Abstract
PURPOSE
To further elucidate the cataract phenotype, and identify the gene and mutation for autosomal dominant cataract (ADC) in an American family of European descent (ADC2) by sequencing the major intrinsic protein gene (MIP), a candidate based on linkage to chromosome 12q13.
DESIGN
Observational case series and laboratory experimental study.
METHODS
We examined two at-risk individuals in ADC2. We PCR-amplified and sequenced all four exons and all intron-exon boundaries of the MIP gene from genomic and cloned DNA in affected members to confirm one variant as the putative mutation.
RESULTS
We found a novel single deletion of nucleotide (nt) 3223 (within codon 235) in exon four, causing a frameshift that alters 41 of 45 subsequent amino acids and creates a premature stop codon.
CONCLUSIONS
We identified a novel single base pair deletion in the MIP gene and conclude that it is a pathogenic sequence alteration.
Autosomal dominant cataracts (adc) are genetically heterogeneous; 24 chromosomal regions and 15 genes have been identified. We examined two additional members of a family of European descent (ADC2) with ADC and added a different cataract morphology and location within the lens to the phenotype. We identified the putative mutation in the major intrinsic protein gene (MIP).
We studied an American family (ADC2) (Figure 1) in which the gene had been mapped to 12q13.1,2 Informed consent, in accordance with the Declaration of Helsinki and approved by both the University of California, Los Angeles (UCLA) and University of Colorado institutional review boards, was obtained.
FIGURE 1.
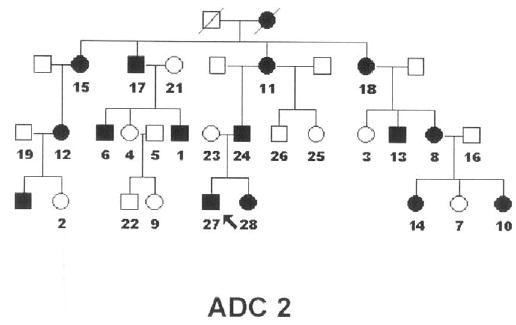
Pedigree of family ADC2 with autosomal dominant cataracts. The arrow marks the proband, affected individuals are identified by solid squares (male) and circles (female), and the studied individuals are numbered.
Two individuals originally categorized as unknown were examined.
On the basis of published sequences,3,4 we designed PCR primers for amplification and sequencing in both directions of genomic and cloned DNA for all four exons of MIP including intron-exon boundaries and 2.84 kb of the 5′ untranslated region (Table). We sequenced products with the PE-ABI Prism FS dye terminator kit (Applied Biosystems, Foster City, California, USA) with an ABI 373 DNA sequencer. Chromatogram alignments were performed by Sequencher (Gene Codes, Ann Arbor, Michigan, USA), and DNA sequence analyses and protein effect of the putative mutation were performed by DNA Strider (Cedex, France).
TABLE.
Primers used for the amplification and sequencing of Major Intrinsic Protein Gene (MIP)
| Region | Sequence | Fragment size | |
|---|---|---|---|
| Exon 1 | F | 5′ -AAGGGGACTGTCCACCCAG- 3′ | 270 bp |
| R | 5′ -TGGGCTCCACTGATGTGGC- 3′ | ||
| Exon 1 | F | 5′ -GGCTATGGCATTTGGCTTGG- 3′ | 306 bp |
| R | 5′ -CTGCACCAGTCAGGGAGTC- 3′ | ||
| Exon 1 | F | 5′ -TCCTCTATAAAGGGGACTGTC- 3′ | 216 bp |
| R | 5′ -GCCACCTGCAGAACATGCAG- 3′ | ||
| Exon 1 and Promoter | F | 5′ -CACTTCTCGTAGTCTCTCTTGCTG- 3′ | 300 bp |
| R | 5′ -GCTGATCGCAGTTCCCACATGGC- 3′ | ||
| Exon 2 | F | 5′ -AGGAGGTAACACTGTGGCAG- 3′ | 263 bp |
| R | 5′ -GAATCCTTGAATGAGAAGTTGC- 3′ | ||
| Exon 3 | F | 5′ -AAGCTGGGGTGCAGTAGGG- 3′ | 169 bp |
| R | 5′ -GAGTGCTGGTACAGCAGCC- 3′ | ||
| Exon 4 | F | 5′ -CAGCGTTGCTGCTCTGTCC- 3′ | 258 bp |
| R | 5′ -TCAGCTGGAGCTTCTACAGG- 3′ | ||
| Exon 4 | F | 5′ -GGGAACCTGTTGAACTGAACA- 3′ | 254 bp |
| R | 5′ -TGGGGAGGAAGGGAAGTTTG- 3′ | ||
| Exon 4 | F | 5′ -CTCAAGAGTATTTCTGAGAGAC- 3′ | 201 bp |
| R | 5′ -ACAGTCTCTTTCTTCATCTAGG- 3′ | ||
| Intron 3 and | F | 5′ -GAATATACACATGCTAAGGTGTGG- 3′ | 279 bp |
| Exon 4 | R | 5′ -AGTCTCTCAAGAAATACTCTTGAGC- 3′ | |
| 5′ UTR | F | 5′ -GGGATTACAGGCATGAGCCATTGCC- 3′ | 359 bp |
| R | 5′ -GTGCAGTGGCACAATCAGCTCACTG- 3′ | ||
| 5′ UTR | F | 5′ -TACGAGTCCCTTTCTTATTCCTATCCC- 3′ | 410 bp |
| R | 5′ -CCATCAAGCTGAGCTCACTTCATGGC- 3′ | ||
| 5′ UTR | F | 5′ -CCTCTCCTTGATCGGCTGTTTCCCC- 3′ | 301 bp |
| R | 5′ -GGGACCGACAGAGAGAAGACACTATC | ||
| 5′ UTR | F | 5′ -CACCATGATGTGCTACCCTCTCTCTT- 3′ | 760 bp |
| R | 5′ -CTTTAATACAGCCATGATCAGCACACC- 3′ | ||
| 5′ UTR | F | 5′ -CACCATGATGTGCTACCCTCTCTCTT- 3′ | 516 bp |
| R | 5′ -AAATAGTGGCAAGTCATACCGGCCAC- 3′ | ||
| 5′ UTR | F | 5′ -CAGTTTATACTCCCACCAGCAGTTGTA- 3′ | 696 bp |
| R | 5′ -CCTATCCACTGGCAGGAAGAGTTGC- 3′ | ||
| 5′ UTR | F | 5′ -TACGAGTCCCTTTCTTATTCCTATCCC- 3′ | 526 bp |
| R | 5′ -TGGGGGTGAAGAACAGCAGGACTG- 3′ |
F = forward, R = reverse
Patients 1 and 6 had mild opacities and a different type of cataract as compared with other affected family members. Patient 1 had fine punctate opacities in the posterior cortex and Y sutures, and a few punctuate opacities in the anterior cortex bilaterally. Patient 6 had fine white punctate opacities in the cortex, larger opacities inferiorly, and in the posterior cortical region bilaterally.
We found a single base pair deletion that cosegregated with the disease at nt 3223 (guanine) (codon 235) of exon 4 of the MIP gene in all 12 affected individuals (Figure 2). None of the 11 unaffected individuals tested had the change.
FIGURE 2.
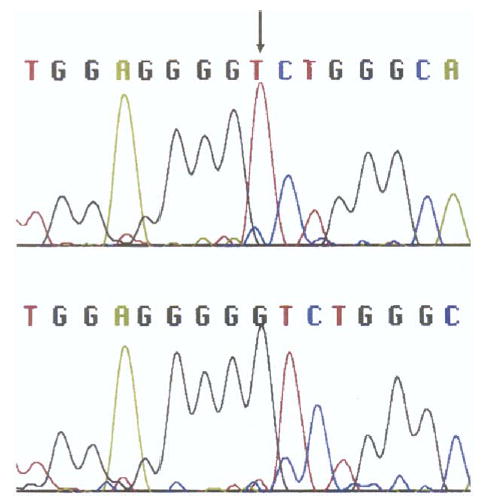
Sequence analysis of exon 4 in the major intrinsic protein (MIP) gene from one individual in the family ADC2 affected with autosomal dominant cataracts (patient 10). The guanine deletion is detected at nucleotide 3223 (arrow) (Bottom) (within codon 235) as compared with the normal sequence from an unaffected individual in the family (Top).
Analyses of the deletion predict a frameshift and the creation of a premature stop codon, resulting in substitutions of 41 of the subsequent 45 amino acids preceding the truncation (Figure S1). The shortened protein would be 257 amino acids as compared with the normal size of 263.
FIGURE S1.
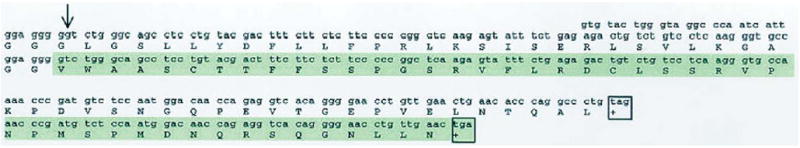
Predicted translational effects of the guanine deletion at position 3223 in exon 4 (arrow), resulting in a substitution of glutamic acid for glycine. The shaded region shows the resultant frameshift and consequent amino acid changes in the mutant allele. Stop codons are boxed, illustrating the introduction of a premature stop codon (nonsense mutation) in the mutant allele.
MIP is an aquaporin and contains four exons; the 5′ -flanking sequence contains an active promoter region for lens expression. It is the predominant lens membrane protein; has water channels, considered essential for lens microcirculation; and is present in membrane junctions, possibly functioning as an adhesion molecule. The C-terminus is located in the cytoplasmic side of the plasma membrane and is cleaved in aging cataractogenesis.
Missense mutations in exon 2 of MIP have been reported in two families.5 One had a C→G transition resulting in a threonine to arginine substitution at codon 138. This family had “progressive, bilateral, punctuate opacities in the mid- and peripheral lamellae… common to all affected individuals”; some individuals had “symmetric anterior and posterior” cataracts, and one had a cortical cataract.5,6 The second family had an A→G transversion resulting in a glutamic acid substitution for glycine at codon 134 and fine, nonprogressive lamellar and sutural opacities.5
Affected ADC2 individuals exhibited variable expressivity of cataract with radiating, vacuolar, or dense opacities in the embryonal nucleus.1,2 The two newly examined members had milder and different cataracts with fine punctate cortical opacities; one had punctate opacities in the posterior Y sutures, and the other had small inferior posterior cortical opacities in the inferior posterior cortex.
We found a single base pair deletion at nt 3223 (codon 235; exon 4) in MIP. This deletion is likely to have functional significance because it is within the DNA sequence encoding the sixth and last transmembrane region and the C-terminus of the protein. This mutation results in a shortened protein that may alter voltage dependence and calmodulin-binding properties. The clinical variability of the cataract may be similar to the effects of C-terminus truncation on the aging human lens.
Footnotes
Supported in part by National Eye Institute RO1 Grant EY 08282 (J.B.B.).
Supplemental Material available at AJO.com.
Eleanor Roosevelt Institute affiliated with the University of Denver after this research project.
References
- 1.Bateman JB, Spence MA, Marazita ML, Sparkes RS. Genetic linkage analysis of autosomal dominant congenital cataracts. Am J Ophthalmol. 1986;101:218–225. doi: 10.1016/0002-9394(86)90599-4. [DOI] [PubMed] [Google Scholar]
- 2.Bateman JB, Johannes M, Flodman P, et al. A new locus for autosomal dominant cataract on chromosome 12q13. Invest Ophthalmol Vis Sci. 2000;41:2665–2670. [PubMed] [Google Scholar]
- 3.Pisano MM, Chepelinsky AB. Genomic cloning, complete nucleotide sequence, and structure of the human gene encoding the major intrinsic protein (MIP) of the lens. Genomics. 1991;11:981–990. doi: 10.1016/0888-7543(91)90023-8. [DOI] [PubMed] [Google Scholar]
- 4.Wang XY, Ohtaka-Maruyama C, Pisano MM, Jaworski CJ, Chepelinsky AB. Isolation and characterization of the 5′ -flanking sequence of the human ocular lens MIP gene. Gene. 1995;167:321–325. doi: 10.1016/0378-1119(95)00637-0. [DOI] [PubMed] [Google Scholar]
- 5.Berry V, Francis P, Kaushal S, Moore A, Bhattacharya S. Missense mutations in MIP underlie autosomal dominant “polymorphic” and lamellar cataracts linked to 12q. Nat Genet. 2000;25:15–17. doi: 10.1038/75538. [DOI] [PubMed] [Google Scholar]
- 6.Francis P, Berry V, Bhattacharya S, Moore A. Congenital progressive polymorphic cataract caused by a mutation in the major intrinsic protein of the lens, MIP (AQP0) Br J Ophthalmol. 2000;84:1376–1379. doi: 10.1136/bjo.84.12.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]


