Abstract
Eg5 or KSP is a homotetrameric Kinesin-5 involved in centrosome separation and assembly of the bipolar mitotic spindle. Analytical gel filtration of purified protein and cryo-electron microscopy (cryo-EM) of unidirectional shadowed microtubule–Eg5 complexes have been used to identify the stable dimer Eg5-513. The motility assays show that Eg5-513 promotes robust plus-end-directed microtubule gliding at a rate similar to that of homotetrameric Eg5 in vitro. Eg5-513 exhibits slow ATP turnover, high affinity for ATP, and a weakened affinity for microtubules when compared to monomeric Eg5. We show here that the Eg5-513 dimer binds microtubules with both heads to two adjacent tubulin heterodimers along the same microtubule protofilament. Under all nucleotide conditions tested, there were no visible structural changes in the monomeric Eg5–microtubule complexes with monastrol treatment. In contrast, there was a substantial monastrol effect on dimeric Eg5-513, which reduced microtubule lattice decoration. Comparisons between the X-ray structures of Eg5-ADP and Eg5-ADP-monastrol with rat kinesin-ADP after docking them into cryo-EM 3-D scaffolds revealed structural evidence for the weaker microtubule–Eg5 interaction in the presence of monastrol.
Keywords: cryo-electron microscopy, KSP, microtubule, mitosis, molecular motor
Introduction
Molecular motors are prominent participants in mitosis and not only provide the force necessary to assemble the spindle, but they also strike a delicate balance of forces to generate the synchronized set of movements necessary for chromosome segregation (reviewed by Wittmann et al, 2001; McIntosh et al, 2002; Kwon and Scholey, 2004). One of the mitotic motors, Eg5 or KSP, is a member of the homotetrameric BimC Kinesin-5 subfamily. These N-terminal kinesins are responsible for providing a plus-end-directed force associated with sliding microtubules (MTs) during centrosome separation to assemble the bipolar spindle and for prevention of bipolar spindle collapse before anaphase (Saunders and Hoyt, 1992; Sawin et al, 1992; Cole et al, 1994; Blangy et al, 1995; Kashina et al, 1996; Gordon and Roof, 1999; Mayer et al, 1999; Mountain et al, 1999; Sharp et al, 1999; Kapoor et al, 2000; Goshima and Vale, 2003; Kwok et al, 2004).
Of the Kinesin-5 members, human Eg5 is of particular interest because of its potential as a therapeutic target for cancer treatment. A number of small molecule inhibitors have been identified that can specifically and reversibly inhibit Eg5 function leading to monoaster formation and arrested cell division (Mayer et al, 1999; Duhl and Renhowe, 2005). The best known of these inhibitors, monastrol, has been shown to slow ADP release, significantly alter Eg5 interactions with the MT, and alter the conformation of the switch II region and neck linker of the catalytic core (Maliga et al, 2002, 2006; Cochran et al, 2004, 2005; Crevel et al, 2004; Yan et al, 2004). However, for these initial experiments, a monomeric Eg5 was used. To understand the mechanochemical properties of Eg5 for its mitotic function and the mechanism of inhibition by the monastrol class of small molecule inhibitors, it is critical to define the cooperative interactions that occur between the two motor domains that interact on a single MT and to define the pathway of communication between the catalytic active site and the motor–MT interface.
The study presented here analyzes monomeric and dimeric Eg5 motors comprising the N-terminal 367 (Eg5-367) and 513 (Eg5-513) amino acids of the human gene sequence expressed in Escherichia coli. Analytical gel filtration and cryo-electron microscopy (cryo-EM) in combination with high-resolution surface shadowing (e.g. see Hoenger et al, 2000) indicate that Eg5-513 forms a stable dimer, and the MT gliding assays indicate that the motility promoted by the dimer is robust and comparable to homotetrameric Eg5. Steady-state kinetics, equilibrium binding, and cryo-EM 3-D analyses have been used to compare the monomeric and dimeric motors in the presence and absence of monastrol. The results indicate that in dimeric Eg5-513, the cooperative head–head interactions slow ATP turnover, tighten ATP binding, and weaken binding to MTs. While monomeric Eg5 can bind to MTs in the presence of monastrol, the cryo-EM studies show that monastrol disrupts dimeric Eg5-513 interactions with the MT at every nucleotide condition. Comparisons between X-ray structures of Eg5 head domains in the presence (Yan et al, 2004) and absence of monastrol (Turner et al, 2001) and docking of these structures into EM-derived 3-D maps of Eg5–microtubule complexes suggest that monastrol induces a conformational change into the switch II region that modifies in particular the conformation of helix α4 at the Eg5 motor–MT interface and the position of the neck linker. We propose a model for dimeric Eg5 in which monastrol stabilizes a conformation with the neck linker of each motor domain locked onto its catalytic core, resulting in dissociation of the MT·Eg5 complex or an inhibition of MT binding.
Results
Eg5-513 is dimeric
A new Eg5 truncation was generated that extends from the start methionine until the initial break in the coiled-coil as predicted by PAIRCOIL (Berger et al, 1995). Eg5-513, containing the N-terminal 513 amino acids, was compared to dimeric conventional kinesin K401 (Gilbert and Johnson, 1993; Correia et al, 1995) and monomeric Eg5-367 (Turner et al, 2001; Maliga et al, 2002; Cochran et al, 2004; Yan et al, 2004). As shown in Figure 1B, Eg5-513 and Eg5-513-5His elute from the gel filtration column at similar retention times, preceding dimeric K401 and monomeric Eg5-367. A plot of Kav versus log Mr yielded an apparent molecular mass of 182 000 for K401, 189 688 for Eg5-513, and 181 036 for the Eg5-513-5His. Additional evidence of oligomeric state was provided by the unidirectional shadowing of both monomeric Eg5-367 and the prospective dimeric Eg5-513 on MTs and tubulin sheets (Figure 2F–I). Using a substoichiometric 1:8 ratio of Eg5-367 to tubulin, individual motor domains were visible and spaced stochastically along the MT lattice (Figure 2F and G). The same ratio of Eg5-513 to tubulin also produced a random distribution of motors (Figure 2H and I); however, the density of one motor domain was predominantly accompanied by a companion (see red and yellow dots in Figure 2I). In most cases, the second motor head was located in the axial direction 8 nm apart to the next tubulin dimer on the same protofilament, which is indicative of the physical attachment expected for a dimeric motor. A few isolated motor domains were also seen (Figure 2I, marked in blue), but it is not apparent whether they represent the overlay of two motor domains or are monomeric motors.
Figure 1.
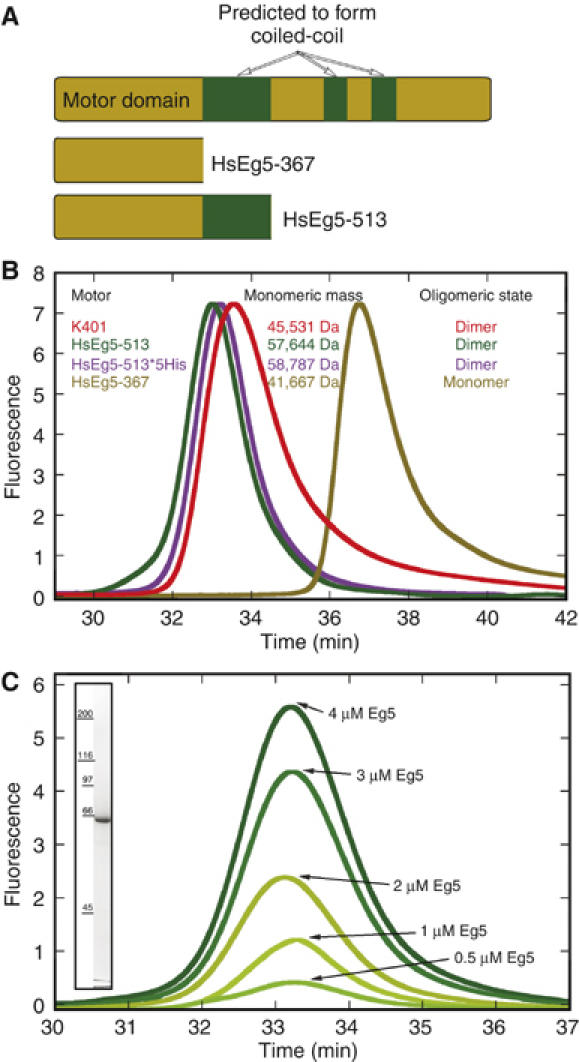
Eg5 motor proteins. (A) The domain structure of the human gene as predicted by PAIR-COIL with Eg5-367 and Eg5-513. (B) Analytical gel filtration was carried out to compare Eg5-513, Eg5-513-5His, dimeric Kinesin-1 K401, and monomeric Eg5-367. The void volume eluted at 16 min and the included volume at 51.5 min. (C) Gel filtration of Eg5-513 at the protein concentration range used for the reported experiments indicates that the proposed dimeric state of Eg5-513 remained stable over the range of protein concentrations used in this study (0.5–4 μM). The inset shows an SDS–Coomassie blue stained gel of Eg5-513.
Figure 2.

Shadowgraphs of freeze-dried MTs decorated with Eg5 motors. Unidirectional shadowing as shown here reveals specifically the surface features of MTs in the absence of Eg5 (A) and MTs complexed with monomeric Eg5 (Eg5-367) and dimeric Eg5 (Eg5-513), either attempting full saturation (B–E) or undersaturation (F–I). Full saturation was achieved with Eg5-367 in the presence of MgAMPPNP (B) and MgAMPPNP+S-monastrol (C). Dimeric Eg5 allows complete surface decoration in the presence of MgAMPPNP (D) but not in the presence of S-monastrol+MgAMPPNP (E) owing to a strikingly reduced affinity of motors to MTs. The repetitive surface pattern in panel D indicates that Eg5-513 dimers bind with both heads to MTs and do not leave one head unbound and tethered (e.g. as seen with Ncd). At subsaturating conditions (F, G), monomeric Eg5-367 binds randomly along the MT lattice, indicated by the red dots in (G) to mark the motor head position. (H, I) Eg5-513 also shows stochastic binding, but here the motors are clearly visible as double-densities that mostly align along protofilaments. The Eg5-513 head positions are marked in red with potential dimers as double-red dots connected by a yellow marker (I).
Eg5-513 promotes robust MT gliding at a rate comparable to homotetrameric Eg5
Preformed MT·Eg5-513-5His complexes were flowed into a microscopy perfusion chamber, followed by the addition of MgATP to initiate MT motility. As expected for a plus-end-directed motor, polarity marked MTs were observed to move with their brightly labeled minus-ends leading when the Eg5 motor was present (Figure 3 and Supplementary Movie). MT gliding was not observed in the absence of either Eg5 or MgATP. The average rate at which Eg5-513-5His promoted MT gliding was 2.83±0.06 μm/min (47±1 nm/s) with a range of 1.6–5.2 μm/min (27–87 nm/s).
Figure 3.
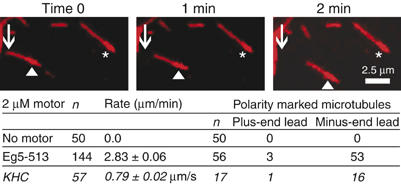
Eg5-513-5His promotes plus-end-directed MT gliding in the presence of 1.5 mM MgATP. Polarity-marked MTs exhibit a more highly fluorescent MT minus-end. ▴ indicates a MT that is changing its position over time in comparison to the stationary MT denoted by ★. Conventional Kinesin-1 (KHC) was used to assess the polarity of the MTs. See Supplementary data for a movie.
Eg5-513 exhibits slow steady-state ATP turnover
The steady-state ATPase kinetics for Eg5-513 (Figure 4) were pursued as a function of either ATP concentration or MT concentration in the presence or absence of 150 μM monastrol. In comparison to monomeric Eg5-367, Eg5-513 was 10-fold slower (kcat=0.48 s−1 versus 5.5 s−1 for Eg5-367), exhibited a three-fold tighter affinity for ATP (Km,ATP=7.9 versus 25 μM for Eg5-367), yet ∼3-fold weaker affinity for MTs (K1/2,MT=1.8 versus 0.7 μM for Eg5-367) (Table I). The Eg5-513 steady-state kcat is significantly lower than would be predicted based on the motility assays. At 2.83 μm/min or 47 nm/s with an assumed 8 nm step, the kcat is predicted to be ∼6 s−1, a rate constant more similar to the steady-state kcat of monomeric Eg5-367. However, the motility assays quantify the Eg5 motors only when bound and stepping along the MT. In contrast, the solution kinetics include the steps of motor detachment from the MT and its rebinding, both of which contribute to the overall rate for steady-state ATP turnover. Furthermore, the slower ATPase kinetics observed for the dimer in comparison to the monomer are suggestive of cooperative interactions between the motor domains of the dimer to slow the overall rate of the reaction. This behavior has been well documented for Kinesin-1, Myosin-V, and Myosin-VI (Moyer et al, 1996, 1998; Rosenfeld et al, 1996; De La Cruz et al, 2001; Sakamoto et al, 2003; Cross, 2004).
Figure 4.
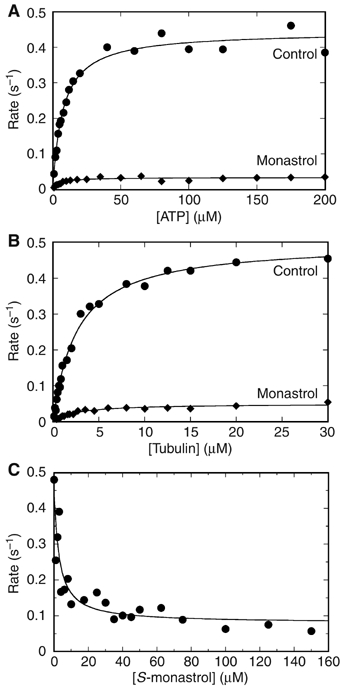
Steady-state ATPase kinetics and monastrol inhibition. A preformed MT·Eg5 (•) or MT·Eg5-monastrol (⧫) complex was rapidly mixed with [α-32P]ATP. (A) Final concentrations: 0.5 μM Eg5-513 motor domain, 20 μM tubulin, 20 μM Taxol, 150 μM S-monastrol, and 1–200 μM MgATP. The fit of the data to the Michaelis–Menten equation provided the steady-state parameters: kcat=0.44±0.01 s−1, Km,ATP=7.5±0.7 μM and kcat-Mon=0.034±0.001 s−1, Km,ATP-Mon=4.5±0.9 μM. (B) Final concentrations: 1 μM Eg5-513 motor domains, 0.1–40 μM tubulin, 20 μM Taxol, 150 μM S-monastrol, and 400 μM MgATP. The data were fit to equation (1): kcat=0.48±0.01 s−1, K1/2,MT=1.8±0.1 μM and kcat-Mon=0.048±0.002 s−1, K1/2,MT -Mon=1.7±0.3 μM. (C) Final concentrations: 0.5 μM Eg5-513 motor domain, 20 μM tubulin, 20 μM Taxol, 150 μM MgATP, and 0–150 μM S-monastrol. The data were fit to equation (2): Kd-Mon=2.88±0.04 μM.
Table 1.
Comparison of Eg5-513 with Eg5-367
| Kinetic parameter | Eg5-513 | Eg5-513 monastrol | Eg5-367a | Eg5-367b monastrol |
|---|---|---|---|---|
| Steady-state kcat | 0.48±0.02 s−1, n=17 | 0.046±0.006 s−1, n=6 | 5.5±0.3 s−1 | 1.2±0.03 s−1 |
| Km,ATP | 7.9±2.4 μM, n=10 | 4.5±0.06 μM, n=3 | 25.1±0.7 μM | 3.6±0.3 μM |
| K1/2,MT | 1.8±0.2 μM, n=7 | 1.94±0.2 μM, n=3 | 0.71±0.15 μM | 6.7±0.4 μM |
| Kd-Mon | 4.8±1.3 μM, n=3 | 13.8±1.0 μM | ||
| aCochran et al (2004). | ||||
| bCochran et al (2005). | ||||
In the presence of monastrol, the steady-state kcat of Eg5-513 was reduced 10-fold to 0.046 s−1 with the affinity for S-monastrol at Kd-Mon=4.8 μM (Figure 4C and Table I). Interestingly, the apparent Kd-Mon determined for dimeric Eg5-513 was at least two-fold tighter than that reported for monomeric Eg5-367 (Maliga et al, 2002; Cochran et al, 2005).
Monastrol disrupts the MT binding properties of dimeric Eg5-513 but not monomeric Eg5-367
Structural studies on MT–motor interactions were performed with cryo-EM and helical 3-D analysis. Owing to symmetry constraints, we used 15-protofilament MTs because this configuration allows for helical 3-D reconstructions. Figure 5 summarizes the results obtained by cryo-EM. Columns from left to right show for each condition a raw-data micrograph of an MT·Eg5 complex, a Fourier-filtered 2-D projection of an averaged data set, and the corresponding 2-D diffraction pattern with the helical layer lines running vertically in accordance with the orientation of the images on their left. The right column shows graphs that are generated by projecting the layer lines along their elongation. This way we receive a summing (total power) of each layer line to better compare the individual power of the 1/8-nm layer line cluster relative to the 1/4-nm layer line cluster. Plain MTs composed of 15 protofilaments generate a cluster of strong layer lines only at 1/4 nm, reflecting the first order of the α-β-α-β tubulin 4-nm monomer repeat, convoluted with the protofilament supertwist, typical for 15-protofilament MTs (explained by Beuron and Hoenger, 2001). Owing to the strong structural similarities of α- and β-tubulin, plain MTs lack a strong 8-nm heterodimer repeat. However, the 1/8 layer line cluster becomes very strong once MTs are complexed with kinesin motor head domains. The binding of one motor head per each αβ-tubulin dimer strongly enhances the visibility of the repetitive 8-nm pattern along the protofilaments. Under ideal conditions, the motors bind stoichiometrically and with low mobility or flexibility in the interface. The states here that came close to such ideal conditions were decoration with monomeric Eg5-367 in the presence of AMPPNP (Figures 2B and 5B) and nucleotide-free (Figure 5A), and with dimeric Eg5-513 in the presence of AMPPNP (Figures 2D and 5E). The full saturation and low-mobility configuration is reflected in the corresponding ratios of total layer line powers as found in these graphs (Figure 5). All other conditions tested revealed different ratios. Eg5-367 in the presence of monastrol reveals a slightly reduced 1/8 nm power. Comparing this result with that of surface shadowing indicates that at the conditions used here, this motor decorates stoichiometrically (see Figure 2C), but the lower power suggests a higher degree of mobility and/or a higher on–off rate that averages out the total mass of motor domains on the MT surface after helical averaging due to variable binding geometries. This effect is also visible in axial sections through helical 3-D reconstructions as shown in Figure 6. Comparison of MT·Eg5-367 complexes in Figure 6A (nucleotide-free) with Figure 6B (nucleotide-free+monastrol) or Figure 6C (AMPPNP) with Figure 6D (AMPPNP+monastrol) reveals a shift in the concentration of motor mass toward the center of each head when monastrol was added (Figure 6B and D) whereas the density in the outer peripheries was reduced.
Figure 5.
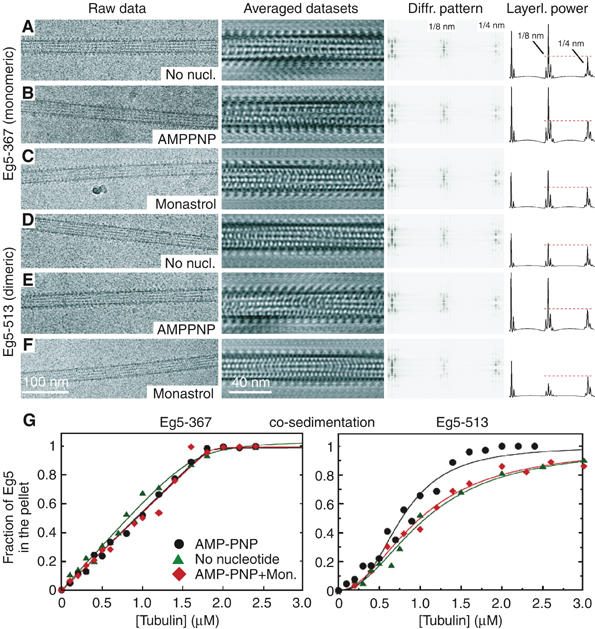
Eg5 motor binding to MTs. Cryo-EM analysis of monomeric MT·Eg5-367 complexes (A–C) and dimeric MT·Eg5-513 complexes (D–F) are shown in different nucleotide states. Diffraction patterns were taken from the averaged data sets. The graphs on the right display the projected total power of each layer line. The leftmost peak is not the equator, but represents the first layer line marking the protofilament supertwist. The 1/4 nm cluster (marked in panel A) relates mainly to the tubulin monomer repeat along the MT axis, whereas 1/8 nm marks the repeat of the αβ-tubulin dimer or a tubulin dimer complexed with one motor head domain. Eg5-367 binds regularly along the MTs in the absence of nucleotides (A), in the presence of MgAMPPNP (B), and in the presence of MgAMPPNP+S-monastrol (C). Dimeric Eg5-513 in the presence of MgAMPPNP also reveals a strong MT affinity (E) that is, however, significantly reduced in the absence of nucleotides (D). AMPPNP+S-monastrol show an even further reduced affinity (F, also see Figure 2E). MT·Eg5 cosedimentation (G) has been performed to compare Eg5-367 (left) and Eg5-513 (right). The MT binding patterns were not significantly different for monomeric Eg5-367 with MgAMPPNP, apyrase-induced nucleotide-free state, or AMPPNP+S-monastrol. In contrast, dimeric Eg5-513 exhibited a sigmoidal MT binding behavior. The AMPPNP state was more tightly bound (KHill=0.49±0.06 μM3, n=2.8±0.3) than the nucleotide-free state (KHill=0.96±0.07 μM2, n=1.7±0.1). Monastrol treatment with AMPPNP (KHill=1.0±0.05 μM2, n=2.0±0.1) resulted in a binding pattern more similar to the apyrase-induced nucleotide-free state.
Figure 6.
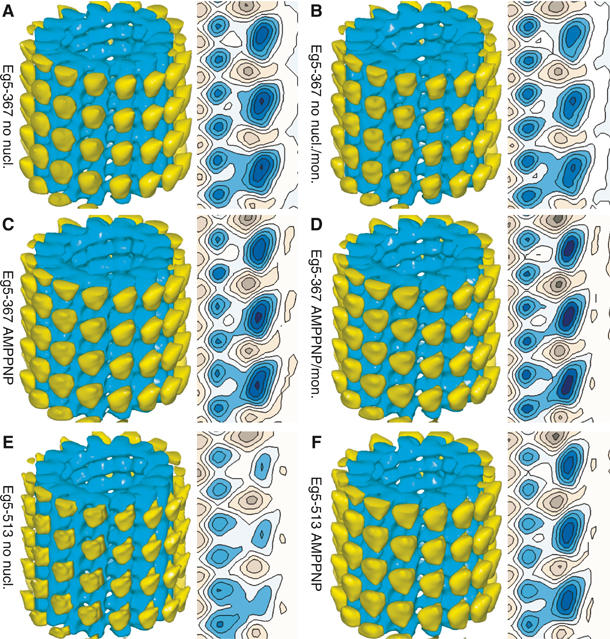
3-D analysis of MT·Eg5 complexes. Here, we show surface-rendered 3-D volumes and a selected axial cross-section of helically reconstructed MTs decorated with monomeric (A–D) and dimeric (E, F) Eg5 under various nucleotide conditions. The MTs are oriented with the plus-end at the top of the reconstruction (tubulin: blue; motors: yellow). The maps are normalized to the MT volume. In contrast to Kinesin-1 (Skiniotis et al, 2003), the structural differences for Eg5 according to nucleotide state and/or presence of S-monastrol were not resolved, and all maps obtained with monomeric Eg5-367 appear identical. Reconstructions of MT·Eg5-513 complexes treated with apyrase (nucleotide-free; (E)) show reduced density in the head portion, indicating incomplete surface decoration due to a lower MT affinity of dimeric motors under these conditions. In the presence of MgAMPPNP (F), the density of the head portion is identical to monomers, whereas addition of monastrol abolishes binding almost completely. Hence, no meaningful 3-D map could be calculated. The head shape on reconstructions with dimeric motor constructs (E, F) reveals an average over trailing and leading heads (see Figure 7), but the close similarity to monomer reconstructions confirms the findings with monomers that these heads do not change configuration upon changes in nucleotide conditions. There was no obvious density in the reconstruction with the dimeric Eg5 that could be attributed to the second head being tethered but detached from the MT.
In contrast to Eg5 monomers, dimeric Eg5-513 reacts very differently to nucleotide-free conditions (Figure 5D) and decorates the MT surface less efficiently. In the presence of monastrol, nucleotide conditions do not seem to matter anymore, and in all cases MT binding is significantly reduced (AMPPNP+monastrol; Figure 5F). In summary, stoichiometric decoration with dimeric Eg5 could only be achieved with AMPPNP in the absence of monastrol (Figure 5E), and monastrol seems to abolish MT decoration of Eg5-513 almost entirely (Figures 2E and 5F).
In the presence of ADP, motor affinity for MTs was very low for both monomers and dimers, and we could not generate sufficiently decorated MT·Eg5 complexes required for a meaningful helical 3-D analysis (data not shown). The equilibrium binding analysis also reflected the weak affinity of Eg5-513 for MTs in the ADP state (Supplementary Figure S1G and H). Note that only 45% of Eg5-513 motors sedimented with MTs in the presence of 2 mM MgADP. With the addition of monastrol+MgADP, this percentage decreased further to 19%.
Cosedimentation agrees well with the cryo-EM results
The pattern observed in the cryo-EM analysis was consistent with the results of the cosedimentation assays. For these experiments, 2 μM Eg5 was incubated with varying concentrations of MTs plus nucleotide or apyrase, allowed to come to equilibrium, and then centrifuged. For each reaction, equal volumes of the supernatant and pellet were analyzed by SDS–PAGE to determine the fraction of Eg5 motor that partitioned with the supernatant or the MT pellet. Sedimentation of monomeric Eg5-367 in the presence of AMPPNP, in the nucleotide-free state achieved with apyrase treatment, and in the presence of AMPPNP+monastrol revealed similar results (Figure 5G, left). Cosedimentation of Eg5-513 with MTs produced two important findings: first, monastrol altered the binding performance of Eg5-513, and second, the data were sigmoid (Figure 5G, right). In the presence of AMPPNP, dimeric Eg5-513 exhibits the tightest binding. However, when the MT·Eg5-513 complex was treated with AMPPNP+monastrol, the data became more similar to the weaker, nucleotide-free binding curve. These results for dimeric Eg5-513 are consistent with the observations in which monastrol treatment results in Eg5 motors that can remain attached to MTs, yet the motors have lost the ability to generate force (Mayer et al, 1999; Kapoor et al, 2000; Crevel et al, 2004).
Discussion
Monastrol induces an increase in monomeric Eg5 head domain mobility
At the resolution achieved in our cryo-EM 3-D reconstructions, monomeric Eg5-367 motor domains bind to MTs with a very similar binding geometry at both the nucleotide-free and the AMPPNP states with and without monastrol (compare Figure 6A and B, and Figure 6C and 6D). However, there are indications that in the presence of monastrol+AMPPNP, the heads bind with a higher mobility. This is reflected in the Fourier-transformed structural image data after averaging. First, there is a lower ratio of the total power in the layer line clusters at 1/8 nm (reflecting mostly the motor binding to each αβ-tubulin heterodimer) versus 1/4 nm (reflecting the axial α-β-α-β tubulin subunit repeat of the microtubule template) (Figure 5C). Second, the tighter concentration of the head density into a smaller central region (Figure 6D) indicates mobility because there is a smaller overlap of averaged positions when the heads are wobbling. These cryo-EM results are consistent with previous solution cosedimentation studies of Eg5-367 in the presence of monastrol (Cochran et al, 2005). Monastrol-treated Eg5-367 remained effectively bound to the MT in the presence of AMPPNP although the motor could no longer generate force.
Nucleotide state and monastrol alter MT binding properties of Eg5-513
The MT binding properties of dimeric Eg5-513 were clearly more affected by nucleotide conditions and monastrol than monomeric Eg5-367, suggestive of cooperative interactions within the dimer. AMPPNP induced the only strong MT binding condition for Eg5-513 (Figures 5E and 6F). Cryo-EM revealed a strong 1/8 nm layer line cluster (Figure 5E) indicating saturated conditions with low head mobility within the motor–tubulin complex. High-resolution surface shadowing confirmed that Eg5-513 was capable of fully saturating the MT surface in a very ordered way (Figure 2D) that rendered the corresponding images very comparable to these of monomeric Eg5-367 (Figure 2B). The regularity of the surface pattern found in Figure 2D indicates that both heads of the dimer are bound to the MT. The double-binding and dimeric nature of Eg5-513 could indeed be confirmed by comparing surface shadowgraphs of intentionally undersaturated MT·Eg5-367 complexes (Figure 2F and G; identical image with heads marked as red dots) with MT·Eg5-513 complexes (Figure 2H and I: identical image with heads marked as red dots in (H) and potential dimers with yellow dots). It is evident in these images that monomeric Eg5-367 heads scatter randomly on the MT surface (Figure 2F and G), whereas Eg5-513 heads form doublets that bind mostly along one protofilament (Figure 2H and I). Hence, our data contradict the findings of Hirose et al (2000), who reported that dimeric Eg5·AMPPNP binds with one head only. In our hands, Eg5-513 resembles the binding pattern of dimeric Kinesin-1 (Skiniotis et al, 2003), while being clearly different from Drosophila Ncd (Wendt et al, 2002) or Caenorhabditis elegans Zen-4 (Kinesin-6; Hizlan et al, 2006).
Compared to AMPPNP conditions, nucleotide-free conditions showed a significantly weaker MT-binding affinity of at least one of the two heads in Eg5-513 (Figures 5D and 6E), and the presence of monastrol abolished MT-binding almost entirely, no matter what nucleotide conditions were chosen (Figures 2E and 5F; data shown only in the presence of AMPPNP). The different MT-binding patterns for monomeric versus dimeric Eg5 motors as presented here are indicative of a strong communication between the two heads in the dimer. The relatively weak binding pattern observed under nucleotide-free conditions can be explained by a binding inhibition of the second head once the first one touched the MT surface. The second head cannot bind until ATP or AMPPNP enters the first head, but its presence as the tethered head blocks the adjacent tubulin binding site from occupation by the motor head of another Eg5 dimer, thereby resulting in an incomplete decoration of the MT surface as confirmed by cryo-EM and helical averaging (Figures 5D and 6E). Averaged motor heads appear on every tubulin binding site owing to the averaging procedure, but the averaged density of heads represents only ∼50–60% partial decoration on the MT (compare Figure 6E (nucleotide-free: low decoration) with Figure 6F (AMPPNP: full decoration)).
The Eg5 structural data presented here were also consistent with the cosedimentation results. For Eg5-513, hyperbolic MT binding was observed in the presence of MgADP (Supplementary Figure S1G and H) and in the absence of apyrase (Supplementary Figure S1A and B); however, Eg5 never reached complete saturation for either condition. In contrast, the cosedimentation experiments with either AMPPNP or apyrase to induce the nucleotide-free state revealed complete partitioning of Eg5 to the MT (Figure 5G and Supplementary Figure S1C–F). The difference in results may reflect the conditions of the structural versus solution studies. For the structural studies, the MTs were first applied to the grid to obtain individual MTs, followed by the addition of the Eg5 and other additives, before rapid freezing. In contrast, the solution studies assess the partitioning of Eg5 motors at equilibrium and at higher concentrations. Therefore, the Eg5 motors partition on and off the MT until the equilibrium is established, and the centrifugation sediments Eg5 dimers that are bound by either one or both heads. In contrast, the cryo-EM is technically more demanding to achieve full decoration because of the experimental design, and probably underestimates the relative affinity. Regardless, both methods indicate that there is a significant difference in the MT binding properties of dimeric Eg5-513 in comparison to monomeric Eg5-367.
Neither Eg5-367 nor Eg5-513 decorates MTs in a significantly cooperative manner as previously observed for Ncd, where free MTs were observed next to fully decorated MTs (Wendt et al, 2002). However, there must be communication between Eg5 dimers. If it were completely absent, the regularity of the surface pattern would most likely be disrupted as found for dimeric Kinesin-1 (see Figure 4 in Hoenger et al, 2000). The cosedimentation studies with AMPPNP and apyrase (Figure 5G) produced sigmoidal binding profiles suggestive of cooperativity. In fact, a recent study with Kinesin-1 revealed cooperative binding toward the plus-end of the MT during ATP turnover, yet absent with the addition of ADP or AMPPNP (Muto et al, 2005). Possibly, the interplay between the Eg5 motor domains and the MT causes a more subtle global effect preventing binding cooperativity from being easily detected by our structural analysis of dimeric Eg5 in comparison to dimeric Ncd.
A structural model for monastrol inhibition of dimeric Eg5-513
How can the effect of monastrol on dimeric Eg5-513 be explained? The reason for the observation may be found in the EM docking models of, and within, the corresponding X-ray structures as illustrated in Figure 7. According to the X-ray crystallography studies (Turner et al, 2001; Yan et al, 2004), monastrol affects the switch II region and the relay helix α4 that modulates the MT-binding interface. It also seems to alter the configuration of the neck linker as outlined below.
Figure 7.
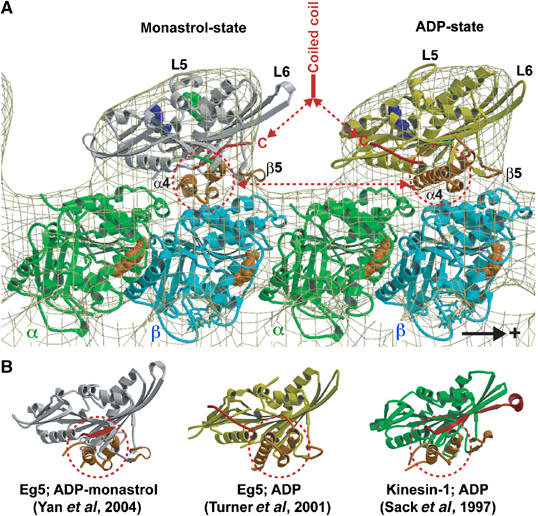
Molecular docking of the atomic resolution X-ray structures of monomeric Eg5 in the presence of ADP and monastrol (left; Yan et al, 2004) and ADP (right; Turner et al, 2001) into our EM-derived 3-D map of the MT·Eg5-513·AMPPNP complex. The plus-end of the MT is on the right. One major structural difference in the two heads that is relevant to MT binding is the position of helix α4 that is rotated by about 20° from the monastrol to ADP structure. Monastrol (green in (A)) binds close to the ATP pocket. The other relevant feature is the locked neck linker (red) in the monastrol structure. Hence, the arrangement as shown here could mimic a dimer with the trailing head in an ATP state and the leading head ready to release ADP and assume a nucleotide-free state. (B) Crystal structures of monomeric Eg5 in the presence of monastrol and ADP, Eg5 with ADP, and rat kinesin with ADP displayed in aligned orientations according to their internal β-sheet.
According to the cosedimentation results in Figure 5G, the MT binding affinity of Eg5-513 is relatively strong, yet decreases to that of the nucleotide-free conditions upon addition of monastrol. The cryo-EM in Figure 5D–F suggests, however, that the affinity may actually drop even more. Fourier analysis revealed that in the nucleotide-free state, the power of the 1/8 nm layer line in MT·Eg5-513 complexes (Figure 5D, right column) was substantially lower than that in the presence of AMPPNP (Figure 5E) and that of the rigor state of the MT·Eg5-367 monomer (Figure 5A). This relative decrease in the power of the lower 1/8 nm layer line with respect to the 1/4 nm line indicates partial MT decoration by Eg5-513 motors. Furthermore, even in the presence of AMPPNP, monastrol treatment caused the relative power of the 1/8 nm layer line to drop even further, indicating an even lower affinity for MTs (Figure 5F), which again is in sharp contrast to what we observed with monomeric Eg5-367. For the Eg5 monomer, monastrol appears to induce somewhat more mobility into the motor–MT interface, but the decoration of the MT surface was effectively achieved. The solution studies reported previously (Cochran et al, 2005) also show that monastrol-treated Eg5-367 cosediments with MTs even though monastrol treatment inhibits ATP turnover and the ability to generate force.
These Eg5 results were somewhat surprising because the atomic resolution structure of monomeric Eg5 motor domains in an ADP state (Turner et al, 2001) compared to that in the presence of monastrol (Yan et al, 2004) revealed striking conformational differences in the switch II region and also in the neck linker (see Figure 7). In particular, Eg5 relay helix α4 shows a very different orientation in the presence of monastrol, manifested through a rotation in the motor–tubulin interface plane (illustrated in Figure 7). Docking the X-ray structures of Eg5 monomers into our cryo-EM 3-D maps illustrates clearly how much the MT interface must be affected by monastrol. Figure 7B illustrates that the monastrol structure shows a helix α4 orientation resembling that of Kinesin-1·ADP (Kozielski et al, 1997; Sack et al, 1997; Hoenger et al, 1998). However, both structures have been suggested to be closer to an ATP state (Vale and Milligan, 2000). The Eg5·ADP structure shows helix α4 rotated by about 20° within the motor–MT interface plane. Hence, for monomeric Eg5-367, we also expected to see a head rotation as demonstrated for KIF1A (Kikkawa et al, 2001) and Kinesin-1 (Skiniotis et al, 2003), but this was clearly not the case (Figure 6A–D). Possibly, binding of Eg5 to the MT lattice induces different conformational changes into the interface region than the ones previously observed with either monomeric KIF1A or Kinesin-1.
A similar pattern as described for the switch II region can also be seen for the position of the neck linker such that the Eg5·ADP-monastrol structure shows a so-called locked neck linker in contact with the motor domain as it was described for an ATP state (Rice et al, 1999; Kikkawa et al, 2001; Skiniotis et al, 2003). These structural observations are consistent with the presteady-state ATP binding kinetics of Eg5-367. Monastrol did not alter ATP binding and appeared to stabilize an ATP conformation (Cochran and Gilbert, 2005; Cochran et al, 2005).
The Eg5·ADP X-ray structure (Turner et al, 2001), however, shows a free neck linker (Figure 7A, yellow head domain) that mimics the predicted nucleotide-free configuration of Kinesin-1 (Rice et al, 1999; Skiniotis et al, 2003). Hence, when projected into a walking dimer that momentarily has both heads attached to two adjacent tubulin heterodimers (Figure 7A), the Eg5·ADP·monastrol crystal structure (Figure 7, gray head domain) shows a trailing head conformation (ATP-like state of Kinesin-1) whereas the Eg5·ADP crystal structure (Figure 7, yellow head domain) resembles a leading head as in a nucleotide-free state of Kinesin-1 (Skiniotis et al, 2003).
Under the various experimental conditions applied here, we see a clear difference in how monastrol affects Eg5 in different oligomeric states: for AMPPNP conditions, monastrol interferes slightly with monomeric Eg5-367 (compare Figure 5B and C) but still allows for fully decorated MT·Eg5 complexes. In contrast, with dimers (Eg5-513·AMPPNP), monastrol interferes strongly (compare Figure 5E and F) and reduces MT affinity. At the resolution achieved in the cryo-EM reconstructions, we obviously cannot see directly what happens to the relevant structural elements (switch II region, helix α4) at the motor–tubulin interface upon Eg5 binding to the MT lattice. However, the effect of monastrol on the neck linker region suggests that the absence of head–head communication in Eg5 monomers makes monomers less affected by monastrol. This observation indicates that monastrol interferes with the communication between the heads in the dimer through their neck linker and neck regions rather than simply altering the relative affinity for the MT through individual head domains. Hence, rather than strongly influencing the motor–tubulin interface through the switch II region, the effects of monastrol on the neck linker and neck region may be even more effective by inducing a sterically unfavorable dimer configuration for microtubule binding.
MT gliding by Eg5-513 suggests that dimeric Eg5 is mechanistically similar to the homotetramer
The rate of Eg5-513-promoted MT gliding was comparable to what has been observed in vitro for Xenopus homotetrameric Eg5 with translocation on MT asters (Wilde et al, 2001) and for a Baculovirus-expressed and purified Eg5 homotetramer sliding MTs against axonemes (Kapitein et al, 2005). We used a similar in vitro motility assay, and the results show that dimeric Eg5-513 was able to generate MT gliding as fast as homotetrameric Eg5 motors. This commonality between the results for dimeric Eg5-513 and homotetrameric Eg5 motors strongly supports the idea that mechanistic data obtained using Eg5-513 will be applicable in defining the cooperative interactions and mechanistic basis of homotetrameric Eg5 function in vivo.
Materials and methods
Experimental conditions
The biochemical analysis of Eg5-513 was performed in ATPase buffer: 20 mM Hepes, pH 7.2 with KOH, 5 mM magnesium acetate, 0.1 mM EDTA, 0.1 mM EGTA, 50 mM potassium acetate, and 1 mM dithiothreitol at 25°C. Paclitaxel in DMSO was used throughout to stabilize the MT polymer, and the more active enantiomer S-monastrol was used. The concentrations reported are final concentrations after mixing.
Cloning, expression, and protein purification
The necessary coding region was PCR amplified out of a pBluescript plasmid containing the full-length human Eg5 cDNA (the generous gift of Dr Anne Blangy, Centre de Recherches de Biochimie Macromoléculaire). Amplification used 5′-GCGTATGGCCAAACTGGCAC as the N-terminal primer, and 5′-ATCCAGCTCGAGTTAGAGACCAGATACATC (Eg5-513) and 5′-ATCCAGCTCGAGTTAGTGGTGGTGGTGGTGCA TGGCCTTGTGGAGACCAGATACATC (Eg5-513-5His) as the C-terminal primers. Both C-terminal primers engineered a TAA stop codon and an Xho1 restriction site at the end of the desired coding region. The primer specific for Eg5-513-5His also inserted a short amino-acid linker (HKAM) between the native sequence and the 5His sequence. The amplified region was then ligated into a pRSETa plasmid containing an Eg5 construct of the first 405 residues of the human protein (Maliga et al, 2002). BstEII and Xho1 were the restriction enzymes used. The ligation mix was then transformed into the Nova Blue cell line (Novagen, Madison, WI) for amplification and storage. Plasmids from the clones were verified by restriction digest with BglI and sequenced before transformation into the BL21-CodonPlus (DE3)-RIL cell line (Stratagene) for protein expression. Eg5-513 and Eg5-513-5His were subsequently purified by MT affinity followed by MgATP release (see Supplementary data). The Eg5-513 concentrations reported represent monomer motor domain, that is, single Eg5 active site concentrations.
Analytical gel filtration
Gel filtration was performed using a Superose-6 HR 10/30 gel filtration column with the System Gold high-pressure liquid chromatography system (Beckman Coulter Inc.). Elution was monitored using intrinsic protein fluorescence. All proteins were eluted in ATPase buffer at a flow rate of 0.5 ml/min at room temperature. The protein standards used were aldolase, catalase, ferritin, and thyroglobin.
Motility assays
Polarity marked MTs were polymerized as described previously and stabilized with 20 μM Taxol in 10 mM PIPES, 5 mM MgCl2, and 1 mM EGTA (Sproul et al, 2005). The resulting MTs exhibited a highly fluorescent short minus-end and a less fluorescent, extended plus-end. A preformed MT·Eg5·AMPPNP complex (2 μM Eg5-513-5His, 300 nM tubulin, 20 μM Taxol, 1 mM MgAMPPNP) was flowed into the perfusion chamber followed by buffer plus 1 mM MgAMPPNP to remove unattached MTs. The reaction was initiated by 1.5 mM MgATP plus an ATP regeneration system (Sproul et al, 2005). Eg5-513 was not as effective for the MT gliding assays as Eg5-513-5His. We suspect that the 5His sequence of the protein enhances the interaction with the glass coverslip to establish motor orientation for MT gliding. Native squid conventional kinesin (KHC), a well-described plus-end-directed kinesin, was used to evaluate the polarity of the MTs.
Steady-state ATPase kinetics
Steady-state kinetics were determined by monitoring the hydrolysis of [α-32P]ATP (Gilbert and Mackey, 2000). In Figure 4, the observed rate of ATP turnover as a function of MgATP concentration was fit to the Michaelis–Menten equation, and the ATPase rate as a function of MTs (tubulin concentration) was fit to following quadratic equation 1:
![]()
Rate is the concentration of product formed per second per active site, and kcat is the maximum rate constant for product formation at saturating MT and MgATP conditions. The Km,ATP and K1/2,MT are the constants at which the concentration of substrate yields half the maximal velocity. E0 is the concentration of motor active sites in the reaction, and MT0 is the tubulin concentration as MT polymer.
For the inhibition of Eg5 steady-state ATPase as a function of S-monastrol concentration (Figure 4C), the data were fit to the following quadratic equation:
![]()
Ainh is the amplitude of S-monastrol inhibition defined by kmax (kcat in the absence of S-monastrol) minus kmin (kcat at saturating S-monastrol), Kd-Mon is the apparent dissociation constant for S-monastrol, and S0 is the S-monastrol concentration.
Cosedimentation assays
The cosedimentation assays were performed as previously described (Cochran et al, 2004). Briefly, 2 μM Eg5-513 was incubated with varying concentrations of MTs for 10 min before the addition of nucleotide. The MT·Eg5·AXP complex was incubated at room temperature for 30 min to reach equilibrium and centrifuged. In the apyrase-treated experiments, the MT·Eg5 complex was incubated with 0.1 U/ml apyrase (Grade VII, Sigma-Aldrich) for 1 h before centrifugation. For each reaction, the supernatant was collected, and the MT pellet was resuspended to an equal volume with ATPase buffer. Laemmli 5 × sample buffer was added to both supernatant and pellet samples and subjected to SDS–PAGE followed by staining with Coomassie brilliant blue R-250 and analysis. For Eg5-513 (Figure 5G and Supplementary Figure S1) where sigmoid data were obtained, the Hill equation was used to estimate the degree of cooperativity:
![]()
where Ys is the fraction of motor sedimenting with the MTs, [MT] is the tubulin concentration as MT polymer, n is the Hill coefficient, and Kh is the product of the Kd values for the cooperative sites.
Decoration of MTs with Eg5 for cryo-EM
MTs were polymerized for 30 min at 37°C in BRB80 (80 mM PIPES, pH 6.8, 2 mM MgCl2) at a concentration of 5 mg/ml and in the presence of 10% (v/v) DMSO, 2 mM MgGTP, and 20 μM paclitaxel and left overnight at room temperature to stabilize. Decoration of polymerized MTs with monomeric and dimeric Eg5 was performed in solution at a final tubulin concentration of 0.5 mg/ml (∼4.5 μM αβ-tubulin heterodimers) and varying concentrations of Eg5. The stoichiometry of Eg5 heads to αβ-tubulin heterodimers ranged from 4:1 to 10:1.
Preparation of cryo-EM samples
Experiments were performed either in the presence of 2 mM MgAMPPNP or after incubation with apyrase to achieve the nucleotide-free state. Samples of monomeric Eg5-367 were incubated for 2 min and subsequently adsorbed to holey carbon grids for 1 min and quick-frozen in liquid ethane by using a plunger (Dubochet et al, 1988). With dimeric Eg5, decoration of MTs was performed directly on the grid to avoid bundling of MTs. In these cases, MTs at a concentration of 0.5 mg/ml were adsorbed on holey carbon grids for 1 min, incubated for 2 min with an Eg5 solution, and quick-frozen as described above. To ensure complete decoration, conditions with a high motor to tubulin ratio of 4:1 were used.
Cryo-EM, image processing, and 3-D reconstruction
Cryo-EM was performed on a Philips CM200-FEG microscope, using a GATAN-626 cryo-holder. Images were recorded at × 38 000 nominal magnification on Kodak SO-163 EM film at a defocus range of −1.5 to −2.0 μm.
The 3-D reconstructions were performed using 15-protofilament/2-start helical MTs (Beuron and Hoenger, 2001). Micrographs were digitized using a Zeiss-SCAI scanner at a step size of 21 μm corresponding to 0.526 nm on the original object. Suitable MTs were helically reconstructed by using the program suite PHOELIX (Whittaker et al, 1995) and SUPRIM (Schroeter and Bretaudiere, 1996). All data sets were truncated to a maximum resolution of 25 Å. 3-D maps were visualized by AVS (advanced visualization software). The reconstruction statistics are shown in Table II. Data sets (20–40) were included in the reconstruction giving a total of 20 000–40 000 asymmetric units per reconstruction. Docking of the atomic coordinates into the electron density maps was carried out with the program suite O (Jones et al, 1991). Figures of the model were prepared using Bobscript 2.3 (Esnouf, 1997).
Table 2.
Cryo-EM reconstruction data
| Sample | Nucleotide state | Averaged asymmetrical units |
|---|---|---|
| Eg5-367 | Nucleotide free | ∼36 200 |
| Eg5-367 | AMPPNP | ∼21 000 |
| Eg5-367 | AMPPNP+S-monastrol | ∼40 700 |
| Eg5-513 | Nucleotide free | ∼20 700 |
| Eg5-513 | AMPPNP | ∼37 000 |
Unidirectional shadowing
MTs were assembled in BRB80 as described above. Eg5 motors were mixed with MTs to a final concentration of 1 μM (saturating conditions) or 0.1 μM (undersaturating conditions) Eg5 and MTs at 0.5 μM tubulin. Samples were adjusted to 5 mM MgAMPPNP±150 μM S-monastrol. The grids were quick-frozen in liquid nitrogen and transferred into the freeze-drying/shadowing device (MIDILAB; Gross et al, 1990). After freeze-drying for 2 h at 10–7 bar/140 K, samples were unidirectionally shadowed at an elevation angle of 45°, and subsequently transferred into the microscope for imaging under constant vacuum conditions. Images were recorded with a Gatan-794 Multiscan CCD camera at an electron dose of 500–1000 electrons/nm2 and a nominal magnification of × 35 000 on the microscope resulting in a pixel size of 0.534 nm on the camera.
Note added in proof:
Dimeric Eg5-513 has been shown to be a processive motor. Valentine MT, Fordyce PM, Krzysiak TC, Gilbert SP, Block SM (2006) Individual dimers of the mitotic kinesin motor Eg5 step processively and support substantial loads in vitro. Nat Cell Biol Apr2; [ePub ahead of print].
Supplementary Material
Supplementary Materials and Methods
Supplementary Figure 1
Supplementary Movie Legend
Supplementary Movie
Acknowledgments
We thank Anne Blangy (Centre de Recherches de Biochimie Macromoléculaire, Montpellier, France) for providing human Eg5 cDNA, Wasyl Halczenko and George Hartman (Merck Research Laboratories, West Point, PA) for the gift of S-monastrol, Jared Cochran for Eg5-367 protein and helpful discussion, Steven Block for native squid Kinesin-1 protein, William Saunders for use of his microscope, and Valerie Oke for her assistance during cloning. This work was supported by grants from The European Molecular Biology Laboratory and the German Research Society to AH and grant GM54141 and Career Development Award K02-AR47841 from the National Institutes of Health, U.S. Department of Health and Human Services to SPG.
References
- Berger B, Wilson DB, Wolf E, Tonchev T, Milla M, Kim PS (1995) Predicting coiled coils by use of pairwise residue correlations. Proc Natl Acad Sci USA 92: 8259–8263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuron F, Hoenger A (2001) Structural analysis of the microtubule–kinesin complex by cryo-electron microscopy. Methods Mol Biol 164: 235–254 [DOI] [PubMed] [Google Scholar]
- Blangy A, Lane HA, d'Herin P, Harper M, Kress M, Nigg EA (1995) Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell 83: 1159–1169 [DOI] [PubMed] [Google Scholar]
- Cochran J, Gatial JI, Kapoor T, Gilbert SP (2005) Monastrol inhibition of mitotic kinesin Eg5. J Biol Chem 280: 12658–12667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran JC, Gilbert SP (2005) ATPase mechanism of Eg5 in the absence of microtubules: insight into microtubule activation and allosteric inhibition by monastrol. Biochemistry 44: 16633–16648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran JC, Sontag CA, Maliga Z, Kapoor TM, Correia JJ, Gilbert SP (2004) Mechanistic analysis of the mitotic kinesin Eg5. J Biol Chem 279: 38861–38870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole DG, Saxton WM, Sheehan KB, Scholey JM (1994) A ‘slow' homotetrameric kinesin-related motor protein purified from Drosophila embryos. J Biol Chem 269: 22913–22916 [PMC free article] [PubMed] [Google Scholar]
- Correia JJ, Gilbert SP, Moyer ML, Johnson KA (1995) Sedimentation studies on the kinesin motor domain constructs K401, K366, and K341. Biochemistry 34: 4898–4907 [DOI] [PubMed] [Google Scholar]
- Crevel IM, Alonso MC, Cross RA (2004) Monastrol stabilises an attached low-friction mode of Eg5. Curr Biol 14: R411–R412 [DOI] [PubMed] [Google Scholar]
- Cross RA (2004) The kinetic mechanism of kinesin. Trends Biochem Sci 29: 301–309 [DOI] [PubMed] [Google Scholar]
- De La Cruz EM, Ostap EM, Sweeney HL (2001) Kinetic mechanism and regulation of myosin VI. J Biol Chem 276: 32373–32381 [DOI] [PubMed] [Google Scholar]
- Dubochet J, Adrian M, Chang JJ, Homo JC, Lepault J, McDowall AW, Schultz P (1988) Cryo-electron microscopy of vitrified specimens. Q Rev Biophys 21: 129–228 [DOI] [PubMed] [Google Scholar]
- Duhl DM, Renhowe PA (2005) Inhibitors of kinesin motor proteins – research and clinical progress. Curr Opin Drug Discov Dev 8: 431–436 [PubMed] [Google Scholar]
- Esnouf RM (1997) An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J Mol Graph Model 15: 132–133 [DOI] [PubMed] [Google Scholar]
- Gilbert SP, Johnson KA (1993) Expression, purification, and characterization of the Drosophila kinesin motor domain produced in Escherichia coli. Biochemistry 32: 4677–4684 [DOI] [PubMed] [Google Scholar]
- Gilbert SP, Mackey AT (2000) Kinetics: a tool to study molecular motors. Methods 22: 337–354 [DOI] [PubMed] [Google Scholar]
- Gordon DM, Roof DM (1999) The kinesin-related protein Kip1p of Saccharomyces cerevisiae is bipolar. J Biol Chem 274: 28779–28786 [DOI] [PubMed] [Google Scholar]
- Goshima G, Vale RD (2003) The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J Cell Biol 162: 1003–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross H, Krusche K, Tittmann P (1990) Recent progress in high resolution shadowing for biological TEM. In XIIth International Congress for Electron Microscopy, Peachey LD, Williams DB (eds) pp 510–511. Seattle, WA: San Francisco Press Inc [Google Scholar]
- Hirose K, Henningsen U, Schliwa M, Toyoshima C, Shimizu T, Alonso M, Cross RA, Amos LA (2000) Structural comparison of dimeric Eg5, Neurospora kinesin (Nkin) and Ncd head–Nkin neck chimera with conventional kinesin. EMBO J 19: 5308–5314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hizlan D, Mishima M, Tittmann P, Gross H, Glotzer M, Hoenger A (2006) Structural analysis of the ZEN-4/CeMKLP1 motor domain and its interaction with microtubules. J Struct Biol 153: 73–84 [DOI] [PubMed] [Google Scholar]
- Hoenger A, Doerhoefer M, Woehlke G, Tittmann P, Gross H, Song YH, Mandelkow E (2000) Surface topography of microtubule walls decorated with monomeric and dimeric kinesin constructs. Biol Chem 381: 1001–1011 [DOI] [PubMed] [Google Scholar]
- Hoenger A, Sack S, Thormählen M, Marx A, Müller J, Gross H, Mandelkow E (1998) Image reconstructions of microtubules decorated with monomeric and dimeric kinesins: comparison with x-ray structure and implications for motility. J Cell Biol 141: 419–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Kapitein LC, Peterman EJ, Kwok BH, Kim JH, Kapoor TM, Schmidt CF (2005) The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature 435: 114–118 [DOI] [PubMed] [Google Scholar]
- Kapoor TM, Mayer TU, Coughlin ML, Mitchison TJ (2000) Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J Cell Biol 150: 975–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashina A, Baskin RJ, Cole D, Wedaman K, Saxton W, Scholey J (1996) A bipolar kinesin. Nature 379: 270–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkawa M, Sablin EP, Okada Y, Yajima H, Fletterick RJ, Hirokawa N (2001) Switch-based mechanism of kinesin motors. Nature 411: 439–445 [DOI] [PubMed] [Google Scholar]
- Kozielski F, Sack S, Marx A, Thormählen M, Schönbrunn E, Biou V, Thompson A, Mandelkow EM, Mandelkow E (1997) The crystal structure of dimeric kinesin and implications for microtubule-dependent motility. Cell 91: 985–994 [DOI] [PubMed] [Google Scholar]
- Kwok BH, Yang JG, Kapoor TM (2004) The rate of bipolar spindle assembly depends on the microtubule-gliding velocity of the mitotic kinesin Eg5. Curr Biol 14: 1783–1788 [DOI] [PubMed] [Google Scholar]
- Kwon M, Scholey J (2004) Spindle mechanics and dynamics during mitosis in Drosophila. Trends Cell Biol 14: 194–205 [DOI] [PubMed] [Google Scholar]
- Maliga Z, Kapoor TM, Mitchison TJ (2002) Evidence that monastrol is an allosteric inhibitor of the mitotic kinesin Eg5. Chem Biol 9: 989–996 [DOI] [PubMed] [Google Scholar]
- Maliga Z, Xing J, Cheung H, Juszczak LJ, Friedman JM, Rosenfeld SS (2006) A pathway of structural changes produced by monastrol binding to Eg5. J Biol Chem 281: 7977–7982 [DOI] [PubMed] [Google Scholar]
- Mayer TU, Kapoor TM, Haggarty SJ, King RW, Schreiber SL, Mitchison TJ (1999) Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science 286: 971–974 [DOI] [PubMed] [Google Scholar]
- McIntosh JR, Grishchuk EL, West RR (2002) Chromosome–microtubule interactions during mitosis. Annu Rev Cell Dev Biol 18: 193–219 [DOI] [PubMed] [Google Scholar]
- Mountain V, Simerly C, Howard L, Ando A, Schatten G, Compton DA (1999) The Kinesin-related Protein, HSET, opposes the activity of Eg5 and cross-links microtubules in the mammalian mitotic spindle. J Cell Biol 147: 351–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer ML, Gilbert SP, Johnson KA (1996) Purification and characterization of two monomeric kinesin constructs. Biochemistry 35: 6321–6329 [DOI] [PubMed] [Google Scholar]
- Moyer ML, Gilbert SP, Johnson KA (1998) Pathway of ATP hydrolysis by monomeric and dimeric kinesin. Biochemistry 37: 800–813 [DOI] [PubMed] [Google Scholar]
- Muto E, Sakai H, Kaseda K (2005) Long-range cooperative binding of kinesin to a microtubule in the presence of ATP. J Cell Biol 168: 691–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice S, Lin AW, Safer D, Hart CL, Naber N, Carragher BO, Cain SM, Pechatnikova E, Wilson-Kubalek EM, Whittaker M, Pate E, Cooke R, Taylor EW, Milligan RA, Vale RD (1999) A structural change in the kinesin motor protein that drives motility. Nature 402: 778–784 [DOI] [PubMed] [Google Scholar]
- Rosenfeld SS, Rener B, Correia JJ, Mayo MS, Cheung HC (1996) Equilibrium studies of kinesin–nucleotide intermediates. J Biol Chem 271: 9473–9482 [DOI] [PubMed] [Google Scholar]
- Sack S, Müller A, Marx M, Thormählen M, Mandelkow E-M, Brady ST, Mandelkow E (1997) X-ray structure of motor and neck domains from rat brain kinesin. Biochemistry 36: 16155–16165 [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Wang F, Schmitz S, Xu Y, Xu Q, Molloy JE, Veigel C, Sellers JR (2003) Neck length and processivity of myosin V. J Biol Chem 278: 29201–29207 [DOI] [PubMed] [Google Scholar]
- Saunders WS, Hoyt MA (1992) Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell 70: 451–458 [DOI] [PubMed] [Google Scholar]
- Sawin KE, LeGuellec K, Philippe M, Mitchison TJ (1992) Mitotic spindle organization by a plus-end-directed microtubule motor. Nature 359: 540–543 [DOI] [PubMed] [Google Scholar]
- Schroeter JP, Bretaudiere JP (1996) SUPRIM: easily modified image processing software. J Struct Biol 116: 131–137 [DOI] [PubMed] [Google Scholar]
- Sharp DJ, McDonald KL, Brown HM, Matthies HJ, Walczak C, Vale R, Mitchison TJ, Scholey JM (1999) The bipolar kinesin, KLP61F, cross-links microtubules within interpolar microtubule bundles of drosophila embryonic mitotic spindles. J Cell Biol 144: 125–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skiniotis G, Surrey T, Altmann S, Gross H, Song YH, Mandelkow E, Hoenger A (2003) Nucleotide-induced conformations in the neck region of dimeric kinesin. EMBO J 22: 1518–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sproul LR, Anderson DJ, Mackey AT, Saunders WS, Gilbert SP (2005) Cik1 targets the minus-end kinesin depolymerase kar3 to microtubule plus ends. Curr Biol 15: 1420–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J, Anderson R, Guo J, Beraud C, Fletterick R, Sakowicz R (2001) Crystal structure of the mitotic spindle kinesin Eg5 reveals a novel conformation of the neck-linker. J Biol Chem 276: 25496–25502 [DOI] [PubMed] [Google Scholar]
- Vale RD, Milligan RA (2000) The way things move: looking under the hood of molecular motor proteins. Science 288: 88–95 [DOI] [PubMed] [Google Scholar]
- Wendt TG, Volkmann N, Skiniotis G, Goldie KN, Muller J, Mandelkow E, Hoenger A (2002) Microscopic evidence for a minus-end-directed power stroke in the kinesin motor ncd. EMBO J 21: 5969–5978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker M, Carragher BO, Milligan RA (1995) PHOELIX: a package for semi-automated helical reconstruction. Ultramicroscopy 58: 245–259 [DOI] [PubMed] [Google Scholar]
- Wilde A, Lizarraga SB, Zhang L, Wiese C, Gliksman NR, Walczak CE, Zheng Y (2001) Ran stimulates spindle assembly by altering microtubule dynamics and the balance of motor activities. Nat Cell Biol 3: 221–227 [DOI] [PubMed] [Google Scholar]
- Wittmann T, Hyman A, Desai A (2001) The spindle: a dynamic assembly of microtubules and motors. Nat Cell Biol 3: E28–E34 [DOI] [PubMed] [Google Scholar]
- Yan Y, Sardana V, Xu B, Homnick C, Halczenko W, Buser CA, Schaber M, Hartman GD, Huber HE, Kuo LC (2004) Inhibition of a mitotic motor protein: where, how, and conformational consequences. J Mol Biol 335: 547–554 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials and Methods
Supplementary Figure 1
Supplementary Movie Legend
Supplementary Movie


