Abstract
The BRCA1 tumour suppressor and its heterodimeric partner BARD1 constitute an E3-ubiquitin (Ub) ligase and function in DNA repair by unknown mechanisms. We show here that the Caenorhabditis elegans BRCA1/BARD1 (CeBCD) complex possesses an E3-Ub ligase responsible for ubiquitylation at DNA damage sites following ionizing radiation (IR). The DNA damage checkpoint promotes the association of the CeBCD complex with E2-Ub conjugating enzyme, Ubc5(LET-70), leading to the formation of an active E3-Ub ligase on chromatin following IR. Correspondingly, defects in Ubc5(let-70) or the DNA damage checkpoint genes atl-1 or mre-11 abolish CeBCD-dependent ubiquitylation in vivo. Extending these findings to human cells reveals a requirement for UbcH5c, the MRN complex, γ-H2AX and a co-dependence for ATM and ATR kinases for BRCA1-dependent ubiquitylation at DNA damage sites. Furthermore, we demonstrate that the DNA damage checkpoint promotes the association between BRCA1 and UbcH5c to form an active E3-Ub ligase on chromatin after IR. These data reveal that BRCA1-dependent ubiquitylation is activated at sites of DNA repair by the checkpoint as part of a conserved DNA damage response.
Keywords: BRCA1, C. elegans, checkpoint, DNA damage, ubiquitylation
Introduction
The BRCA1 tumour suppressor gene and its structurally related partner, BARD1, form a heterodimeric complex that functions in a variety of cellular processes including transcriptional regulation, cell cycle progression and maintenance of X chromosome inactivation (Scully and Livingston, 2000; Ganesan et al, 2002). Several lines of evidence also strongly implicate BRCA1 and BARD1 in DNA repair (Kerr and Ashworth, 2001; Jasin, 2002; Venkitaraman, 2002). Mutant cell lines defective for BRCA1 or BARD1 exhibit genome instability, are exquisitely sensitive to DNA damaging agents and display defects in DNA double-strand break (DSB) repair by homologous recombination (Moynahan et al, 1999; Scully et al, 1999). In S-phase cells or following exposure to DNA damaging agents, BRCA1 and BARD1 form discrete nuclear foci at sites of DNA damage where they colocalize with other DNA repair proteins such as the DNA strand exchange protein, RAD51 (Scully et al, 1997b). BRCA1 is also phosphorylated during the cell cycle and following treatment with genotoxic agents by the DNA damage checkpoint kinases ATM and ATR (Tibbetts et al, 2000). Despite these observations, the role(s) of BRCA1 and BARD1 in DNA repair processes remains elusive.
BRCA1 and BARD1 both possess RING and BRCT domains, and BARD1 also contains ankyrin repeats. Recent studies suggest that the BRCT motifs may function as a phosphopeptide-binding domain that may be required for mediating protein–protein interactions with phospho-proteins (Manke et al, 2003; Yu et al, 2003). The N-terminal RING domains form a dimerization interface responsible for tight association of the two proteins (Brzovic et al, 2001b; Morris et al, 2002). This motif also confers E3-ubiquitin (Ub) ligase activity (Hashizume et al, 2001; Ruffner et al, 2001) raising the possibility that BRCA1/BARD1 heterodimer may specifically ubiquitylate proteins required for transcription, cell cycle and/or DNA repair.
Ubiquitylation is the stepwise process by which a target protein is modified by covalent attachment of mono-Ub or poly-Ub chains (Weissman, 2001). This process occurs in an ATP-dependent manner and is initiated by the formation of a thiol-ester bond between a cysteine residue in the Ub activating enzyme (E1) and the C-terminal glycine of Ub. The second step involves the transfer of Ub from the E1 enzyme to a conjugating enzyme (E2) with the formation of a new thio-ester bond. Finally, an E3-Ub ligase catalyzes the transfer of Ub from the E2 to a lysine residue in the target protein. Accumulating evidence from in vitro experiments suggest that the E3-Ub ligase activity of the BRCA1/BARD-1 heterodimer requires the E2 conjugating enzyme, UbcH5c, that has been shown to directly interact with the first and second Zn2+ loops and central α-helix of the BRCA1 RING (Brzovic et al, 2001b, 2003; Mallery et al, 2002; Dong et al, 2003). The biological importance of the RING domain is highlighted by the observation that a tumour-derived mutation in BRCA1 (C61G) abolishes E3 ligase activity in vitro (Brzovic et al, 2001a; Ruffner et al, 2001). Autoubiquitylation of BRCA1/BARD1 results in a 20-fold stimulation in E3 ligase activity in vitro, but how this activity is regulated in vivo is unknown (Mallery et al, 2002). It has also been shown that BRCA1/BARD1 autoubiquitylation occurs through K6 in Ub suggesting that this enzyme may conjugate Ub through less conventional linkages in vivo (Wu-Baer et al, 2003; Morris and Solomon, 2004). Although the BRCA1/BARD1 E3-Ub ligase is able to mono-ubiquitylate p53, H2AX, RNA polymerase II and the nucleosome core histones H2A, H2B, H3 and H4 in vitro, bona fide targets in vivo remain elusive (Mallery et al, 2002; Dong et al, 2003). Moreover, the functional relevance of BRCA1/BARD1 E3-Ub ligase activity during DNA repair processes is not known.
We have previously described Caenorhabditis elegans BRCA1 and BARD1 orthologues (Cebrc-1 and Cebrd-1, respectively) that possess many of the functional domains present in their human counterparts, including RING, ankyrin and BRCT domains (Boulton et al, 2004). Consistent with conserved roles in DNA repair, CeBRC-1 and CeBRD-1 interact to form a heterodimer via their respective RING domains and animals depleted for Cebrc-1 or Cebrd-1 by RNAi display p53-dependent germ cell death, radiation sensitivity and chromosome fragmentation (Boulton et al, 2004). To explore the mechanistic role of CeBRC-1 and CeBRD-1 in DNA repair processes, we have characterized a CeBRC-1/CeBRD-1 complex (CeBCD) purified by tandem immunoaffinity before and at different time points after IR-treatment. This approach is a first for C. elegans and demonstrates that protein complexes purified in this manner are amenable to biochemical analysis and can be used in combination with genetics and cell biology to accelerate functional discoveries. We present evidence that the CeBCD complex possesses an E3-Ub ligase that is activated on chromatin in response to IR-treatment, and further demonstrate that the DNA damage checkpoint promotes association of the CeBCD complex with E2-Ub conjugating enzyme, Ubc5(LET-70), to form an active E3-Ub ligase in response to DNA damage. We also show that ubiquitylation events at DNA damage sites require Cebrc-1, Cebrd-1, ubc5(let-70), mre-11 and atl-1, thus providing in vivo evidence to support our biochemical analysis. Finally, we extended our findings in C. elegans to human cells and show that BRCA1-dependent ubiquitylation at DNA damage sites is abolished by UbcH5c siRNA and in checkpoint mutant cells. Moreover, our studies reveal that BRCA1 and UbcH5c associate in a checkpoint-dependent manner to form an active E3-Ub ligase on chromatin after IR in human cells. These results define a conserved checkpoint-dependent mechanism in C. elegans and human cells responsible for activating BRCA1-dependent ubiquitylation at DNA damage sites.
Results
RAD-51 associates with the CeBCD complex after DNA damage
Functional orthologues of BRCA1 and BARD1 exist in C. elegans (Cebrc-1 and Cebrd-1, respectively) and share many functional similarities with their human counterparts (Boulton et al, 2004). To examine the role of CeBRC-1 and CeBRD-1 in DNA repair processes, we have biochemically characterized CeBRD-1 associated proteins (CeBCD complexes) purified by tandem immuno-affinity (Polanowska et al, 2004) (Figure 1A) from soluble and chromatin bound fractions (Supplementary Figure S1A and B), under normal growth conditions or at different time points after exposure to IR. This purification strategy is performed on whole animals at all developmental stages so has the potential to identify CeBRD-1 association proteins from all cell types that express CeBRD-1, including cycling cells in the mitotic compartment of the germline and during embryogenesis, and in cells at progressive stages of meiotic prophase (Supplementary Figure S3 and data not shown). Using this approach up to 20 discrete bands reproducibly co-purify with CeBRD-1 from soluble and chromatin bound fractions before IR-treatment with a number of additional bands found exclusively in the soluble or chromatin bound fractions (Figure 1B, lanes 2 and 4). The vast majority of these specific bands are absent from the mock purification from an untagged strain (Figure 1B, lane 1) (Polanowska et al, 2004). We have so far identified four previously uncharacterized proteins in the CeBCD complex by mass spectrometry that are the subject of ongoing work to determine their biological relevance to the CeBCD pathway. We have also identified six characterized proteins with established meiosis specific roles that have clear functional connections with CeBRC-1/CeBRD-1 during meiotic prophase that will be described elsewhere. The CeBCD complex also contains UBC-9 that was identified previously as a yeast-two hybrid interacting partner with CeBRD-1 (Boulton et al, 2004) (data not shown), Ubc5 (E2 conjugating enzyme) and the RAD-51 recombinase (see below).
Figure 1.
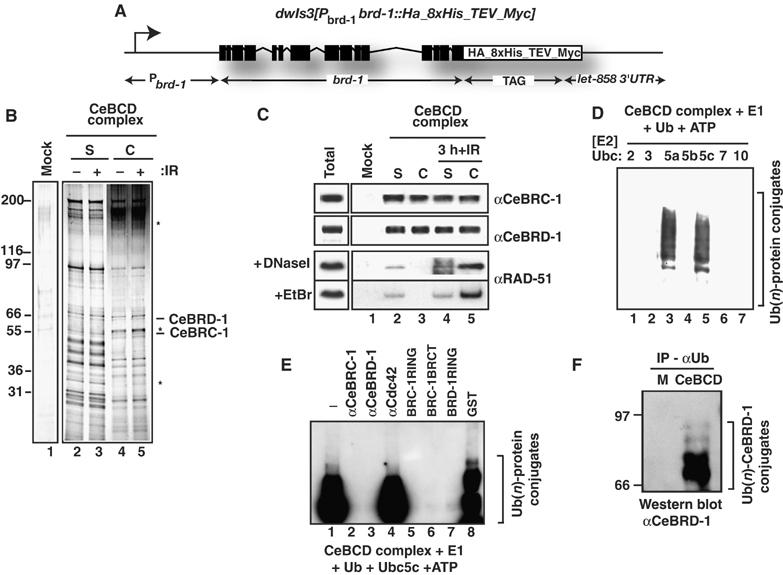
CeBCD complex contains CeBRC-1, CeBRD-1 and RAD-51 and possess E3-Ub ligase activity. (A) The CeBRD-1 transgene used to purify the CeBCD complex. (B) Silver stain of the mock purification and CeBCD complexes following tandem immunoaffinity purification. Complexes shown have been normalized for CeBRC-1 and CeBRD-1. (S) Soluble and (C) chromatin bound, before and after IR-treatment. Asterisks mark bands enriched in the CeBCD complex (C) after IR-treatment. (C) Western blots of the mock purification and CeBCD complexes for CeBRC-1 and CeBRD-1. CeBCD complexes purified from extracts treated with DNaseI or ethidium bromide (EtBr) and Western blotted for RAD-51. (D) The CeBCD complex possesses E3-Ub ligase activity when incubated with recombinant E1 activating enzyme, ATP, Ub and recombinant UbcH5a or UbcH5c, but not with recombinant UbcH2, 3, 7 or 10. Poly-Ub conjugates were resolved on 10% PAGE gels and subjected to western blotting with anti-Ub antibodies. (E) E3-Ub ligase activity of the CeBCD complex was performed as in (D) following incubation with antibodies to CeBRC-1, CeBRD-1 or Cdc42 (lanes 2–4) or GST-fusions of the CeBRC-1-RING, CeBRC-1-BRCT, CeBRD-1-RING or GST alone (lanes 5–8). Poly-Ub conjugates were resolved on 10% PAGE gels and subjected to Western blotting with anti-Ub antibodies. (F) Western blot with αCeBRD-1 antibodies of mock (M) and CeBCD complexes following immunoprecipitation with αUb antibodies to enrich for ubiquitylated proteins.
After IR-treatment with 75 Gy (an intermediate dose for C. elegans (Gartner et al, 2000; Boulton et al, 2002)), a number of bands are enriched in the CeBCD complex purified from the chromatin bound fraction (Figure 1B, lane 5) with no detectable changes in composition of the CeBCD complex purified from the soluble fraction (Figure 1B, lane 3). Western blotting reveals that CeBRC-1 and CeBRD-1 are present in soluble and chromatin bound CeBCD complexes both before and after IR-treatment (Figure 1C, lanes 2–5). In contrast, the RAD-51 recombinase is only detectable as a stable component of the chromatin bound CeBCD complex after IR-treatment (Figure 1C, lane 5) (Dong et al, 2003). Association of RAD-51 with the CeBCD complex is resistant to DNaseI and ethidium bromide treatment, suggesting that this interaction is direct and does not require bridging through DNA (Figure 1C). Taken together with previous observations that the human BRCC complex is also enriched for Rad51 after DNA damage, our findings suggest further functional similarities between C. elegans CeBCD and human BRCC complexes (Dong et al, 2003).
The CeBCD complex possess E3-Ub ligase activity
To determine if the CeBCD complex possesses an E3-Ub ligase activity (Hashizume et al, 2001; Ruffner et al, 2001; Mallery et al, 2002; Brzovic et al, 2003), the purified CeBCD complex was incubated with recombinant Ub activating enzyme Uba1 (E1), Ub, ATP and a panel of different recombinant E2 conjugating enzymes (see Materials and methods for details). Reactions containing the purified CeBCD complex supplemented with either UbcH5a or UbcH5c possess potent E3-Ub ligase activity (Figure 1D, lanes 3 and 5), whereas reactions containing UbcH2, 3, 5b, 7, 10, methyl-Ub, Ub mutated for all seven lysines or a mutant form of UbcH5c (C85A) defective for Ub-conjugation give no activity (Figure 1D, lane, 1, 2, 4, 6 and 7; Supplementary Figure S1C). Furthermore, the E3-Ub ligase activity of the CeBCD complex is inhibited by incubation with CeBRC-1 and CeBRD-1 antibodies that recognize their respective RING domains but not by Cdc42 antibodies (Figure 1E). Also, GST fusions to the CeBRC-1-RING, CeBRC-1-BRCT or the CeBRD-1-RING act as dominant negatives to inhibit E3-Ub ligase activity of the CeBCD complex whereas GST alone had no effect (Figure 1E). These data strongly suggest that the CeBRC-1/CeBRD-1 heterodimer is the E3-Ub ligase in the CeBCD complex and it exhibits the same specificity for the E2-Ub conjugating enzyme Ubc5(LET-70) (orthologue of human UbcH5c) as the human BRCA1/BARD1 heterodimer (Hashizume et al, 2001; Ruffner et al, 2001; Mallery et al, 2002; Brzovic et al, 2003; Dong et al, 2003). Moreover, we have found by mass-spectrometry and confirmed by Western blotting that CeBRD-1 is ubiquitylated in the CeBCD complex (Figure 1F), indicating that at least part of the ubiquitylation activity observed in the CeBCD complex corresponds to CeBRD-1 autoubiquitylation similar to that previously described for its human counterpart (Mallery et al, 2002; Brzovic et al, 2003).
E3-Ub ligase activity of the CeBCD complex is activated on chromatin after DNA damage
To determine if the CeBCD E3-Ub ligase activity is regulated in response to DNA damage, we compared E3-Ub ligase activity in CeBCD complexes purified from soluble or chromatin bound extracts at different time points after IR-treatment. Under normal growth conditions, the vast majority of the E3-Ub ligase activity is present in soluble CeBCD complexes (Figure 2A, lane 3), an activity that may correspond to CeBCD-dependent ubiquitylation events that occur during normal meiosis (see below). No detectable E3-Ub ligase activity is found associated with chromatin bound CeBCD complexes purified under normal growth conditions (Figure 2A, lane 9). In contrast, in response to IR-treatment, the E3-Ub ligase activity associated with soluble CeBCD complexes is rapidly abolished and remains inactive for at least 16 h post-IR treatment (Figure 2A, lanes 4–8). The mechanism by which this inhibition occurs is unknown and is the subject of ongoing investigation. Conversely, chromatin bound CeBCD complexes become activated as an E3-Ub ligase approximately 3 h after IR-treatment, and remain active for at least 16 h post-treatment (Figure 2A, lanes 10–14). These data suggest that chromatin bound CeBCD complexes are activated as an E3-Ub ligase in response to DNA damage.
Figure 2.
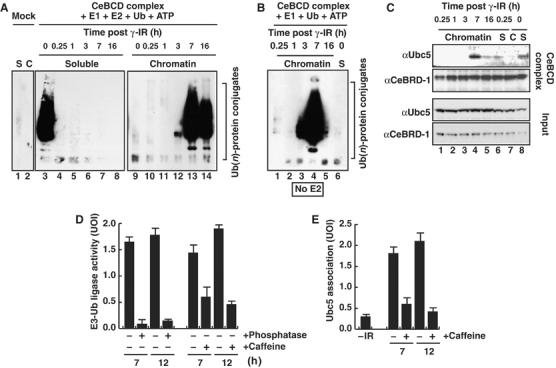
CeBCD E3-Ub ligase is activated on chromatin in response to IR through a checkpoint-dependent interaction with Ubc5(LET-70). (A) E3-Ub ligase activity of CeBCD complexes purified from (S) Soluble and (C) chromatin bound extracts at the indicated times after IR. Assays were performed as in Figure 1D. (B) E3-Ub ligase activity of chromatin bound CeBCD complexes without addition of recombinant Ubc5 (E2) as a cofactor. (C) CeBCD complexes normalized for CeBRD-1 and Western blotted for Ubc5(LET-70) and CeBRD-1. (D) E3-Ub ligase activity of chromatin bound CeBCD complexes normalized with respect to CeBRC-1 and CeBRD-1, without addition of recombinant Ubc5 as a cofactor, 7 and 12 h after 75 Gy IR, without and with alkaline phosphatase or caffeine treatment. (E) Ubc5(LET-70) associated with chromatin bound CeBCD complexes normalized with respect to CeBRC-1 and CeBRD-1, 7 and 12 h after IR, without and with caffeine treatment. UOI, units of intensity. The averages (columns) and standard error of mean (s.e.m.: bars) of three independent experiments are presented. E3-Ub ligase activity assays were performed as in Figure 1D.
Ubc5(LET-70) associates with chromatin bound CeBCD complexes after IR
To determine how chromatin bound CeBCD complexes are activated as an E3-Ub ligase following IR-treatment, we next analyzed CeBCD complexes for their E1 and E2 cofactor requirements in E3-Ub ligase assays in vitro. CeBCD complexes purified before and at all time points after IR-treatment show no E3-Ub ligase activity when recombinant E1 activating enzyme is omitted from the reaction (data not shown), indicating that endogenous C. elegans E1 activating enzyme is not a stable component of CeBCD complexes. Similarly, omitting recombinant UbcH5c (E2) from the reaction abolishes E3-Ub ligase activity in soluble CeBCD complexes purified under normal growth conditions (Figure 2B, lane 6). In contrast, chromatin bound CeBCD complexes purified following IR-treatment still possess potent E3-Ub ligase activity even when the recombinant UbcH5c (E2) is omitted from the reaction (Figure 2B, lanes 3–5). This result suggests that after DNA damage, chromatin bound CeBCD complexes are enriched for a factor that is able to compensate when recombinant UbcH5c (E2) is omitted from the reaction. Indeed, mass-spectrometry identified Ubc5(LET-70) as a component of the chromatin-bound CeBCD complexes, but only after DNA damage. This was confirmed by Western blotting with Ubc5(LET-70) antibodies that revealed the presence of endogenous Ubc5(LET-70) as a stable component of chromatin bound CeBCD complex only after IR-treatment (Figure 2C, lane 4). These data suggest that CeBCD complexes and Ubc5(LET-70) associate to form an active E3-Ub ligase on chromatin in response to IR.
Activation of E3-Ub ligase activity of the CeBCD complex following IR requires the DNA damage checkpoint
Human BRCA1 is phosphorylated by the checkpoint kinases ATM and ATR in response to DNA damage raising the intriguing possibility that activation of BRCA1-dependent ubiquitylation may depend on phosphorylation by the DNA damage checkpoint (Tibbetts et al, 2000). To determine if phosphorylation is important for activating the E3-Ub ligase of CeBCD complexes, we first treated purified complexes with alkaline phosphatase prior to performing the E3-Ub ligase assay in the presence of an excess of phosphatase inhibitors (see Supplementary data and Supplementary Figure S2A and B). Phosphatase treatment of the soluble and chromatin bound CeBCD complexes abolishes E3-Ub ligase activity (Figure 2D and Supplementary Figure S2A). Moreover, silver staining of CeBCD complexes following phosphatase treatment indicates that the majority of CeBCD complex components are unaffected by phosphatase treatment. However, the association of a subset of components is reduced or abolished following phosphatase treatment (Supplementary Figure S2C). Together, these results indicate that phosphorylation of the CeBCD complex is important for E3-Ub ligase activity, as well as for stable association of a subset of components with the complex.
Next, we treated animals with caffeine, an inhibitor of ATM and ATR kinase activity (Sarkaria et al, 1999). Caffeine treatment had no effect on the E3-Ub ligase activity of soluble CeBCD complexes indicating that ATM and ATR are dispensable for this activity (data not shown). However, chromatin bound CeBCD complexes purified after DNA damage exhibit severely reduced E3-Ub ligase activity after caffeine treatment (Figure 2D and Supplementary Figure S2D). Indeed, the DNA damage dependent association between chromatin bound CeBCD complexes and endogenous Ubc5(LET-70) is severely impaired following caffeine treatment (Figure 2E and Supplementary Figure S2E). These results raise the possibility that the DNA damage checkpoint is required for activation of the E3-Ub ligase of chromatin bound CeBCD complexes and may do so by regulating the association between the CeBCD complex and Ubc5(LET-70).
The CeBCD complex is required for ubiquitylation events at DNA damage sites in vivo
To further these studies in an in vivo setting, we took advantage of recent observations showing that ubiquitylation events at DNA damage sites in human cells can be visualized with an antibody to Ub (FK2 antibody) that has been extensively characterized and shown to recognize the conjugated Ub chains but not the free form of Ub (Morris and Solomon, 2004). Morris and Solomon demonstrated that ubiquitylation events at DNA damage sites are abolished by BRCA1 siRNA and by overexpression of a K6R-Ub mutant that prevents autoactivation of BRCA1 activity (Mallery et al, 2002; Morris and Solomon, 2004). Immunostaining of the C. elegans germline with the FK2 antibody reveals ubiquitylation events in the later stages of meiotic prophase under normal growth conditions (Supplementary Figure S3) that are rapidly abolished following IR-treatment with similar kinetics to that observed for the loss of E3-Ub ligase activity in soluble CeBCD complexes (Figure 2A; data not shown). It is therefore possible that these ubiquitylation events in late meiotic prophase correspond to the E3-Ub ligase activity in soluble CeBCD complexes purified under normal growth conditions. Although Cebrc-1 and Cebrd-1 mutants exhibit a meiotic phenotype (Boulton et al, 2004), the function of the ubiquitylation events in late meiosis I and the nature of the E3-Ub ligase inhibitory mechanism following DNA damage remains to be determined and is the subject of ongoing studies.
In nonirradiated mitotic cells within the C. elegans germline, a small number of dispersed cytoplasmic conjugated-Ub foci are detectable, with few if any concordant with chromatin (Figure 3B, panel 1). After IR-treatment, multiple conjugated-Ub foci are detectable at DNA damage sites in wild-type (N2) mitotic nuclei between 3 and 16 h after treatment (Figure 3B, panel 2; data not shown). In contrast, Cebrc-1 and Cebrd-1 mutants are defective for conjugated-Ub focus formation after IR (Figure 3B, panels 3 and 4), indicating that ubiquitylation at DNA damage sites in mitotic cells requires the action of a functional CeBCD complex, similar to that shown in human cells (Morris and Solomon, 2004). Thus, ubiquitylation at DNA damage sites is a conserved function of BRCA1 orthologues in C. elegans and human cells.
Figure 3.
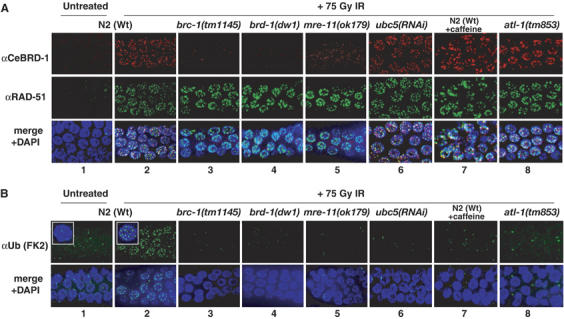
Conjugated Ub focus formation after IR is abolished in Cebrc-1, Cebrd-1, mre-11, atl-1 and Ubc5/let-70 mutants and after caffeine treatment. Representative images of fixed mitotic nuclei at the distal end of the germline stained for (A) CeBRD-1 and RAD-51, (B) conjugated-Ub (FK2) before (N2 wild type) and 3 h after 75 Gy IR-treatment for the indicated genotypes. brc-1(tm1145), brd-1(dw1), mre-11(ok179) and atl-1(tm853) are deletion mutants in C. elegans BRCA1, BARD1, MRE11 and ATR, respectively.
CeBCD-dependent ubiquitylation at DNA damage sites is abolished in ubc5(let-70), mre-11 and atl-1 mutants
To examine the genetic requirements for CeBCD-dependent ubiquitylation events at DNA damage sites, we next analyzed CeBRD-1, RAD-51 and conjugated-Ub focus formation in various mutant backgrounds following IR-treatment. Cebrc-1 mutants or a mutant in the mre-11 component of the MRN complex (Chin and Villeneuve, 2001) are severely impaired for IR-induced CeBRD-1 focus formation, but remain competent for RAD-51 focus formation after IR (Figure 3A, panels 2–5). Unlike Cebrc-1 mutants that lack any detectable CeBRD-1 staining due to protein instability (data not shown), the vast majority of CeBRD-1 protein detected in mre-11 mutants after IR treatment is defuse within nuclei indicative of a defect in recruitment of CeBRD-1 to DNA damage sites (Figure 3A, panel 5; data not shown). In contrast, RAD-51 and CeBRD-1 focus formation in response to IR-treatment is unaffected by caffeine treatment of wild-type (N2) animals, by a deletion mutant in the C. elegans orthologue of the ataxia-telangiectasia related (ATR) DNA damage checkpoint gene atl-l (Garcia-Muse and Boulton, 2005), or in animals subjected to RNAi depletion of Ubc5(let-70), the E2 conjugating enzyme that confers ubiquitylation activity to the CeBCD complex in vitro (Figure 3A, panels 6–8). These data indicate that efficient CeBRC-1/CeBRD-1 recruitment to DNA damage sites is dependent upon mre-11, but occurs independently of ubc5(let-70) and atl-1. Immunostaining with the FK2 antibody reveals that wild-type (N2) animals treated with caffeine, mre-11, atl-1 or ubc5(let-70) RNAi depleted animals are all compromised for ubiquitylation at DNA damage sites in vivo (Figure 3B, panels 5–8). Collectively, these results suggest that a failure to recruit the CeBCD complex to sites of DNA damage (mre-11 dependent) or an inability to activate the E3-Ub ligase of the CeBCD complex (ubc5(let-70) and atl-1 dependent) abolishes CeBCD-dependent ubiquitylation at DNA damage sites.
Collectively, these data suggest that the DNA damage checkpoint promotes association of the CeBCD complex and Ubc5(LET-70) to form an active E3-Ub ligase on chromatin following IR-treatment that is responsible for ubiquitylation events at DNA damage sites.
BRCA1 and UbcH5c interact in a checkpoint dependent manner to from an active E3-Ub ligase on chromatin in response to DNA damage
To determine if our findings in C. elegans are conserved in human cells, we first assessed whether BRCA1 and UbcH5c interact to from an active E3-Ub ligase on chromatin in response to DNA damage. Under normal growth conditions, BRCA1 immuno-precipitates from soluble and chromatin bound MCF7 cell extracts contain very low levels of UbcH5c (Figure 4A, lanes 1 and 3, respectively). In contrast, following IR-treatment, UbcH5c is significantly increased in BRCA1 immuno-precipitates from chromatin bound MCF7 extracts (Figure 4A, lane 4). The increased level of association between BRCA1 and UbcH5c is not seen in soluble extracts after IR (Figure 4A, lane 2) and is abolished in chromatin bound extracts following caffeine treatment (Figure 4A, lane 8). Correspondingly, BRCA1 immuno-precipitates from chromatin bound extracts following IR-treatment possess ubiquitylation activity when incubated with Ub, ATP and E1 activating enzyme (Figure 4B, lane 4). In contrast, ubiquitylation activity is not observed in BRCA1 immunoprecipitates incubated with Ub, ATP and E1 activating enzyme from soluble or chromatin bound extracts before IR treatment and from soluble or chromatin bound extracts treated with caffeine before or after IR (Figure 4B). This result indicates that the increased levels of endogenous UbcH5c present in BRCA1 immunoprecipitates from the chromatin fraction after IR-treatment is able to compensate for the absence of recombinant UbcH5c in the in vitro E3-Ub ligase assay. Finally, BRCA1 immuno-precipitates from MCF7 cells treated with caffeine or from Nbs1−/−, A-T or ATR-Seckel cells do not exhibit the increased levels of UbcH5c after IR treatment seen in MCF7 cells (Figure 4C and D). Collectively, these data indicate that the DNA damage checkpoint somehow promotes association between BRCA1 and UbcH5c to form an active E3-Ub ligase on chromatin following DNA damage, similar to our observation in C. elegans.
Figure 4.
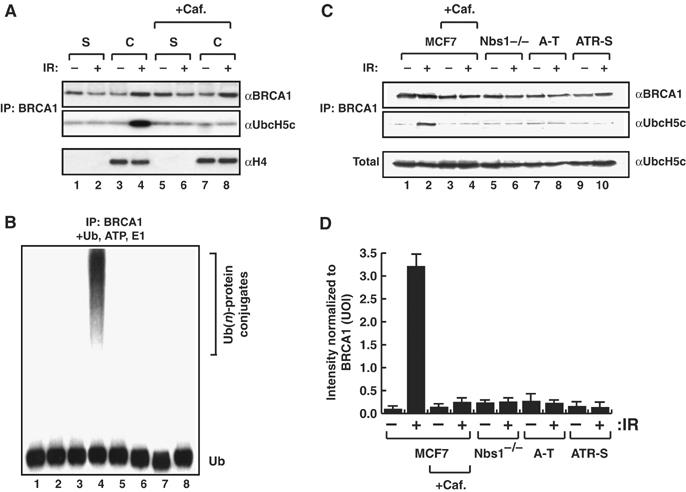
Human BRCA1 and UbcH5c associate in a checkpoint dependent manner to form an active E3-Ub ligase on chromatin. (A) UbcH5c and BRCA1 immunoblots of BRCA1 immunoprecipitates from MCF7 soluble (S) or chromatin bound (C) extracts before and 7 h after IR-treatment±caffeine treatment. The bottom panel is an immunoblot of the inputs with Histone H4 (chromatin marker) antibodies. (B) E3-Ub ligase assay performed on BRCA1 immunoprecipitates described in (A) in the presence of Ub, ATP and recombinant E1 activating enzyme (note: recombinant UbcH5c is not added to these reactions). Poly-Ub conjugates were resolved on 10% PAGE gels and subjected to western blotting with anti-Ub antibodies. The Ub-protein conjugates in lane 4 likely reflect autoubiquitylation of BRCA1. (C) UbcH5c and BRCA1 immunoblots of BRCA1 immunoprecipitates from MCF7 whole-cell extracts before and 7 h after IR-treatment±caffeine treatment, and from Nbs1−/−, A-T and ATR-Seckel whole-cell extracts before and 7 h after IR-treatment. Total is UbcH5c immunoblot of whole-cell extracts for each time point and genotype. (D) Quantification of UbcH5c association in BRCA1 immunoprecipitates shown in (C) normalized with respect to BRCA1. The averages (columns) and s.e.m. (bars) of two independent experiments are shown.
BRCA1-dependent ubiquitylation at DNA damage sites is abolished by BRCA1 and UbcH5c siRNA, and in ATLD1/D, NBS1−/−, A-T and ATR-Seckel cell lines
To examine the genetic requirements for BRCA1-dependent ubiquitylation at DNA damage sites in human cells, we next correlated γ-H2AX, an early marker of DNA breaks (Fernandez-Capetillo et al, 2004), with BRCA1-mediated ubiquitylation events (Morris and Solomon, 2004) after IR-treatment in MCF7 cells and in cell lines deficient for Atm, Atr, DNA-PKcs, Mre11, Nbs1, Chk2 and γ-H2AX (reviewed in Bradbury and Jackson, 2003; Shiloh, 2003; Kastan and Bartek, 2004). Representative images of γ-H2AX and conjugated-Ub staining (Figure 5A) and quantification of conjugated-Ub focus formation (Figure 5B) are shown. In untreated MCF7 cells, γ-H2AX and conjugated-Ub foci are detectable at very low levels (Figure 5A, panel 1 and B). In contrast, multiple γ-H2AX and conjugated-Ub foci are detectable at sites of DNA damage following IR-treatment (Figure 5A, panel 2 and B) (Fernandez-Capetillo et al, 2004). Consistent with previous observations and our findings in C. elegans, BRCA1 siRNA (Figure 5C) abolishes IR-induced conjugated-Ub foci, but has no effect on the accumulation of γ-H2AX foci (Figure 5A, panel 3 and B) (Morris and Solomon, 2004). Furthermore, siRNA to UbcH5c, but not to UbcH2 or Ube2K, abrogates BRCA1-dependent ubiquitylation at DNA damage sites, thus confirming UbcH5c as the bona fide E2-conjugating enzyme for BRCA1 in vivo (Figure 5A, panel 4 and B and Supplementary Figure S4). Also, MCF7 cells treated with caffeine (Sarkaria et al, 1999), as well as ATLD1/2, A-T, ATR-Seckel and NBS1−/− cell lines (deficient for Mre11, ATM, ATR and Nbs1, respectively) accumulate γ-H2AX but are defective for conjugated-Ub focus formation after IR (Figure 5A, panels 5, 7, 8, 9 and 11, and B). Similar to our findings in C. elegans, these results indicate that a functional checkpoint is required for BRCA1-dependent ubiquitylation events at DNA damage sites in human cells.
Figure 5.
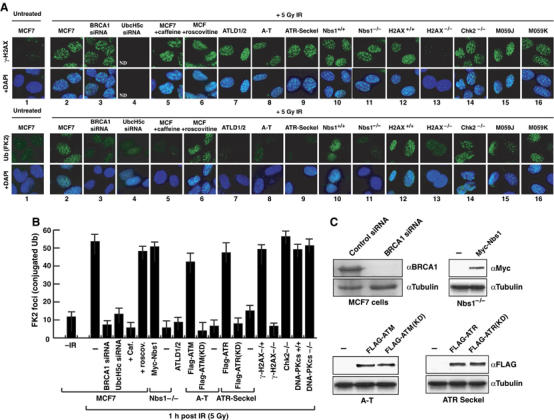
Genetic requirements for conjugated Ub focus formation after IR in mammalian cells. (A) Representative images of γ-H2AX, conjugated-Ub (FK2)±DAPI staining of DNA 1 h after 5 Gy of IR-treatment in cells of the indicated genotypes. ATLD1/2 cells are defective for Mre11; A-T cells are defective for Atm; ATR-Seckel cells carry a hypomorphic mutation in Atr; M059J (DNA-PKcs−/−), M059K (DNA-PKcs+/+). MCF7 cells were treated with 5 mM caffeine or 10 μM roscovitine for 6 h prior to IR-treatment. (B) Quantification of conjugated-Ub (FK2) focus formation 1 h post-treatment with 5 Gy of IR in cells of the indicated genotype. NBS1−/−, A-T and ATR-Seckel cells were complemented with the indicated plasmids: KD corresponds to kinase dead versions of ATM and ATR. The number of conjugated-Ub (FK2) foci was counted in at least 25 cells of each genotype. The averages (columns) and s.e.m. (bars) of three independent experiments are presented. (C) BRCA1 and Tubulin western blots of MCF7 extracts following control or BRCA1 siRNA. Western blots for Myc (Nbs1), Flag (ATM, ATM(KD), ATR, ATR(KD) and tubulin following correction of NBS1−/−, A-T and ATR-Seckel cell lines with the indicated plasmids. KD=kinase dead.
BRCA1-dependent ubiquitylation at DNA damage sites requires ATM and ATR kinase activity and γ-H2AX
To further explore the role of the DNA damage checkpoint in the activation of BRCA1-dependent ubiquitylation at DNA damage sites, we corrected Nbs1−/−, A-T and ATR-Seckel cell lines by expression of wild-type Myc-Nbs1, Flag-ATM and Flag-ATR, respectively (Figure 5C). Correction of Nbs1−/−, A-T and ATR-Seckel cell lines with their respective wild-type cDNA restores BRCA1-dependent ubiquitylation events (Figure 5B). However, expression of kinase dead versions of ATM and ATR fails to restore this response in A-T and ATR-Seckel cells (Figure 5B). These results indicate that the kinase activities of both ATM and ATR are required for BRCA1-dependent ubiquitylation events at DNA damage sites. Consistent with previous observations that γ-H2AX phosphorylation by ATM/ATR is required for retention of BRCA1 at DNA damage sites (Celeste et al, 2003), H2AX−/− cells that lack γ-H2AX foci are compromised for conjugated-Ub focus formation after IR (Figure 5A, panel 13 and B). Recent studies have shown that BARD1 is also a substrate for phosphorylation by cyclin-dependent kinases (Hayami et al, 2005). However, MCF7 cells treated with roscovitine, a potent and selective inhibitor of cyclin-dependent kinases (Mailand and Diffley, 2005), has no effect on BRCA1-dependent ubiquitylation events after IR (Figure 5A, panel 6 and B). BRCA1-dependent ubiquitylation at DNA damage sites is also unaffected in NBS+/+, H2AX+/+, Chk2−/−, DNA-PKcs−/−(M059J) and DNA-PKcs+/+(M059K) cells as shown by the accumulation of both γ-H2AX and conjugated-Ub foci after IR (Figure 5A, panels 10, 12, 14, 15 and 16 and Figure 5B). Collectively, these results reveal that the DNA damage checkpoint, including the MRN complex, the kinase activities of ATM and ATR, and γ-H2AX, are required for BRCA1-dependent ubiquitylation at DNA damage sites. However, DNA-PKcs, a critical component of the non-homologous end-joining pathway, Chk2, an effector kinase downstream of ATM, and cyclin-dependent kinases are dispensable for this response.
Discussion
Our previous report revealed the intriguing possibility that BRCA1 and BARD1 homologs exist in C. elegans (Boulton et al, 2004). This notion was based on extensive sequence and domain conservation between the C. elegans and human proteins, the formation of a RING-dependent heterodimer between CeBRC-1 and CeBRD-1, and DNA repair defects observed in worms RNAi depleted for Cebrc-1 or Cebrd-1 (Boulton et al, 2004). In this study, we have purified the first native protein complex from C. elegans amenable to extensive biochemical analyses. Our studies of CeBCD complexes have revealed extensive functional similarities with the human BRCC complex, as well as providing new insight into the function and regulation of BRCA1. First, we have shown that CeBCD complexes contain RAD-51, but only after DNA damage, similar to the human BRCC complex (Dong et al, 2003). Second, CeBCD complexes possess E3-Ub ligase activity and exhibit the same specificity for the E2-conjugating enzyme Ubc5/LET-70 (human UbcH5c) as reported for its human counterpart (Brzovic et al, 2001a; Ruffner et al, 2001; Mallery et al, 2002; Dong et al, 2003). Third, CeBRD-1 is subjected to autoubiquitylation, similar to its human counterpart (Mallery et al, 2002). Fourth, like BRCA1/BARD1 in human cells, CeBRC-1/CeBRD-1 are recruited to sites of DNA damage (Scully et al, 1997a). Fifth, Cebrc-1 or Cebrd-1 mutants are defective for ubiquitylation events at DNA damage sites in vivo, consistent with recent studies in human cells (Morris and Solomon, 2004). Finally, we have also shown for the first time that Ubc5/UbcH5c is the bona fide E2-conjugating enzyme for CeBCD/BRCA1 in C. elegans and in human cells, respectively, and present evidence that the DNA damage checkpoint promotes association between CeBCD/BRCA1 and Ubc5/UbcH5c leading to activation of BRCA1-dependent ubiquitylation events at DNA damage sites.
Morris and Solomon (2004) previously demonstrated that BRCA1 is responsible for ubiquitylation events at DNA damage sites in human cells. Our findings that Cebrc-1 and Cebrd-1 are required for ubiquitylation events at DNA damage sites in C. elegans substantiates the findings of Morris and Solomon and demonstrates that this response to DNA damage is conserved in a simple metazoan. A number of reports have shown that the human BRCA1/BARD1 proteins preferentially use UbcH5c as the E2-Ub conjugating enzyme in E3-Ub ligase reactions in vitro (Brzovic et al, 2001a, 2003; Ruffner et al, 2001; Mallery et al, 2002; Dong et al, 2003). Studies have also shown that UbcH5c interacts with the first and second Zn2+ loops and central α-helix of the BRCA1 RING (Brzovic et al, 2003). However, the importance of UbcH5c in BRCA1-dependent ubiquitylation reactions in vivo has not been explored. We present two lines of evidence that implicate Ubc5(LET-70)/UbcH5c as the E2-conjugating enzyme that functions with CeBCD/BRCA1 at DNA damage sites in vivo. First, we show in C. elegans and in human cells that CeBCD/BRCA1 and Ubc5(LET-70)/UbcH5c co-immunoprecipitate from chromatin following IR-treatment, respectively. Second, we find that RNAi depletion of Ubc5(let-70) in C. elegans and siRNA to UbcH5c in human cells abolishes ubiquitylation events at DNA damage sites. It has not been possible to assess whether Ubc5/let-70 and UbcH5c depletion confer DNA damage sensitivity similar to CeBCD/BRCA1 mutants as both Ubc5(let-70) and UbcH5c function as the E2-Ub conjugating enzyme in other cellular processes and are essential (Frazier et al, 2004).
It was not clear from previous studies how BRCA1-dependent ubiquitylation events are regulated and activated in cells. The data presented here provide new insight into the regulation of BRCA1-dependent ubiquitylation events at DNA damage sites. We have shown first in C. elegans and then in human cells that chromatin bound CeBCD/BRCA1 complexes associate with Ubc5(LET-70)/UbcH5c to form an active E3-Ub ligase in response to DNA damage. The fact that this association was blocked by phosphatase or caffeine treatment raised the possibility that checkpoint dependent phosphorylation could be required for CeBCD/BRCA1 complexes to associate with Ubc5(LET-70)/UbcH5c, in C. elegans and human cells, respectively, Indeed, caffeine treatment was also found to significantly reduce E3-Ub ligase activity in chromatin bound CeBCD/BRCA1 complexes after IR-treatment. The importance of the DNA damage checkpoint in activating CeBCD/BRCA1 dependent ubiquitylation was confirmed by our findings that (1) atl-1 (C. elegans ATR), mre-11 mutants and caffeine treated wild-type (N2) animals are compromised for CeBCD-dependent ubiquitylation at DNA damage sites; (2) Nbs1−/−, A-T and ATR-Seckel cells fail to exhibit increased association between BRCA1 and UbcH5c on chromatin, after IR-treatment; and (3) caffeine treated MCF7 cells, as well as ATLD1/2, Nbs1−/−, A-T, ATR-Seckel and H2AX−/− cell lines are defective for BRCA1-dependent ubiquitylation at DNA damage sites. Together, these data indicate that the checkpoint promotes association between BRCA1 and UbcH5c leading to the formation of an active E3-Ub ligase on chromatin after IR.
It is perhaps surprising that BRCA1-dependent ubiquitylation events have a co-requirement for both ATM and ATR checkpoint kinases, as it was conventionally thought that these kinases respond primarily to DSBs and replication stress, respectively. However, recent work in C. elegans and mammalian cells has revealed that checkpoint responses to DSBs require both ATM and ATR kinases (Garcia-Muse and Boulton, 2005; Jazayeri et al, 2006). Thus, BRCA1-dependent ubiquitylation is one of the first DNA damage responses reported to have a co-dependence on ATM and ATR. In future studies, it will be important to determine how checkpoint-dependent phosphorylation regulates association between Ubc5/UbcH5c and CeBCD/BRCA1, respectively. It is unlikely that that phosphorylation is a determinant of the interaction surface between BRCA1 and UbcHc5, as recombinant BRCA1 and UbcH5c associate in vitro to form an active E3-Ub ligase in the absence of any detectable phosphorylation and BRCA1 and UbcH5 can interact in the yeast-two hybrid in the absence of DNA damage (Brzovic et al, 2003). Rather, checkpoint dependent phosphorylation is more likely to regulate the co-recruitment or retention of BRCA1 and UbcH5c as an active complex at DNA damage sites. Our observation that H2AX−/− cells are defective for BRCA1-dependent ubiquitylation supports this possibility as previous work has demonstrated a role for γ-H2AX in retaining BRCA1 at DNA damage sites (Celeste et al, 2003; Nakamura et al, 2004). It is also plausible that the DNA damage checkpoint may target/mark a substrate protein for CeBCD/BRCA1 dependent ubiquitylation at DNA damage sites through phosphorylation.
In summary, these results reveal that CeBCD/BRCA1-dependent ubiquitylation is a conserved response to DNA damage that is activated by the checkpoint (Figure 6). Our data implicate Ubc5(LET-70)/UbcH5c, MRE-11/MRN complex, ATM, ATL-1/ATR and H2AX in the activation of CeBCD/BRCA1-dependent ubiquitylation at DNA damage sites. Checkpoint activation following IR-treatment promotes the association between CeBCD/BRCA1 and its E2-Ub conjugating enzyme Ubc5(LET-70)/UbcH5c on chromatin. This triggers the formation of an active E3 Ub-ligase complex that is responsible for ubiquitylation events at sites of DNA damage (Figure 6). It is important to note that the timing of CeBCD/BRCA1-dependent ubiquitylation at DNA damage sites is relatively late compared with initial checkpoint activation and γH2AX phosphorylation, suggesting that this response may play a late role in DNA repair processes and may occur at complex DNA lesions that take a longer time to repair. While convincing evidence for in vivo substrates for the E3-Ub ligase of CeBCD/BRCA1 remains elusive, our data and those of Morris and Solomon strongly suggest that bona fide substrates reside at sites of DNA damage and may therefore function in DNA repair and/or regulation of chromatin dynamics.
Figure 6.
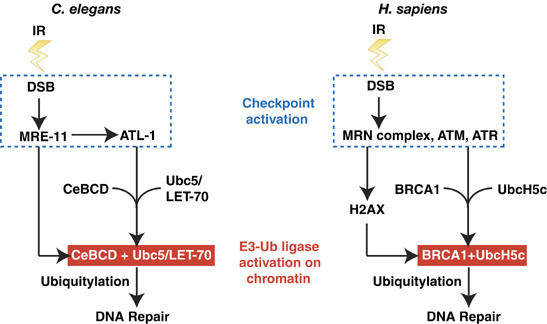
A conserved DNA damage-signalling pathway required for activation of BRCA1-dependent ubiquitylation during DNA repair. Following IR-induced damage, checkpoint activation in C. elegans and human cells promotes the association of CeBCD/BRCA1 with Ubc5/UbcH5c, respectively, and results in E3-Ub ligase activation. Once active, the E3-Ub ligase is responsible for ubiquitylation events at DNA damage sites. The sensitivity of Cebrc-1/BRCA1 mutants to DNA damaging agents suggest that their role in ubiquitylation at DNA damage sites may be required for efficient DNA repair.
Materials and methods
Worm strains
C. elegans strains were cultured as described previously(Brenner, 1974). For caffeine experiments, C. elegans were cultured in standard growth media supplemented with 20 mM caffeine for at least 6 h prior to purification of the CeBCD complex. The following strains were kindly provided by the Caenorhabditis Genetics Centre (University of Minnesota, St Paul, MN), including wild-type Bristol N2, mre-11(ok179)(Chin and Villeneuve, 2001), rad-51 (lg08701)(Alpi et al, 2003). brd-1(dw1) will be described in detail elsewhere. brc-1(tm1145) and atl-1(tm853) were generated and kindly provided by Shoehi Mitani of the National Bioresource Project for the nematode, Japan.
Cell culture
Human HeLa ohio cells (ATCC), human breast carcinoma MCF7 cells (ATCC), human fibroblasts F02-98 (ATR-Seckel), M059J (DNA-PKcs−/−), M059K (DNA-PKcs+/+), GM02052 (ATM−/−) and GM15989 (NBS1−/−), human colon tumour cells Chk2−/− and mouse embryonic fibroblast H2AX+/+ and H2AX−/− were maintained in minimum essential media (MEM) supplemented with 20% fetal calf serum, L-glutamine and penicillin–streptomycin (100 μg/ml and 100 U/ml, respectively). ATLD1/2 cells carry a truncating mutation in Mre11 and were grown in MEM with 15% fetal calf serum, L-glutamine and penicillin–streptomycin. Cells were kindly provided by Penny Jeggo (ATR-Seckel, ATLD1/2), Andre Nussenzweig (H2AX+/+ and H2AX−/−) Fred Bunz and Bert Vogelstein (Chk2−/−) and Ted DeWeese (ATM−/−, M059J and M059K). All cell lines were maintained at 37°C in a 5% CO2/95% air incubator. BRCA1, UbcH5c, UbcH2 and Ube2K siRNA (Dharmacon) was performed using Dharmafect transfection reagent and analyzed for FK2 focus formation following IR-treatment 48 h post-transfection. Caffeine and roscovitine were purchased from Sigma and Calbiochem, respectively. Plasmids for correcting NBS1−/− (Myc-Nbs1), A-T (Flag-ATM and Flag-ATM-KD) and ATR-Seckel (Flag-ATR and Flag-ATR-KD) were kindly provided by Matt Weitzman, Yossi Shiloh, Jacob Falck and Steve Jackson. Transiently transfected cells and stably selected clones gave similar results when assessed for correction of BRCA1-dependent ubiquitylation.
In vitro E3-Ub ligase assay
To measure E3-Ub ligase activity, purified CeBCD complexes were incubated at 30°C for 60 min in 15 μl of assay buffer containing 0.1 μg Uba1 E1, 0.04 μg UbcH5c E2, 5 μg Ub, 50 mM Tris–HCl pH 8.0, 0.2 mM CaCl2, 5 mM MgCl2, 1 mM DTT, 4 mM ATP. The reaction was resolved on SDS–PAGE and transferred onto nitrocellulose membrane. After preincubation in denaturating buffer (6 M guanidine–HCl, 20 mM Tris–HCl pH7.5, 5 mM β-mercaptoethanol, 1 mM PMSF for 30 min 4°C) and extensive washing in PBS, the activity was revealed by Western blotting with mouse anti-Ub antibody (1:300, Zymed Laboratories). Reactions were performed in the presence of 0.04 μg of recombinant E2 enzymes: UbcH2, UbcH3, UbcH5a, UbcH5b, Ubch5c, UbcH6, UbcH7, UbcH10 (Boston Biohem). Reactions in Figure 1E were performed following a 30 min incubation of the CeBCD complex with 0.04 μg of CeBRC-1, CeBRD-1 or Cdc42 antibodies or with recombinant GST-BRC-1-RING (1–150 aa), GST-BRC-1-BRCT (350–547 aa), GST-BRD-1-RING (1–150aa) or GST alone. C. elegans Ubc5(LET-70) was detected with rabbit anti-Ubc5 antibody (Boston Biochem).
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Methods
Acknowledgments
We thank Shohei Mitani (Bioresource Project for the nematode) for kindly providing brc-1(tm1145); Penny Jeggo, Andre Nussenzweig, Fred Bunz, Bert Vogelstein and Ted DeWeese for kindly providing cell lines; Matt Weitzman, Yossi Shiloh, Jacob Falck and Steve Jackson for plasmids; Nicola O'Reilly for peptide synthesis, Ruth Peat and Rachel Horton-Harpin for cell culture. Thanks to Jo Morris for helpful discussion and to Juliet Reid, Phil Zegerman, Jesper Svejstrup, Helle Ulrich and John Diffley for comments on the manuscript. This work was funded by Breast Cancer Campaign (GA3221) and Cancer Research UK.
References
- Alpi A, Pasierbek P, Gartner A, Loidl J (2003) Genetic and cytological characterization of the recombination protein RAD-51 in Caenorhabditis elegans. Chromosoma 112: 6–16 [DOI] [PubMed] [Google Scholar]
- Boulton SJ, Gartner A, Reboul J, Vaglio P, Dyson N, Hill DE, Vidal M (2002) Combined functional genomic maps of the C. elegans DNA damage response. Science 295: 127–131 [DOI] [PubMed] [Google Scholar]
- Boulton SJ, Martin JS, Polanowska J, Hill DE, Gartner A, Vidal M (2004) BRCA1/BARD1 orthologs required for DNA repair in Caenorhabditis elegans. Curr Biol 14: 33–39 [DOI] [PubMed] [Google Scholar]
- Bradbury JM, Jackson SP (2003) ATM and ATR. Curr Biol 13: R468. [DOI] [PubMed] [Google Scholar]
- Brenner S (1974) The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzovic PS, Keeffe JR, Nishikawa H, Miyamoto K, Fox D III, Fukuda M, Ohta T, Klevit R (2003) Binding and recognition in the assembly of an active BRCA1/BARD1 ubiquitin–ligase complex. Proc Natl Acad Sci USA 100: 5646–5651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzovic PS, Meza JE, King MC, Klevit RE (2001a) BRCA1 RING domain cancer-predisposing mutations. Structural consequences and effects on protein–protein interactions. J Biol Chem 276: 41399–41406 [DOI] [PubMed] [Google Scholar]
- Brzovic PS, Rajagopal P, Hoyt DW, King MC, Klevit RE (2001b) Structure of a BRCA1-BARD1 heterodimeric RING–RING complex. Nat Struct Biol 8: 833–837 [DOI] [PubMed] [Google Scholar]
- Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A (2003) Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol 5: 675–679 [DOI] [PubMed] [Google Scholar]
- Chin GM, Villeneuve AM (2001) C. elegans mre-11 is required for meiotic recombination and DNA repair but is dispensable for the meiotic G(2) DNA damage checkpoint. Genes Dev 15: 522–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Hakimi MA, Chen X, Kumaraswamy E, Cooch NS, Godwin AK, Shiekhattar R (2003) Regulation of BRCC, a holoenzyme complex containing BRCA1 and BRCA2, by a signalosome-like subunit and its role in DNA repair. Mol Cell 12: 1087–1099 [DOI] [PubMed] [Google Scholar]
- Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A (2004) H2AX: the histone guardian of the genome. DNA Repair (Amst) 3: 959–967 [DOI] [PubMed] [Google Scholar]
- Frazier T, Shakes D, Hota U, Boyd L (2004) Caenorhabditis elegans UBC-2 functions with the anaphase-promoting complex but also has other activities. J Cell Sci 117: 5427–5435 [DOI] [PubMed] [Google Scholar]
- Ganesan S, Silver DP, Greenberg RA, Avni D, Drapkin R, Miron A, Mok SC, Randrianarison V, Brodie S, Salstrom J, Rasmussen TP, Klimke A, Marrese C, Marahrens Y, Deng CX, Feunteun J, Livingston DM (2002) BRCA1 supports XIST RNA concentration on the inactive X chromosome. Cell 111: 393–405 [DOI] [PubMed] [Google Scholar]
- Garcia-Muse T, Boulton SJ (2005) Distinct modes of ATR activation after replication stress and DNA double-strand breaks in Caenorhabditis elegans. EMBO J 24: 4345–4355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner A, Milstein S, Ahmed S, Hodgkin J, Hengartner MO (2000) A conserved checkpoint pathway mediates DNA damage-induced apoptosis and cell cycle arrest in C. elegans. Mol Cell 5: 435–443 [DOI] [PubMed] [Google Scholar]
- Hashizume R, Fukuda M, Maeda I, Nishikawa H, Oyake D, Yabuki Y, Ogata H, Ohta T (2001) The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J Biol Chem 276: 14537–14540 [DOI] [PubMed] [Google Scholar]
- Hayami R, Sato K, Wu W, Nishikawa T, Hiroi J, Ohtani-Kaneko R, Fukuda M, Ohta T (2005) Down-regulation of BRCA1-BARD1 ubiquitin ligase by CDK2. Cancer Res 65: 6–10 [PubMed] [Google Scholar]
- Jasin M (2002) Homologous repair of DNA damage and tumorigenesis: the BRCA connection. Oncogene 21: 8981–8993 [DOI] [PubMed] [Google Scholar]
- Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP (2006) ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol 8: 37–45 [DOI] [PubMed] [Google Scholar]
- Kastan MB, Bartek J (2004) Cell-cycle checkpoints and cancer. Nature 432: 316–323 [DOI] [PubMed] [Google Scholar]
- Kerr P, Ashworth A (2001) New complexities for BRCA1 and BRCA2. Curr Biol 11: R668–R676 [DOI] [PubMed] [Google Scholar]
- Mailand N, Diffley JF (2005) CDKs promote DNA replication origin licensing in human cells by protecting Cdc6 from APC/C-dependent proteolysis. Cell 122: 915–926 [DOI] [PubMed] [Google Scholar]
- Mallery DL, Vandenberg CJ, Hiom K (2002) Activation of the E3 ligase function of the BRCA1/BARD1 complex by polyubiquitin chains. EMBO J 21: 6755–6762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manke IA, Lowery DM, Nguyen A, Yaffe MB (2003) BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science 302: 636–639 [DOI] [PubMed] [Google Scholar]
- Morris JR, Keep NH, Solomon E (2002) Identification of residues required for the interaction of BARD1 with BRCA1. J Biol Chem 277: 9382–9386 [DOI] [PubMed] [Google Scholar]
- Morris JR, Solomon E (2004) BRCA1: BARD1 induces the formation of conjugated ubiquitin structures, dependent on K6 of ubiquitin, in cells during DNA replication and repair. Hum Mol Genet 13: 807–817 [DOI] [PubMed] [Google Scholar]
- Moynahan ME, Chiu JW, Koller BH, Jasin M (1999) Brca1 controls homology-directed DNA repair. Mol Cell 4: 511–518 [DOI] [PubMed] [Google Scholar]
- Nakamura TM, Du LL, Redon C, Russell P (2004) Histone H2A phosphorylation controls Crb2 recruitment at DNA breaks, maintains checkpoint arrest, and influences DNA repair in fission yeast. Mol Cell Biol 24: 6215–6230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polanowska J, Martin JS, Fisher R, Scopa T, Rae I, Boulton SJ (2004) Tandem immunoaffinity purification of protein complexes from Caenorhabditis elegans. Biotechniques 36: 778–780, 782 [DOI] [PubMed] [Google Scholar]
- Ruffner H, Joazeiro CA, Hemmati D, Hunter T, Verma IM (2001) Cancer-predisposing mutations within the RING domain of BRCA1: loss of ubiquitin protein ligase activity and protection from radiation hypersensitivity. Proc Natl Acad Sci USA 98: 5134–5139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkaria JN, Busby EC, Tibbetts RS, Roos P, Taya Y, Karnitz LM, Abraham RT (1999) Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res 59: 4375–4382 [PubMed] [Google Scholar]
- Scully R, Chen J, Ochs RL, Keegan K, Hoekstra M, Feunteun J, Livingston DM (1997a) Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell 90: 425–435 [DOI] [PubMed] [Google Scholar]
- Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T, Livingston DM (1997b) Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell 88: 265–275 [DOI] [PubMed] [Google Scholar]
- Scully R, Ganesan S, Vlasakova K, Chen J, Socolovsky M, Livingston DM (1999) Genetic analysis of BRCA1 function in a defined tumor cell line. Mol Cell 4: 1093–1099 [DOI] [PubMed] [Google Scholar]
- Scully R, Livingston DM (2000) In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature 408: 429–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh Y (2003) ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer 3: 155–168 [DOI] [PubMed] [Google Scholar]
- Tibbetts RS, Cortez D, Brumbaugh KM, Scully R, Livingston D, Elledge SJ, Abraham RT (2000) Functional interactions between BRCA1 and the checkpoint kinase ATR during genotoxic stress. Genes Dev 14: 2989–3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkitaraman AR (2002) Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell 108: 171–182 [DOI] [PubMed] [Google Scholar]
- Weissman AM (2001) Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol 2: 169–178 [DOI] [PubMed] [Google Scholar]
- Wu-Baer F, Lagrazon K, Yuan W, Baer R (2003) The BRCA1/BARD1 heterodimer assembles polyubiquitin chains through an unconventional linkage involving lysine residue K6 of ubiquitin. J Biol Chem 278: 34743–34746 [DOI] [PubMed] [Google Scholar]
- Yu X, Chini CC, He M, Mer G, Chen J (2003) The BRCT domain is a phospho-protein binding domain. Science 302: 639–642 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Methods


