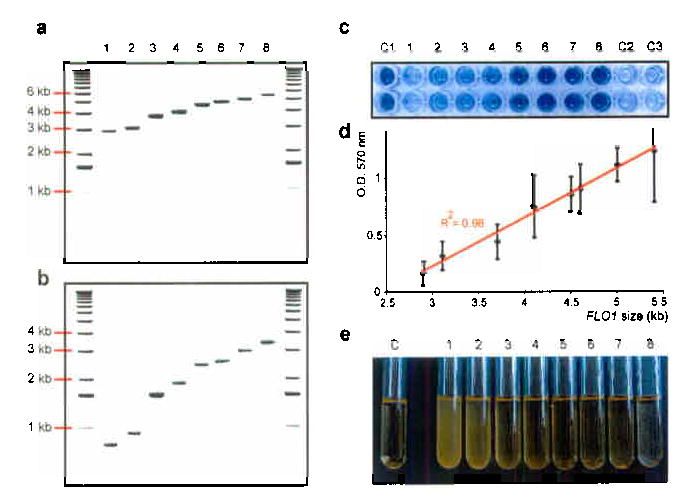
S. cerevisiae strain KV133 (FLO1::URA3) was plated onto 5-FOA medium to select Ura− segregants. (a) The Ura− segregants harbor FLO1 alleles of different lengths, ranging from 2.9 kb to 5.4 kb. (b) PCR amplification of the FLO1 repetitive domains shows that the different lengths of the alleles are due to corresponding differences in the lengths of the FLO1 repeat region. Lane 1: strain KV298 (FLO1 ORF = 2.9 kb); lane 2: KV308 (3.1 kb), lane 3: KV220 (3.7 kb), lane 4: KV219 (4.1 kb), lane 5: KV224 (4.5 kb), lane 6: KV211 (4.6 kb), lane 7: KV312 (5.0 kb), lane 8: KV311 (5.4 kb). (c) Expansion of FLO1 repeats leads to increased adherence to polystyrene. The FLO1 genes of the S288C strain BY4742 (KV210, lane C1) and of 8 strains containing different-sized FLO1 alleles (see panel a) were fused to the GAL1 promoter. Cells were grown on galactose medium and subsequently tested for adhesion to polystyrene by staining with crystal violet29. Cells expressing a long allele of FLO1 showed strong adhesion to polystyrene, whereas cells expressing shorter alleles do not adhere. When grown on glucose, strain KV311 fails to adhere (lane C2). Strain KV306, which contains the same FLO1 allele as strain KV311, but lacks the GAL1 promoter, fails to adhere when grown on galactose medium (lane C3). (d). Linear relationship between adherence to polystyrene and number of repeats. The error bars represent the standard deviation between three independent experiments. (e). Expansion of FLO1 repeats results in stronger cell-cell adhesion. The pGAL1-FLO1 fusion strains (tubes 1-8, see panel b) were tested for flocculation. Cells expressing a long allele of FLO1 show extremely strong cell-cell adhesion (all cells have sedimented on the bottom of the test tube). Tube C contains a strain (KV210) carrying the wild-type FLO1 allele of S. cerevisiae strain BY4742 fused to the GAL1 promoter.
To analyze the mechanism involved in the recombination of intragenic FLO1 repeats, the stability of the repeats in various key DNA repair and recombination mutants was measured (Table 1). Elegant studies on recombination of intergenic repeats have shown that in most cases, replication slippage and/or the repair of doubled-stranded breaks during DNA replication are the main mechanisms for repeat expansion and contraction9–13. These studies also identified various RAD genes that influence mutation frequencies in repeats. We found that loss of the RAD27-encoded flap endonuclease, which causes the formation of double-stranded breaks during replication12-15, increases the instability of FLO1 repeats almost 40-fold. The increased recombination frequency in rad27Δ mutants suggests that FLO1 repeat instability is associated with the occurrence of double-stranded breaks due to defective DNA replication12,14,15. Deletion of RAD52 and RAD50 severely reduces the frequency of rearrangements, whereas deletion of the RecA homologue RAD51 does not affect the frequency. Rad51 is required for ATP-dependent strand invasion during conservative DNA repair and recombination processes16. The absence of an effect in rad51Δ mutants suggests that FLO1 recombination does not require strand invasion and thus gene conversion, break-induced replication and crossing over are unlikely recombination mechanisms. Instead, the decrease in recombination observed in rad50Δ, rad52Δ and rad1Δ rad52Δ mutants suggests that the process depends on break repair by single-strand annealing11, a conclusion further supported by the decrease in FLO1 recombination in the rad59Δ mutant, which is known to be deficient in this type of DNA repair17. Moreover, in contrast to many other possible models, the proposed model also accounts for the expansion in the number of repeats found in some of the Ura− segregants. Taken together, the recombination frequencies observed in the various mutants indicate that recombination between the FLO1 repeats is caused by a replication slippage process similar to that observed in intergenic repeats (Supplementary Fig. 5 online).
Table 1. Frequency of recombination between intragenic repeats in selected DNA repair and recombination mutants.
The frequency of recombination in the intragenic repeats in FLO1 was determined by measuring the frequency of Ura− segregants (see Methods). The frequency measured in various DNA recombination and repair mutants was compared to that of an otherwise isogenic Rad+ strain (KV133). The recombination frequency in strain KV133 is 1 ×10−5 (see Fig. 3c). Values marked with an * indicate significant differences from the Rad+ control as measured by the student t-test at a 99% confidence level. All measurements were repeated at least three times.
| Relevant Genotype | Ratio compared to WT |
|---|---|
| FLO1::URA3 (WT) | 1 |
| FLO1::URA3 Δrad1 | 0.7 |
| FLO1::URA3 Δrad27 | 37* |
| FLO1::URA3 Δrad50 | 0.09* |
| FLO1::URA3 Δrad51 | 0.94 |
| FLO1::URA3 Δrad52 | 0.05* |
| FLO1::URA3 Δrad59 | 0.24* |
| FLO1::URA3 Δrad1 Δrad52 | 0.01* |
| FLO1::URA3 Δdnl4 | 0.9 |
| URA3 inserted at URA3 locus | < 0.03* |
Our data show that expansion and contraction of repeats results in gradual, quantitative and fully reversible functional changes that permit existing features of the organism to be rapidly attuned to a particular environment. The presence of repeats in the FLO adhesins, for example, enables Saccharomyces to adapt its adhesion behavior, finding an optimal balance between adherent cells and free cells that can escape from the mass and explore new surfaces. For pathogenic fungi like Candida albicans and Candida glabrata, such recombination events in their adhesin genes (ALS and EPA genes, respective homologs of FLO1) could enable the cells to adhere to novel host tissues. Variability at the cell surface of these pathogens may also permit evasion of the host immune system2. Interestingly, intragenic repeats are also present in cell-surface genes of non-fungal pathogens, including Haemophilus influenzae18, Bacillus anthracis19, Leishmania infantum20, and various Plasmodium species21. Hence, recombination of intragenic repeats may be a widespread mechanism among microorganisms to generate cell surface diversity from a single gene. This mechanism differs from that in Trypanosomes, where diversity arises from the expression of different, unlinked members of a large library of genes22.
In multicellular eukaryotes, repeat expansion and contraction may have significance for the generation of variability in genes other than those that function in the cell surface. For example, the rapid yet topologically conservative evolution of canine skeletal morphology has been attributed to the expansion and contraction of intragenic repeats within developmental genes23. In humans, the mucin (MUC) genes, which are homologues of the S. cerevisiae FLO genes, contain variable numbers of a 60 bp intragenic tandem repeat. Elevated expression of MUC genes induces tumorigenesis24 and is currently used as a marker for malignant tumors. Extensive size differences in MUC genes have been reported25, but the relationship of this variation to malignancy is not yet known.
METHODS
Bioinformatics
To find intragenic repeats, the EMBOSS ETANDEM software26 was used to screen the sequences of all S. cerevisiae ORFs. Two separate screens identified the short (3-39 nt) or long (>40 nt) repeats. ETANDEM threshold score was set at 20. This first screen identified 323 ORFs with long repeats and 859 ORFs with short repeats (see Supplementary Table 1 online). A secondary screen further refined the results of these two initial screens by excluding dubious ORFs and ORFs with poorly conserved (degenerated) repeats. Intragenic repeats were considered significant if three conditions were fulfilled: (1) the ORF was not a dubious or hypothetical ORF according to the Saccharomyces genome database (http://www.yeastgenome.org/); (2) repeat conservation was at least 85% and (3) the number of repeats was at least 20 for trinucleotide repeats, 16 copies for repeats between 4 and 10 nucleotides, 10 for repeats with a length between 11 and 39 nucleotides and 3 copies for repeats of at least 40 nucleotides.
Strains and Molecular Biology
All yeast strains used are listed in Supplementary Table 2 online. Yeast cultures were grown as described before27. YPGal medium contained 2% raffinose, 2% galactose (Sigma Chemical Co.), 2% peptone (Difco) and 1% yeast extract (Difco). Standard procedures and reagents for molecular biology were used. The URA3 marker was inserted into the intragenic repeats in FLO1 by transformation. A URA3 cassette was PCR amplified using primers containing 5′ tails with sequence homologous to the consensus repeated motif found in FLO1 and the plasmid pRS30628 as a template; these constructs were transformed into a Ura— recipient. Due to the similarity between the repeats, the construct integrated at various positions in the FLO1 repeats, thereby replacing a variable number of repeats. In some cases, insertion of URA3 led to an increase in repeat units. Real-time PCR using the ABI 7500 system (Applied Biosystems) was carried out with the appropriate enzymes and chemicals from Applied Biosystems as recommended by the supplier. All PCR primers are listed in Supplementary Table 3 online. CHEF chromosome separation was performed using a BioRad CHEF-DRII using the protocol supplied by BioRad. Flocculation and adhesion to polystyrene were tested as described previously6,29.
Recombination analysis
In order to measure the recombination frequency in the various FLO1::URA3 strains, single colonies growing on SD −Ura plates were inoculated in SD −Ura medium, and used to inoculate a 50 ml culture with an initial cell concentration of 1× 106 cells ml−1. This culture was shaken for 14 h at 28 °C, after which cells were harvested, washed with sterile distilled water and resuspended in water to a concentration of 5× 108 cells ml−1. This cell suspension was used to make a dilution series of which 150 microliter was plated onto SD plates containing 1 g l−1 5-fluoroorotic acid (5-FOA) to select for Ura− segregants. Since there is no growth on non-selective medium, frequencies measured by this method provide a good estimate of actual recombination rates (number of events per cell division). Loss of the URA3 marker was confirmed by PCR. All experiments were repeated at least 3 times and the average number of colonies was used to calculate the recombination frequency. Statistical significance was estimated using the student t-test.
GenBank Accession numbers
Short FLO1 alleles: AY949845-48; FLO1-YAR062W fusion genes: DQ029324-DQ029325.
Acknowledgments
The authors thank Anthony Sinskey, Angelica Amon and all Fink lab members for the valuable discussions, Kimberly Walker for assistance with the bioinformatics and Tom DiCesare for help with the graphics. K.J.V. is a post-doctoral fellow of the Fund for Scientific Research Flanders (F.W.O. Vlaanderen) and a D. Collen Fellow of the Belgian American Educational Foundation (B.A.E.F.). G.R.F. is an American Cancer Society Professor of Genetics. This research was supported by NIH grant 5RO1 GM35010 to G.R.F.
References
- 1.Hartl DL. Molecular melodies in high and low C. Nature Rev Genet. 2000;1:145–149. doi: 10.1038/35038580. [DOI] [PubMed] [Google Scholar]
- 2.Verstrepen KJ, Reynolds TB, Fink GR. Origins of variation in the fungal cell surface. Nat Rev Microbiol. 2004;2:533–540. doi: 10.1038/nrmicro927. [DOI] [PubMed] [Google Scholar]
- 3.Li YC, Korol AB, Fahima T, Nevo E. Microsatellites within genes: structure, function and evolution. Mol Biol Evol. 2004;21:991–1007. doi: 10.1093/molbev/msh073. [DOI] [PubMed] [Google Scholar]
- 4.Jin P, Alisch RS, Warren ST. RNA and microRNAs in fragile X mental retardation. Nat Cell Biol. 2004;6:1048–53. doi: 10.1038/ncb1104-1048. [DOI] [PubMed] [Google Scholar]
- 5.Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–54. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 6.Guo B, Styles CA, Feng Q, Fink G. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc Natl Acad Sci USA. 2000;97:12158–12163. doi: 10.1073/pnas.220420397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harrison P, et al. A small reservoir of disabled ORFs in the yeast genome and its implications for the dynamics of proteome evolution. J Mol Biol. 2002;316:409–419. doi: 10.1006/jmbi.2001.5343. [DOI] [PubMed] [Google Scholar]
- 8.Liu HP, Styles CA, Fink GR. Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth. Genetics. 1996;144:967–978. doi: 10.1093/genetics/144.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lovett ST. Encoded errors: mutations and rearrangements mediated by misalignment at repetitive DNA sequences. Mol Microbiol. 2004;52:1243–1253. doi: 10.1111/j.1365-2958.2004.04076.x. [DOI] [PubMed] [Google Scholar]
- 10.Viguera E, Canceill D, Ehrlich SD. Replication slippage involves DNA polymerase pausing and dissociation. EMBO J. 2001;20:2587–2595. doi: 10.1093/emboj/20.10.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richard GF, Dujon B, Haber JE. Double-strand break repair can lead to high frequencies of deletions within short CAG CTG trinucleotide repeats. Mol Gen Genet. 1999;261:871–882. doi: 10.1007/s004380050031. [DOI] [PubMed] [Google Scholar]
- 13.Kokoska RJ, et al. Destabilization of yeast micro- and minisatellite DNA sequences by mutations affecting a nuclease involved in Okazaki fragment processing (rad27) and DNA polymerase delta (pol3-t) Mol Cell Biol. 1998;18:2779–2788. doi: 10.1128/mcb.18.5.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Callahan JL, Andrews KJ, Zakian VA, Freudenreich CH. Mutations in yeast replication proteins that increase CAG/CTG expansions also increase repeat fragility. Mol Cell Biol. 2003;23:7849–7860. doi: 10.1128/MCB.23.21.7849-7860.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Debrauwère H, Loeillet S, Lin W, Nicolas A. Links between replication and recombination in Saccharomyces cerevisiae: a hypersensitive requirement for homologous recombination in the absence of Rad27 activity. Proc Natl Acad Sci USA. 2001;98:8263–8269. doi: 10.1073/pnas.121075598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krogh BO, Symington LS. Recombination proteins in yeast. Annu Rev Genet. 2004;38:233–71. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 17.Davis AP, Symington LS. The yeast recombinational repair protein Rad59 interacts with Rad52 and stimulates single-strand annealing. Genetics. 2001;159:515–525. doi: 10.1093/genetics/159.2.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hood DW, et al. DNA repeats identify novel virulence genes in Haemophilus influenzae. Proc Natl Acad Sci USA. 1996;93:11121–11125. doi: 10.1073/pnas.93.20.11121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keim P, et al. Molecular diversity in Bacillus anthracis. J Appl Microbiol. 1999;87:215–7. doi: 10.1046/j.1365-2672.1999.00873.x. [DOI] [PubMed] [Google Scholar]
- 20.Boceta C, Alonso C, Jimenez-Ruiz A. Leucine rich repeats are the main epitopes in Leishmania infantum PSA during canine and human visceral leishmaniasis. Parasite Immunol. 2000;22:55–62. doi: 10.1046/j.1365-3024.2000.00269.x. [DOI] [PubMed] [Google Scholar]
- 21.Sakihama N, et al. Relative frequencies of polymorphisms of variation in Block 2 repeats and 5′ recombinant types of Plasmodium falciparum msp1 alleles. Parasitol Int. 2004;53:59–67. doi: 10.1016/j.parint.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 22.Pays E, Vanhamme L, Berberof M. Genetic controls for the expression of surface antigens in African trypanosomes. Annu Rev Microbiol. 1994;48:25–52. doi: 10.1146/annurev.mi.48.100194.000325. [DOI] [PubMed] [Google Scholar]
- 23.Fondon JW, 3rd, Garner HR. Molecular origins of rapid and continuous morphological evolution. Proc Natl Acad Sci USA. 2004;101:18058–63. doi: 10.1073/pnas.0408118101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schroeder JA, et al. MUC1 overexpression results in mammary gland tumorigenesis and prolonged alveolar differentiation. Oncogene. 2004;23:5739–47. doi: 10.1038/sj.onc.1207713. [DOI] [PubMed] [Google Scholar]
- 25.Patton S, Gendler SJ, Spicer AP. The epithelial mucin, MUC1, of milk, mammary gland and other tissues. Biochim Biophys Acta. 1995;1241:407–23. doi: 10.1016/0304-4157(95)00014-3. [DOI] [PubMed] [Google Scholar]
- 26.Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 27.Sherman, F., Fink, G.R. & Hicks, J. Methods in yeast genetics, 263 (Cold Spring Harbour Laboratory Press, Cold Spring Harbour, NY, 1991).
- 28.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reynolds TB, Fink GR. Bakers’ yeast, a model for fungal biofilm formation. Science. 2001;291:878–881. doi: 10.1126/science.291.5505.878. [DOI] [PubMed] [Google Scholar]
- 30.Marinangeli P, Angelozii D, Ciani M, Clementi F, Mannazzu M. Minisatellites in Saccharomyces cerevisiae genes encoding cel wall proteins: a new way towards wine strain characterisation. FEMS Yeast Res. 2004;4:427–435. doi: 10.1016/S1567-1356(03)00172-7. [DOI] [PubMed] [Google Scholar]


