Abstract
We describe here a system for the rapid identification, assay development, and characterization of gene-based single nucleotide polymorphisms (SNPs). This system couples informatics tools that mine candidate SNPs from public expressed sequence tag resources and automatically designs assay reagents with detection by a chip-based matrix-assisted laser desorption/ionization time-of-flight mass spectrometry platform. As a proof of concept of this system, a genomewide collection of reagents for 9,115 gene-based SNP genetic markers was rapidly developed and validated. These data provide preliminary insights into patterns of polymorphism in a genomewide collection of gene-based polymorphisms.
Genomewide genetic analysis of gene-based single nucleotide polymorphisms (SNPs) may be an efficient paradigm for the discovery of genes important in complex traits in humans (1). Genetic analysis would focus on variation in the transcribed portion of the genome. This approach, however, has been practically limited by the lack of availability of suitable reagents and experimental platforms for high-throughput SNP evaluation. Moreover, little is known about genomewide patterns of variation in transcript-based SNPs. We have begun to address these issues by developing a set of genomewide, gene-based genetic analysis reagents. Using the SNPpipeline, a set of publicly accessible sequence analysis tools (http://cgap.nci.nih.gov/GAI and ref. 2), we identified 10,243 high probability gene-based SNPs among sequences contained in the UniGene (3) release. Candidates were derived from 6,458 UniGene clusters, for which four or more chromatographs were available from Washington University (St. Louis) (http://genome.wustl.edu/est/esthmpg.html and ref. 4). For this study, only candidate SNPs with scores of 0.99 or greater were considered. A score of this magnitude indicates that an alternative nucleotide is observed at the given location in the sequence assembly with 99% certainty.
The candidate SNPs were evaluated by chip-based matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (DNA MassARRAY) of primer extension products generated from PCR-generated sequences amplified from pooled DNA samples (Fig. 1). Previous studies have shown this approach to be a highly accurate means for uniplex or multiplex individual SNP determination (5–7). Pilot studies have suggested that this approach can be used to estimate SNP allele frequencies in pooled samples. These studies indicate that allele frequencies can be estimated with an average accuracy of ±1.6% for SNPs where the rare allele is present in the sample pool at a frequency of 10% or greater.
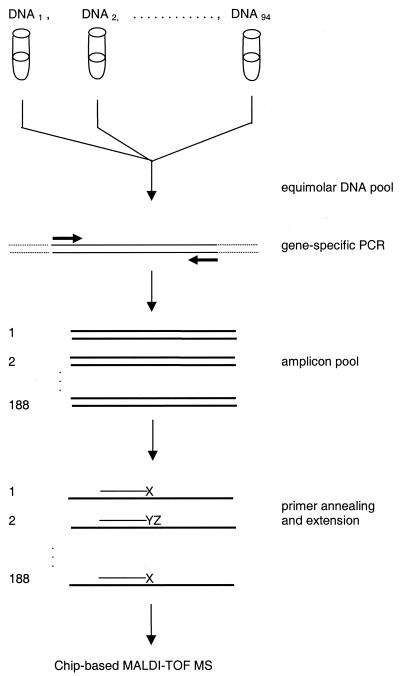
Figure 1.
Scheme of the analytical process to confirm and validate SNPs using chip-based mass spectrometry. After setup of an equimolar mixture of 94 individual Centre d'Étude du Polymorphisme Humain DNA samples, gene-specific uniplex PCRs were performed. The amplicon pool was subjected to a post-PCR primer extension reaction (massextend). Nanoliter amounts of the extension products were loaded onto SpectroCHIPs and subsequently analyzed by an array mass spectrometer.
Materials and Methods
PCR Primer Design Algorithm.
Primers were designed based on our assembly consensus sequences. Primers were designed by using primer3 version 0.6 (http://www.genome.wi.mit.edu/genome_software/other/primer3.html). These assays were constrained to produce products of minimal size (80–120 bp) to maximize the PCR success rate of assays designed from cDNA sequences. At least 10 bp (and a maximum of 35 bp) were reserved on either side of the SNP site (for probe primer design).
Extension Primer Design.
The massextend program is used to design a primer that is extended across the SNP site by using a sequencing polymerase reaction. The extension reaction is controlled by a mixture of dideoxy-terminated nucleotides, such that one single-base extension product is created and one double-base extension product is created. This scheme creates two peaks in the mass spectrometer that are separated by about 300 Da, which is optimal for peak area calculations. The choice of which strand to use for this extendable primer is based on the typical criteria related to hybridization and polymerase priming, specifically %G + C (40–70%), complexity of the primers (at least three of the four bases present), distribution of bases in the primers (maximum four of the same base in a row), and the absence of any unknown base (N, R, Y, etc.).
DNA Quantitation.
Pico green was purchased from Molecular Probes. The DNA concentration was determined according to the manufacturer's protocol. The fluorescence was measured by using an Ascent Flouroscan plate reader (excitation 485 nm; emission 538 nm). The final concentration of equimolar mixture of 94 DNA samples was adjusted to 5 ng/μl; 5 μl (25 ng) of this DNA pool was used for PCR. The equimolarity of the pool was checked by testing the factor VII polymorphism (R353Q) (8).
PCR Protocol.
The PCR was performed under standard conditions for all assays. The volume was 50 μl including 1 unit of Taq-polymerase (Hotstar, Qiagen, Chatsworth, CA), 200 μmol of dNTP, 25 pmol of PCR primer 1, 2 pmol of PCR primer 2 with universal sequence tag, and 10 pmol of biotinylated universal sequence primer (5′-bio-agcggataacaatttcacacagg-3′). PCR primer 2 produced DNA strand complementary to the massextend primer. Temperature conditions were initial denaturation at 95°C for 15 min followed by 50 cycles of 95°C for 5 s, 56°C for 20 s, and 72°C for 30 s.
Primer Extension Protocol.
massextend reactions were performed in four groups of different terminations according to the design rational (ddACG, ddACT, ddAGT, and ddCGT, respectively). The reaction volume was 10 μl including 1 unit of Thermosequenase (Amersham Pharmacia), 50 μmol of respective termination mix (e.g., dTTP, ddATP, ddCTP, ddGTP), and 10 pmol of test-specific massextend primer. A universal temperature program was applied (80° for 30 s followed by 3 cycles of 40°C for 15 s and 60°C for 1 min). After denaturation of primer extension products, approximately 7 nl was loaded onto four positions of SpectroCHIPs preloaded with 7 nl of matrix. SpectroCHIPs were analyzed in fully automated mode by a MassARRAY mass spectrometer (Bruker-Sequenom).
MALDI-TOF MS Analysis.
PCR products generated from the pooled DNA sample were bound via biotin–streptavidin coupling to paramagnetic beads. After denaturation of the double-stranded template, the post-PCR extension reaction was performed. The resulting ≈20-mer single-stranded extension products then were denatured into ammonium hydroxide solution before serial piezoelectric pipette transfer of low nanoliter aliquots onto individual 200-μm elements of a 96-element silicon chip, which had been preloaded with nanoliter volumes of crystalline matrix (3-hydroxy-picolinic acid). In preparation for MALDI-TOF MS analysis, the analyte solution was used to dissolve the matrix patch, and, upon solvent evaporation, analyte was incorporated into newly formed crystals. Under high vacuum conditions, the matrix crystals, which absorb strongly in the UV, were irradiated with nanosecond duration 337-nm laser pulses, leading to formation of a plume of volatilized matrix and analyte as well as charge transfer from matrix ions to analyte molecules. After electric field-induced acceleration in the mass spectrometer source region, the gas-phase ions travel through a ≈1-m field-free region at a velocity inversely proportional to their mass-to-charge ratios.
The resulting time-resolved spectrum is translated into a mass spectrum upon calibration. These mass spectra were further processed and analyzed by proprietary software (spectrotyper) for baseline correction, peak identification, and peak area calculations. Whereas conventional (0.1–0.5 μl matrix and analyte) MALDI preparations result in crystals with total surface area ≈100× that of the laser irradiation area, uneven analyte incorporation into matrix crystals (9), and thus nonstatistical sampling of relative analyte ratios, we have shown previously that uniformly arrayed low nanoliter preparations enhance reproducibility and automated serial spectrum acquisition. Nearly the entire sample is irradiated; this improved sampling efficiency enables determination of relative concentrations of similarly sized DNA molecules in a mixture by comparing the respective areas under each mass spectral peak.
Genotype Generation.
The spectrotyper software uses a set of digital filters optimized for mass spectra of DNA. The overriding design constraint is to have a digital processing algorithm that achieves maximum noise reduction with minimal resolution degradation and processing artifacts. These processes are automated and completely integrated as a postacquisition application for data entry into a relational database. Special care was taken in the development of the algorithm for peak area integration to ensure reproducibility and robustness against noise and baseline errors. Normalized areas are computed as individual peak areas divided by the sum of all peak area.
Results
An automated procedure was used to design the large collection of PCR primers required to amplify the region surrounding each candidate SNP. A suitable set of PCR reagents could be designed for 9,484 of the candidate SNPs (93% of the total candidate set). The primer extension assays were designed so that the binary SNP alleles would terminate after extension by either one or two nucleotides. The postamplification extendable primers also were selected in silico by an automated procedure designed to minimize the total number of extension experimental conditions. Suitable primers could be designed for 9,893 (97% of the total candidate set). Combining the PCR and post-PCR in silico designs, an overall automated design efficiency of 89% (9,115 of 10,243) was achieved. Inspection and manual redesign of failed assays may improve this further.
The candidate SNPs for which such automated assay design was successful (9,115 assays; 1 set of 90 assays and 95 sets of 95 assays) were experimentally evaluated by using a pool of DNA samples obtained from 94 independent individuals of European ancestry from the Centre d'Étude du Polymorphisme Humain (Paris) panel (10). The Centre d'Étude du Polymorphisme Humain panel was used in these studies to permit the evaluation of Mendelian transmission in families and to assess the accuracy of genotyping by examining individual SNP genotypes in the context of other markers in high-density genetic maps. DNA was obtained from the Coriell Cell Repositories, Camden, NJ. DNA concentrations for samples used in the pool were confirmed by testing with pico green.
A total of 7,666 (84% of those for which an assay was developed) candidate SNPs could be amplified and successfully analyzed under universal reaction conditions. Among the 1,449 failures (i.e., those that produced no mass spectra after the entire analytical process was performed), 803 (55%) produced no PCR product, 457 (31%) produced a PCR product similar in size of the one predicted from the sequence assembly, and 189 (14%) showed PCR products with single or multiple bands of different size, all as determined by agarose gel electrophoresis. Re-evaluation of a randomly chosen subset of 185 failed PCRs showed that 72 (39%) of the set recovered the appropriate PCR products and subsequent primer extension reactions simply by reordering (resynthesizing) the same PCR primers.
A total of 6,404 of the 7,666 experimentally proven candidates produced outcomes that could be unequivocally scored as polymorphic (defined as containing a minor allele for frequency >0.1) or nonpolymorphic (no alternative mass spectral peak corresponding to the second allele detectable in four independent measurements). Among these, 3,646 (57%) were classified as SNPs as defined above. Data from an additional 690 (9%) of the 7,666 candidates were indicative of rarer minor alleles, with relative frequencies in the range of 0.01 to 0.1.
To assess the validity of these results, individual samples from the pools were genotyped to test for the presence of alternative genotypes. A total of 41 loci were evaluated on a minimum of 10 individuals for the set of SNPs estimated to have frequencies of greater than 10%. Of these, 16 loci were selected from the highly polymorphic subset (rare allele frequency greater than or equal to 0.4). All loci tested demonstrated the presence of a minimum of two alternative genotypes; 84 loci were tested from the subset with frequencies greater than zero but less than 10%. Within this set, only one locus did not show evidence of a polymorphism within the 23 or 46 individuals, respectively. A subset (153 loci) of the SNPs described above has been previously characterized using restriction fragment length polymorphism analysis of PCR products. All but seven (11%) loci that were observed to be polymorphic in restriction fragment length polymorphism tests were detected as polymorphic in the pooled MALDI-TOF analysis. All polymorphisms tested demonstrated Mendelian transmission patterns in Centre d'Étude du Polymorphisme Humain families and could be placed in genetic reference maps.
In 81 assays (1%), unexpected peaks were observed. These additional products could indicate tri-allelic SNPs, secondary SNPs, sequence errors, or simply nonspecificity of the amplification reaction. The analysis of 41 tests using individual samples demonstrated these peaks to be unspecific products. The most likely explanation is the coamplification of homologous DNA sequences or pseudogenes by PCR.
In high-throughput SNP studies, the quality of the assays is important for the final successful analysis. Indicators for the robustness of assays are peak areas and the signal-to-noise ratio. In this study, the average signal-to-noise ratio was 83.1. Example spectra are shown in Fig. 2.
Figure 2.
Example spectra representing the range of assay qualities. Allelic peaks are indicated at the top of each spectrum. (A) Low-end assay quality, total peak area of both alleles (pa; arbitrary units) = 768, signal-to-noise ratio (snr) = 24.7, relative frequency for C allele = 0.23, T allele = 0.77, standard deviation (SD) of four individual spottings onto SpectroCHIPs followed by mass spectrometric analysis = 0.034. (B) pa = 1,995, snr = 33.3, C allele = 0.37, T allele = 0.63, SD = 0.012. (C) Average assay quality, pa = 9,957, snr = 77.4, C allele = 0.55, T allele = 0.45, SD = 0.009. (D) High-end assay quality, pa = 48,679, snr = 199, C allele = 0.77, T allele = 0.23, SD = 0.006.
The accuracy of the allele frequency estimates was evaluated by contrasting the frequencies observed from the pooled sample with those observed from typing the individual samples from which the pool was constituted. For 10 loci, all 95 individuals that constituted the pool were examined individually. The rare allele frequency observed in the individual sample ranged from 0.42 to 0.19. The estimate obtained from the pooled samples for these 10 loci differed by an average of only 0.007 (range 0.0023 to 0.0096). The allele frequencies also were obtained from the smaller subset of individuals typed for the 41 loci used to confirm the presence of SNPs described above. The mean difference observed was 0.07615 (SD 0.06128). Note that this larger difference is attributable to sampling error in the estimate obtained from individuals caused by examination only of subset of individuals from the pool.
To determine the precision of the frequency estimates determined by the assay, 81 of the MALDI-TOF assays were performed in two sets of replicates of four. In one set of replicates, the same primer extension product was spotted four times. The median standard deviation for these evaluations was 1.6%. An alternative set of replicates examined the outcome of the entire experiment when repeated four times (for the same locus). For this set of experiments, the median standard deviation was 1.7%.
Using publicly available expressed sequence tag mapping data (11), it is possible to place 2,377 of the SNPs into a human genome map. These 2,377 SNPs describe 1,924 locations (UniGene clusters). The average interlocus distance observed is 5.70 centirays/1.74 centimorgans. The average genetic distance was calculated by assuming a uniform relationship between centirays and centimorgans. The distribution of markers also is given in a more realistic genetic context by placing them on a map scaled by genetic distances estimated from microsatellite markers that have been placed on the physical map for reference. These maps are available at http://cgap.nci.nih.gov/GAI. As would be expected, the distribution of gene-based SNPs mirrors the distribution of genes in the genome. Therefore, chromosomes observed to have higher or lower gene densities in GeneMap'99 are observed to have comparable variations in their marker densities. More specifically, chromosome 19, which had previously been shown to have the highest gene density, is observed to have one of the highest SNP marker densities (3.22 centirays/0.98 centimorgan spacing). Similarly, chromosome 18 is observed to have both the lowest autosomal gene density and the lowest SNP density in these maps (25.41 centirays/6.57 centimorgan spacing). The average heterozygosity for the set is 0.39 (range 0.18–0.50), making them of utility for linkage studies.
Discussion
The above study demonstrates that it is currently feasible to perform large-scale gene-based SNP studies. Thousands of SNP assays can be rapidly and cost-efficiently designed from transcript data currently available in public databases. Studies considering thousands of SNPs can be cost-efficiently performed with analytic platforms that are currently commercially available. The validated SNPs described in this pilot study represent a collection of reagents of immediate utility to the human genetics community. They are one of the largest collections of validated gene-based SNPs described and genotyped using technology that is already available to the genetics community.
Whereas the pilot SNP collection is of insufficient density to allow genomewide studies of linkage disequilibrium, it does provide a reagent of utility for larger-scale candidate locus studies than are currently feasible. The 3,646 validated SNPs are distributed among 2,987 UniGene clusters (1.2 SNPs/transcript). Of these, 3,148 (86%) have not been previously described in the National Center for Biotechnology Information's dbSNP. By definition, all of the SNPs are in transcribed portions of the genome. Therefore, the information generated by this study is of immediate, practical utility to investigators interested in performing large-scale gene-based association studies. A factor critical to this is the observed accuracy of the pooled allele frequency estimates obtained by chip-based MALDI-TOF MS. It should, therefore, be practical to routinely assess the role of hundreds to thousands of gene-based markers by using population rather than family designs.
The above pilot is only a first step in obtaining a comprehensive set of gene-based SNPs. Based on efforts such as the National Cancer Institute's Cancer Genome Anatomy Project (CGAP), the number of transcripts and genes available in the public domain continues to grow by thousands per week. The results of the above study can be used to inform future SNP mining efforts. It is projected that the overall yield can be improved by incorporating additional information when determining which candidate SNPs to validate. The current SNP discovery algorithm uses only information in the sequences themselves. Therefore, the algorithm used to nominate candidate SNPs says nothing about their frequency in the population. It is practical to also include information on the diversity of libraries and the distribution of variants within the libraries. Using standard logistic regression methods, one can derive a model that classifies a candidate SNP as polymorphic with 86% accuracy. These modifications are being incorporated into the tools provided at http://cgap.nci.nih.gov/GAI.
It is, in fact, pragmatic to consider the construction of a genomewide collection of gene-based SNPs using this approach. The automated design of reagents is performed in hours. The success rate of these design protocols will be improved with the availability of genomic sequence and high-quality state-of-the-art primer synthesis. Indeed, preliminary analysis of PCR failures where draft genome sequence is available indicates a substantial amount of the failures can be attributed to intron spanning. More provocatively, given the designed reagents, pooled validation experiments were performed for the 9,115 candidates in less then 4 weeks.
Given the manner by which the expressed sequence tags are ascertained, caution must be exercised in interpreting what this data set concludes about the distribution of variation in genes. The sources used to identify the variation are heterogeneous with respect to number of individuals examined, geographic origin of populations, and disease state. However, the population used in validation was uniform in number and constitution. Therefore, several observations are of note. Interestingly, the observed rare allele frequency distribution for the 3,646 unequivocal SNP loci was uniform (Fig. 3). Given the very small number of chromosomes and limited number of libraries that went into making SNP predictions, it is not unexpected that the observed frequency of variation in a defined population varies considerably. Indeed, although there was a significant correlation between the observed heterozygosity in the defined sample and heterozygosity estimated from data mining (P < 0.0001), the data mining estimate explained only 9% of the variation in observed heterozygosity.
Figure 3.
Allelic frequency range of 3,646 SNP genetic markers with a frequency in the Centre d'Étude du Polymorphisme Humain population greater than 10%.
The clusters contain 1,851 named genes (genes present in GenBank that have an assigned gene symbol). One can, therefore, make further observations concerning the distribution and magnitude of gene-based variation throughout the genome. A total of 373 of the SNPs are observed in the described coding regions of these genes. A total of 148 of the validated SNPs is predicted to result in an amino acid substitution within a named gene. Interestingly, the frequency distribution of the rare allele is uniform in both. No significant difference (t test: −1.0166, df = 1839, P = 0.3094) in mean frequency of the rare allele is observed whether the SNP results in an amino acid substitution (0.30) or not (0.31). The distribution of the frequency of the rare allele is not observed to differ whether or not the SNP resulted in an amino acid substitution (χ2(df:7) = 3.73, P = 0.81). Whereas it is premature to draw conclusions, the uniformity of the rare allele distributions suggests that the amino acid substituting variants have not been selected against. Complete information on the distribution of SNPs in each transcript and whether they result in amino acid substitutions is available at http://cgap.nci.nih.gov/GAI.
Finally, it is worth noting that failure to validate a candidate SNP using the Centre d'Étude du Polymorphisme Humain panel does not necessarily mean that it is not polymorphic. Whereas a significant fraction of failures may be attributable to experimental artifacts such as reverse transcriptase–PCR error, it is also likely that the candidates are polymorphic in other populations or at a frequency below that which is detectable in this sample.
Acknowledgments
We thank the members of the Sequenom team for their excellent support. This study was supported in part by a Small Business Innovation Research grant (phase I) granted to Sequenom (IR43CA83429-01).
Abbreviations
- SNP
single nucleotide polymorphism
- MALDI-TOF
matrix-assisted laser desorption/ionization time-of-flight
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.021506298.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.021506298
References
- 1.Risch N, Merikangas K. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 2.Buetow K H, Edmonson M N, Cassidy A B. Nat Genet. 1999;21:323–325. doi: 10.1038/6851. [DOI] [PubMed] [Google Scholar]
- 3.Schuler G D, Boguski M S, Stewart E A, Stein L D, Gyapay G, Rice K, White R E, Rodriguez-Tome P, Aggarwal A, Bajorek E, et al. Science. 1996;274:540–546. [PubMed] [Google Scholar]
- 4.Hillier L D, Lennon G, Becker M, Bonaldo M F, Chiapelli B, Chissoe S, Dietrich N, DuBuque T, Favello A, Gish W, et al. Genome Res. 1996;6:807–828. doi: 10.1101/gr.6.9.807. [DOI] [PubMed] [Google Scholar]
- 5.Braun A, Little D P, Koster H. Clin Chem (Washington, DC) 1997;43:1151–1158. [PubMed] [Google Scholar]
- 6.Little D P, Braun A, Darnhofer-Demar B, Koster H. Eur J Clin Chem Clin Biochem. 1997;35:545–548. [PubMed] [Google Scholar]
- 7.Little D P, Braun A, O'Donnell M J, Koster H. Nat Med. 1997;3:1413–1416. doi: 10.1038/nm1297-1413. [DOI] [PubMed] [Google Scholar]
- 8.Iacoviello L, Di Castelnuovo A, De Knijff P, D'Orazio A, Amore C, Arboretti R, Kluft C, Benedetta Donati M. N Engl J Med. 1998;338:79–85. doi: 10.1056/NEJM199801083380202. [DOI] [PubMed] [Google Scholar]
- 9.Dai Y, Whittal R M, Li L, Weinberger S R. Rapid Commun Mass Spectrom. 1996;10:1792–1796. doi: 10.1002/(SICI)1097-0231(199611)10:14<1792::AID-RCM753>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 10.Dausset J, Cann H, Cohen D, Lathrop M, Lalouel J M, White R. Genomics. 1990;6:575–577. doi: 10.1016/0888-7543(90)90491-c. [DOI] [PubMed] [Google Scholar]
- 11.Deloukas P, Schuler G D, Gyapay G, Beasley E M, Soderlund C, Rodriguez-Tome P, Hui L, Matise T C, McKusick K B, Beckmann J S, et al. Science. 1998;282:744–746. doi: 10.1126/science.282.5389.744. [DOI] [PubMed] [Google Scholar]