Abstract
The presence of large number of false lesion classification on segmented brain MR images is a major problem in the accurate determination of lesion volumes in multiple sclerosis (MS) brains. In order to minimize the false lesion classifications, a strategy that combines parametric and nonparametric techniques is developed and implemented. This approach uses the information from the proton density (PD)- and T2- weighted and fluid attenuation inversion recovery (FLAIR) images. This strategy involves CSF and lesion classification using the Parzen window classifier. Image processing, morphological operations and ratio maps of PD and T2 weighted images are used for minimizing false positives. Contextual information is exploited for minimizing the false negative lesion classifications using hidden Markov random field – expectation maximization (HMRF-EM) algorithm. Lesions are delineated using fuzzy connectivity. The performance of this algorithm is quantitatively evaluated on 23 MS patients. Similarity index, percentages of over, under and correct- estimations of lesions are computed by spatially comparing the results of present procedure with expert manual segmentation. The automated processing scheme detected 80% of the manually segmented lesions in the case of low-lesion load and 93% of the lesions in those cases with high lesion load.
Keywords: Segmentation, Feature classification, Multiple Sclerosis, Expectation maximization, Hidden Markov random field, MRI
INTRODUCTION
Magnetic resonance imaging (MRI) is a sensitive modality for visualizing MS lesions. MRI-determined lesion burden is used as a secondary outcome measure in a number of multicenter clinical trials on MS.18, 12 Accurate tissue segmentation is a prerequisite for robust estimation of lesion volumes. Independent of the segmentation technique used, various factors such as noise, intensity inhomogeneities, and partial volume effects introduce false tissue classifications. These false classifications can be reduced to some extent by applying image preprocessing techniques aimed at reducing noise and shading.13, 4 However, these steps only partially reduce the false classifications. Elimination of these false classifications requires considerable human intervention. Such intervention is impractical when dealing with large number of images that are typically encountered in multicenter clinical trials. Therefore, the image processing and segmentation techniques need to be either fully automatic or involve minimum human intervention.
While the emphasis of the current studies is on lesion segmentation, as indicated in the methods section below, minimization of false lesion classifications requires segmentation of other brain tissues. Wells et al.17 have proposed expectation maximization (EM) approach for iteratively and simultaneously correcting for the bias field and classifying tissues. Zhang et al.19 used hidden Markov random field (HMRF) model along with EM algorithm to incorporate contextual information into segmentation. These parametric methods assume the tissue classes to be modeled by some known distribution. While this appears to be true for gray matter (GM) and white matter (WM) classes which follow Gaussian distribution, lesions and CSF do not follow any known distribution. For example, Van Leemput et al.11 have identified MS lesions as outliers that are not well handled by this model. In contrast, nonparametric methods such as Parzen window classifier6 do not assume any distribution for tissues. These methods segment images using the feature space generated from the feature vectors comprised of the seed points for different classes.4 However, nonparametric techniques such as Parzen classifier are also prone to false classifications.
Both parametric and nonparametric techniques have their own strengths and weaknesses. For example, parametric techniques are robust but assume a known intensity distribution. This is not satisfied for lesions and to some extent CSF. On the other hand, nonparametric techniques do not assume any distribution. However, nonparametric techniques also introduce false tissue classifications. Therefore, by combining these two techniques, it is possible to improve the segmentation quality. This is the approach that we have followed in the current studies. This method was quantitatively validated using similarity measures against the results of manual segmentation performed by an expert neuroradiologist using MR images acquired on 23 MS patients.
METHODS
Patients
Twenty three patients (18 females and 5 males) with clinically definite MS were included in this study. Their mean age was 36 years (mean ± SD: 36.42 ± 9.9, range: 20-51 years). Their expanded disability status scale (EDSS) ranged from 0 to 5 (median score of 1.3). Written informed consent was obtained from all patients. These studies were approved by our Institutional Committee for the Protection of Human Subjects.
MRI acquisition
MR images were acquired on a 1.5 T, General Electric scanner. Subjects in the present study were recruited and scanned over a period of time. During this period of time, the scanner software was upgraded from 8.x to 9.x. Therefore data on some patients was acquired under 8.x while data on others was acquired under 9.x operating system. A quadrature birdcage resonator was used for radiofrequency transmission and signal reception. Dual echo images were acquired using the fast spin echo (FSE) sequence with the following parameters: TR = 6800 ms, TE1/TE2 = 12 ms/ 86 ms. Fast Fluid Attenuated Inversion Recovery (FLAIR) images were acquired with TR = 10002 ms, TI = 2200 ms, and TE = 91 ms. A total of 42 contiguous and interleaved slices, each of 3 mm thick, were acquired using the FSE and FLAIR sequences with the following parameters: FOV of 240 mm × 240 mm, and image matrix of 256 × 256.
Segmentation of Lesions, CSF, and Parenchyma
FLAIR images were registered with the interleaved dual echo images using the 3D rigid body registration technique.9 This multi-resolution technique maximizes the mutual information using the global optimization technique that is based on the genetic algorithm in continuous space and dividing rectangle method. Bias field correction was applied on these three sets of images using the module provided in statistical parametric mapping (SPM2).2 The extrameningeal tissues from the MR images were removed (image stripping) using semi-automated, in-house developed software. Anisotropic diffusion filter was applied to reduce noise in the images without concomitant image blurring.15, 7 While the filtration operation can be performed either before or following the image stripping, in these studies we chose to filter the post-stripped images. Intensity standardization14 was applied to all data sets so that the same feature maps could be applied to images acquired on different patients for Parzen window classification.13 Initially, lesions, CSF and parenchyma were classified on the late-echo FSE (T2-weighted or T2 images) and FLAIR images based on the two-dimensional feature map generated using the nonparametric Parzen window algorithm.6 The training points used for the generation of the feature map were identified by an individual with expertise in neuroanatomy. The two-dimensional feature map was generated using Parzen estimator with a Gaussian kernel:
| [1] |
The n samples of the training set in the two-dimensional feature space is denoted by ξi, i ∈ {1, ..., n}. Here, and the parameter h1 was chosen as:
| [2] |
where R is the rejection radius. Based on our experience, a value of 90 was used for R.
Removal of Surface Lesions
Small errors in the image registration and partial volume effects tend to produce false lesion classifications around the brain edges. Careful observation on a large image database obtained from multiple patients indicated that most of these false positives occurred within 2 to 3 pixels from the brain surface. Using this criterion and employing the morphological erosion operation with 2D kernel of size 3×3, all false lesions at the surface were eliminated.
False Positive Minimization (FPM) Inside Brain
False classifications inside the brain were eliminated using the ratio maps that were generated by taking the ratio of PD and T2 weighted images.10 A single threshold value, based on careful observation of a number of images, was used for all data sets to obtain a binary ratio mask of brain, excluding CSF and lesions (See Table 1). Negation of this mask was multiplied by the lesions obtained with Parzen classifier. Sizes of lesions obtained by the masking were usually smaller than the sizes seen on Parzen classifier. These sizes were matched with the Parzen classification by considering all the connected pixels of lesion. This procedure was observed to significantly minimize false positives. These identified false positive regions were reclassified as parenchyma which was later segmented into GM and WM.
Table 1.
Similarity measures calculated at different FPM thresholds on one patient data (Manual segmented lesion volume: 29.41 cc).
| S. No. | Threshold to eliminate false positive lesions | Segmented Lesion volume (in cc) | POE | PUE | PCE | SI |
|---|---|---|---|---|---|---|
| 1. | 1.5 | 19.28 | 13.03 | 47.48 | 52.52 | 0.63 |
| 2. | 1.55 | 21.94 | 15.10 | 40.49 | 59.51 | 0.68 |
| 3. | 1.6 | 25.75 | 21.50 | 33.96 | 66.04 | 0.70 |
| 4. | 1.65 | 31.57 | 28.99 | 21.64 | 78.36 | 0.76 |
| 5. | 1.7 | 33.03 | 31.22 | 18.93 | 81.07 | 0.76 |
| 6. | 1.75 | 34.52 | 34.19 | 16.82 | 83.18 | 0.77 |
| 7. | 1.8 | 35.78 | 37.27 | 15.64 | 84.36 | 0.76 |
| 8. | 1.85 | 37.87 | 43.23 | 14.49 | 85.51 | 0.75 |
| 9. | 1.9 | 39.24 | 46.97 | 13.55 | 86.45 | 0.74 |
| 10. | 1.95 | 40.67 | 50.95 | 12.69 | 87.31 | 0.73 |
| 11 | 2.0 | 41.65 | 53.98 | 12.36 | 87.64 | 0.73 |
Classification of GM and WM
Usually, GM and WM classes, but not lesions and CSF, follow a Gaussian distribution. Therefore, in the present study we have classified CSF and lesions using the Parzen classifier. Then the remaining brain parenchyma, excluding CSF and lesions, was classified into GM and WM using the proton density (PD or short echo FSE) and T2 images (long echo FSE). A parametric method, the HMRF-EM algorithm19 that is applicable to multispectral case was used for GM and WM classification.
A d-dimensional HMRF model with a Gaussian distribution can be specified as:
| [3] |
where , with Φ = {μ, Σ}. L represents the set of all class labels. yi is a feature vector in d-dimensional space and xNi is the neighborhood configuration of xi determined from the local characteristics of Markov random fields. Estimation of the model parameters for tissue classification simultaneously with bias field correction using expectation maximization (EM) approach is discussed in detail elsewhere.19 This method incorporates the contextual information into segmentation through MRF theory.
False Negative Minimization (FNM)
During the minimization of false positives, some true lesions, particularly subtle lesions, were eliminated from lesion classification. Following GM and WM segmentation, these regions were mostly classified as GM. More than 95% of MRI-observed MS lesions reside within WM. So, all the GM islands surrounded by WM or lesion were re-examined for possible reclassification as lesions. This was done by first labeling all disjoint GM segments by the connected component analysis (blob coloring3) that assigns every disconnected island a distinct numerical value (identifier/flag). For each GM segment, G, the outer boundary β(G) was obtained by
| [4] |
where is the dilation of G by the 2D structure element S of size 3×3. If the boundary β(G) had pixels of WM and/or lesion only, then the associated GM region, G, was classified as lesion if it was originally segmented as lesion on the Parzen classifier but was deleted during the FPM. Though this procedure recovered most of the lesions inside the WM, it did not recover the deleted lesions located in the GM and WM mixture regions and cortical gray matter.
Following the minimization of false classifications, lesions were delineated using fuzzy connectivity16 on the FLAIR images. In the present application, the degree of fuzzy adjacency between two spatial elements was assigned a value of unity if they differed in exactly one of the coordinates by one, and zero otherwise. The strength of fuzzy affinity, μκ (c,d), between the spatial elements c and d was described by the relation
| [5] |
Here, μα (c, d) is the degree of adjacency assigned to the spatial elements (c, d), g1 and g2 are multivariate Gaussian functions and ω1 and ω2 are non-negative weights such that ω1 + ω2 = 1. All the terms in equation [5] have the same meaning and definitions as described by Udupa et al.16 The values for ω1 and ω2 were chosen as 0.7 and 0.3 respectively. The threshold for fuzzy connectivity was set to 0.5. All thresholds and weights used for the delineation of lesion using fuzzy connectedness were optimized and fixed following a careful examination of a large number of data sets.
The final segmentation results, as judged by the expert neuroradiologist, have demonstrated more accurate lesion sizes with significantly reduced false lesion classifications.
The complete segmentation procedure is summarized in Fig. 1. The software was implemented on a PC under the Interactive Data Language (IDL, Research Systems, Inc., Boulder, CO) environment. SPM2 module for bias field correction and HMRF-EM algorithm were re-implemented in IDL.
FIGURE 1.
Schematic representation of segmentation procedure.
Manual Segmentation
In the absence of histologic confirmation, the true lesions and true lesion volumes are not known. Therefore, we relied on the expertise of a neuroradiologist (RKG) to establish the “true” lesions and their volumes by manually segmenting the lesions using the in-house developed software. All lesions identified on the MR images of the 23 MS patients were manually segmented. The expert classification was based on interpretation of the registered PD, T2 and FLAIR images and considered to be the “gold standard” or “reference” in these studies. Regions with high intensity in WM and GM on PD, T2 and FLAIR were defined as lesions. Hyperintense regions at tissue interfaces with the vasculature and the hyperintense ventricular lining seen on FLAIR were not considered as lesions. Periaqueductal hyperintensity was also not labeled as lesion. We arbitrarily classified patients into two categories: category I if the total lesion volume in the whole brain was less than 10 cc (N = 8), and category II if the total lesion burden was greater than or equal to 10 cc (N = 15).
Evaluation
The performance of the automatic segmentation results were quantitatively compared against the “gold standard” using four different similarity measures (Eqs. [6a]-[6d]): similarity index (SI), percentage of correct estimation (PCE), percentage of over estimation (POE), and percentage of under estimation (PUE). The SI is a measure of agreement in lesion volume between the reference and the segmented results. The PCE measures the percentage of correctly classified segmented lesion volumes relative to the reference. The POE measures the percentage of false positive classification relative to the reference while the PUE measures the percentage of missed lesion classifications. While the performance of the segmentation is evaluated based on the value of SI, the other three similarity measures provide an insight into the effect of each processing step on the lesion segmentation. The four similarity measures are formally defined as:
| [6a] |
| [6b] |
| 6c |
| 6d |
In these definitions, Ref and Seg denote the volumes based on the manual (expert classification) and the segmentation algorithm described above, respectively. The intersection of Ref and Seg represents the volume of the correctly classified voxels. The volume Seg corresponds to the false positives while Seg represents the false negatives.1 In all these measures the spatial correspondence between the automatic and manual segmentations was considered.
RESULTS
Segmentation
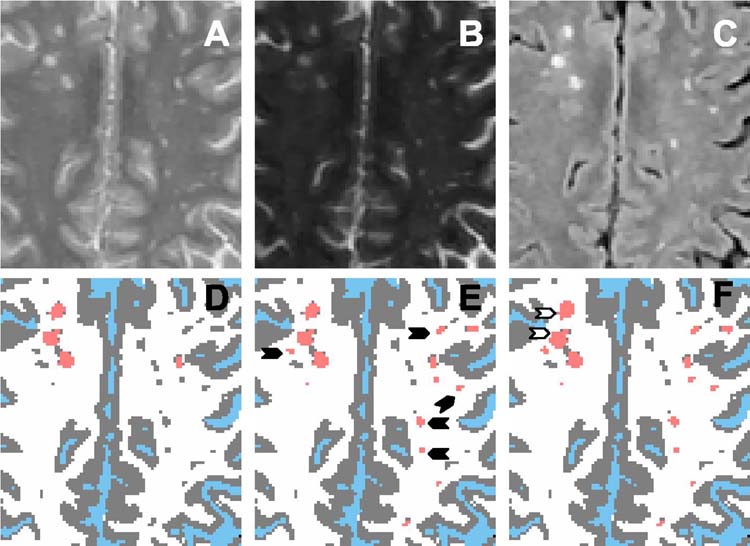
Figure 2 shows PD (A), T2 (B) and FLAIR (C) images of an MS brain. Figure 2D shows the segmented image in which CSF (blue), parenchyma (gray), and lesions (salmon) were classified. The segmented image was based on the FLAIR and T2 images using the Parzen window classifier. The presence of large number of false lesion classifications, particularly those close to the surface, can easily be appreciated on this image. Some of these false classifications are indicated by the open arrow heads. The same image following the removal of surface false positive lesions, as described earlier, is shown in Fig. 2E. However, this step did not completely eliminate all of the false positives within the brain parenchyma as some of them are shown by the solid arrow heads. These remaining false positives were removed using the ratio mask obtained with a threshold of 1.75, as described earlier. The value of the threshold was determined following a careful examination of a large number of images on different patients. The same threshold was used for all images with consistent results obtained. As an example Table 1 shows various similarity measures calculated at different thresholds on one patient data. As can be seen from this table, a threshold value of 1.75 yielded the best results. This value was employed in the segmentation of all the patient data. On the ratio mask, lesions and CSF appear with zero intensity. Application of the ratio mask eliminated the majority of the remaining false positive lesions (Fig. 2F).
FIGURE 2.

A-C: PD, T2 and FLAIR images respectively. D: Parzen classification: CSF (blue), brain parenchyma (gray) and lesions (salmon). False positives are clearly seen on the surface brain parenchyma (open arrow heads). E: After removing surface false positive lesions. The presence of false positives inside the parenchyma is shown by solid arrow heads. F: Minimization of false positives after applying the ratio mask to E. G: Binary mask of brain parenchyma excluding lesions and CSF. H: Gray and white matter classification (HMRF-EM). I: Merging CSF (blue), lesions (salmon), gray matter (gray) and white matter (white) segments.
Identification and elimination of the false negatives that occur in the segmentation process require contextual or neighborhood information. Therefore, prior to eliminating false negative classifications, brain parenchyma was classified into GM and WM using the HMRF-EM algorithm as described earlier. For this purpose, the brain mask excluding the lesions and CSF was generated (Fig. 2G). This mask was applied to the T2 and PD images and using the HMRF-EM algorithm, the parenchyma was classified into GM and WM (Fig. 2H). The segmented images shown in Figs. 2F and 2H were merged to generate the final segmented image (Fig. 2I) in which all the tissues were classified.
Figure 3 demonstrates the false negative minimization. In the prior example, we observed few false negative lesions and the effectiveness of false negative minimization could not be clearly demonstrated. Figures 3A - 3C show the magnified regions on PD, T2 and FLAIR images respectively. Figure 3D is the segmented image obtained as described above. A comparison of the FLAIR and segmented images shows a number of false negative lesions. The gray matter islands surrounded by WM or connected to lesions only were identified as described in the previous section. By comparing with the original Parzen classification, some of these GM islands were reclassified as lesions (Fig. 3E), as indicated by solid arrow heads. However, the sizes of these identified lesions were generally smaller than the actual lesions. Fuzzy connectivity was used for complete delineation of the lesions (Fig. 3F), shown by open arrow heads.
FIGURE 3.
A-C: Magnified regions on PD, T2 and FLAIR images respectively. D: Gray (gray) and white (white) matter segmented image after false positive minimization, but with some lesions seen on C classified as gray matter. E: Some of the lesions recovered during the FNM shown by solid arrow heads. F: Delineated lesions (open arrow heads).
Quantitative Analysis
Since the main purpose of these studies was to evaluate the quality of lesion segmentation, we quantitatively evaluated the performance of the segmentation results on the brain images of 23 MS patients using the evaluation metrics described above (Eqs. [6a] - [6d]). The same Parzen map and threshold values were used in segmenting all of the images on all of the patients.
In order to determine the extent of improvement in the segmentation quality following each step (segmentation based on the Parzen classification, FPM and FNM), we computed the similarity measures at each stage. It should be pointed out that FNM also includes lesion delineation using the fuzzy connectivity. The scatter plots of SI, over- and underestimation of lesion volumes against reference lesion volume for all subjects are shown in Figs 4A–4C.
FIGURE 4:
Scatter plots of similarity measures against reference lesion volume. Similarity indices (Plot A), Overestimated (Plot B) and Underestimated (Plot C) lesion volumes are computed at Parzen, FPM and FNM stages of the segmentation procedure on all 23 patients. In all the three plots, X-axis represents the reference lesion volume.
The similarity index using the Parzen classification without the application of FPM and FNM varied from 0.05 to 0.7 depending on the patient. However, this value increased from 0.3 to 0.9 following FPM, suggesting the importance of this step in accurate lesion segmentation. Significant improvement in the similarity indices in all the 23 subjects is evident from this plot [Fig. 4A]. FNM resulted in a smaller increment in improvement compared to FPM. These results also indicate that the process of FPM did not introduce large numbers or volumes of false negative lesions. This behavior was observed in all the 23 patients.
As can be seen from Fig. 4B, Parzen classification alone overestimated the total lesion volume by as much as 60 cc. This number was dramatically reduced by the FPM step. The effect of various processing steps on the underestimated lesion volume is shown in Fig. 4C. In this analysis, Fig. 4C suggests that Parzen window classifier alone did not underestimate the lesions. Reduction in underestimated lesion volume after FNM step indicates that most of the lesions deleted during FPM were recovered.
The effects of various segmentation steps, averaged over all the patients, are summarized in Tables 2 - 4. The values in these tables indicate the mean ± sd and are shown separately for the category I and category II patients. The results suggest that FNM has provided an overall improvement in all the similarity measures. This effect seems to be particularly impressive for patients with low lesion load (category I). The POE values are particularly high for small lesion load (category I) compared to large lesion load (category II). The values of PCE and SI were higher for category II compared to category I. Overall, these results suggest the importance of both FPM and FNM in maintaining the segmentation fidelity.
Table 2.
Various Similarity measures between manual segmentation and automatic segmentation following Parzen window classification.
| Parzen | POE | PUE | PCE | SI |
|---|---|---|---|---|
| Category I (8) | 1735.77 ± 1646.24 | 15.25 ± 1.94 | 84.74 ± 1.94 | 0.15 ± 0.10 |
| Category II (15) | 176.82 ± 112.52 | 9.34 ± 2.55 | 90.65 ± 2.55 | 0.53 ± 0.13 |
| Category I + II (23) | 719.06 ± 1202.80 | 11.39 ± 3.69 | 88.60 ± 3.69 | 0.39 ± 0.21 |
Table 4.
Various Similarity measures between manual segmentation and automatic segmentation following false negative minimization and fuzzy delineation.
| FNM | POE | PUE | PCE | SI |
|---|---|---|---|---|
| Category I (8) | 59.36 ± 34.34 | 19.74 ± 20.29 | 80.25 ± 20.29 | 0.67 ± 0.14 |
| Category II (15) | 27.18 ± 15.98 | 6.94 ± 2.70 | 93.05 ± 2.70 | 0.84 ± 0.05 |
| Category I + II (23) | 38.38 ± 27.98 | 11.37 ± 13.21 | 88.62 ± 13.21 | 0.78 ± 0.12 |
Quantitative analysis indicates high similarity measures between manual and the proposed segmentation procedure. For instance the percentage of correctly estimated lesion volumes is 93% for high lesion load (≥ 10 cc; category II) patients and 80% for patients with smaller lesion loads (<10 cc; category I). When both categories are combined, the percentage agreement is about 88%. Perhaps a better index that reflects the quality of segmentation is the similarity index that accounts for both false and correct classification. The similarity index in the current studies is 0.84 for category II patients and drops to 0.67 for category I patients. However, the overall value of 0.78 is one of the highest reported values in white matter lesion classification.
Bland-Altman Plot
Bland-Altman analysis was used for an objective evaluation of the agreement between the manual and segmentation results.5 Bland-Altman method is a statistical technique for assessing the agreement between two imperfect measures of the same variable. In this method the difference between the two measurements of the same variable (also referred to as bias) is plotted against the estimate of the true value (mean of the two measurements). In the present analysis, difference was computed by subtracting the lesion volume obtained by our method from the manually segmented volume. Generally the mean and mean ± 2 sd values of the differences are shown on these plots to provide a visual estimation of both random and systematic differences between the two measurements. The Bland-Altman plot for the lesion volumes is shown in Fig. 5. The plot demonstrates a close agreement between the two segmentation methods. This analysis also shows a bias that is below the zero mean, indicating that the automatic analysis systematically overestimated the total lesion volume. However, the bias is well within the two standard deviations.
FIGURE 5.
Bland Altman plot for comparing the bias in manual and automatic lesion segmentations. Average volume (X-axis) is the mean lesion volume of automatic and manual methods for each patient. Bias is computed by subtracting the lesion volume obtained by the present method from the reference lesion volume.
DISCUSSION
In spite of significant advances in image segmentation, lesion classification in MS continues to be problematic. To some extent this can be alleviated by manually editing the segmentation results. However, this becomes impractical when handling large amounts of data. Automatic detection of false negative classifications is particularly problematic, and to the best of our knowledge has not been addressed in any formal publications. In the present study, a unified approach combining both parametric and nonparametric segmentation methods with morphological operations is used to minimize these false classifications. Such an approach overcomes the limitations of the parametric methods that require tissue distribution to follow a well defined distribution. Since lesions and CSF do not obey this requirement, as powerful as they may be, parametric techniques do not perform well in the classification of lesions. The nonparametric techniques, in contrast, do not require any type of distribution for tissues. In the present studies, the minimization of false negative lesion classification after FPM utilizes the knowledge of the tissue type in the neighborhood of lesions and classification of GM and WM. Previous studies have also shown that both of these tissues follow a Gaussian distribution. Therefore, in the current studies WM and GM were classified using the HMRF-EM algorithm, which is a parametric technique that has been shown to be robust in classifying tissues that obey a well defined distribution. As can be appreciated from our results, such an approach appears to have been reasonably successful.
Automatic identification and minimization of false lesion classifications is an important part of the current studies. This aspect of segmentation received relatively little attention in the literature. As can be seen from our results, without these steps, the lesion volumes are overestimated by as much as 60 cc. The concept of using ratio images for lesion segmentation was originally proposed by Krishnan and Atkins.10 These authors used this technique as a part of the overall segmentation, but have not explicitly used the ratio images for minimization of false positives. In the current studies, removal of false positives, in some instances, resulted in the elimination of true lesions, particularly subtle ones. The GM and WM classification and the “blob coloring” technique used in these studies for automatic identification of the false negatives appear to have reduced this problem, as can be judged from the results [Table 4]. This step does remove intracortical lesions. It is worth pointing out that MRI is not very sensitive in demonstrating cerebral cortex lesions. For example, in a recent study Geurts et al.8 have shown that even on the FLAIR images only about 5% of cortical lesions appear on MR imaging. In all the 23 patients, we did not see on MR any lesion within 2 pixels distance from brain surface.
In these studies we quantitatively evaluated the performance of our segmentation using the similarity measures. These metrics were originally applied by Leemput et al.11, Zijdenbos et al.20, and Anbeek et al.1 to measure the concordance of their results with manual segmentation of white matter lesions. With their segmentation techniques, Leemput et al.11 and Zijdenbos et al.20, achieved similarity indices of 0.51 and 0.68 respectively, while Anbeek et al.1, achieved a value of 0.7 for SI for all of their study patients. In our present approach this value is 0.78 suggesting the superior performance of our method. It is difficult to quantitatively compare our results with other published results since very few publications evaluated the performance of the segmentation using the metrics of the current studies.
As indicated earlier, one of our main objectives is to automate all the processing steps in the lesion segmentation. Of these various steps, image registration, bias field correction, filtration, intensity normalization, gray and white matter classification, identification and elimination of false lesion classification and lesion delineation are fully automatic. The necessary thresholds in the ratio images and lesion delineation do involve manual intervention. However, once fixed, the same values were used for fully automated processing of all the subsequent images. Similarly, identification of training feature vectors for feature map generation needs to be performed only once. The only processing step that involves some manual intervention is in image stripping which was performed on the T2 set of images on each subject. However, this procedure is significantly simplified by providing various tools such as connectivity and island removal. The same mask was then automatically applied to the PD and FLAIR images. In our studies, stripping of all 42 images corresponding to 42 cross-sections of one brain volume typically took between 5 and 10 minutes, depending on the experience of the operator.
The HMRF-EM algorithm includes the bias field correction as a part of the tissue classification. When we tried the HMRF-EM algorithm with and without bias field corrected (by SPM2) input data for a fixed number of iterations with the same initial approximations, we observed better classification with the bias field corrected input data. Therefore, SPM2 bias field correction module was used before Parzen classification.
The Bland-Altman analysis indicates that the automatic segmentation yields values that are in close agreement with manual segmentation, but consistently overestimates the total lesion load. Like the majority of segmentation procedures, our technique classifies the ependymal lining around the ventricles which appears bright on FLAIR and T2 weighted images as a lesion, but our neuroradiologist did not classify this as a lesion. This perhaps is the major reason why the present technique consistently overestimated the lesion load.
Similarity index is better suited for category II subjects compared to category I subjects. This is because even a few false lesion classifications would have a higher impact on smaller lesion loads. Similar behavior was also observed by Anbeek et al.1
The multi-spectral segmentation used in these studies requires that the dual FSE and FLAIR data be in exact registration. Otherwise segmentation results in a large number of false lesion classifications. Even though both of these image sets were acquired in the same session, the inevitable movement of the subject between two different image series requires retrospective registration with sub-voxel accuracy. In these studies we used the global optimization of mutual information for image registration. This is shown to be a robust technique9 and in the current studies it provided excellent results in 95% of cases.
While this method has been applied to the detection and quantification of MS lesions, it should be applicable, with a few modifications, for automatically detecting and quantifying anatomical lesions that are present in other pathologies such as cancer.
In conclusion, we implemented a segmentation technique that combines parametric and nonparametric techniques for lesion classification in MS with minimal human intervention. This method was quantitatively evaluated using similarity measures. The percentage of correct estimation of lesions between our method and manual segmentation was found to be better than 88%.
Table 3.
Various Similarity measures between manual segmentation and automatic segmentation following false positive minimization.
| FPM | POE | PUE | PCE | SI |
|---|---|---|---|---|
| Category I (8) | 47.20 ± 28.13 | 50.88 ± 24.73 | 49.11 ± 24.73 | 0.47 ± 0.18 |
| Category II (15) | 22.91 ± 13.46 | 13.45 ± 3.06 | 86.54 ± 3.06 | 0.82 ± 0.05 |
| Category I + II (23) | 31.36 ± 22.52 | 26.47 ± 23.08 | 73.52 ± 23.08 | 0.70 ± 0.20 |
ACKNOWLEDGMENT
This work is supported by National Institutes of Health Grant EB002095 to PAN.
Glossary of Terms
- CSF
Cerebrospinal fluid
- EDSS
Expanded disability status scale
- EM
Expectation maximization
- FLAIR
Fluid attenuation inversion recovery
- FNM
False negative minimization
- FPM
False positive minimization
- FSE
Fast spin echo
- GM
Gray matter
- HMRF
Hidden Markov random field
- HMRF-EM
Hidden Markov random field-expectation maximization
- IDL
Interactive data language
- MR
Magnetic Resonance
- MRF
Markov random field
- MRI
Magnetic Resonance Imaging
- MS
Multiple sclerosis
- PCE
Percentage of correct estimation
- PD
Proton density
- POE
Percentage of overestimation
- PUE
Percentage of underestimation
- SI
Similarity index
- SPM
Statistical parametric mapping
- WM
White matter
REFERENCES
- 1.Anbeek P, Vincken KL, van Osch MJ, Bisschops RH, van der Grond J. Probabilistic segmentation of white matter lesions in MR imaging. NeuroImage. 2004;21:1037–1044. doi: 10.1016/j.neuroimage.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 2.Ashburner J, Friston K. MRI sensitivity correction and tissue classification. NeuroImage. 1998;7:S706. [Google Scholar]
- 3.Ballard DH, Brown CM. Computer Vision. Prentice-Hall; New Jersey: 1982. [Google Scholar]
- 4.Bedell BJ, Narayana PA, Wolinsky JS. A dual approach for minimizing false lesion classifications on magnetic resonance images. Magn. Reson. Med. 1997;37:94–102. doi: 10.1002/mrm.1910370114. [DOI] [PubMed] [Google Scholar]
- 5.Bland JM, Altman DG. Comparing methods of measurement: why plotting difference against standard method is misleading. Lancet. 1995;346:1085–1087. doi: 10.1016/s0140-6736(95)91748-9. [DOI] [PubMed] [Google Scholar]
- 6.Duda RO, Hart PE, Stork DG. Pattern Classification. John Wiley & Sons; New York: 2001. [Google Scholar]
- 7.Gerig G, Kubler O, Kikinis R, Jolesz FA. Nonlinear anisotropic Filtering of MRI data. IEEE Trans. Med. Imaging. 1992;11:221–232. doi: 10.1109/42.141646. [DOI] [PubMed] [Google Scholar]
- 8.Geurts JJG, Bö L, Pouwels PJW, Castelijns JA, Polman CH, Barkhof F. Cortical Lesions in Multiple Sclerois: Combined Postmortem MR Imaging and Histopathology. AJNR Am J Neuroradiol. 2005;26:572–577. [PMC free article] [PubMed] [Google Scholar]
- 9.He R, Narayana PA. Global optimization of mutual information: application of three dimensional retrospective registration of magnetic resonance images. Comput. Med. Imag. Graph. 2002;26:277–292. doi: 10.1016/s0895-6111(02)00019-8. [DOI] [PubMed] [Google Scholar]
- 10.Krishnan K, Atkins MS. Segmentation of Multiple Sclerosis lesions in MRI - an image analysis approach. Proc SPIE Med. Imaging. 1998;3338:1106–1116. [Google Scholar]
- 11.Leemput KV, Maes F, Vandermeulen D, Colchester A, Suetens P. Automated segmentation of Multiple Sclerosis lesions by model outlier detection. IEEE Trans. Med. Imaging. 2001;20:677–688. doi: 10.1109/42.938237. [DOI] [PubMed] [Google Scholar]
- 12.Miller DH, Filippi M, Fazekas F, Frederiksen JL, Matthews PM, Montalban X, Polman CH. Role of magnetic resonance imaging within diagnostic criteria for multiple sclerosis. Ann. Neurol. 2004;56:273–278. doi: 10.1002/ana.20156. [DOI] [PubMed] [Google Scholar]
- 13.Narayana PA, Borthakur A. Effect of radio frequency inhomogeneity correction on the reproducibility of intra-cranial volumes using MR image data. Magn. Reson. Med. 1995;33:396–400. doi: 10.1002/mrm.1910330312. [DOI] [PubMed] [Google Scholar]
- 14.Nyul LG, Udupa JK, Zhang X. New variants of a method of MRI scale standardization. IEEE Trans. Med. Imaging. 2000;19:143–150. doi: 10.1109/42.836373. [DOI] [PubMed] [Google Scholar]
- 15.Perona P, Malik J. Scale-space and edge detection using anisotropic diffusion. IEEE Trans. Pattern. Anal. Mach. Intell. 1990;12:629–639. [Google Scholar]
- 16.Udupa JK, Wei L, Samarasekera S, Miki Y, van Buchem MA, Grossman RI. Multiple Sclerosis lesion quantitation using fuzzy-connectedness principles. IEEE Trans. Med. Imaging. 1997;16:598–609. doi: 10.1109/42.640750. [DOI] [PubMed] [Google Scholar]
- 17.Wells WM, III, Grimson WEL. Adaptive segmentation of MRI data. IEEE Trans. Med. Imaging. 1996;15:429–442. doi: 10.1109/42.511747. [DOI] [PubMed] [Google Scholar]
- 18.Wolinsky JS, Narayana PA, Johnson KP. Multiple Sclerosis Study Group and the MRI Analysis Center. United States open-label glatiramer acetate extension trial for relapsing multiple sclerosis: MRI and clinical correlates. Multiple Sclerosis Study Group and the MRI Analysis Center. Mult. Scler. 2001;7:33–41. doi: 10.1177/135245850100700107. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization. IEEE Trans. Med. Imaging. 2001;20:45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- 20.Zijdenbos AP, Forghani R, Evans AC. Automatic “pipeline” analysis of 3-D MRI data for clinical trials: application to multiple sclerosis. IEEE Trans. Med. Imaging. 2002;21:1280–1291. doi: 10.1109/TMI.2002.806283. [DOI] [PubMed] [Google Scholar]